Introduction
As a chronic joint disease, osteoarthritis (OA) has
seriously threatened the health of millions of individuals globally
and has led to disability (1).
Multiple factors are considered to be involved in the etiology of
OA, including intracellular metabolism, inflammation and
extracellular matrix (ECM) degeneration (2-4).
Moreover, obesity and aging increase the risk of developing OA
(5). The current treatments for OA,
such as drug, electromagnetic therapy and acupuncture therapy, are
mainly aimed at pain relief (6-8),
thus OA remains difficult to cure. Further investigation of the
pathogenesis of OA is required to elucidate effective therapies to
prevent OA and reduce its severity.
Long non-coding RNAs (lncRNAs) are RNA molecules
with >200 nucleotides that do not encode proteins (9). A multitude of studies have manifested
that lncRNAs are involved in multiple diseases (10-12).
For example, lncRNA AT-rich interactive domain 2-IR aggravates
renal inflammation by activating the IL-1β-induced NF-κB signaling
pathway, as well as inducing the production of inflammatory
cytokines (13). LncRNA urothelial
cancer-associated 1 promotes the development of Parkinson's disease
by aggravating the inflammatory response and exacerbating the
damage of dopaminergic neurons (14). LncRNA metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) inhibits ECM degradation and
cell apoptosis by targeting miR-150-5p/AKT3 axis (15). Moreover, previous studies have
revealed that long intergenic non-protein coding RNA 473
(LINC00473) serves a notable role in various diseases. For example,
LINC00473 has been identified as a notable biomarker in ischemic
stroke through the subpathway-LNCE method, which was designed to
identify lncRNAs that competitively modulated functions in diseases
(16). LINC00473 downregulation
contributes to the development of preeclampsia (17). Additionally, a previous study
demonstrated that LINC00473 was highly expressed in patients with
severe pain associated with OA (18). However, the importance of LINC00473
in OA pathology requires further elucidation.
It has been proposed that competing endogenous RNAs
(ceRNAs) could function as molecular sponges for microRNAs
(miRNAs/miRs) and then modulate the expression of target genes
(19). In previous years, an
increasing number of studies have demonstrated that lncRNAs
function as ceRNAs during the pathogenesis of multiple diseases,
including OA. For example, upregulation of lncRNA RNA component of
mitochondrial RNA processing endoribonuclease aggravates the
development of coronary heart disease via the
miR-206/autophagy-related 3 axis (20). LncRNA MALAT1 facilitates renal
tubular epithelial pyroptosis by sponging miR-23c and targeting
ELAV-like protein 1 in diabetic nephropathy (21). LncRNA small nucleolar RNA host gene
5 promotes chondrocyte proliferation by targeting the miR-26a/sex
determining region Y-box 2 axis in OA (22). Additionally, recent research has
indicated that LINC00473 may participate in OA pathogenesis via
dysregulated ceRNA interactions (18). However, the specific mechanism of
LINC00473 in OA has not been fully explored. The present study
aimed to investigate the effects of LINC00473 on the pathogenesis
of OA.
Materials and methods
Clinical samples
OA cartilage tissues (n=24) were obtained from 24
patients (11 males and 13 females; age, 51-70 years) undergoing
total knee replacement surgery and 24 healthy cartilage tissue
samples were collected from patients (10 males and 14 females; age,
33-42 years) undergoing traumatic amputations at the Children's
Hospital of Soochow University (Jiangsu, China) between January
2014 and December 2019. Additionally, normal cartilage tissues
(n=24) were collected from donors who underwent amputation with no
diagnostic history of rheumatoid arthritis or OA at the same
hospital. All donors signed informed consent forms. The present
study was approved by The Human Ethics Committee of Children's
Hospital of Soochow University. All tissue samples were stored at
-80˚C. Inclusion criteria were listed as follows: i) Patients who
met clinical manifestations of knee osteoarthritis (23); ii) patients with complete medical
records; iii) patients who had not received any antibiotic
treatment within 3 months before admission; iv) patients who were
willing to cooperate and participate in the investigation.
Exclusion criteria were also listed as follows: i) Patients with
tumors, organ dysfunction, infectious diseases, autoimmune
diseases, neurological disorders, cardiovascular diseases or
cerebrovascular diseases; ii) patients who were allergic to drugs
used in the present study; iii) patients with physical disabilities
and unable to take care of themselves; iv) patients who were
transferred to other hospitals.
Rat model of OA
The animal experiments were approved by The Attitude
of the Animal Care & Welfare Committee of Children's Hospital
of Soochow University (permit no. 2019-044). A total of 20 male
rats (age, 6 weeks; weight, 230-260 g; Beijing Vital River
Laboratory Animal Technology Co., Ltd.) were randomly divided into
the following four groups (n=5 rats/group) for subsequent
experiments: i) Sham; ii) OA; iii) OA + adeno-associated virus
(AAV) small interfering RNA (si/siRNA)-negative control (NC); and
iv) and OA+AAV si-LINC00473. All procedures were conducted
following recommendations of The Guide for the Care and Use of
Laboratory Animals (24).
The OA rat model was established by anterior
cruciate ligament (ACL) transection and partial medial
meniscectomy. In brief, after anesthetization by the
intraperitoneal injection of pentobarbital (35 mg/kg weight), a
1-cm longitudinal incision on the medial side of the right knee
joint was made and the medial ligament was exposed. After the ACL
and medial meniscus were removed, the joint capsule and the skin
was sutured and bandaged under a sterile environment. For the sham
group, the same incisions were made in rats but without meniscus or
ACL resection. For AAV-injected groups, OA model rats were
intra-articularly injected with recombinant (r)AAV5 expressing
si-NC or si-LINC00473 (1.65x1010 vg/joint) using
33-gauge needles and 25-µl gastight syringes (both Hamilton
Company) after modeling. rAAV5 was obtained from Han heng
Biotechnology (Shanghai) Co., Ltd. All groups of rats (sham, OA
model and AAV injected) were kept in separate cages and provided
with 12/12 h light/dark cycle and ad libitum food and water.
Humidity was 50-55% and feeding temperature was set to 20-23˚C. At
4 weeks after surgery, under intraperitoneal anesthesia using 1,000
mg/kg urethane, rats were sacrificed by cervical dislocation and
death was confirmed by absence of corneal reflex, spontaneous
breathing and heartbeat for a period of >5 min. The cartilage
tissues of rats in each group were collected for subsequent
experiments.
Cell culture and treatment
Human cartilage C28/I2 cells were provided by
American Type Culture Collection. The articular cartilage specimens
of rats were minced into small sections and then digested with 0.2%
type II collagenase (MilliporeSigma; Merck KGaA) in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) at 37˚C for 5-6 h. To separate the
rat chondrocytes, the cell suspension was centrifuged at 12,000 x g
for 10 min at 4˚C. C28/I2 cells and rat chondrocytes were incubated
in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS, 0.1
mg/ml streptomycin and 100 U/ml penicillin (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C with 5% CO2. To induce the
pathological condition of OA in cells, C28/I2 cells were treated
with 10 ng/ml IL-1β (Sigma-Aldrich; Merck KGaA) for 24 h at 37˚C.
Non-treated cells served as a control.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cartilage tissues
of patients with OA, control donors, cultured C28/I2 cells and the
cartilage tissues of rats in each group using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). High-Capacity
cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used to reverse transcribe total RNA to cDNA
according to the manufacturer's recommendations. After reverse
transcription, SYBR® Green Mix (Takara Bio, Inc.) was
used for qPCR with the Biosystems 7300 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR was
conducted at 95˚C for 10 min followed by 40 cycles of 95˚C for 30
sec and 60˚C for 1 min. The 2-ΔΔCt method was applied to
calculate the relative gene expression (25). Internal references were U6 and
GAPDH. The relative primer sequences are provided in Table I.
 | Table ISequences of primers used for reverse
transcription-quantitative PCR. |
Table I
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5'→3') |
|---|
| LINC00473 | F:
GGGAGCTTGAGCTGAGATGG |
| | R:
TTCGCAGTTTCCTAGTGGGAC |
| miR-424-5p | F:
CAGCAGCAATTCATGTTTTGAA |
| | R:
CTCTACAGCTATATTGCCAGCCAC |
| miR-15b-5p | F:
TAGCAGCACATCATGGTTTACA |
| | R:
CTCTACAGCTATATTGCCAGCCAC |
| miR-497-5p | F:
CAGCAGCACACTGTGGTTTG |
| | R:
CTCTACAGCTATATTGCCAGCCAC |
| miR-16-5p | F:
TAGCAGCACGTAAATATTGGCG |
| | R:
CTCTACAGCTATATTGCCAGCCAC |
| miR-6838-5p | F:
TAATCTCAGCTGGCAACTGTG |
| | R:
CTCTACAGCTATATTGCCAGCCAC |
| miR-195-5p | F:
TAGCAGCACAGAAATATTGGCG |
| | R:
CTCTACAGCTATATTGCCAGCCAC |
| miR-15a-5p | F:
TAGCAGCACATAATGGTTTGTGC |
| | R:
CTCTACAGCTATATTGCCAGCCAC |
| LY6E | F:
CTGCGTGACTGTGTCTGCTA |
| | R:
CCTGCATGGGAAATGAGGCT |
| GAPDH | F:
GAAGGTGAAGGTCGGAGTC |
| | R:
GAAGATGGTGATGGGATTTC |
| U6 | F:
GCTTCGGCAGCACATATACTAAAAT |
| | R:
CGCTTCACGAATTTGCGTGTCAT |
Cell transfection
Small interfering RNA (siRNA) against LINC00473
(si-LINC00473) and lymphocyte antigen 6 locus E (LY6E; si-LY6E)
with si-negative control (NC) as the control and pcDNA3.1/LY6E with
empty pcDNA3.1 vector as the control were synthesized by Shanghai
GenePharma Co., Ltd. miR-424-5p mimics, miR-424-5p inhibitor and
their corresponding controls (NC mimics and NC inhibitor) were
purchased from Shanghai GenePharma Co., Ltd. Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.) was
utilized to transfect these plasmids or vectors into normal or
IL-1β-stimulated C28/I2 cells (5x106 cells/well) for 48
h following the manufacturer's instructions. The concentration of
miR-424-5p mimics/inhibitors or NC mimics/inhibitors was 50 nM, and
that of siRNAs and vectors were 40 and 10 nM, respectively. The
transfection efficiency was examined by RT-qPCR after 48 h. The
sequences of all molecules used for cell transfection are presented
in Table II.
 | Table IISequences of molecules used for cell
transfection. |
Table II
Sequences of molecules used for cell
transfection.
| Molecule | Sense (5'-3') | Antisense
(5'-3') |
|---|
| si-LINC00473 |
CAAGCACUCAUGUUCUAAAGA |
UUUAGAACAUGAGUGCUUGUG |
| si-NC (for
si-LINC00473) |
GGACCGCUAAUCUAAUCAAUA |
UAUUGAUUAGAUUAGCGGUCC |
| si-LY6E |
GCUUGAACCAGAAGAGCAAUC |
UUGCUCUUCUGGUUCAAGCAG |
| si-NC (for
si-LY6E) |
AACGACCAUGAGAGUAGUCAC |
GUGACUACUCUCAUGGUCGUU |
| miR-424-5p
inhibitor |
UUCAAAACAUGAAUUGCUGCUG | - |
| NC inhibitor |
CAGUACUUUUGUGUAGCACAAA | - |
| miR-424-5p
mimics |
CAGCAGCAAUUCAUGUUUUGAA | - |
| NC mimics |
CAAAACAUGAAUUGCUGCUGUU | - |
Fluorescence in situ hybridization
(FISH)
LINC00473 probes were designed and synthesized by
Guangzhou RiboBio Co., Ltd. Probe signals were detected using an
RNA FISH Kit (Guangzhou RiboBio Co., Ltd.) following the
manufacturer's protocols. In brief, IL-1β-stimulated C28/I2 cells
were fixed in 4% formalin for 10 min at room temperature. After
permeabilization in PBS with 0.5% Triton X-100 at 4˚C for 20 min,
cells were dehydrated using 100% ethanol for 15 min at room
temperature. Subsequently, cells were incubated in 10 µl
hybridization solution (900 mM NaCI, 20 mM Tris/HCI, 20% formamide,
distilled H2O and 0.01% SDS) at 37˚C for 30 min and then
further incubated with LINC00473 probe (300 ng/ml) overnight at
4˚C. Afterwards, cells were washed twice with saline sodium for 10
min at 25˚C. The slides were incubated with tyramide signal
amplification (TSA) fluorescent signal reaction solution (cat. no.
NEL701001KT; PerkinElmer, Inc.) for 30 min and counterstained with
DAPI (Beyotime Institute of Biotechnology) at room temperature for
5 min following the manufacturer's protocol. Fluorescence
microscopy (Leica Microsystems GmbH) was used to capture images at
x200 magnification.
Luciferase reporter assay
The binding sites between LINC00473 and miR-424-5p
were predicted using starBase version 3.0 (http://starbase.sysu.edu.cn/). The sequences of
LINC00473 were cloned to the pmirGLO reporter plasmids (Promega
Corporation) to form wild-type (Wt; pmirGLO-LINC00473-Wt-1/2)
LINC00473. Mutated (Mut; pmirGLO-LINC00473-Mut-1/2) LINC00473
luciferase vectors were constructed using Phusion Site-Directed
Mutagenesis Kit (Thermo Fisher Scientific, Inc.). The pmirGLO
luciferase reporter vectors of the LY6E 3'untranslated region
(3'UTR) were obtained in the same way. C28/I2 cells were seeded
into 24-well plates (5x104 cells/well) at 37˚C
overnight. Subsequently, in the first test, the
pmirGLO-LINC00473-Wt-1/2 or pmirGLO-LINC00473-Mut-1/2 vectors were
co-transfected with NC mimics or miR-424-5p mimics into C28/I2
cells using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.). In the second test, the pmirGLO-LY6E
3'UTR-Wt or pmirGLO-LY6E 3'UTR-Mut vectors (Generay Biotech Co.,
Ltd.) were co-transfected with NC mimics or miR-424-5p mimics into
C28/I2 cells using Lipofectamine® 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.). The concentration of plasmids was
50 ng/well and that of miR-424-5p mimics or NC mimics was 20 nM.
After 48 h of transfection, the Dual-Luciferase Reporter Assay
system (Promega Corporation) was used to detect the luciferase
activity. The firefly luciferase activity was normalized to that of
Renilla luciferase.
RNA immunoprecipitation (RIP)
assay
The RIP assay was performed using the EZ-Magna RIP
Kit (MilliporeSigma) following the manufacturer's recommendations.
In brief, C28/I2 cells were lysed in RIP lysis buffer
(MilliporeSigma) containing protease inhibitor cocktail and RNase
inhibitor for 5 min of incubation on ice followed by centrifugation
at 10,000 x g for 10 min at 4˚C. Cell lysate was incubated with RIP
buffer containing Protein G magnetic beads (cat. no. 88848; Thermo
Fisher Scientific, Inc.) coated with Ago2 antibody (1:50; cat. no.
ab186733; Abcam) or the control IgG antibody (1:1,000; cat. no.
ab6702; Abcam) overnight at 4˚C according to the manufacturer's
recommendations. Next, the magnetic beads were washed three times
using RIP wash buffer and treated with proteinase K (Thermo Fisher
Scientific, Inc.) at 55˚C for 30 min. Finally, the co-precipitated
RNAs were measured using RT-qPCR.
Hematoxylin and eosin (H&E)
staining assay
The cartilage tissues from rats in the sham or OA
groups were fixed with 4% paraformaldehyde (Wuhan Boster Biological
Technology, Ltd.) at room temperature for 24 h, embedded in
paraffin, cut into 4-µm thick sections and visualized under light
microscope (Leica Microsystems GmbH) at x200 magnification after
H&E staining.
Flow cytometry analysis
Early and late apoptosis was evaluated by flow
cytometry using the Annexin V-Fluorescein Isothiocyanate (FITC)
Apoptosis Detection kit (MilliporeSigma). Briefly, transfected
C28/I2 cells (1x105 cells/well) were washed twice with
cold PBS and resuspended in binding buffer. Next, cells were
incubated with Annexin V-FITC and propidium iodide in the dark at
room temperature for 10 min. Stained cells were identified by a
FACScalibur flow cytometer (Becton, Dickinson and Company) analyzed
using FlowJo v.10 (FlowJo LLC).
Western blotting
Proteins were extracted from cartilage tissues of
rats in each group and cultured C28/I2 cells using RIPA lysis
buffer (Beyotime Institute of Biotechnology) containing
phenylmethylsulfonyl fluoride. The protein concentration was
quantified using a Bicinchoninic Acid Protein Assay kit (Beyotime
Institute of Biotechnology). After 25 µg protein/lane was separated
by 10% SDS-PAGE, proteins were transferred onto PVDF membranes
(MilliporeSigma). Next, the membranes were blocked at room
temperature for 2 h with 5% non-fat milk and then incubated
overnight at 4˚C with primary antibodies (all Abcam) targeted
against the following: Caspase-3 (cat. no. ab13847; 1:500), Bax
(cat. no. ab32503; 1:1,000), Bcl-2 (cat. no. ab196495; 1:1,000),
β-actin (cat. no. ab8227; 1:1,000), collagen II (cat. no. ab239007;
1:5,000), MMP1 (cat. no. ab134184; 1:1,000), MMP13 (cat. no.
ab39012; 1:3,000) and LY6E (cat. no. ab201098; 1:1,000). β-actin
was used as the internal control. After washing three times with
TBST containing 0.1% Tween 20, the membranes were further incubated
with goat anti-rabbit IgG H&L (HRP) secondary antibodies (cat.
no. ab97051; 1:10,000; Abcam) at room temperature for 2 h. Finally,
the immunoreactive bands were visualized using an ECL
chemiluminescent detection system (Thermo Fisher Scientific, Inc.)
and GeneGenius Gel Light Imaging system (Syngene) and analyzed
using ImageJ software (version 1.49; National Institutes of
Health).
ELISA
Blood was collected from the rats of each group and
sera was obtained after centrifugation at 6,000 x g for 15 min at
4˚C. The concentrations of inflammatory cytokines, including IL-6
(cat. no. ab234570; Abcam), IL-8 (cat. no. SEKR-0071; Solarbio) and
TNF-α (cat. no. ab236712; Abcam), were measured using ELISA kits
according to the manufacturer's protocols.
Statistical analysis
All experiments were repeated three times
independently. Experimental data are expressed as the mean ±
standard deviation (unless otherwise indicated). Statistical
analyses were performed using SPSS v19.0 software (IBM Corp.). The
independent Student's t-test was used to analyze the significant
differences between two groups. Statistical significance among
three or more groups was assessed using one-way ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Bioinformatics analysis
Seven candidate miRNAs (miR-424-5p, miR-15b-5p,
miR-497-5p, miR-16-5p, miR-6838-5p, miR-195-5p and miR-15a-5p) were
predicted using starBase v3.0 software (http://starbase.sysu.edu.cn/) under the condition of
cross linking and immunoprecipitation-sequence data (high
stringency ≥3). The downstream target gene of miR-424-5p, LY6E, was
also predicted from the starBase v3.0 website under the condition
of degradome data (low strigency ≥1) and the prediction program
tools PITA, RNA22 and PicTar.
Results
LINC00473 is upregulated in cartilage
tissues of patients with OA and IL-1β-treated chondrocytes
It has been previously reported that LINC00473 is
one of the candidate biomarkers for OA (18). To explore the role of LINC00473 in
the pathogenesis of OA, RT-qPCR analysis was conducted to examine
LINC00473 expression levels in the cartilage tissues of patients
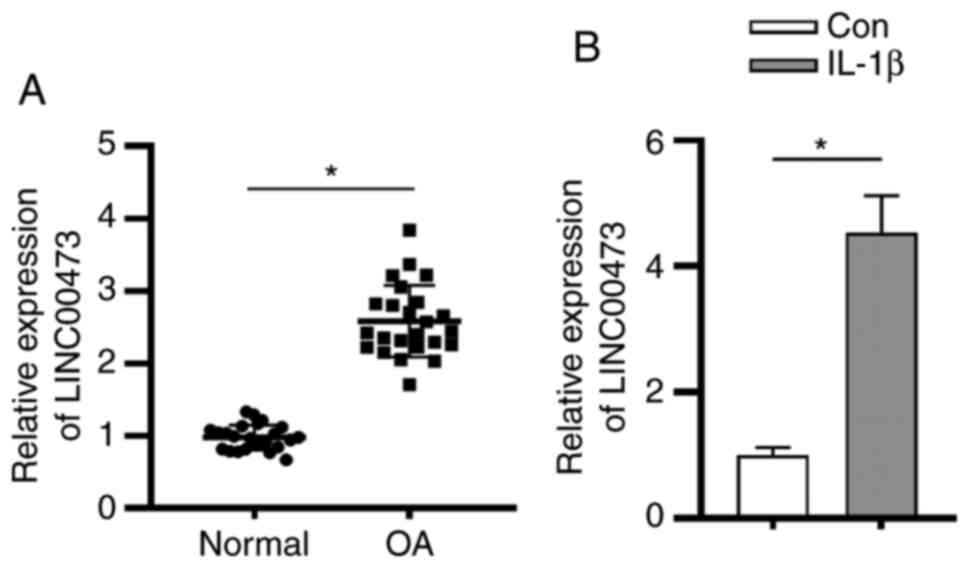
with OA. As presented in Fig. 1A,
LINC00473 expression was significantly upregulated in the cartilage
tissues of patients with OA compared with individuals without OA.
In accordance with previous studies (26,27),
the OA cell model was constructed using IL-1β to induce normal
human chondrocytes C28/I2. The present study revealed that the
expression level of LINC00473 was significantly higher in
IL-1β-stimulated C28/I2 cells compared with that in control C28/I2
cells (Fig. 1B).
LINC00473 knockdown reduces
chondrocyte apoptosis and the inflammatory response in
IL-1β-treated C28/I2 cells
Apoptosis and inflammation serve notable roles in OA
development (28). Based on the
aforementioned results, the effects of LINC00473 on cell injury in
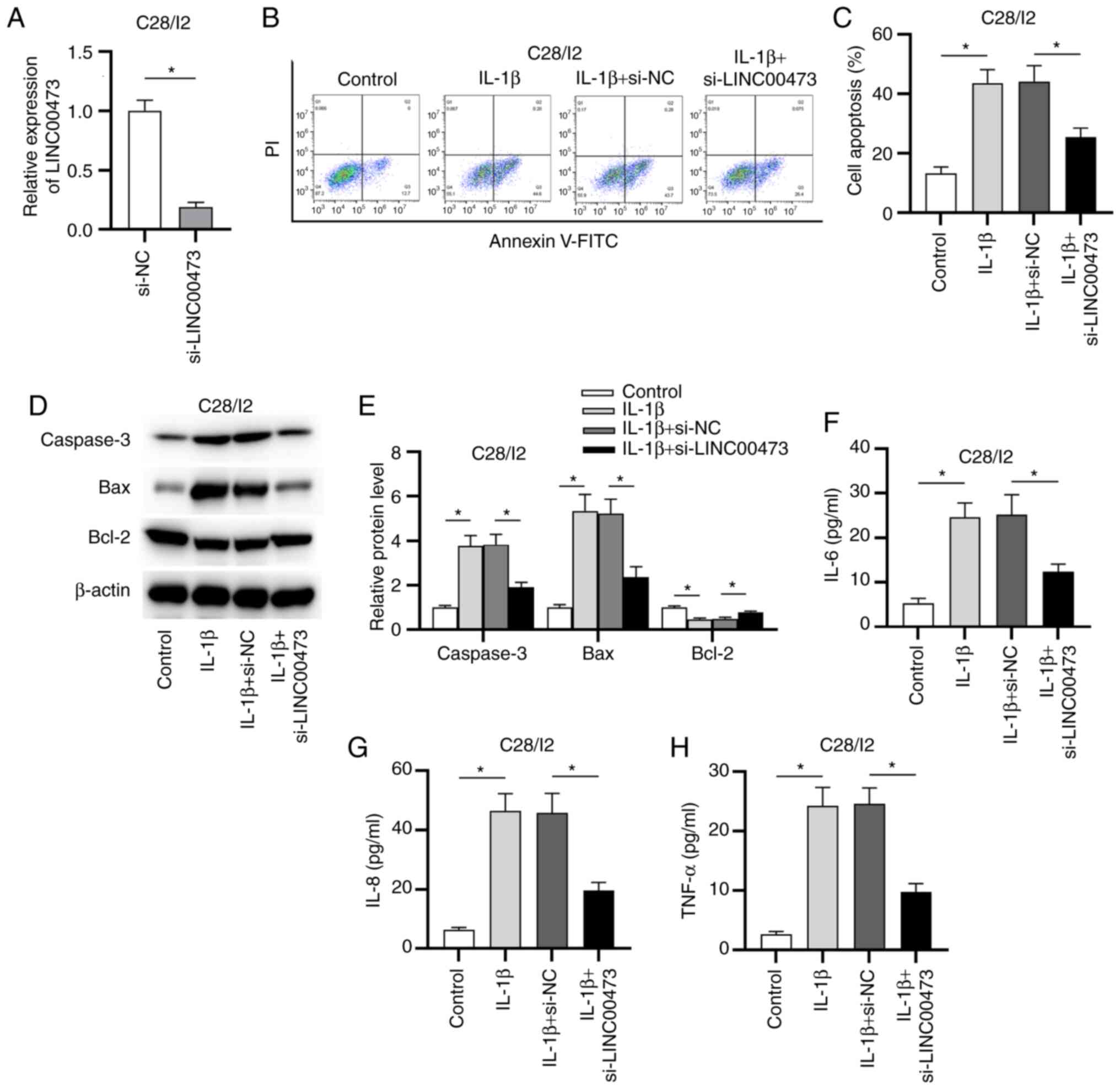
OA were investigated. As presented in Fig. 2A, the expression level of LINC00473
was significantly decreased in C28/I2 cells transfected with
si-LINC00473 compared with those transfected with si-NC. In the
subsequent experiments, C28/I2 cells were treated with 10 ng/ml
IL-1β to induce the pathological effects of OA, including
apoptosis. The flow cytometry results revealed that LINC00473
knockdown significantly inhibited the apoptosis of IL-1β-stimulated
C28/I2 cells compared with the IL-1β + si-NC group (Fig. 2B and C). Moreover, western blotting revealed
that the protein levels of caspase-3 and Bax were significantly
increased while Bcl-2 protein level was decreased after IL-1β
treatment. Compared with the IL-1β + si-NC group, transfection of
si-LINC00473 partially reversed IL-1β-induced promotive or
suppressive effect on protein levels of these apoptotic markers in
C28/I2 cells (Fig. 2D and E). These data further indicated that
LINC00473 knockdown decreased apoptosis in IL-1β-treated C28/I2
cells. Increasing evidence has demonstrated that inflammation
exacerbates the development of OA by producing excessive
proinflammatory cytokines in chondrocytes (29-31).
To verify whether LINC00473 influenced the release of
proinflammatory cytokines, ELISAs were performed to determine the
concentrations of IL-6, IL-8 and TNF-α released by IL-1β-treated
C28/I2 cells. As presented in Fig.
2F-H, the levels of these proinflammatory cytokines were
significantly increased in C28/I2 cells treated with IL-1β compared
with these in the control group. Furthermore, these effects were
partially inhibited by LINC00473 knockdown, as seen by the
significant decrease in the levels of these proteins in the IL-1β +
si-LINC00473 group compared with those in the IL-1β + si-NC
group.
LINC00473 knockdown alleviates OA
development in vivo
To further investigate the effect of LINC00473 on OA
development in vivo, an OA model was successfully
established in rats. H&E staining was conducted to determine
the pathological alterations to the knee joint in OA model rats. As
presented in Fig. 3A, sham rats
presented a smoothed cartilage surface, no cartilage sclerosis and
synovial hyperplasia. However, OA model rats manifested clear
hyperplasia, sclerosis and joint wear. Subsequently, RT-qPCR was
conducted to detect the expression of LINC00473 in vivo. As
presented in Fig. 3B, the
expression levels of LINC00473 were significantly higher in the
cartilage tissues of rats with OA compared with those of sham rats.
Compared with the OA + AAV si-NC group, expression of LINC00473 was
significantly decreased by the injection of AAV si-LINC00473. It
has been previously reported that the degradation of ECM was
closely associated with the destruction of joints in OA (32). To evaluate whether LINC00473
affected OA progression by promoting ECM degradation, the
expression levels of collagen II, MMP1 and MMP13 in the cartilage
tissue of rats in each group were measured (Fig. 3C and D). The data revealed that the protein
expression levels of MMP1 and MMP13 were significantly upregulated
in the cartilage tissues of OA model rats compared with the sham
group. In contrast to the OA + AAV si-NC group, the injection of
AAV si-LINC00473 significantly downregulated the protein expression
levels of MMP1 and MMP13 in the cartilage tissues of rats with OA.
However, the protein expression levels of collagen II indicated the
opposite trend. In addition, it was revealed that the
concentrations of proinflammatory cytokines, including IL-6, IL-8
and TNF-α, were significantly increased in the cartilage tissues of
OA model rats compared with sham rats. Moreover, LINC00473
knockdown in the OA + AAV si-LINC00473 group induced a significant
decrease in proinflammatory cytokine concentrations in the
cartilage tissues of rats with OA compared with the OA + AAV si-NC
group (Fig. 3E-G).
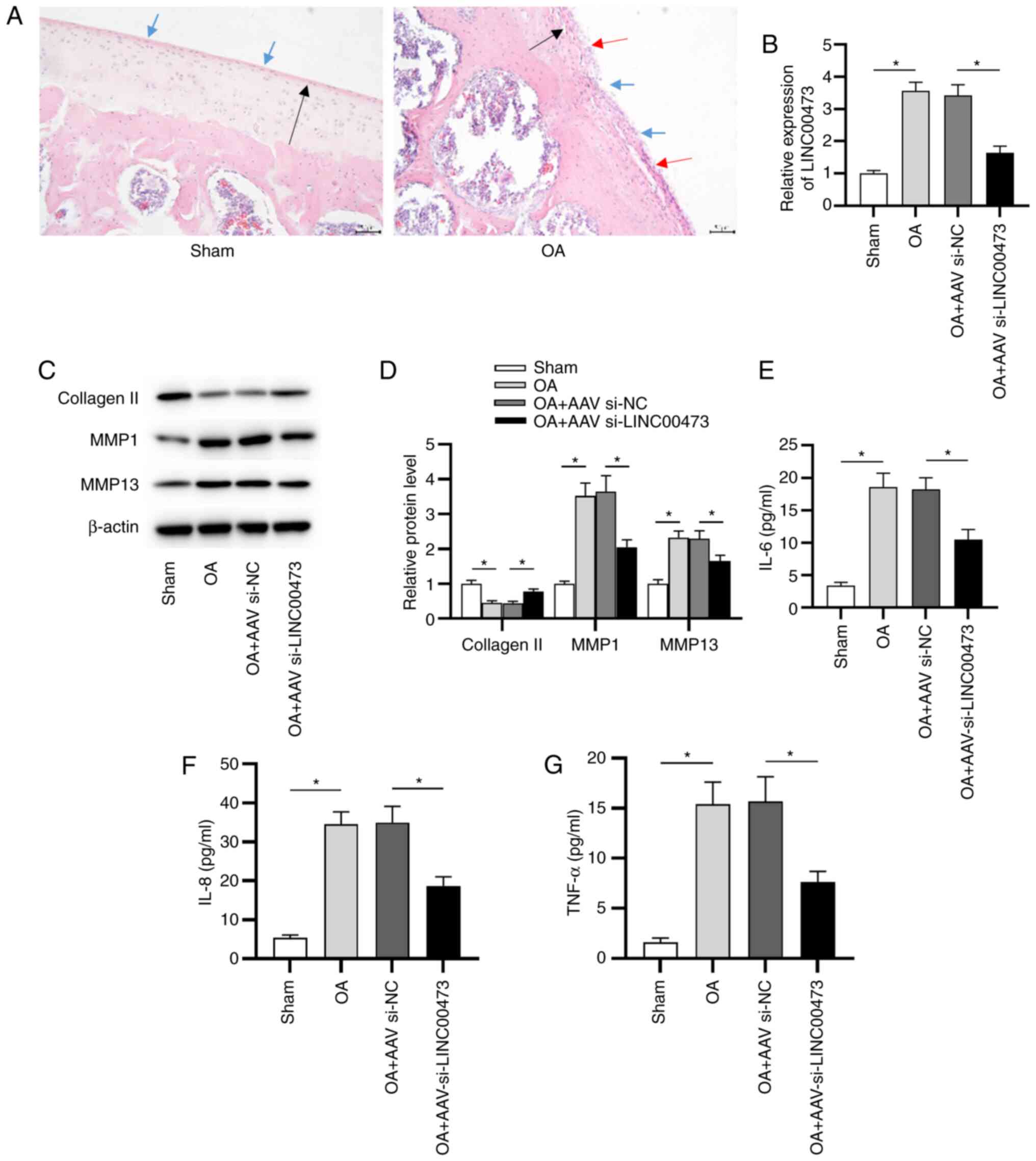 | Figure 3LINC00473 knockdown alleviates OA
development in vivo. (A) Pathological changes in the knee
joints of rats were observed using hematoxylin and eosin staining
(magnification, x200; scale bar, 50 µm). Black arrow indicates
thinning of articular cartilage tissue; blue arrow indicates the
damage of cartilage surface and the existence of cartilage debris;
red arrow indicates the formation of chondrocyte clusters around
the bare cartilage. (B) Expression of LINC00473 was detected in rat
cartilage tissues using reverse transcription-quantitative PCR.
Protein expression levels of collagen II, MMP1 and MMP13 were (C)
detected using western blotting and (D) semi-quantified after the
injection of AAV si-LINC00473. ELISAs were used to determine the
concentrations of proinflammatory cytokines (E) IL-6, (F) IL-8 and
(G) TNF-α in the cartilage tissues of rats from each group.
*P<0.05. LINC00473, long intergenic non-protein
coding RNA 473; OA, osteoarthritis; MMP, matrix metallopeptidase;
AAV, adeno-associated virus; si, small interfering RNA; NC,
negative control. |
LINC00473 acts as a sponge for
miR-424-5p
Previous studies have indicated that lncRNAs
participate in the pathogenesis of OA via the ceRNA pattern
(33,34). As a typical post-transcriptional
mechanism, the ceRNA pattern refers to the lncRNAs that compete
with mRNAs to bind with miRNAs, thus antagonizing the suppressive
effects of miRNAs on mRNAs (35).
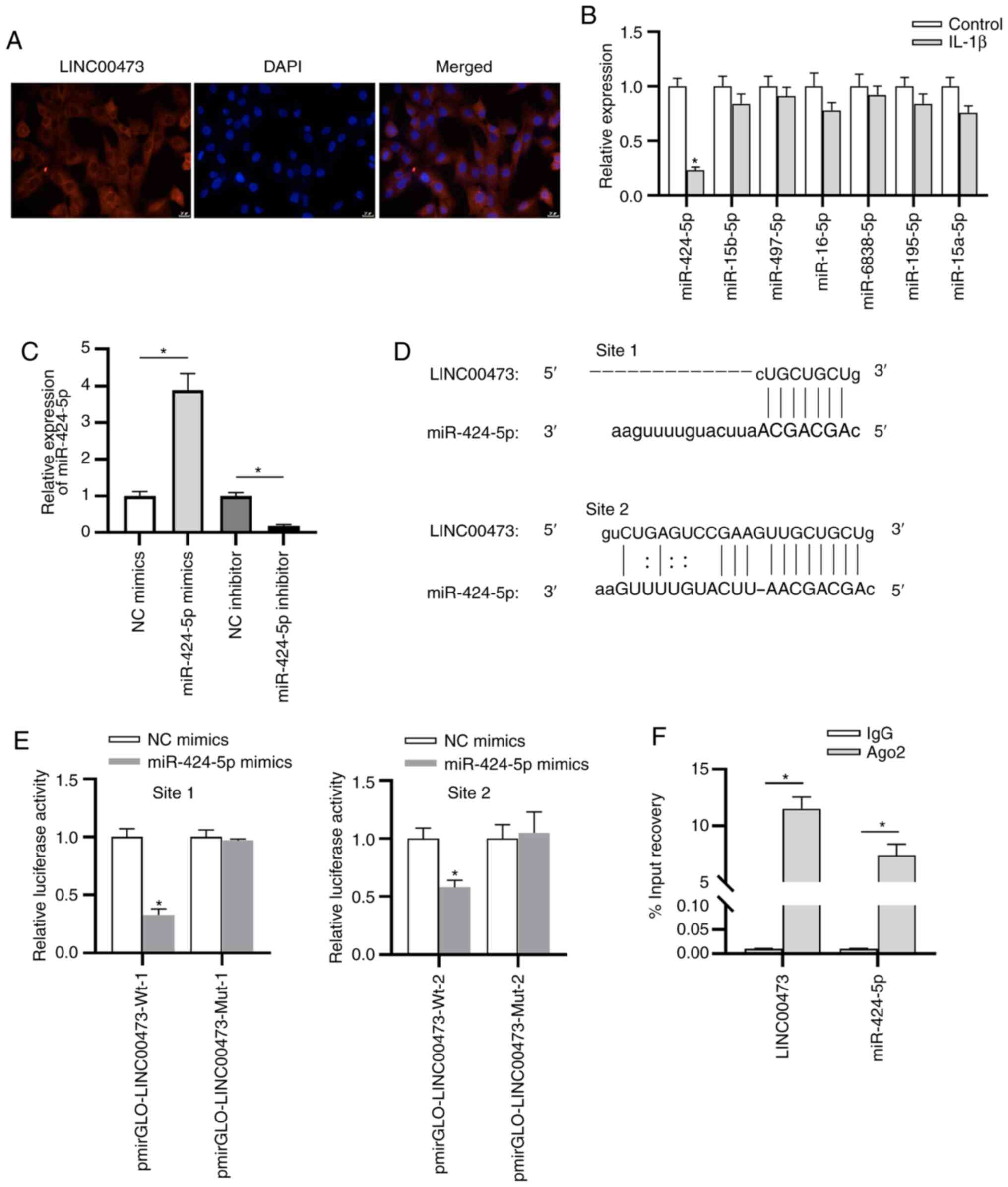
As presented in Fig. 4A, LINC00473
was primarily localized in the cytoplasm of cells, indicating the
post-transcriptional control of LINC00473 on gene expression in
C28/I2 cells. To further investigate the potential mechanism of
LINC00473 in C28/I2 cells, bioinformatics analysis was performed.
According to the prediction of starBase, seven candidate miRNAs
(miR-424-5p, miR-15b-5p, miR-497-5p, miR-16-5p, miR-6838-5p,
miR-195-5p and miR-15a-5p) were revealed to share potential a
binding site with LINC00473. Subsequently, RT-qPCR was performed to
estimate the expressions of these candidate miRNAs, the results of
which revealed that miR-424-5p demonstrated the most significant
downregulation in IL-1β-stimulated C28/I2 cells compared with the
control cells (Fig. 4B). Whether
LINC00473 interacted with miR-424-5p in C28/I2 cells was
subsequently investigated. As presented in Fig. 4C, compared with the NC groups,
miR-424-5p expression was significantly increased and decreased in
C28/I2 cells following transfection with miR-424-5p mimics or
miR-424-5p inhibitor, respectively. The two binding sites between
LINC00473 and miR-424-5p were predicted using the starBase database
(Fig. 4D). The luciferase reporter
assay revealed that the luciferase activity of pmirGLO-LINC00473-Wt
vectors at both site 1 and site 2 was significantly reduced by
miR-424-5p mimics compared with the NC mimic (Fig. 4E). By contrast, no difference was
identified in the LINC00473-Mut-1/2 group between the NC mimics and
miR-424-5p mimics groups, indicating that LINC00473 bound to
miR-424-5p at the predicted sites in C28/I2 cells. The results
implied that LINC00473 could bind with the two binding sites of
miR-424-5p in C28/I2 cells. In addition, the RIP assay elucidated
that LINC00473 and miR-424-5p were significantly enriched in
RNA-induced silencing complexes immunoprecipitated by anti-Ago2
compared with those immunoprecipitated by anti-IgG in C28/I2 cells
(Fig. 4F).
LY6E is a direct target of
miR-424-5p
Subsequently, the downstream target of miR-424-5p,
i.e., LY6E was identified according to the prediction from the
starBase website under the condition of degradome data Low
strigency (≥1) and the prediction program tools PITA, RNA22 and
PicTar (miRNA-targets predicting software). As determined by
RT-qPCR, the expression of LY6E was significantly increased in
C28/I2 cells stimulated with IL-1β compared with control cells
(Fig. 5A). As demonstrated in
Fig. 5B, miR-424-5p contained the
binding site on the 3'UTR of LY6E in C28/I2 cells. The luciferase
reporter assay revealed that the luciferase activity of the
pmirGLO-LY6E 3'UTR-Wt vector was significantly decreased by
miR-424-5p mimics transfection in C28/I2 cells compared with
transfection with NC mimics. However, no difference was revealed in
the luciferase activity of the pmirGLO-LY6E 3'UTR-Mut vector
between the NC mimics and miR-424-5p mimics groups in C28/I2 cells
(Fig. 5C). Additionally, both
miR-424-5p and LY6E were revealed to have a significantly increased
level of co-immunoprecipitation by Ago2 antibody compared with IgG
antibody in C28/I2 cells (Fig. 5D).
Moreover, the mRNA and protein expression levels of LY6E in C28/I2
cells were significantly decreased by miR-424-5p mimics
transfection or LINC00473 knockdown compared with the corresponding
NC groups (Fig. 5E-H). As presented
in Fig. 5I, LY6E expression in
C28/I2 cells was significantly decreased by the transfection of
si-LY6E compared with that of si-NC. The flow cytometry results
indicated that IL-1β promoted the apoptosis of C28/I2 cells
compared with the control group, and cell apoptosis was
significantly suppressed in IL-1β + si-LY6E group compared with
that in IL-1β + si-NC group (Fig.
5J and K). Furthermore, the
concentrations of proinflammatory cytokines (IL-6, IL-8 and TNF-α)
were significantly elevated in IL-1β-stimulated C28/I2 cells
compared with the control cells; whereas these effects were
partially reversed by LY6E knockdown (Fig. 5L-N).
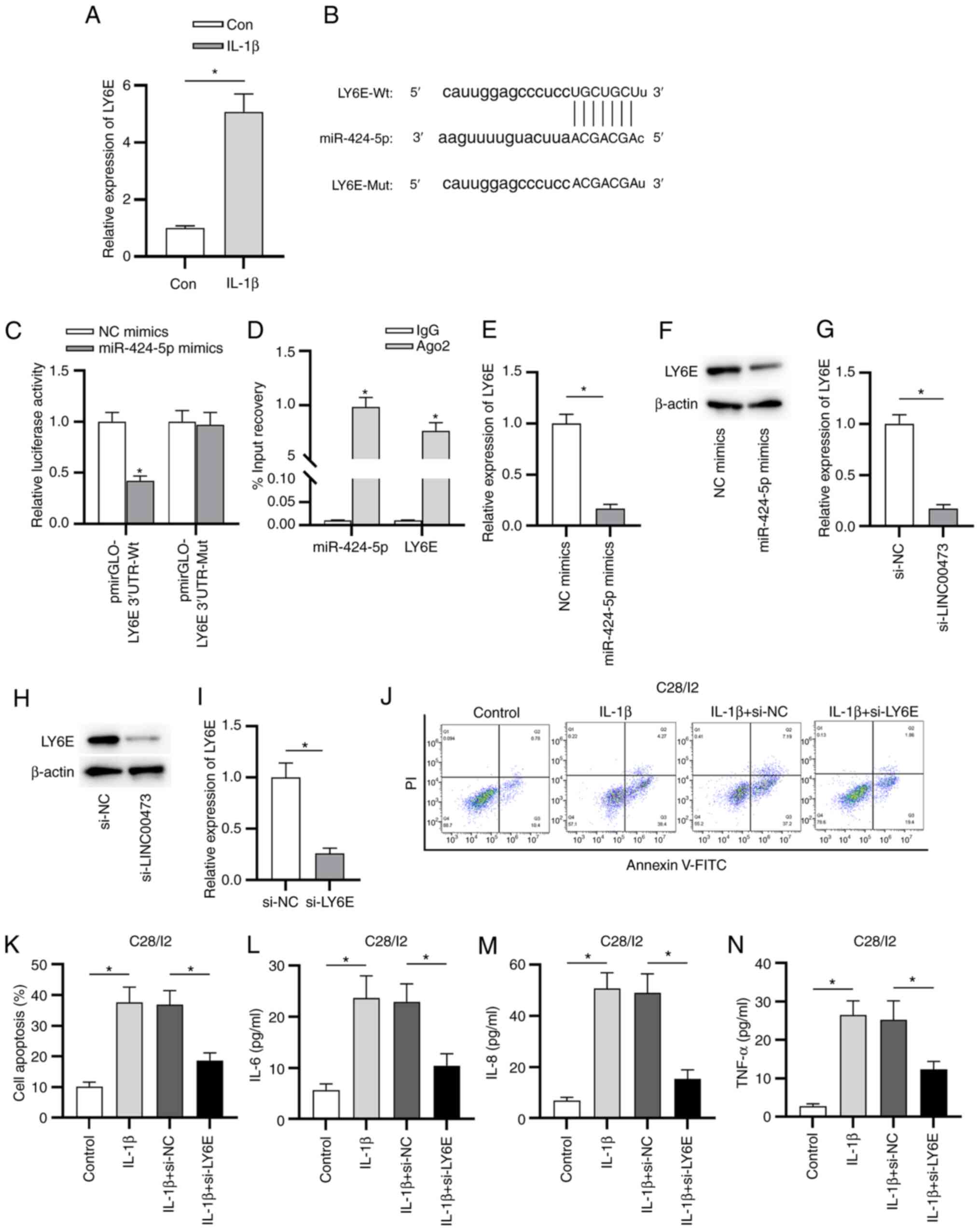 | Figure 5LY6E is a direct target of
miR-424-5p. (A) RT-qPCR analysis was used to evaluate LY6E
expression in IL-1β-stimulated C28/I2 cells. The binding capacity
between miR-424-5p and LY6E in C28/I2 cells was (B) predicted via
starBase and (C) verified by performing the luciferase reporter
assay. (D) Enrichment of miR-424-5p and LY6E in C28/I2 cells was
measured using an RNA immunoprecipitation assay. Effects of
miR-424-5p-overexpression on the (E) mRNA and (F) protein
expression levels of LY6E in C28/I2 cells. (G) mRNA and (H) protein
expression levels of LY6E were detected in si-LINC00473- or
si-NC-transfected C28/I2 cells using RT-qPCR and western blotting,
respectively. (I) RT-qPCR analysis was conducted to evaluate the
knockdown efficacy of si-LY6E in C28/I2 cells. (J and K) Effect of
LY6E knockdown on IL-1β-induced apoptosis in C28/I2 cells was
measured. ELISAs were used to determine the concentrations of
inflammatory cytokines (L) IL-6, (M) IL-8 and (N) TNF-α in
IL-1β-stimulated C28/I2 cells. *P<0.05. LY6E,
lymphocyte antigen 6 locus E; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; si, small interfering RNA;
LINC00473, long intergenic non-protein coding RNA 473; NC, negative
control; Wt, wild-type; Mut, mutant; Con, control; PI, propidium
iodide. |
LINC00473 exacerbates OA progression
by regulating LY6E
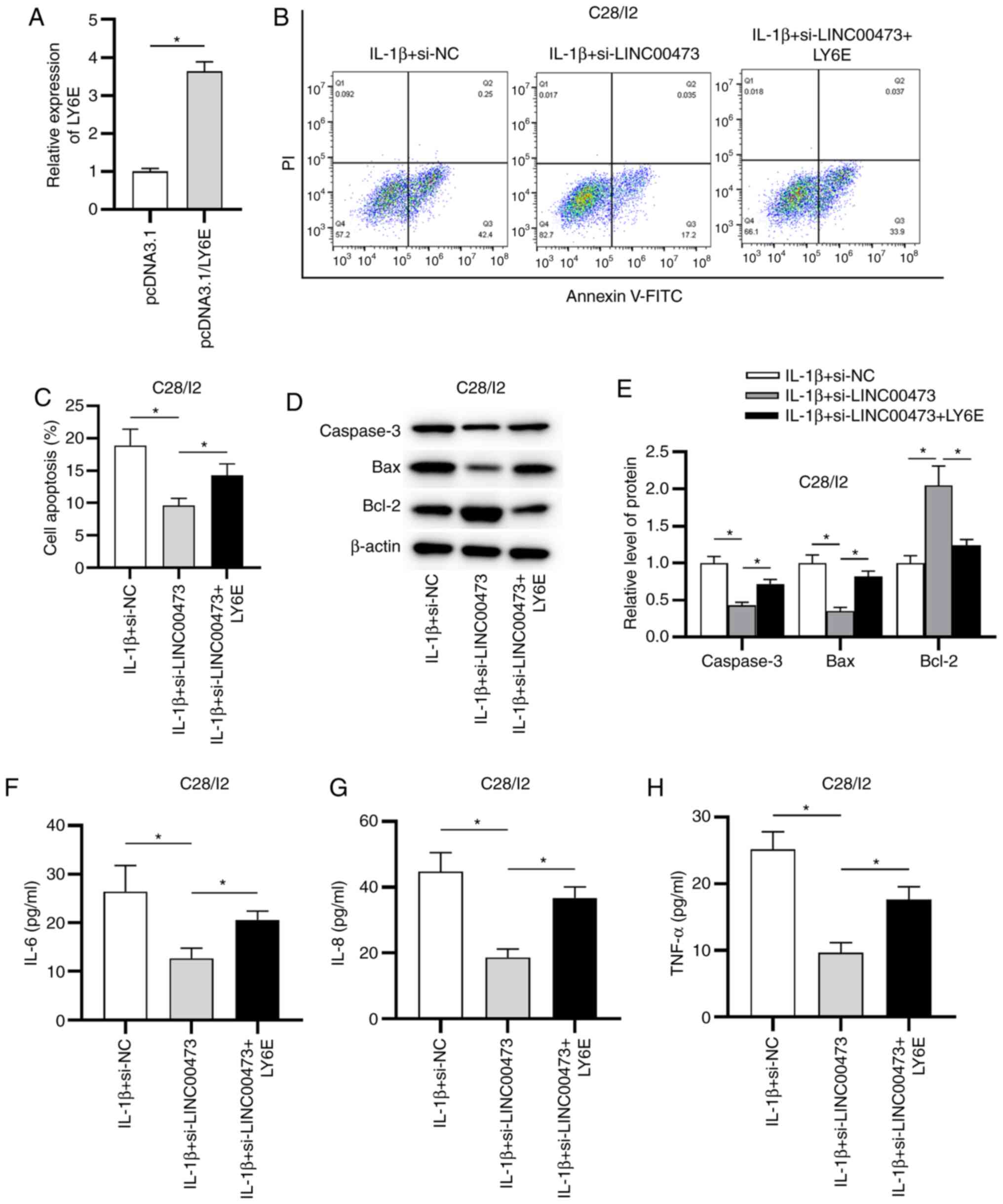
The follow-up rescue assays were performed to
determine whether LINC00473 regulated LY6E to induce OA
development. The overexpression efficacy of LY6E in
IL-1β-stimulated C28/I2 cells was verified by RT-qPCR. The results
revealed that the expression of LY6E was significantly elevated in
C28/I2 cells transfected with pcDNA3.1/LY6E compared with the empty
vector (Fig. 6A). As presented in
Fig. 6B and C, LINC00473 knockdown suppressed the
apoptosis of IL-1β-treated C28/I2 cells compared with the IL-1β +
si-NC group, while LY6E overexpression partially reversed LINC00473
knockdown-mediated inhibitory effect on the apoptosis of
IL-1β-treated C28/I2 cells. In addition, the protein expression
levels of caspase-3 and Bax were significantly decreased in the
IL-1β + si-LINC00473 group compared with the si-NC, whereas Bcl-2
expression was significantly increased. However, these effects were
significantly offset by LY6E overexpression in IL-1β-stimulated
C28/I2 cells (Fig. 6D and E). Furthermore, the results demonstrated
that LY6E overexpression partially reversed the suppressive effects
of LINC00473 knockdown on the production of inflammatory cytokines
(IL-6, IL-8 and TNF-α) by IL-1β-stimulated C28/I2 cells (Fig. 6F-H).
Discussion
OA is a chronic disease that is characterized by
several features, such as cartilage degradation and synovial
inflammation (30). The cumulative
incidence of symptomatic knee OA among middle-aged and older
Chinese adults was 8.5%, and the incidence among females (11.2%)
was higher than that among males (5.6%) based on data collected
from China Health and Retirement Longitudinal Study, which is a
national population survey with a 4-year follow-up, between 2011
and 2015(36). Over the next few
decades, the incidence of OA is expected to continuously rise,
which may have a significant impact on the daily activities of
individuals (37). Current medical
therapy of OA primarily focuses on pain relief (38). However, there are still a number of
challenges, including the lack of disease-modifying drugs,
therapeutic efficacy of anti-inflammatory drugs and the safety of
new therapies, in the treatment of OA (39).
It is known that lncRNAs are non-coding segments of
RNAs that have notable regulatory functions in the
pathophysiological processes of multiple diseases, including OA
(13-15).
LINC00473, a novel lncRNA, has been revealed to be upregulated in
several diseases, such as ischemic stroke and preeclampsia
(16,17). Moreover, LINC00473 is upregulated in
patients with severe pain associated with OA (18). Consistent with the high expression
of LINC00473 in ischemic stroke (16), preeclampsia (17) and patients diagnosed with OA
(18), the current study revealed
that LINC00473 was highly expressed in the cartilage tissue of
patients with OA, in IL-1β-treated C28/I2 cells and in the
cartilage tissues of OA model rats.
Accumulating evidence has suggested that the
pathogenesis of OA is complex and is usually associated with
chondrocyte apoptosis, inflammation and ECM degeneration (40,41).
For example, hypoxia-inducible factor-2α aggravated cartilage
destruction by enhancing chondrocyte apoptosis in OA (42). Furthermore, cartilage injury-related
lncRNAs facilitated the degradation of ECM in OA progression
(43). MALAT1 also regulated the
inflammation of synovial fibroblasts in obese patients and those
with OA (44). Thus, further
determining the mechanism of LINC00473 may be useful to elucidate a
novel therapeutic strategy for OA. The present study revealed that
LINC00473 knockdown efficiently alleviated OA development in
vitro and in vivo by decreasing chondrocyte apoptosis,
degradation of the ECM and production of proinflammatory
cytokines.
miRNAs are a series of endogenous small non-coding
RNAs that have been reported to be involved in numerous diseases
(45). For example, miR-155
exacerbated pulmonary inflammation induced by cigarette smoke and
the development of chronic obstructive pulmonary disease (46). miR-182 served a protective role in
hypoxia-induced cardiomyocytes by targeting hes family bHLH
transcription factor 1(47).
Additionally, miR-30a served as a virulence miRNA in OA development
by promoting ECM degradation (48).
Emerging evidence has revealed that miRNAs exert their biological
functions by interacting with lncRNAs in OA (49-51).
In the present study, two binding sites between miR-424-5p and
LINC00473 were predicted by bioinformatics analysis. The binding
ability between them was subsequently verified, indicating that
LINC00473 functioned as a sponge for miR-424-5p in C28/I2 cells.
Furthermore, a number of studies have demonstrated that miRNAs
modulate the expression levels of specific genes in diseases
(47,52,53).
As a key member of the lymphostromal cell membrane
Ly6 protein superfamily, LY6E serves a notable role in
immunological regulation and oncogenesis (54). Previous studies have suggested that
LY6E participates in the progression of several diseases (55-57).
For example, LY6E expression could function as a biomarker in the
diagnosis of systemic lupus erythematosus (55,56).
Using RNA sequencing, LY6E was also revealed to be involved in
ischemic stroke (57). However, to
the best of our knowledge, the role of LY6E has not been identified
in OA. The present study revealed that miR-424-5p had binding sites
on the 3'UTR of LY6E and further proved the binding capability
between miR-424-5p and LY6E. Moreover, LY6E knockdown significantly
inhibited IL-1β-induced apoptosis and proinflammatory cytokine
production in C28/I2 cells. In addition, rescue experiments
revealed that LY6E overexpression significantly reversed the
inhibitory effects of LINC00473 knockdown on chondrocyte apoptosis
and proinflammatory cytokine production. These data indicated that
LINC00473 may be involved in OA pathogenesis via dysregulated ceRNA
interactions.
In summary, the present study revealed that
LINC00473 knockdown suppressed IL-1β-induced apoptosis and
production of proinflammatory cytokines in C28/I2 cells via the
miR-424-5p/LY6E axis. The results may provide a novel perspective
for the improved understanding of the pathogenesis of OA, and may
also improve the treatment of OA.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GF and XG designed the research. GF and JL performed
the research. GF and YZ analyzed the data. GF and XG wrote the
paper. GF and XG confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by The Human Ethics
Committee of Children's Hospital of Soochow University (Jiangsu,
China; approval number, KS20190036). Informed consent was obtained
from all participants included in the current study. All procedures
performed in studies involving human participants were in
accordance with the ethical standards of the institutional and/or
national research committee and with the 1964 Declaration of
Helsinki and its later amendments or comparable ethical standards.
The animal experiments were approved by The Attitude of the Animal
Care & Welfare Committee of Children's Hospital of Soochow
University (Jiangsu, China; approval no. 2019-044).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Palazzo C, Nguyen C, Lefevre-Colau MM,
Rannou F and Poiraudeau S: Risk factors and burden of
osteoarthritis. Ann Phys Rehabil Med. 59:134–138. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Liu-Bryan R: Inflammation and
intracellular metabolism: New targets in OA. Osteoarthritis
Cartilage. 23:1835–1842. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Maldonado M and Nam J: The role of changes
in extracellular matrix of cartilage in the presence of
inflammation on the pathology of osteoarthritis. Biomed Res Int.
2013(284873)2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Perez Martin A: Symptoms. Localizations:
Knee, hip, hands, spine, other localizations. Aten Primaria. 46
(Suppl 1):S11–S17. 2014.PubMed/NCBI View Article : Google Scholar : (In Spanish).
|
|
5
|
Loeser RF, Goldring SR, Scanzello CR and
Goldring MB: Osteoarthritis: A disease of the joint as an organ.
Arthritis Rheum. 64:1697–1707. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Majeed MH, Sherazi SAA, Bacon D and Bajwa
ZH: Pharmacological treatment of pain in osteoarthritis: A
descriptive review. Curr Rheumatol Rep. 20(88)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu YK, Zhu FG, Feng EY, Fan CL, Yu GS, Wei
YZ and Xie LH: External therapies of traditional Chinese medicine
combined with sodium hyaluronate injected in articular cavity
therapy on knee osteoarthritis: Meta-analysis. Zhongguo Zhong Yao
Za Zhi. 43:1934–1939. 2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
8
|
Richards MM, Maxwell JS, Weng L, Angelos
MG and Golzarian J: Intra-articular treatment of knee
osteoarthritis: From anti-inflammatories to products of
regenerative medicine. Phys Sportsmed. 44:101–108. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA-LncRNA interactions. Methods Mol Biol.
1402:271–286. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kazemzadeh M, Safaralizadeh R and Orang
AV: LncRNAs: Emerging players in gene regulation and disease
pathogenesis. J Genet. 94:771–784. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mathieu EL, Belhocine M, Dao LT, Puthier D
and Spicuglia S: Functions of lncRNA in development and diseases.
Med Sci (Paris). 30:790–796. 2014.PubMed/NCBI View Article : Google Scholar : (In French).
|
|
12
|
Xie F, Liu YL, Chen XY, Li Q, Zhong J, Dai
BY, Shao XF and Wu GB: Role of MicroRNA, LncRNA, and exosomes in
the progression of osteoarthritis: A review of recent literature.
Orthop Surg. 12:708–716. 2020.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Zhou Q, Huang XR, Yu J, Yu X and Lan HY:
Long noncoding RNA Arid2-IR is a novel therapeutic target for renal
inflammation. Mol Ther. 23:1034–1043. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cai L, Tu L, Li T, Yang X, Ren Y, Gu R,
Zhang Q, Yao H, Qu X, Wang Q and Tian J: Downregulation of lncRNA
UCA1 ameliorates the damage of dopaminergic neurons, reduces
oxidative stress and inflammation in Parkinson's disease through
the inhibition of the PI3K/Akt signaling pathway. Int
Immunopharmacol. 75(105734)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhang Y, Wang F, Chen G, He R and Yang L:
LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3
axis. Cell Biosci. 9(54)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zheng Y, Sun S, Yu M and Fu X:
Identification of potential hub-lncRNAs in ischemic stroke based on
Subpathway-LNCE method. J Cell Biochemistry. 120:12832–12842.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu D, Xu Y, Zou Y, Zuo Q, Huang S, Wang S,
Lu X, He X, Wang J, Wang T and Sun L: Long noncoding RNA 00473 is
involved in preeclampsia by LSD1 binding-regulated TFPI2
transcription in trophoblast cells. Mol Ther Nucleic Acids.
12:381–392. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen Y, Lin Y, Bai Y, Cheng D and Bi Z: A
long noncoding RNA (lncRNA)-associated competing endogenous RNA
(ceRNA) network identifies eight lncRNA biomarkers in patients with
osteoarthritis of the knee. Med Sci Monit. 25:2058–2065.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kong F, Jin J, Lv X, Han Y, Liang X, Gao Y
and Duan X: Long noncoding RNA RMRP upregulation aggravates
myocardial ischemia-reperfusion injury by sponging miR-206 to
target ATG3 expression. Biomed Pharmacother. 109:716–725.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li X, Zeng L, Cao C, Lu C, Lian W, Han J,
Zhang X, Zhang J, Tang T and Li M: Long noncoding RNA MALAT1
regulates renal tubular epithelial pyroptosis by modulated miR-23c
targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res.
350:327–335. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shen H, Wang Y, Shi W, Sun G, Hong L and
Zhang Y: LncRNA SNHG5/miR-26a/SOX2 signal axis enhances
proliferation of chondrocyte in osteoarthritis. Acta Biochim
Biophys Sin (Shanghai). 50:191–198. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Glyn-Jones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals. 8th
edition. Washington, DC, National Academies Press (US), 2011.
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang Y, Shen S, Li Z, Li W and Weng X:
MIR-140-5p affects chondrocyte proliferation, apoptosis, and
inflammation by targeting HMGB1 in osteoarthritis. Inflamm Res.
69:63–73. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cheng C, Shan W, Huang W, Ding Z, Cui G,
Liu F, Lu W, Xu J, He W and Yin Z: ACY-1215 exhibits
anti-inflammatory and chondroprotective effects in human
osteoarthritis chondrocytes via inhibition of STAT3 and NF-κB
signaling pathways. Biomed Pharmacother. 109:2464–2471.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang Y, Yu T, Jin H, Zhao C and Wang Y:
Knockdown MiR-302b alleviates LPS-induced injury by targeting Smad3
in C28/I2 chondrocytic cells. Cell Physiol Biochem. 45:733–743.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fernandes JC, Martel-Pelletier J and
Pelletier JP: The role of cytokines in osteoarthritis
pathophysiology. Biorheology. 39:237–246. 2002.PubMed/NCBI
|
|
30
|
Charlier E, Deroyer C, Ciregia F, Malaise
O, Neuville S, Plener Z, Malaise M and de Seny D: Chondrocyte
dedifferentiation and osteoarthritis (OA). Biochem Pharmacol.
165:49–65. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Goldring MB and Otero M: Inflammation in
osteoarthritis. Curr Opin Rheumatol. 23:471–478. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang Y, Lin J, Zhou X, Chen X, Chen AC,
Pi B, Pan G, Pei M, Yang H, Liu T and He F: Melatonin prevents
osteoarthritis-induced cartilage degradation via targeting
MicroRNA-140. Oxid Med Cell Longev. 2019(9705929)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang P, Sun J, Liang C, Gu B, Xu Y, Lu H,
Cao B and Xu H: lncRNA IGHC γ Acts as a ceRNA to regulate
macrophage inflammation via the miR-6891-3p/TLR4 axis in
osteoarthritis. Mediators Inflamm. 2020(9743037)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen H and Chen L: An integrated analysis
of the competing endogenous RNA network and co-expression network
revealed seven hub long non-coding RNAs in osteoarthritis. Bone
Joint Res. 9:90–98. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tay Y, Rinn J and Pandolfi PJN: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ren Y, Hu J, Tan J, Tang X, Li Q, Yang H,
Liu C, He Q, Zou K, Sun X and Tan B: Incidence and risk factors of
symptomatic knee osteoarthritis among the Chinese population:
Analysis from a nationwide longitudinal study. BMC Public Health.
20(1491)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Callahan LF, Ambrose KR, Albright AL,
Altpeter M, Golightly YM, Huffman KF, Nelson AE and Weisner SE:
Public Health interventions for osteoarthritis-updates on the
osteoarthritis Action Alliance's efforts to address the 2010 OA
Public Health Agenda Recommendations. Clin Exp Rheumatol. 37 (Suppl
120):S31–S39. 2019.PubMed/NCBI
|
|
38
|
Vargas Negrin F, Medina Abellan MD,
Hermosa Hernan JC and de Felipe Medina R: Treatment of patients
with osteoarthritis. Aten Primaria. 46 (Suppl 1):S39–S61.
2014.PubMed/NCBI View Article : Google Scholar : (In Spanish).
|
|
39
|
Hermann W, Lambova S and Muller-Ladner U:
Current treatment options for osteoarthritis. Curr Rheumatol Rev.
14:108–116. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hwang HS and Kim HA: Chondrocyte apoptosis
in the pathogenesis of osteoarthritis. Int J Mol Sci.
16:26035–26054. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Minguzzi M, Cetrullo S, D'Adamo S,
Silvestri Y, Flamigni F and Borzi RM: Emerging players at the
intersection of chondrocyte loss of maturational arrest, oxidative
stress, senescence and low-grade inflammation in osteoarthritis.
Oxid Med Cell Longev. 2018(3075293)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Ryu JH, Shin Y, Huh YH, Yang S, Chun CH
and Chun JS: Hypoxia-inducible factor-2alpha regulates Fas-mediated
chondrocyte apoptosis during osteoarthritic cartilage destruction.
Cell Death Differ. 19:440–450. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L,
Zhou C and Ao Y: Long noncoding RNA related to cartilage injury
promotes chondrocyte extracellular matrix degradation in
osteoarthritis. Arthritis Rheumatol. 66:969–978. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Nanus DE, Wijesinghe SN, Pearson MJ,
Hadjicharalambous MR, Rosser A, Davis ET, Lindsay MA and Jones SW:
Obese osteoarthritis patients exhibit an inflammatory synovial
fibroblast phenotype, which is regulated by the long non coding RNA
MALAT1. Arthritis Rheumatol. 72:609–619. 2019.
|
|
45
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
De Smet EG, Van Eeckhoutte HP, Avila Cobos
F, Blomme E, Verhamme FM, Provoost S, Verleden SE, Venken K, Maes
T, Joos GF, et al: The role of miR-155 in cigarette smoke-induced
pulmonary inflammation and COPD. Mucosal Immunol. 13:423–436.
2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang Y, Peng B and Han Y: MiR-182
alleviates the development of cyanotic congenital heart disease by
suppressing HES1. Eur J Pharmacol. 836:18–24. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chang T, Xie J, Li H, Li D, Liu P and Hu
Y: MicroRNA-30a promotes extracellular matrix degradation in
articular cartilage via downregulation of Sox9. Cell Prolif.
49:207–218. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zhang G, Wu Y, Xu D and Yan X: Long
Noncoding RNA UFC1 promotes proliferation of chondrocyte in
osteoarthritis by acting as a sponge for miR-34a. DNA Cell Biol.
35:691–695. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li Y, Li S, Luo Y, Liu Y and Yu N: LncRNA
PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as
a sponge for miR-488-3p. DNA Cell Biol. 36:571–580. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Li YF, Li SH, Liu Y and Luo YT: Long
noncoding RNA CIR promotes chondrocyte extracellular matrix
degradation in osteoarthritis by acting as a sponge For Mir-27b.
Cell Physiol Biochem. 43:602–610. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Pan W, Yu H, Zheng B, Gao Y, Li P, Huang
Q, Xie C and Ge X: Upregulation of MiR-369-3p suppresses cell
migration and proliferation by targeting SOX4 in Hirschsprung's
disease. J Pediatr Surg. 52:1363–1370. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen L, Li Q, Wang J, Jin S, Zheng H, Lin
J, He F, Zhang H, Ma S, Mei J and Yu J: MiR-29b-3p promotes
chondrocyte apoptosis and facilitates the occurrence and
development of osteoarthritis by targeting PGRN. J Cell Mol Med.
21:3347–3359. 2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Yu J and Liu SL: Emerging role of LY6E in
virus-host interactions. Viruses. 11(1020)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Feng X, Huang J, Liu Y, Xiao L, Wang D,
Hua B, Tsao BP and Sun L: Identification of interferon-inducible
genes as diagnostic biomarker for systemic lupus erythematosus.
Clin Rheumatol. 34:71–79. 2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Komatsuda A, Wakui H, Iwamoto K, Ozawa M,
Togashi M, Masai R, Maki N, Hatakeyama T and Sawada K: Up-regulated
expression of Toll-like receptors mRNAs in peripheral blood
mononuclear cells from patients with systemic lupus erythematosus.
Clin Exp Immunol. 152:482–487. 2008.PubMed/NCBI View Article : Google Scholar
|
|
57
|
He W, Wei D, Cai Chen S, Li S and Chen W:
Altered long non-coding RNA Transcriptomic Profiles in Ischemic
Stroke. Hum Gene Ther. 29:719–732. 2018.PubMed/NCBI View Article : Google Scholar
|