Introduction
In December 2019, an unexplained viral pneumonia,
now known to be part of the pathology of coronavirus disease 2019
(COVID-19), emerged in Wuhan (China) (1,2). The
common clinical manifestations were fever, cough and regions of
ground-glass opacity on chest computed tomography (CT) scans
(3).
No effective medical treatment exists for the early
stages of COVID-19 (4-6).
Supportive care has been indicated to be the most effective
strategy during the COVID-19 outbreak. Since 90% of cases of severe
acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection are
asymptomatic, it is important to evaluate the risk of patients with
SARS-CoV-2 infection regarding the progression to severe forms for
prompt individual treatment and medical resource management, which
may prevent imposing restrictions on whole populations and
facilitate the identification of high-risk populations (7,8).
Therefore, the approach employed towards the risk assessment of
severe SARS-CoV-2 infection is of critical significance for the
effective treatment of COVID-19.
Patients were reported to have recovered from
COVID-19, but in numerous cases, a subsequent PCR test indicated
SARS-CoV-2 nucleic acid-positive results (9-11).
Re-positive patients usually have no or mild clinical symptoms;
however, their health status, infectivity and the mechanisms of
acquiring re-positivity remain elusive (12,13).
In the course of COVID-19 treatment, numerous patients test
false-negative, but there appear to be many and complex influencing
factors interfering with these results (14,15).
Clinical practice guidelines recommend repeated PCR testing to
confirm the clinical diagnosis (16). Research on COVID-19 patient
populations with re-positive or false-negative test results is
still limited and no relevant reference clinical risk assessment
indicators exist for re-positive and false-negative patients.
Furthermore, the possible infection and replication patterns of
SARS-CoV-2 in humans have remained elusive. To explore the clinical
characteristics and risk factors associated with acquiring
re-positive or false-negative SARS-CoV-2 test results, a
cross-sectional observational study of hospitalized patients with
COVID-19 was performed.
Materials and methods
Study design and participants
A cross-sectional observational study was performed
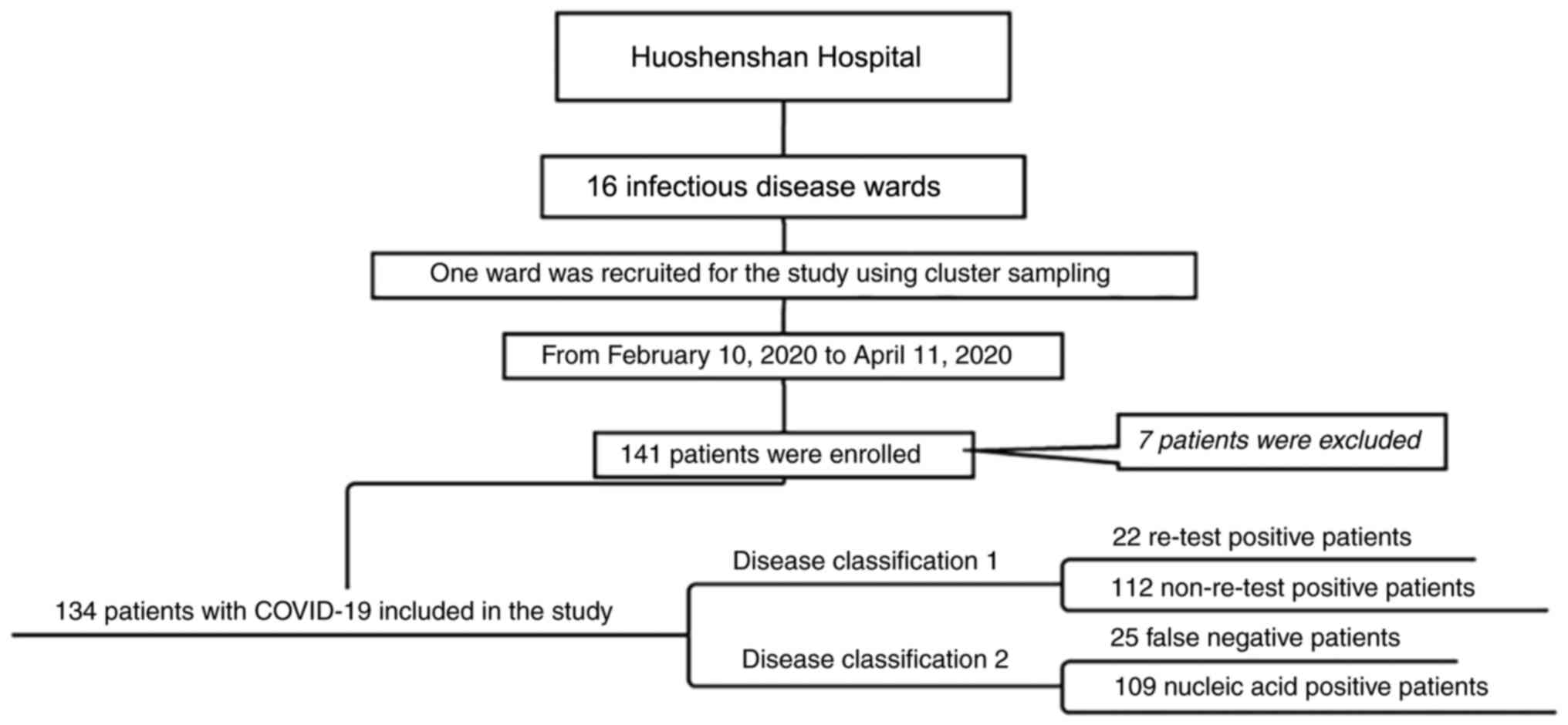
at Wuhan Huoshenshan Hospital (Wuhan, China). The flowchart of the
study is provided in Fig. 1. The
Wuhan Municipal Government assigned COVID-19 patients to
Huoshenshan Hospital (Wuhan, China) and this Hospital then randomly
assigned these patients to 16 infectious disease wards. To study
the disease characteristics of patients with COVID-19, one specific
ward was selected. The present study included patients hospitalized
between February 2020 and April 2020 with fever, respiratory
symptoms, and chest CT scans indicating pneumonia (Fig. S1) (17). According to the Chinese Management
Guidelines for COVID-19 (version 7.0) (17), suspected cases with one of the
following etiological or serological forms of evidence were
diagnosed as having COVID-19 infection: i) Positive real-time
fluorescent quantitative PCR (qPCR) detection of SARS-CoV-2 nucleic
acids; ii) viral gene sequencing indicating high homology with
SARS-CoV-2; iii) positive detection of SARS-CoV-2-specific IgM/IgG
antibodies in serum on admission. When none of these three
conditions was met, the patient was excluded. The hospital's
workflow pattern is outlined in Data
S1 and Fig. S2, Fig. S3, Fig.
S4 and Fig. S5. A total number
of 134 patients in the ward were diagnosed with COVID-19 and were
finally enrolled in the present study.
The present retrospective study was reviewed and
approved by the Ethics Committee of Huoshenshan Hospital (Wuhan,
China; approval no. HSSLL032) and written informed consent was
obtained from each enrolled patient or a first-degree relative. The
nucleic acid test of patients with COVID-19 who were admitted for
the first time was positive. After systematic treatment, their
clinical symptoms improved and at least two nucleic acid tests were
negative. However, after a few days, their clinical symptoms
worsened and at least two or more nucleic acid tests were
SARS-CoV-2-positive; such patients were defined as re-positive
patients. Patients were considered COVID-19 false-negative if they
had COVID-19-related symptoms and typical imaging manifestations of
COVID-19 pneumonia and multiple COVID-19 nucleic acid tests had
been previously negative but they obtained a positive result in a
recent nucleic acid test.
Data collection
The clinical records, clinical classification, chest
CT scans, laboratory test results, treatment details and outcome
data were collected from the electronic medical records of the
patients. The information for all patients was collated in a
standardized form. The data were then independently reviewed by two
physicians.
The blood test results of the first medical
evaluation after admission were collected. White blood cells (WBC),
red blood cells (RBC), hemoglobin concentration (HBC), hematocrit
(HCT), lymphocytes and platelets were detected using a BC-6800 Auto
Hematology Analyzer and the original matching reagent (Shenzhen
Mindray Bio-Medical Electronics Co., Ltd.). The levels of
C-reactive protein (CRP), high-sensitivity CRP (hs-CRP), alanine
aminotransferase, aspartate aminotransferase, procalcitonin,
creatine kinase isoenzyme, lactate dehydrogenase (LDH),
α-hydroxybutyrate dehydrogenase (α-HBDH), albumin (ALB), cystatin C
and urea nitrogen were determined using an SAL9000 fully automatic
biochemical analyzer and the original matching reagent (Shenzhen
Mindray Bio-Medical Electronics Co., Ltd.). Blood coagulation
parameters were detected using an EXC810 fully automatic
coagulation analyzer (Shenzhen Mindray Bio-Medical Electronics Co.,
Ltd.). SARS-CoV-2 IgM/IgG antibodies were detected by an iFlash
3000 fully automatic chemiluminescence immunoassay analyzer and the
original matching reagent (Shenzhen YHLO Biotech Co., Ltd.). IL-6
was detected using a Cobas e411 analyzer and original matching
reagent (Roche Diagnostics). SARS-CoV-2 nucleic acids, open reading
frame 1ab (ORF1ab) and nucleocapsid (N) sequences were detected in
pharyngeal swab samples via the SLAN-96P Real-Time PCR System
(Shanghai Hongshi Medical Technology Co., Ltd.) and detection
reagents (Hunan Shengxiang Biotechnology Co., Ltd.). All specimens
were processed in the tent-type biosafety tertiary laboratory of
Huoshenshan Hospital (Wuhan, China) and all operations were in
strict compliance with the instructions provided by the
manufacturers of the equipment and reagents. All specimens were
transported and tested following the WHO Laboratory Testing
Guidelines (18).
All patients underwent 128-slice CT scans using the
uCT 760 CT X-ray system (United Imaging). The scans ranged from the
thoracic inlet to the bottom of the lung and the scanning type was
helical. The main scanning conditions were as follows: 120 kV;
automatic MAs control; rotation time, 0.5 sec; field of view, 350
mm; matrix, 512x512; pitch, 1.21; and slice thickness/gap,
0.625/0.625 mm. Chest CT results were divided into three stages:
Early (grade 1), advanced (grade 2) and late (grade 3) based on the
Chinese Management Guidelines for COVID-19 (version 7.0) (17).
Statistical analysis
Descriptive statistical methods were used to
summarize and analyze the data obtained. Categorical variables were
expressed as n (%), continuous variables as the median and
interquartile range, as appropriate. When the data of two
independent samples did not follow a normal distribution, the
Mann-Whitney U test was used; multiple groups of samples were
compared with the Kruskal-Wallis test. Proportional data were
compared with categorical variables using the Pearson χ2
test. Continuity correction was adopted when continuous random
variables approached discrete random variables. However, in the
case of limited data volumes, comparison was performed using
Fisher's exact test. Next, receiver operating characteristic (ROC)
curves were generated to analyze the predictive accuracy of various
risk factors. Furthermore, multivariate logistic regression
analysis was utilized to assess the risk factors related to
false-negative detection of viral nucleic acids. P<0.05 was
considered to indicate statistical significance. All statistical
analyses were performed using SPSS software, version 25.0 (IBM
Corp.).
Results
Baseline characteristics
The data of 134 patients (67 females and 67 males)
with COVID-19 who were hospitalized at Huoshenshan Hospital (Wuhan,
China) were analyzed in the present study. The median age of the
patients was 63 (51.69) years, with a minimum age of 21 years and a
maximum age of 91 years. Of these 134 patients, 33 (24.6%) had a
history of epidemic exposure, including direct exposure to the
epidemic site and family collective infection; 103 (76.9%) had
underlying comorbidities [44 (32.8%) had hypertension, 10 (7.5%)
had coronary heart disease, 28 (20.9%) had diabetes and 41 (30.6%)
had other comorbidities (20 patients with hypertension or coronary
heart disease or diabetes; furthermore, chronic bronchitis,
Parkinson's disease, hepatitis B virus (HBV), sequelae of cerebral
infarction, prostatic cancer, breast cancer, gallstones, old
pulmonary tuberculosis, lumbar disc herniation, nephrolithiasis,
rheumatoid arthritis, duodenal ulcer, glaucoma, hepatic cysts,
cataract, schizophrenia and hypothyroidism were present)]. A total
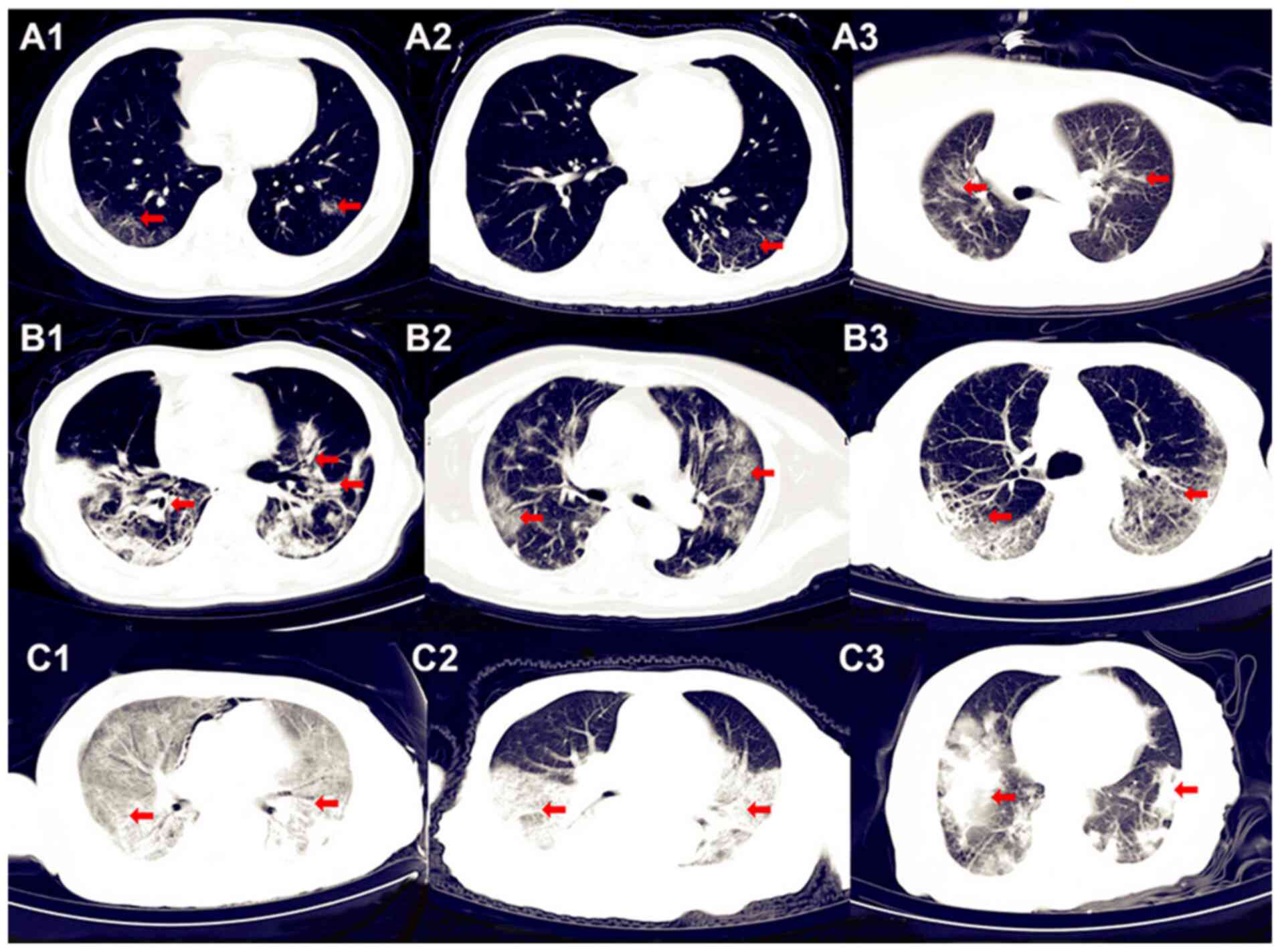
of 38 patients were in the early stage according to their chest CT
images, as a small number of localized patchy or ground-glass
opacities were visible (Fig. 2A).
Furthermore, 90 patients were in the advanced stage of the disease,
the diagnosis of which was established based on their chest CT
images exhibiting multiple bilateral lung ground-glass and patchy
opacities (Fig. 2B). A total of six
patients were in the late stage of the disease according to their
chest CT images, in which large areas of consolidated opacities
were observed (Fig. 2C). According
to the results of the laboratory tests, 68 (50.7%; 37 males and 31
females) patients had reduced numbers of RBC, 55 (41%; 33 males and
22 females) patients had decreased HBC; 73 (54.5%; 43 males and 30
females) patients had lower HCT; 42 (31.3%) patients had a
neutrophil-to-lymphocyte ratio >3.13; and 105 (78.4%) patients
had varying degrees of ALB reduction (data not shown). The chest CT
grades of the patients were as follows: 38 (28.3%) patients were
classified as grade 1 (early stage); 90 (67.2%) as grade 2
(advanced stage); and six (4.5%) as grade 3 (late stage) (Table I). Representative CT images of the
different stages of the disease are provided in Fig. 2. The common symptoms of these
patients were recorded: Median fever maximum temperature of 38.0
(37.6, 38.8)˚C; symptoms of myalgia or fatigue in 75 (56%); dry
cough in 105 (78.4%); dyspnea in 82 (61.2%); diarrhea in 15
(11.2%); and acute respiratory distress syndrome in 12 (9%) of the
patients (Table I). The major
therapeutic methods were antiviral treatment (n=96, 71.6%),
antibiotic treatment (n=81, 60.4%), use of hormones (n=47, 35.1%),
use of immunostimulant drugs (n=38, 28.4%) and traditional Chinese
medicine (Lotus Qingwen capsules; Shijiazhuang Yiling
Pharmaceutical Co., Ltd.) antiviral therapy (n=82, 61.2%). As of
April 11, 2020, 73 (54.5%) of the patients had been discharged, 54
(40.3%) remained in hospital and seven (5.2%) had died (Table I).
 | Table IClinical and laboratory data,
treatments and outcomes for re-positive and non-re-positive
patients. |
Table I
Clinical and laboratory data,
treatments and outcomes for re-positive and non-re-positive
patients.
| Item | Total (n=134) | Re-positive
patients (n=22) | Non-re-positive
patients (n=112) | P-value |
|---|
| Clinical
characteristics | | | | |
|
Sex
(M/F) | 67/67 | 11/11 | 56/56 | 0.652 |
|
Age
(years) | 63 (51, 69) | 60 (52.5,
67.3) | 63.5 (51, 70) | 0.663 |
|
Epidemiological
history | 33 (24.6) | 7 (31.8) | 26 (23.2) | 0.569 |
|
False-negative | 25 (18.7) | 0 (0) | 25 (22.3) | 0.010 |
| Comorbidities | | | | |
|
Hypertension | 44 (32.8) | 10 (45.5) | 34 (30.4) | 0.309 |
|
Coronary
heart disease | 10 (7.5) | 2 (9.1) | 8 (7.1) | 0.858 |
|
Diabetes | 28 (20.9) | 3 (13.6) | 25 (22.3) | 0.264 |
|
Other | 41 (30.6) | 7 (31.8) | 34 (30.4) | 0.867 |
| Signs and
symptoms | | | | |
|
Tmax,˚C | 38.0 (37.6,
38.8) | 38.3 (37.6,
39.0) | 38 (37.6,
38.7) | 0.319 |
|
Dry
cough | 105 (78.4) | 15 (68.2) | 90 (80.4) | 0.054 |
|
Myalgia or
fatigue | 75(56) | 11(50) | 64 (57.1) | 0.27 |
|
Dyspnea | 82 (61.2) | 12 (54.5) | 70 (62.5) | 0.214 |
|
Diarrhea | 15 (11.2) | 3 (13.6) | 12 (10.7) | 0.823 |
|
ARDS | 12(9) | 1 (4.5) | 11 (9.8) | 0.365 |
| Disease severity
classification | | | | 0.051 |
|
Common | 39 (29.1) | 10 (45.5) | 29 (25.9) | |
|
Severe | 85 (63.4) | 11(50) | 74 (66.1) | |
|
Critical | 10 (7.5) | 1 (4.5) | 9(8) | |
| Stages of chest
CT | | | | 0.366 |
|
1 | 38 (28.3) | 7 (31.8) | 31 (27.6) | |
|
2 | 90 (67.2) | 15 (68.2) | 75(67) | |
|
3 | 6 (4.5) | 0 (0) | 6 (5.4) | |
| Laboratory
parameters (reference range) | | | | |
|
RBC,
x1012/l (3.8-5.8) | 4.0 (3.7, 4.5) | 3.7 (3.3, 4.0) | 4.0 (3.7, 4.6) | 0.002 |
|
HBC, g/l
(115-175) | 124 (115,
135.3) | 117 (102.8,
125) | 126 (116.5,
139) | 0.005 |
|
HCT, %
(35-50) | 37.1 (34,
39.8) | 34.8 (31,
37.2) | 37.6 (34.8,
41) | 0.003 |
|
WBC,
x109/l (3.5-9.5) | 5.7 (4.4, 7.0) | 4.9 (3.9, 6.1) | 5.9 (4.6, 7.1) | 0.025 |
|
NEUT, %
(40-75) | 60.8 (54.4,
71.3) | 59.9 (52.2,
68) | 61.3 (54.9,
72.4) | 0.36 |
|
MONO, %
(3-10) | 7.6 (6.3, 9.0) | 7.4 (6.6, 8.9) | 7.6 (6.3, 9.2) | 0.68 |
|
LYM, %
(20-50) | 27.1 (19.7,
33.3) | 29.9 (23.2,
34.8) | 26.4 (17,
32.6) | 0.96 |
|
ALC,
x109/l (1.1-3.2) | 1.4 (1.0, 1.9) | 1.3 (1.1, 1.8) | 1.5 (1.0, 1.9) | 0.77 |
|
NLR | 2.3 (1.6, 3.6) | 2.0 (1.5, 2.9) | 2.3 (1.7, 4.2) | 0.18 |
|
MCV, fl
(82-100) | 92.6 (89.5,
95.4) | 93.6 (91.8,
95.8) | 92 (89, 95.4) | 0.056 |
|
RDW, %
(10.9-15.4) | 12.8 (12.3,
13.3) | 12.8 (12.2,
13.6) | 12.8 (12.3,
13.3) | 0.993 |
|
MCH, pg
(27-34) | 31.2 (30, 32) | 31.5 (30.2,
32.1) | 31.1 (29.9,
32) | 0.412 |
|
MCHC, g/l
(316-354) | 336 (331, 342) | 334 (328.8,
340.3) | 336 (331.3,
342.8) | 0.223 |
|
PLT,
x109/l (125-350) | 222.5 (178.5,
283.5) | 208.5 (162.8,
253.3) | 225.5 (182,
302.5) | 0.08 |
|
FIB, g/l
(2-4) | 3.0 (2.6, 3.5) | 2.9 (2.7, 3.1) | 3.1 (2.6, 3.6) | 0.333 |
|
D-D, mg/l
(<0.5) | 0.5 (0.2, 1.4) | 0.4 (0.2, 1.8) | 0.5 (0.2, 1.4) | 0.81 |
|
CK
isoenzyme, IU/l (0-24) | 8.5 (6.7,
11.2) | 8.2 (7.0, 9.3) | 8.7 (6.7,
11.7) | 0.529 |
|
ALT, IU/l
(7-40) | 22.1 (16.1,
42.1) | 20.4 (12.0,
40.1) | 22.2 (16.3,
43.2) | 0.347 |
|
AST, IU/l
(7-45) | 19.1 (15.3,
30.5) | 17.4 (15.5,
30.7) | 19.2 (15.2,
30.3) | 0.764 |
|
LDH, IU/l
(120-250) | 171.8 (148,
249.6) | 171.3 (151.6,
238.9) | 171.8 (146,
252.6) | 0.871 |
|
α-HBDH, IU/l
(72-182) | 141.4 (120.5,
202.2) | 138.9 (126,
199.8) | 141.5 (120,
203.1) | 0.925 |
| Cystatin C, mg/l
(0.54-1.15) | 0.9 (0.8, 1.1) | 0.9 (0.8, 1.1) | 0.9 (0.8, 1.1) | 0.698 |
|
Urea
nitrogen, mmol/l (2.6-7.5) | 4.2 (3.5, 5.5) | 4.5 (3.7, 5.7) | 4.2 (3.4, 5.4) | 0.378 |
|
Procalcitonin,
ng/ml (<0.15) | 0.04 (0.03,
0.06) | 0.03 (0.03,
0.05) | 0.04 (0.03,
0.06) | 0.717 |
|
IL-6, ng/ml
(<7) | 2.3 (1.5, 5.4) | 2.3 (1.6, 3.1) | 2.4 (1.5, 5.7) | 0.838 |
|
CRP, mg/l
(0-4) | 2.5 (1.0,
18.3) | 2.2 (0.9,
13.6) | 2.8 (1.0,
20.5) | 0.651 |
|
hs-CRP, mg/l
(0-4) | 2.5 (1.0, 10) | 2.2 (0.9, 10) | 2.8 (1.0, 10) | 0.841 |
|
ALB, g/l
(40-55) | 37 (33.4,
39.4) | 36.9 (33.3,
39) | 37.1 (33.3,
39.5) | 0.38 |
|
ORF1ab gene
(+/-) | 12 (8.9) | 9 (40.9) | 3 (2.7) | 0.002 |
|
N gene
(+/-) | 13 (9.7) | 10 (45.5) | 3 (2.7) | 0.002 |
|
SARS-CoV-2
IgM, U/ml (<10) | 27.2 (12.5,
56) | 34.2 (8.9,
61.3) | 23.3 (13.1,
55.5) | 0.83 |
|
SARS-CoV-2
IgG, U/ml (<10) | 89.3 (68.4,
170.7) | 137.3 (72.3,
183.9) | 87.5 (66.1,
170.3) | 0.969 |
| Treatment | | | | |
|
Antiviral
therapy | | | | 0.037 |
|
Arbidol | 69 (51.5) | 11(50) | 58 (51.8) | |
|
Oseltamivir | 13 (9.7) | 2 (9.1) | 11 (9.8) | |
|
Ribavirin | 9 (6.7) | 5 (22.7) | 4 (3.6) | |
|
Ganciclovir | 2 (1.5) | 0 (0) | 2 (1.8) | |
|
Chloroquine
diphosphate | 1 (0.7) | 1 (4.5) | 0 (0) | |
|
Lopinavir/Ritonavir
tablets | 1 (0.7) | 1 (4.5) | 0 (0) | |
|
Interferon | 1 (0.7) | 1 (4.5) | 0 (0) | |
|
Use of
antibiotics | 81 (60.4) | 10 (45.5) | 71 (64.4) | 0.652 |
|
Use of
hormones | 47 (35.1) | 6 (27.3) | 41 (36.6) | 0.844 |
|
Immune
enhancement therapy | 38 (28.4) | 9 (40.9) | 29 (25.9) | 0.091 |
|
Lotus
Qingwen capsules | 82 (61.2) | 10 (45.5) | 72 (64.3) | 0.149 |
| Outcome | | | | 0.204 |
|
Discharged | 73 (54.5) | 8 (36.4) | 65(58) | |
|
Hospitalized | 54 (40.3) | 14 (63.6) | 40 (35.7) | |
|
Died | 7 (5.2) | 0 (0) | 7 (6.3) | |
Analysis of the laboratory findings of
re-positive/non-re-positive patients
Of the 134 patients with COVID-19 included in the
present study, 22 (16.4%) were re-positive and 112 (83.6%) were
non-re-positive (Table I). The WBC
(P<0.05), RBC (P<0.05), HBC (P<0.05) and HCT (P<0.05)
in the re-positive patients were significantly lower than those in
the non-re-positive patients. A total of nine re-positive patients
were positive for the ORF1ab gene and 10 re-positive patients were
positive for the N gene (Table I).
Positivity for the ORF1ab gene (P<0.01) and N gene (P<0.01)
in re-positive patients was significantly more frequent than in
non-re-positive patients. The parameters of erythrocytes in
re-positive patients (11 males and 11 females) and non-re-positive
patients were quantitatively analyzed by sex (Table II). Among the males, the RBC count
of the re-positive patients [4.0 (3.4, 4.2) x1012/l] was
significantly lower (P<0.05) than that of the non-re-positive
patients [4.4 (3.9, 4.8) x1012/l], the HBC of the
re-positive patients [123 (103, 133) g/l] was significantly lower
(P<0.05) than that of the non-re-positive patients [131 (122,
147) g/l] and the HCT of the re-positive patients [36.6 (31.1,
39.2)%] was significantly lower (P<0.05) than that of the
non-re-positive patients [39.5 (36.1, 42.9)%]. Among the female
patients, the RBC count [3.5 (3.1, 3.7) x1012/l] of the
re-positive patients was significantly lower (P<0.05) than that
of the non-re-positive patients [3.9 (3.6, 4.3)
x1012/l], the HBC [115 (102, 118) g/l] of the
re-positive patients was significantly lower (P<0.05) than that
of the non-re-positive patients [122 (110, 130) g/l] and the HCT
[34.2 (28.5, 34.9)%] of the re-positive patients was significantly
lower (P<0.05) than that of the non-re-positive patients [36.4
(33.1, 38.8)%]. The values of the mean corpuscular volume, RBC
distribution width, mean corpuscular hemoglobin (MCH), and MCH
concentration were within the normal range, with no statistically
significant differences between the re-positive and non-re-positive
groups (Table II).
 | Table IIBlood cell parameters in re-positive
and non-re-positive patients by sex. |
Table II
Blood cell parameters in re-positive
and non-re-positive patients by sex.
| Parameter | Reference
range | Re-positive
(n=22) | Non-re-positive
(n=112) | P-value |
|---|
| RBC,
x1012/l | | | | |
|
Males | 4.3-5.8 | 4.0 (3.4, 4.2) | 4.4 (3.9, 4.8) | 0.023 |
|
Females | 3.8-5.1 | 3.5 (3.1, 3.7) | 3.9 (3.6, 4.3) | 0.017 |
| HBC, g/l | | | | |
|
Males | 130-175 | 123 (103, 133) | 131 (122, 147) | 0.32 |
|
Females | 115-150 | 115 (102, 118) | 122 (110, 130) | 0.026 |
| HCT, % | | | | |
|
Males | 40-50 | 36.6 (31.1,
39.2) | 39.5 (36.1,
42.9) | 0.32 |
|
Females | 35-45 | 34.2 (28.5,
34.9) | 36.4 (33.1,
38.8) | 0.011 |
|
MCV, fl | 82-100 | 93.6 (91.8,
95.8) | 92 (89, 95.4) | 0.056 |
|
RDW, % | 10.9-15.4 | 12.8 (12.2,
13.6) | 12.8 (12.3,
13.3) | 0.993 |
|
MCH, pg | 27-34 | 31.5 (30.2,
32.1) | 31.1 (29.9,
32.0) | 0.412 |
|
MCHC,
g/l | 316-354 | 334 (328.8,
340.3) | 336 (331.3,
342.8) | 0.223 |
ROC curves were generated to evaluate the ability of
RBC and WBC counts to distinguish between re-positive and
non-re-positive patients (Fig. 3).
For male re-positive patients, the area under the ROC curve (AUC)
for RBC, HBC and HCT was 0.7175 (95% CI, 0.5758-0.8593), 0.7054
(95% CI, 0.5614-0.8493) and 0.7054 (95% CI, 0.5617-0.8490),
respectively (Fig. 3D). The
predictive values for recurrence of SARS-CoV-2 positivity in males
were as follows: RBC <4.465x1012/l provided a
sensitivity of 100% (95% CI, 74.12-100%), a specificity of 44.64%
(95% CI, 32.39-57.59%) and a likelihood ratio of 1.806; HBC
<135.5 g/l provided a sensitivity of 100% (95% CI, 74.12-100%),
a specificity of 44.64% (95% CI, 32.39-57.59%) and a likelihood
ratio of 1.806; and HCT <40.3% provided a sensitivity of 100%
(95% CI, 74.12-100%), a specificity of 42.86% (95% CI,
30.77-55.86%) and a likelihood ratio of 1.75 (Fig. 3A and D). To distinguish female re-positive from
female non-re-positive patients, the AUC values of the ROC curves
for RBC, HBC and HCT were 0.7297 (95% CI, 0.5799-0.8795), 0.7135
(95% CI, 0.5765-0.8504) and 0.7427 (95% CI, 0.6028-0.8826),
respectively (Fig. 3D). In terms of
the predictive value for SARS-CoV-2 recurrence, RBC
<3.745x1012/l provided a sensitivity of 81.82% (95%
CI, 52.30-96.77%), a specificity of 64.29% (95% CI, 51.19-75.54%)
and a likelihood ratio of 2.291; HBC <118.5 g/l provided a
sensitivity of 90.91% (95% CI, 62.26-99.53%), a specificity of
62.5% (95% CI, 49.41-73.99%) and a likelihood ratio of 2.424; and
HCT <35.25% provided a sensitivity of 90.91% (95% CI,
62.26-99.53%), a specificity of 64.29% (95% CI, 51.19-75.54%) and a
likelihood ratio of 2.545 (Fig. 3B
and D). The AUC value of the ROC
curve for WBC for re-positive patients was 0.6514 (95% CI,
0.5182-0.7845) and in terms of the value for the prediction of
SARS-CoV-2 recurrence, WBC <5.45x109/l provided a
sensitivity of 68.18% (95% CI, 47.32-83.64%), a specificity of
62.5% (95% CI, 53.26-70.91%) and a likelihood ratio of 1.818
(Fig. 3C).
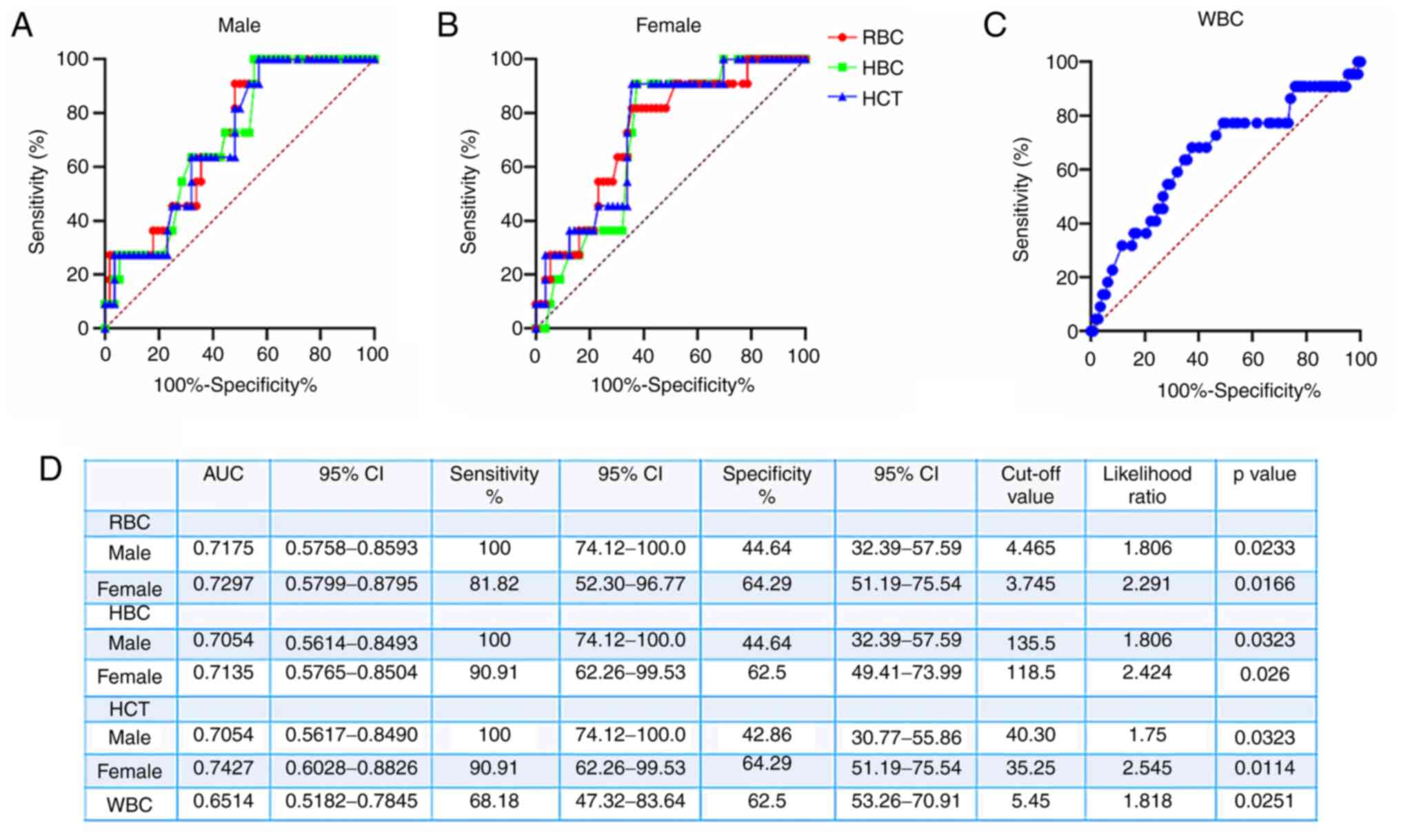 | Figure 3ROC curve analysis of the predictive
performance of RBC, HBC, HCT and WBC for re-positive patients. (A)
ROC curves for RBC, HBC and HCT to distinguish between male
re-positive and male non-re-positive patients. (B) ROC curves for
RBC, HBC and HCT to distinguish between female re-positive and
female non-re-positive patients. (C) ROC curves for WBC to
distinguish between re-positive and non-re-positive patients. (D)
Parameters of the predictive value of RBC, HBC and HCT regarding
virus recurrence obtained from the ROC curves. ROC, receiver
operating characteristic; AUC, area under the ROC curve; RBC, red
blood cells; HBC, hemoglobin concentration; HCT, hematocrit; WBC,
white blood cells. |
Analysis of laboratory findings of
false-negative/nucleic acid-positive patients
As presented in Table
III, of the 134 patients with COVID-19 included in the present
study, 25 (18.7%) had false-negative viral nucleic acid tests and
109 (81.3%) had positive nucleic acid tests. The values of absolute
lymphocyte count (ALC; P<0.01), ALB (P<0.05), RBC
(P<0.05), HBC (P<0.05) and HCT (P<0.05) in the patients
with false-negative SARS-CoV-2 nucleic acid test results were
significantly higher than those of the patients who tested positive
for SARS-CoV-2 nucleic acids. The D-dimer concentration, LDH,
α-HBDH, cystatin C, CRP and hs-CRP in the patients with a
false-negative viral nucleic acid test were significantly lower
than those in the viral nucleic acid-positive patients (P<0.05;
Table III).
 | Table IIIClinical and laboratory parameters,
treatments and outcomes of false-negative/nucleic acid positive
patients. |
Table III
Clinical and laboratory parameters,
treatments and outcomes of false-negative/nucleic acid positive
patients.
| Item | Total (n=134) | False-negative
patients (n=25) | Nucleic acid
positive patients (n=109) | P-value |
|---|
| Clinical
characteristics | | | | |
|
Sex,
M/F | 67/67 | 16/9 | 51/58 | 0.121 |
|
Age,
years | 63 (51, 69) | 53 (43, 65.5) | 64 (55, 70) | 0.008 |
|
Epidemiological
history | 33 (24.6) | 4(16) | 29 (26.6) | 0.267 |
|
Re-positive
patients | 22 (16.4) | 0 (0) | 22 (20.2) | 0.010 |
| Comorbidities | | | | |
|
Hypertension | 44 (32.8) | 5(20) | 39 (35.8) | 0.13 |
|
Coronary
heart disease | 10 (7.5) | 1(4) | 9 (8.3) | 0.465 |
|
Diabetes | 28 (20.9) | 3(12) | 25 (22.9) | 0.225 |
|
Other | 41 (30.6) | 5(20) | 36(33) | 0.202 |
| Signs and
symptoms | | | | |
|
Tmax,˚C | 38.0 (37.6,
38.8) | 38.0 (37.4,
38.4) | 38.2 (37.7,
39.0) | 0.090 |
|
Dry
cough | 105 (78.4) | 17(68) | 88 (80.7) | 0.163 |
|
Myalgia or
fatigue | 75(56) | 14(56) | 61(56) | 0.997 |
|
Dyspnea | 82 (61.2) | 18(72) | 64 (58.7) | 0.219 |
|
Diarrhea | 15 (11.2) | 1(4) | 14 (12.8) | 0.206 |
|
ARDS | 12(9) | 0 (0) | 12(11) | 0.082 |
| Disease severity
classification | | | | 0.031 |
|
Common | 39 (29.1) | 11(44) | 28 (25.7) | |
|
Severe | 85 (63.4) | 14(56) | 71 (65.1) | |
|
Critical | 10 (7.5) | 0 (0) | 10 (9.2) | |
| Stages of chest
CT | | | | 0.095 |
|
1 | 38 (28.3) | 10(40) | 28 (25.7) | |
|
2 | 90 (67.2) | 15(60) | 75 (68.8) | |
|
3 | 6 (4.5) | 0 (0) | 6 (5.5) | |
| Laboratory
parameters | | | | |
|
RBC,
x1012/l | 4.0 (3.7, 4.5) | 4.3 (3.9, 4.8) | 4.0 (3.7, 4.4) | 0.034 |
|
HBC,
g/l | 124 (115,
135.3) | 131 (119.5,
149) | 124 (114, 133) | 0.02 |
|
HCT, % | 37.1 (34,
39.8) | 39 (36, 43.7) | 36.6 (33.4,
39.6) | 0.013 |
|
WBC,
x109/l | 5.7 (4.4, 7.0) | 6.2 (5.0, 7.3) | 5.7 (4.4, 7.0) | 0.202 |
|
NEUT, % | 60.8 (54.4,
71.3) | 57.9 (52,
69.3) | 61.8 (55.3,
72.4) | 0.113 |
|
MONO, % | 7.6 (6.3, 9.0) | 7.7 (6.4, 9.3) | 7.5 (6.3, 9.0) | 0.528 |
|
LYM, % | 27.1 (19.7,
33.3) | 29.4 (21.9,
34.6) | 25.6 (17.1,
33) | 0.143 |
|
ALC,
x109/l | 1.4 (1.0, 1.9) | 1.8 (1.2, 2.3) | 1.3 (1.0, 1.8) | 0.010 |
|
NLR | 2.3 (1.6, 3.6) | 1.9 (1.5, 3.2) | 2.4 (1.7, 4.2) | 0.136 |
|
MCV, fl | 92.6 (89.5,
95.4) | 92.5 (89.3,
94.9) | 92.6 (89.6,
95.5) | 0.728 |
|
RDW, % | 12.8 (12.3,
13.3) | 12.6 (12.3,
13.3) | 12.8 (12.3,
13.3) | 0.71 |
|
MCH, pg | 31.2 (30, 32) | 30.8 (29.8,
32) | 31.2 (30, 32) | 0.495 |
|
MCHC,
g/l | 336 (331, 342) | 335 (331.5,
343.5) | 336 (331,
341.5) | 0.909 |
|
PLT,
x109/l | 222.5 (178.5,
283.5) | 242 (184.5,
289.5) | 219 (177, 284) | 0.515 |
|
FIB,
g/l | 3.0 (2.6, 3.5) | 2.9 (2.4, 3.3) | 3.1 (2.7, 3.6) | 0.726 |
|
D-D,
mg/l | 0.5 (0.2, 1.4) | 0.2 (0.1, 0.5) | 0.6 (0.3, 2.2) | 0.001 |
|
CK
isoenzyme, IU/l | 8.5 (6.7,
11.2) | 7.8 (6.6,
10.0) | 8.8 (7.0,
11.5) | 0.294 |
|
ALT,
IU/l | 22.1 (16.1,
42.1) | 29.4 (17.3,
52.2) | 21.6 (15.5,
40.4) | 0.205 |
|
AST,
IU/l | 19.1 (15.3,
30.5) | 17.6 (14.5,
33.4) | 19.1 (15.4,
30.5) | 0.804 |
|
LDH,
IU/l | 171.8 (148,
249.6) | 151.5 (129.1,
194.7) | 175.8 (152.2,
258.2) | 0.21 |
|
α-HBDH,
IU/l | 141.4 (120.5,
202.2) | 121.8 (104.7,
164.4) | 143.3 (121.9,
208.2) | 0.012 |
|
Cystatin C,
mg/l | 0.9 (0.8, 1.1) | 0.8 (0.8, 1.0) | 1.0 (0.8, 1.1) | 0.025 |
|
Urea
nitrogen, mmol/l | 4.2 (3.5, 5.5) | 4.0 (3.4, 5.0) | 4.2 (3.5, 5.6) | 0.281 |
|
Procalcitonin,
ng/ml | 0.04 (0.03,
0.06) | 0.04 (0.03,
0.04) | 0.04 (0.04,
0.06) | 0.21 |
|
IL-6,
ng/ml | 2.3 (1.5, 5.4) | 3.6 (2.0, 5.2) | 2.2 (1.5, 5.7) | 0.13 |
|
CRP,
mg/l | 2.5 (1.0,
18.3) | 1.4 (0.6, 3.7) | 3.1 (1.0,
27.0) | 0.025 |
|
hs-CRP,
mg/l | 2.5 (1.0, 10) | 1.4 (0.6, 3.7) | 3.1 (1.0, 10) | 0.018 |
|
ALB,
g/l | 37 (33.4,
39.4) | 39 (35.4,
42.2) | 36.7 (32.6,
39) | 0.007 |
|
ORF1ab gene
(+/-) | 12 (8.9) | 0 (0) | 12 (11.0) | 0.055 |
|
N gene
(+/-) | 13 (9.7) | 0 (0) | 13 (11.9) | 0.055 |
|
SARS-CoV-2
IgM, U/ml | 27.2 (12.5,
56) | 16.3 (13.2,
46) | 31.7 (12.2,
61.8) | 0.26 |
|
SARS-CoV-2
IgG, U/ml | 89.3 (68.4,
170.7) | 79.1 (63.7,
163.1) | 93.3 (69.7,
175.8) | 0.132 |
| Treatment | | | | |
|
Antiviral
therapy | | | | 0.187 |
|
Arbidol | 69 (51.5) | 11(44) | 58 (53.2) | |
|
Oseltamivir | 13 (9.7) | 3(12) | 10 (9.2) | |
|
Ribavirin | 9 (6.7) | 1(4) | 8 (7.3) | |
|
Ganciclovir | 2 (1.5) | 1(4) | 1 (0.9) | |
|
Chloroquine
diphosphate | 1 (0.7) | 0 (0) | 1 (0.9) | |
|
Lopinavir/Ritonavir
tablets | 1 (0.7) | 0 (0) | 1 (0.9) | |
|
Interferon | 1 (0.7) | 0 (0) | 1 (0.9) | |
|
Use of
antibiotics | 81 (60.4) | 12(48) | 69 (63.3) | 0.825 |
|
Use of
hormones | 47 (35.1) | 6(24) | 41 (37.6) | 0.198 |
|
Immune
enhancement therapy | 38 (28.4) | 2(8) | 36(33) | 0.032 |
|
Lotus
Qingwen capsules | 82 (61.2) | 16(64) | 66 (60.6) | 0.57 |
| Outcome | | | | 0.027 |
|
Discharged | 73 (54.5) | 20(80) | 53 (48.6) | |
|
Hospitalized | 54 (40.3) | 5(20) | 49(45) | |
|
Died | 7 (5.2) | 0 (0) | 7 (6.4) | |
The variables with statistically significant
differences obtained by grouping patients on the basis of
false-negative/positive viral nucleic acid test results were
analyzed by multivariate logistic regression to identify the
factors that influenced nucleic acid false-negative detection.
Taking a negative viral nucleic acid test result as the dependent
variable, the data with statistically significant differences
(e.g., RBC, HBC and ALC) were used as the independent variables for
logistic regression analysis. The results indicated that RBC [odds
ratio (OR)=0.43, 95% CI: 0.18-0.99], HBC (OR=0.97, 95% CI:
0.94-0.99) and ALC (OR=0.43, 95% CI: 0.20-0.91) were significant
influencing factors for a SARS-CoV-2 false-negative nucleic acid
test result (Fig. 4A). ROC curve
analysis was then applied to evaluate the predictive values of RBC,
HBC and ALC for SARS-CoV-2 false-negative nucleic acid test results
in patients with COVID-19, providing an AUC of 0.7136 (95% CI:
0.6057-0.8215; P=0.0009; Fig. 4B).
Increased RBC, HBC and ALC values led to a greater tendency to
obtain a negative result of the SARS-CoV-2 nucleic acid tests, with
a higher probability of a false-negative SARS-CoV-2 nucleic acid
test result.
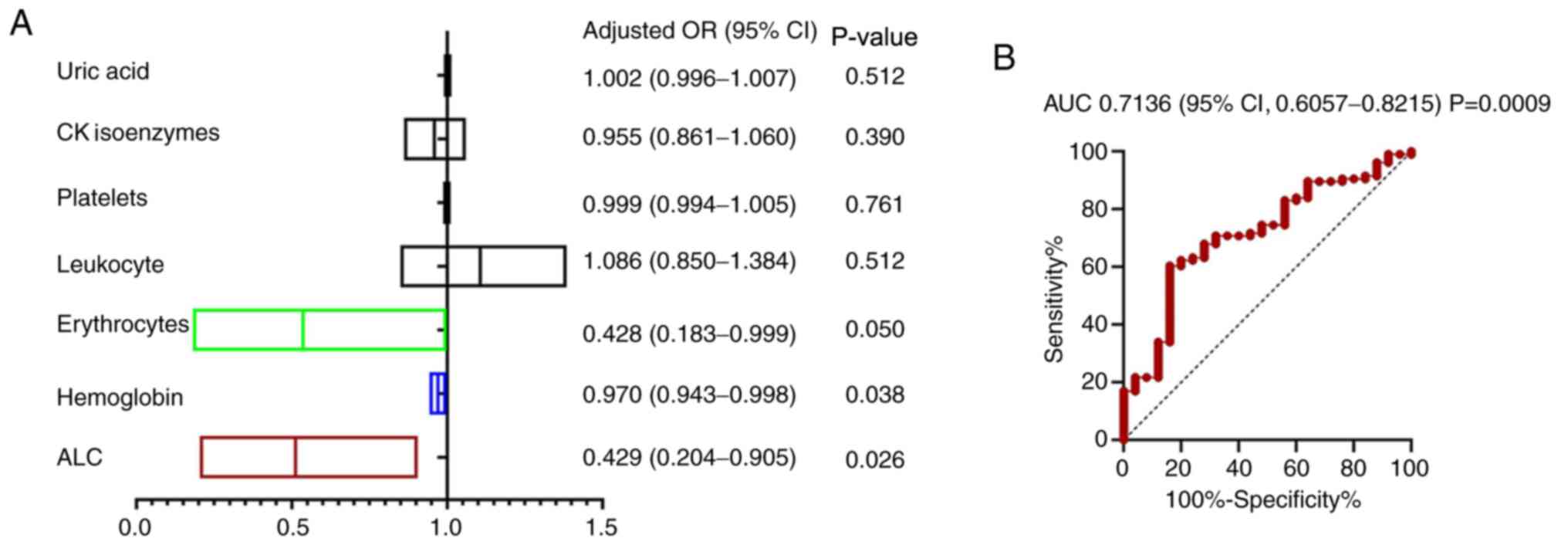 | Figure 4(A) Multivariate logistic regression
to confirm the influencing factors of nucleic acid negative
detection. The probability of the occurrence of a nucleic acid
negative result was predicted by the regression model. According to
the results, RBC, HBC and ALC were significant influencing factors
for a nucleic acid negative test. (B) ROC curve for the predictive
value of combined RBC, HBC and ALC regarding the probability of a
nucleic acid negative result in patients with Coronavirus disease
2019. RBC, red blood cells; HBC, hemoglobin concentration; ALC,
absolute lymphocyte count; HR, hazard ratio; CK, creatine kinase;
ROC, receiver operating characteristic; AUC, area under the ROC
curve. |
Discussion
False-negative results are common during the
treatment process of COVID-19. Cuñarro-López et al (19) reported that 38.7% of 111 obstetric
patients suspected of having COVID-19 had negative PCR results.
False-negatives may occur due to various factors, including the
incubation period of the disease, lack of standardized procedures
for sample collection, poor storage conditions, insufficient
detection and analysis accuracy, and human factors (18). In the present study, the
false-negative rate of SARS-CoV-2 RNA detection in pharyngeal swab
specimens was determined to be 18.7%. The detection of SARS-CoV-2
RNA by PCR in pharyngeal swab specimens is the most commonly used
method for COVID-19 diagnosis in clinical practice. Usually, sputum
specimens are difficult to collect due to the dry cough of most
patients with COVID-19. The false-negative detection results of
SARS-CoV-2 RNA in patients with COVID-19 may be related to various
factors, such as the condition of the sample collection site,
sample quality and laboratory bias (18). Furthermore, Chen et al
(20) indicated that SARS-CoV-2 RNA
was more likely to be detected in blood and anal swabs than in
pharyngeal swabs. Guidelines for laboratory testing of patients
with suspected COVID-19 issued by the World Health Organization in
2020 suggest that SARS-CoV-2-positive detection rates may be higher
in lower respiratory tract specimens (18). An earlier study on SARS also
confirmed that the virus-positive rate of sputum specimens was
higher than that of pharyngeal swabs and there was no significant
association with age, respiratory symptoms and underlying diseases
(21). Considering the high
expression of angiotensin-converting enzyme II, which is utilized
by SARS-CoV-2 as the receptor for entry into type II alveolar cells
(22), it may be recommended that
patients with severe disease and a false-negative SARS-CoV-2 RNA
test should be re-tested using sputum samples from the lower
respiratory tract. In the present study, the age, organ damage
indices (D-D, LDH, α-HBDH and cystatin C) and the immune response
indices (CRP and hs-CRP) of the false-negative patients were
significantly lower than those of the patients who tested positive
for SARS-CoV-2 RNA by PCR. In addition, the values of ALC, ALB,
RBC, HBC and HCT of the false-negative patients were significantly
higher than those of the positive patients and the use of
immunity-enhancing drugs in the false-negative patients was lower
than that in the positive patients. These results indicated that
false-negative patients may have relatively normal immune function
in the early stages of COVID-19 disease. Logistic regression
analysis revealed that the probability of negative viral nucleic
acid detection increases with the elevation of RBC and HBC
values.
In the present cross-sectional study, the rate of
re-positive patients was determined to be 16.4%. It was established
that WBC, RBC, HBC and HCT of the re-positive patients were lower
than those of the non-re-positive patients. Leukopenia has been
previously confirmed in patients with recurrence of hepatitis C
virus (HCV) infection (23,24), which may suggest that these patients
are immunocompromised. In addition to respiratory function, RBCs
also perform multiple immune functions (25,26).
RBCs are the most abundant cell type in the human blood and are
involved in human innate immune responses (27). HBV, HCV, HIV, Epstein-Barr virus,
transfusion-transmitted virus and echovirus have been reported to
cause severe bone marrow aplasia (28,29).
Parvovirus B19 commonly infects pro-erythroblasts and induces
transient RBC aplasia, similar to that observed in patients with
chronic hemolytic anemia (28).
Furthermore, parvovirus B19 infection may also be associated with
pancytopenia, particularly in immunocompromised patients (28). It is noteworthy that viruses have
been postulated to induce lymphocyte activation and eventually lead
to apoptotic death of hematopoietic cells in the bone marrow
(30). In the present study, immune
enhancement therapy of re-positive patients was effective. A total
number of nine patients received immunostimulant drugs, of which
thymosin α 1 was most commonly used.
The present study suggested that ALC, RBC and HBC
were independent predictors of negative viral nucleic acid
detection. Therefore, higher values of RBC, HBC and ALC in patients
with COVID-19 are associated with a higher likelihood of a negative
viral nucleic acid test result. Accumulating evidence has confirmed
the occurrence of lymphopenia in patients with COVID-19 (3,31-33).
However, surprisingly, the present results indicated that higher
RBC was associated with a lower probability of SARS-CoV-2
infection. It was confirmed that re-positive patients had
significantly reduced RBC, HBC and HCT. This result may indicate
that the recurrence of SARS-CoV-2 in re-positive patients leads to
mild normocytic anemia. Studies have confirmed that certain viruses
specifically invade vertebrate RBC, including Orthomyxoviridae
influenza A (34,35), HIV-1(36) and Orthomyxoviridae isavirus
(37). In addition, an earlier
study indicated that salmon erythrocytes were the main Piscine
orthoreovirus (PRV) replicating cells in the early peak phase
of the infection and cytoplasmic inclusions called ‘virus
factories’ were observed in the erythrocytes, which were the
primary sites for the formation of new virus particles (38). Erythrocytes are the main target
cells for PRV in the early infection phase and blood cell infection
precedes myocardial infection (39). The infected erythrocytes provide
further PRV dissemination into various host tissues (40-42).
In the present study, ROC curves were generated to evaluate the
predictive value of the RBC count regarding re-positive patients.
An AUC value of 0.9 is considered to indicate high accuracy,
0.7-0.9 moderate accuracy and 0.5-0.7 low accuracy, while 0.5
indicates a chance result (43,44).
The present results suggested that RBC has moderate accuracy in
predicting virus recurrence in patients with COVID-19.
Based on the present results and literature review,
the following mechanisms may be hypothesized, as outlined in a
schematic in Fig. 5: On the one
hand, false-negative patients have an antiviral response to
SARS-CoV-2 in the early phase of the disease when the viral load is
below the detectable level. At this stage, the virus does not
activate or destroy the erythrocytes, resulting in higher levels of
RBC, HBC and HCT in the false-negative patients than those in
patients who test positive for viral nucleic acids. On the other
hand, immune disorders that induce viral replication in the
erythrocytes may lead to re-positivity of patients, resulting in an
increase in the viral load in damaged erythrocytes. This effect may
account for the lower RBC, HBC and HCT levels in re-positive
patients than those in non-re-positive patients.
The RBC count may be used as an important screening
index to determine whether a patient is COVID-19 false-negative.
Even if the detection of SARS-CoV-2 by PCR is negative, a downward
trend in the RBC count of a suspected patient may indicate that the
virus is being activated and is beginning to destroy RBCs. In this
case, further continuous PCR and chest CT detection are required,
as the aforementioned indications are strongly suggesting that the
patient may have been infected. The changes in the RBC count values
in COVID-19 patients that recovered from hospital treatment and
whose nucleic acid test results were negative may be used to
predict SARS-CoV-2 recurrence. A new decrease in the RBC count
values after their previous gradual return to normal after
treatment may indicate that the increase in the viral load in the
body is causing RBC damage. Based on the changes in the RBC counts,
treatment plans may be prepared in advance and preventive antiviral
treatment may be provided to minimize the virus-induced damage to
patients.
In summary, the RBC values of hospitalized patients
on admission may predict the evolution of COVID-19 disease.
SARS-CoV-2 infection has a relatively lower probability to recur in
patients with high RBC counts and their prognosis is good. This
observation suggests the important role of human erythrocytes in
SARS-CoV-2 infection, which may provide the key to explaining the
subsequent pathogenesis. The present study provided novel insight
into hidden and evasive mechanisms of SARS-COV-2 in the human
body.
Supplementary Material
Hospital workflow pattern
Classification criteria for patients
with COVID-19. COVID-19, coronavirus disease 2019; ICU, intensive
care unit.
Schematic diagram of the hospital
ward. PPE, personal protective equipment.
A step-by-step flow chart for putting
on protective clothing.
A step-by-step flow chart for removing
protective clothing.
Nurses' work schedules and work
content in hospital settings.
Acknowledgements
The authors greatly appreciate the kind assistance
of Professor Ya-juan Yuan (Laboratory, Xijing Hospital, Fourth
Military Medical University, Xi'an, China) in analyzing and
identifying RBC morphological parameters, as well as Ms. Meng-nan
Fan (Epidemiology Biostatistics, First Affiliated Hospital, Henan
University of Science and Technology, Luoyang, China) for her
assistance in statistical analysis.
Funding
Funding: This work was funded by the National Natural Science
Foundation of China (grant nos. 81874156 and 81971206) and the
Natural Science Basic Research Plan in Shaanxi Province of China
(grant no. 2016SF-041).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL and XGZ conceived and designed the study. BL and
XJ confirm the authenticity of all the raw data. BL, YH and XPQ
contributed to writing of the manuscript. XJ, HoJ, HuJ and CML
collected the data. BL, YH, XPQ, GL, DWX, DLW and XJ performed the
statistical analysis. All authors contributed to data acquisition,
data analysis or data interpretation, and have reviewed and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present retrospective study was reviewed and
approved by the Ethics Committee of Huoshenshan Hospital (Wuhan,
China; approval no. HSSLL032) and written informed consent was
obtained from each enrolled patient or a first-degree relative.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, Lu R, et al: A Novel coronavirus from
patients with pneumonia in China, 2019. N Engl J Med. 382:727–733.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong
Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al: Early transmission
dynamics in Wuhan, China, of novel coronavirus-infected pneumonia.
N Engl J Med. 382:1199–1207. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He
JX, Liu L, Shan H, Lei CL, Hui DSC, et al: Clinical characteristics
of coronavirus disease 2019 in China. N Engl J Med. 382:1708–1720.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lai CC, Shih TP, Ko WC, Tang HJ and Hsueh
PR: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
and coronavirus disease-2019 (COVID-19): The epidemic and the
challenges. Int J Antimicrob Agents. 55(105924)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhao W, Zhong Z, Xie X, Yu Q and Liu J:
Relation between chest CT findings and clinical conditions of
coronavirus disease (COVID-19) Pneumonia: A multicenter study. AJR
Am J Roentgenol. 214:1072–1077. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Poland GA: SARS-CoV-2: A time for clear
and immediate action. Lancet Infect Dis. 20:531–532.
2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Calina D, Docea AO, Petrakis D, Egorov AM,
Ishmukhametov AA, Gabibov AG, Shtilman MI, Kostoff R, Carvalho F,
Vinceti M, et al: Towards effective COVID19 vaccines: Updates,
perspectives and challenges (Review). Int J Mol Med. 46:3–16.
2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Stancioiu F, Papadakis GZ, Kteniadakis S,
Izotov BN, Coleman MD, Spandidos DA and Tsatsakis A: A dissection
of SARSCoV2 with clinical implications (Review). Int J Mol Med.
46:489–508. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kang YJ: South Korea's COVID-19 infection
status: From the perspective of re-positive test results after
viral clearance evidenced by negative test results. Disaster Med
Public Health Prep. 14:762–764. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Simon V, van Bakel H and Sordillo EM:
Positive, again! What to make of ‘re-positive’ SARS-CoV-2 molecular
test results. EBioMedicine. 60(103011)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Habibzadeh P, Sajadi MM, Emami A, Karimi
MH, Yadollahie M, Kucheki M, Akbarpoor S and Habibzadeh F: Rate of
re-positive RT-PCR test among patients recovered from COVID-19.
Biochem Med (Zagreb). 30(030401)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wong J, Koh WC, Momin RN, Alikhan MF,
Fadillah N and Naing L: Probable causes and risk factors for
positive SARS-CoV-2 test in recovered patients: Evidence from
Brunei Darussalam. J Med Virol. 92:2847–2851. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
He S, Zhou K, Hu M, Liu C, Xie L, Sun S,
Sun W and Chen L: Clinical characteristics of ‘re-positive’
discharged COVID-19 pneumonia patients in Wuhan, China. Sci Rep.
10(17365)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Pan Y, Long L, Zhang D, Yuan T, Cui S,
Yang P, Wang Q and Ren S: Potential false-negative nucleic acid
testing results for severe acute respiratory syndrome coronavirus 2
from thermal inactivation of samples with low viral loads. Clin
Chem. 66:794–801. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wikramaratna PS, Paton RS, Ghafari M and
Lourenco J: Estimating the false-negative test probability of
SARS-CoV-2 by RT-PCR. Euro Surveill. 25(2000568)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hanson KE, Caliendo AM, Arias CA, Englund
JA, Lee MJ, Loeb M, Patel R, El Alayli A, Kalot MA, Falck-Ytter Y,
et al: Infectious diseases society of America guidelines on the
Diagnosis of COVID-19. Clin Infect Dis. (ciaa760)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
National Health Commission of the People's
Republic of China. Chinese management guideline for COVID-19
(version 7.0). http://www.nhc.gov.cn/xcs/zhengcwj/202003/46c9294a7dfe4cef80dc7f5912eb1989/files/ce3e6945832a438eaae415350a8ce964.pdf.
Accessed March 3, 2020.
|
|
18
|
World Health Organization: Laboratory
testing for 2019 novel coronavirus (2019-nCoV) in suspected human
cases. Interim guidance. https://www.who.int/publications/i/item/10665-331501.
Accessed January 17, 2020.
|
|
19
|
Cunarro-Lopez Y, Cano-Valderrama O,
Pintado-Recarte P, Cueto-Hernández I, González-Garzón B,
García-Tizón S, Bujan J, Asúnsolo Á, Ortega MA and De León-Luis JA:
Maternal and perinatal outcomes in patients with suspected COVID-19
and their relationship with a negative RT-PCR result. J Clin Med.
9(3552)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai
X, Li L, He R, Tan Y, Deng X, et al: Detectable 2019-nCoV viral RNA
in blood is a strong indicator for the further clinical severity.
Emerg Microbes Infect. 9:469–473. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jeong JH, Kim KH, Jeong SH, Park JW, Lee
SM and Seo YH: Comparison of sputum and nasopharyngeal swabs for
detection of respiratory viruses. J Med Virol. 86:2122–2127.
2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zou X, Chen K, Zou J, Han P, Hao J and Han
Z: Single-cell RNA-seq data analysis on the receptor ACE2
expression reveals the potential risk of different human organs
vulnerable to 2019-nCoV infection. Front Med. 14:185–192.
2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Castells L, Vargas V, Allende H, Bilbao I,
Luis Lázaro J, Margarit C, Esteban R and Guardia J: Combined
treatment with pegylated interferon (alpha-2b) and ribavirin in the
acute phase of hepatitis C virus recurrence after liver
transplantation. J Hepatol. 43:53–59. 2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tame M, Buonfiglioli F, Del Gaudio M,
Lisotti A, Cecinato P, Colecchia A, Azzaroli F, D'Errico A, Arena
R, Calvanese C, et al: Long-term leukocyte natural alpha-interferon
and ribavirin treatment in hepatitis C virus recurrence after liver
transplantation. World J Gastroenterol. 19:5278–5285.
2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Siegel I, Liu TL and Gleicher N: The
red-cell immune system. Lancet. 2:556–559. 1981.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Morera D and MacKenzie SA: Is there a
direct role for erythrocytes in the immune response? Vet Res.
42(89)2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Minasyan H: Phagocytosis and oxycytosis:
Two arms of human innate immunity. Immunol Res. 66:271–280.
2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Gonzalez-Casas R, Garcia-Buey L, Jones EA,
Gisbert JP and Moreno-Otero R: Systematic review:
Hepatitis-associated aplastic anaemia-a syndrome associated with
abnormal immunological function. Aliment Pharmacol Ther.
30:436–443. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gonzalez-Casas R, Jones EA and
Moreno-Otero R: Spectrum of anemia associated with chronic liver
disease. World J Gastroenterol. 15:4653–4658. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Young NS, Calado RT and Scheinberg P:
Current concepts in the pathophysiology and treatment of aplastic
anemia. Blood. 108:2509–2519. 2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen T, Wu D, Chen H, Yan W, Yang D, Chen
G, Ma K, Xu D, Yu H, Wang H, et al: Clinical characteristics of 113
deceased patients with coronavirus disease 2019: Retrospective
study. BMJ. 368(m1091)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chen H, Guo J, Wang C, Luo F, Yu X, Zhang
W, Li J, Zhao D, Xu D, Gong Q, et al: Clinical characteristics and
intrauterine vertical transmission potential of COVID-19 infection
in nine pregnant women: A retrospective review of medical records.
Lancet. 395:809–815. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Schoch C, Blumenthal R and Clague MJ: A
long-lived state for influenza virus-erythrocyte complexes
committed to fusion at neutral pH. FEBS Lett. 311:221–225.
1992.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Skehel JJ and Wiley DC: Receptor binding
and membrane fusion in virus entry: The influenza hemagglutinin.
Annu Rev Biochem. 69:531–569. 2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Beck Z, Brown BK, Wieczorek L, Peachman
KK, Matyas GR, Polonis VR, Rao M and Alving CR: Human erythrocytes
selectively bind and enrich infectious HIV-1 virions. PLoS One.
4(e8297)2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Davies AJ and Johnston MR: The biology of
some intraerythrocytic parasites of fishes, amphibia and reptiles.
Adv Parasitol. 45:1–107. 2000.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wessel O, Krasnov A, Timmerhaus G, Rimstad
E and Dahle MK: Antiviral responses and biological concequences of
piscine orthoreovirus infection in salmonid erythrocytes. Front
Immunol. 9(3182)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Finstad OW, Dahle MK, Lindholm TH, Nyman
IB, Løvoll M, Wallace C, Olsen CM, Storset AK and Rimstad E:
Piscine orthoreovirus (PRV) infects Atlantic salmon erythrocytes.
Vet Res. 45(35)2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wessel O, Braaen S, Alarcon M, Haatveit H,
Roos N, Markussen T, Tengs T, Dahle MK and Rimstad E: Infection
with purified Piscine orthoreovirus demonstrates a causal
relationship with heart and skeletal muscle inflammation in
Atlantic salmon. PLoS One. 12(e0183781)2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Takano T, Nawata A, Sakai T, Matsuyama T,
Ito T, Kurita J, Terashima S, Yasuike M, Nakamura Y, Fujiwara A, et
al: Full-genome sequencing and confirmation of the causative agent
of erythrocytic inclusion body syndrome in coho salmon identifies a
new type of piscine orthoreovirus. PLoS One.
11(e0165424)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hauge H, Vendramin N, Taksdal T, Olsen AB,
Wessel Ø, Mikkelsen SS, Alencar ALF, Olesen NJ and Dahle MK:
Infection experiments with novel Piscine orthoreovirus from rainbow
trout (Oncorhynchus mykiss) in salmonids. PLoS One.
12(e0180293)2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Fischer JE, Bachmann LM and Jaeschke R: A
readers' guide to the interpretation of diagnostic test properties:
Clinical example of sepsis. Intensive Care Med. 29:1043–1051.
2003.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Swets JA: Measuring the accuracy of
diagnostic systems. Science. 240:1285–1293. 1988.PubMed/NCBI View Article : Google Scholar
|