Introduction
Hypertrophic scarring is a common complication of
severe trauma, burns and surgical operation, resulting in
dysfunction and deformity of the affected parts, which affects the
quality of life of patients (1-3).
In the formation and development of a hypertrophic scar (HS), the
main features include the abnormal proliferation of fibroblasts and
inhibition of apoptosis, as well as the imbalance of extracellular
matrix collagen synthesis and degradation (4-7).
Fibroblasts in HS tissue aggregate, and their number greatly
increases. The proportion of Collagen I (Col I) and Col III
proteins (also known as type 1 and type 3 collagen, respectively)
is abnormal and their content is increased during HS formation,
which affects the scar appearance and even causes severe deformity
and dysfunction. Currently, the main treatment for hypertrophic
scar is surgical resection plus superficial X-ray radiation and
dot-matrix laser. However, surgical treatment may cause secondary
damage, and radiation therapy may cause radiation dermatitis,
delayed healing of the incision and risk of skin cancer at the
irradiated site (8). Therefore, it
is of great significance to further understand the mechanism
underlying the occurrence of HSs, and to identify new targets
associated with the molecular mechanism for the prevention and
treatment of these scars.
MicroRNAs (miRNAs) are endogenous, non-coding
single-stranded small RNAs of approximately 22 nucleotides in
length (9,10). miRNAs can regulate gene expression
through binding to the 3'-untranslated region (3'-UTR) of target
mRNAs, and have emerged as key players in a wide array of
biological processes, including cell proliferation and apoptosis
(11-13).
An increasing number of studies have reported that miRNAs are
involved in the occurrence and development of HS (14-17).
miR-18a-5p has been studied in several types of cancer, including
osteosarcoma (18,19), glioma (20), breast cancer (21), prostate cancer (22), malignant melanoma (23), esophageal carcinoma (24), and renal cell carcinoma (25), among others. Furthermore, previous
studies have revealed the inhibitory effect of miR-185-5p on
cardiac fibrosis (26) and
sub-pleural pulmonary fibrosis (27). However, to the best of our
knowledge, the expression and functional role of miR-18a-5p in HS
formation remain unclear.
Therefore, the aim of the present study was to
investigate the expression and role of miR-18a-5p in HS formation.
Since fibroblast hyperplasia and extracellular matrix deposition
are the main features of HS formation (28,29),
the effect of miR-18a-5p on scar fibroblast proliferation and
extracellular matrix deposition was further investigated in the
current study in order to explore the underlying mechanism by which
miR-18a-5p is involved in HS formation.
Materials and methods
Clinical samples
A total of 40 HS tissues from 40 patients who were
subjected to scar excision (age range, 27-51 years old; gender
ratio, 1:1) and 40 normal skin tissues from 40 patients subjected
to auto-skin grafting (age range, 28-53 years old; gender ratio,
1:1) were collected at Peking University Shenzhen Hospital
(Shenzhen, China) between February 2015 and February 2017. The
tissues were immediately stored in liquid nitrogen until further
use. For reverse transcription-quantitative PCR (RT-qPCR) analysis,
the tissues were homogenized using an ultrasonic pulverizer (MSE
Soniprep 150 Plus; MSE centrifuges). Written informed consent was
obtained from each patient enrolled in the present study, and the
study was approved by Human Ethical Committee of Peking University
Shenzhen Hospital.
Cell culture
The human embryonic skin fibroblasts CCC-ESF-1 were
obtained from Shanghai Zibo Biological Technology Co., Ltd. (cat.
no. YB-ATCC-3084; Shanghai, China). Human HS fibroblasts (hHSFs)
were provided by Shanghai Guandao Biological Engineering Co., Ltd.
(cat. no. C0618; Shanghai, China). All cells were grown in
Dulbecco's modified Eagle medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated at
37˚C with 5% CO2.
Cell transfection
hHSFs were transfected with 100 nM miR-18a-5p
inhibitor (5'-CUAUCUGCACUAGAUGCACCUUA-3'; Guangzhou RiboBio Co.,
Ltd.), 100 nM inhibitor control (5'-UUCUCCGAACGUGUCACGUTT-3';
Guangzhou RiboBio Co., Ltd.), 100 nM miR-18a-5p mimic (sense,
5'-UAAGGUGCAUCUAGUGCAGAUAG-3' and anti-sense,
5'-AUCUGCACUAGAUGCACCUUAUU-3'; Guangzhou RiboBio Co., Ltd.), 100 nM
mimic control (sense, 5'-UUCUCCGAACGUGUCACGUTT-3' and anti-sense,
5'-ACGUGACACGUUCGGAGAATT-3'; Guangzhou RiboBio Co., Ltd.), 1 µg
Smad2-plasmid (cat. no. sc-421525-ACT; Santa Cruz Biotechnology,
Inc.), 1 µg control-plasmid (cat. no. sc-108083; Santa Cruz
Biotechnology, Inc.) or 100 nM miR-18a-5p mimic + 1 µg
Smad2-plasmid using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) in line with the manufacturer's
protocol. At 48 h after transfection, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
performed to determine transfection efficiency.
RT-qPCR assay
In order to collect the total RNA from tissues or
cells, TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
was used in line with the manufacturer's protocol. RNA
concentration was determined using a NanoDropTM 2000c
Spectrophotometer (Thermo Fisher Scientific, Inc.). For miRNA
detection, reverse transcription and qPCR were performed using
miScript II Reverse Transcription kit (Qiagen GmbH) and miSCRIPT
SYBR Green PCR Kit (Qiagen GmbH), respectively, as per the
manufacturer's protocols. For mRNA detection,
PrimeScriptTM RT reagent kit (Takara Bio, Inc.) was used
for reverse transcription, followed by the SYBR Premix Ex Taq™ II
(Tli RNaseH Plus) kit (Takara Bio, Inc.) was applied for qPCR
analysis, following the manufacturer's protocol. The thermal
cycling conditions were as follows: 35 cycles at 95˚C for 15 sec,
annealing at 60˚C for 1 min, then chain extension at 72˚C for 1 min
and a final extension step at 72˚C for 10 min. U6 and GAPDH were
used as the internal controls for miRNA and mRNA, respectively.
Primer sequences for PCR were listed as follows: GAPDH forward,
5'-TTTGGTATCGTGGAAGGACTC-3' and reverse,
5'-GTAGAGGCAGGGATGATGTTCT-3'; U6 forward,
5'-GCTTCGGCAGCACATATACTAAAAT-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'; miR-18a-5p forward,
5'-ACGTAAGGTGCATCTAGTGCAGATA-3' and reverse,
5'-GTGCAGGGTCCGAGGT-3'; type I collagen forward,
5'-CCCTGAGTGGAAGAGTGGAG-3' and reverse, 5'-GAGGCGTGAGGTCTTCTGTG-3';
Type III collagen forward, 5'-GGAGCTGGCTACTTCTCGC-3' and reverse,
5'-GGGAACATCCTCCTTCAACAG-3' and Smad2 forward,
5'-CGTCCATCTTGCCATTCACG-3' and reverse,
5'-CTCAAGCTCATCTAATCGTCCTG-3'. The relative gene expression was
quantified using the 2-ΔΔCq method (30).
Western blot assay
Radioimmunoprecipitation assay lysis buffer
(Beyotime Institute of Biotechnology) was used to collect the
proteins from cells, and bicinchoninic acid protein assay was then
conducted to quantify the protein concentration. Equal amount of
protein (40 µg/lane) was separated by 12% SDS-PAGE and then
transferred to polyvinylidene difluoride membranes. Next, the
membranes were blocked with 5% non-fat milk at room temperature for
1.5 h, incubated with primary antibodies: Smad2 (1:1,000; cat. no.
5339; Cell Signaling Technology, Inc.), Col I (1:1,000; cat. no.
ab34710; Abcam), Col III (1:1,000; cat. no. Ab7778; Abcam) and
β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.)
at 4˚C overnight. Subsequently, membranes were incubated with
anti-rabbit horseradish peroxidase-conjugated immunoglobulin G
secondary antibody (cat. no. 7074; 1:2,000; Cell Signaling
Technology, Inc.) at room temperature for 2 h. At the end of the
experiment, an enhanced chemiluminescence detection system
(Applygen Technologies, Inc., Beijing, China) was used to observe
the protein bands. For densitometry detection, analysis with ImageJ
1.38X software (National Institutes of Health, Bethesda, MD, USA)
was performed.
Dual-luciferase reporter assay
Bioinformatics analysis using the TargetScanHuman
7.2 tool (www.targetscan.org/vert_72) was performed to predict
the target genes of miR-18a-5p. The results revealed the binding
sites between miR-18a-5p and the 3'-UTR of Smad2. Next, in order to
confirm these binding sites, dual-luciferase reporter assay was
conducted (31). Briefly,
the Smad2-WT vector with the wild-type 3'-UTR of Smad2 mRNA and the
Smad2-MUT vector with the mutated 3'-UTR of Smad2 mRNA were
constructed with the dual-luciferase reporter vector
pmiR-RB-REPORT™ (Guangzhou RiboBio Co., Ltd.). Subsequently, hHSFs
were co-transfected with miR-18a-5p mimic or mimic control, and
with Smad2-WT or Smad2-MUT using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. At 48 h after cell transfection, the
relative luciferase activity was measured using a dual-luciferase
reporter assay system (Promega Corporation) following the
manufacturer's protocols. Luciferase activity was normalized to the
Renilla luciferase activity.
MTT assay
To determine cell proliferation, an MTT assay was
performed. Briefly, hHSFs were seeded into 96-well plates
(1x104 cells per well), and then transfected with
miR-18a-5p inhibitor, inhibitor control, miR-18a-5p mimic, mimic
control or miR-18a-5p mimic + Smad2-plasmid for 48 h. Next, 20 µl
MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) was added to each well and
cultured at 37˚C with 5% CO2 for a further 4 h. The
optical density value at 490 nm was detected using a microplate
reader. Experiments were repeated three times.
Flow cytometry assay
hHSFs were collected in the logarithmic growth phase
and inoculated into 6-well plates at 1x105 cells/well.
hHSFs were then transfected with miR-18a-5p inhibitor, inhibitor
control, miR-18a-5p mimic, mimic control or miR-18a-5p mimic +
Smad2-plasmid for 48 h. Then, cell apoptosis was assessed using the
Annexin V-FITC Apoptosis Detection kit (cat. no. 70-AP101-100;
MultiSciences, Hangzhou, China) and a flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA), according to the
manufacturer's protocol. The data were then analyzed using WinMDI
software (version 2.5; www.cyto.purdue.edu/flowcyt/software/Catalog.htm).
Statistical analysis
Data are expressed as the mean ± standard deviation
of experiments conducted at least in triplicate. The SPSS software,
version 18.0 (IBM Corp., Armonk, NY, USA) was used to perform
statistical analysis. Comparisons between groups were assessed by
Student's t-test, or by one-way analysis of variance and subsequent
Tukey's post-hoc test. P<0.05 was considered to denote a
statistically significant difference.
Results
miR-18a-5p is downregulated in HS
tissues and hHSFs
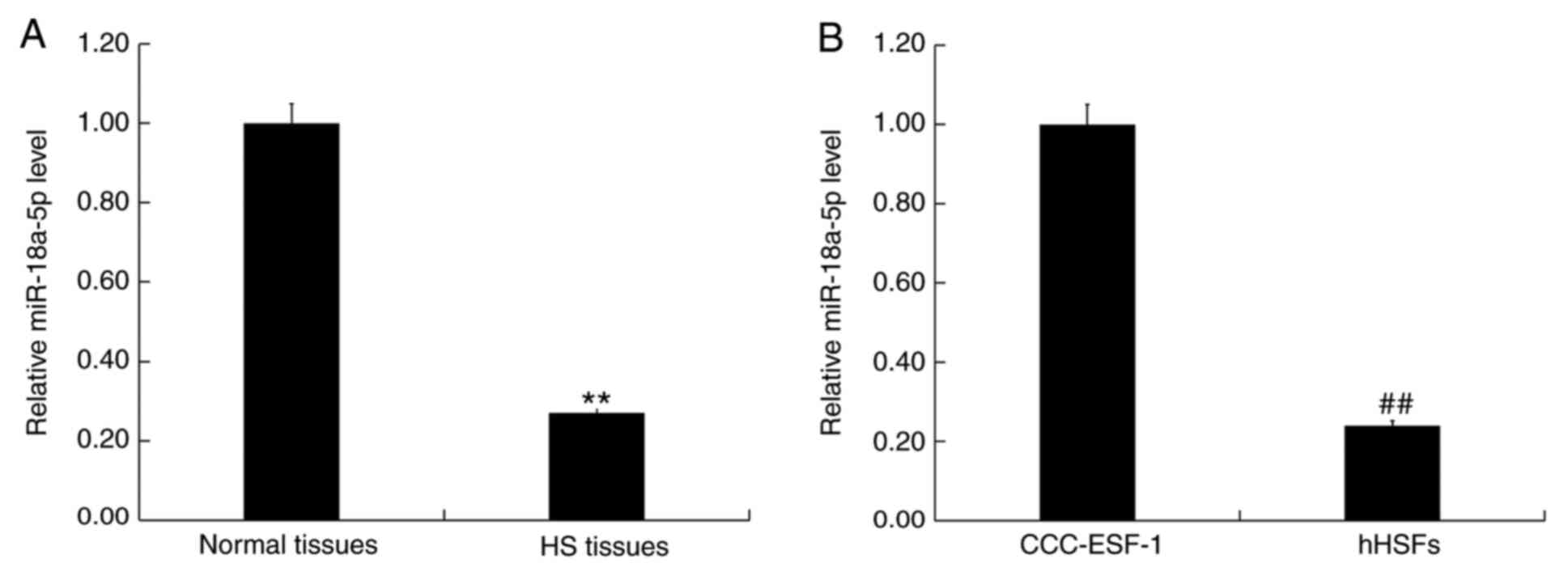
In order to detect the level of miR-18a-5p in HS
tissues and normal skin tissues, as well as in the human embryonic
skin fibroblasts CCC-ESF-1 and hHSFs, RT-qPCR analysis was
performed. The results demonstrated that, compared with the normal
skin tissues, the level of miR-18a-5p was significantly
downregulated in HS tissues (Fig.
1A). Furthermore, compared with the normal fibroblasts
CCC-ESF-1, the level of miR-18a-5p in hHSFs was significantly
decreased (Fig. 1B).
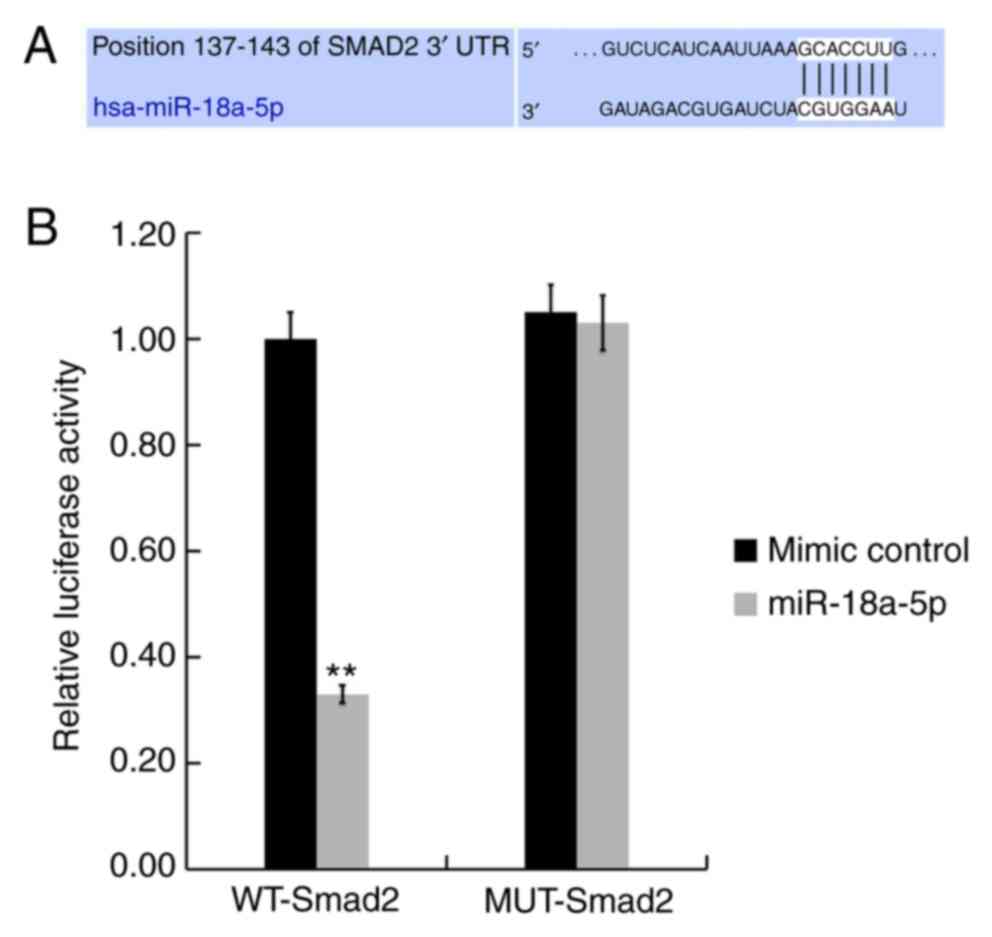
Smad2 is a target of miR-18a-5p
TargetScanHuman 7.2 was used to conduct
bioinformatics analysis, and the binding sites between miR-18a-5p
and the 3'-UTR of Smad2 were identified (Fig. 2A). Next, the findings of the
dual-luciferase reporter assay indicated that, compared with the
mimic control group, miR-18a-5p mimic transfection significantly
decreased the luciferase activity of hHSFs co-transfected with
Smad2-WT. By contrast, no significant difference was observed in
cells co-transfected with Smad2-MUT and miR-18a-5p mimic or mimic
control (Fig. 2B). These results
indicated that miR-18a-5p directly targets Smad2.
miR-18a-5p inhibition promotes
proliferation, inhibits apoptosis and enhances extracellular matrix
deposition in hHSFs
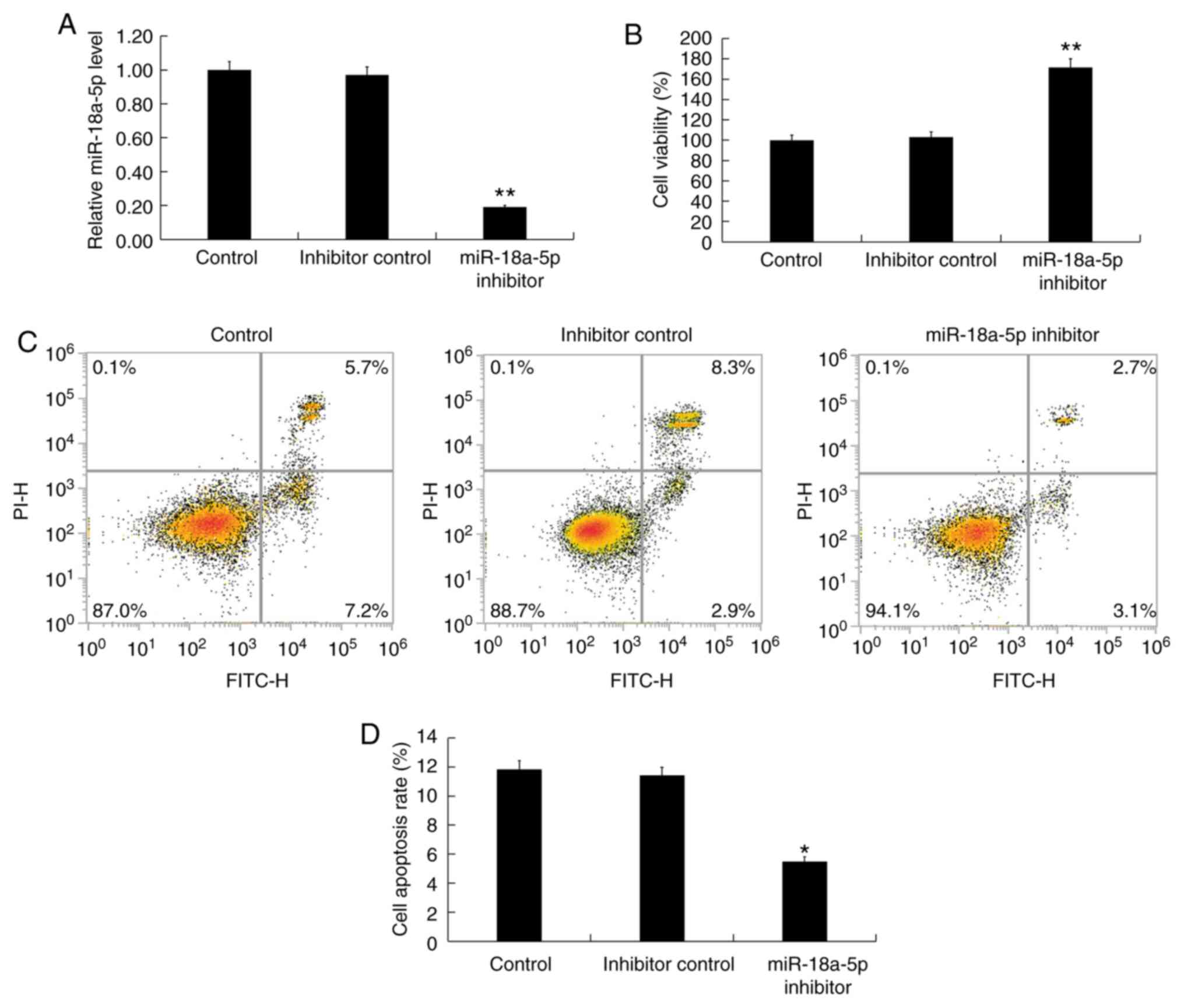
Subsequently, in order to investigate the effect of
miR-18a-5p downregulation on hHSFs, the hHSFs were transfected with
inhibitor control or miR-18a-5p inhibitor for 48 h. The RT-qPCR
results indicated that transfection with miR-18a-5p inhibitor
significantly decreased the expression of miR-18a-5p in hHSFs,
compared with the untransfected control and inhibitor control
groups (Fig. 3A). Next, it was
observed that, compared with the control groups, miR-18a-5p
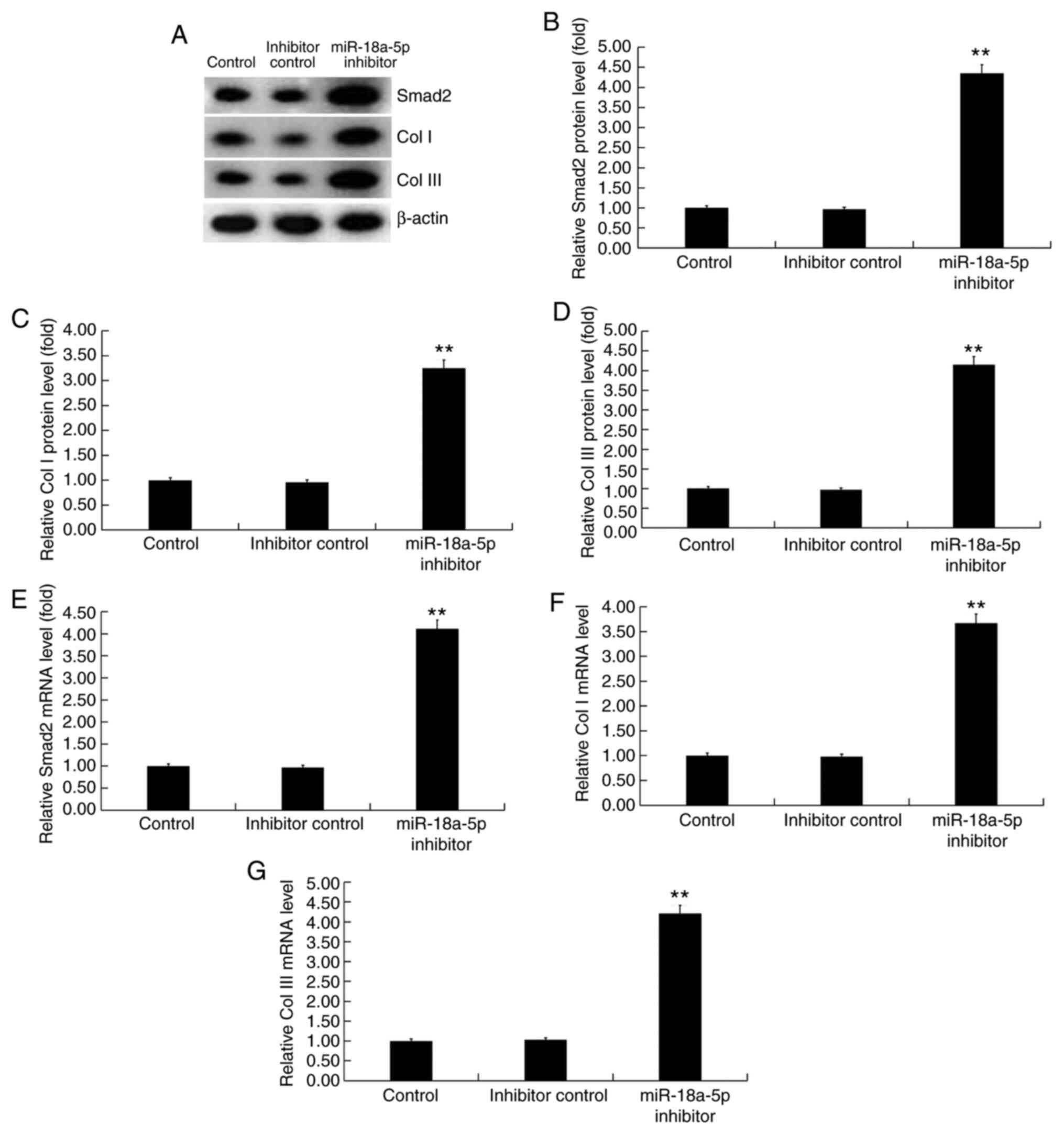
inhibitor significantly promoted cell proliferation (Fig. 3B), decreased cell apoptosis
(Fig. 3C and D), and enhanced the protein (Fig. 4A-D) and mRNA (Fig. 4E-G) expression levels of Smad2
(Fig. 4A, B and E),
Col I (Fig. 4A, C and F),
and Col III (Fig. 4A, D and G) in
hHSFs. These findings suggest that miR-18a-5p inhibitor could
enhance extracellular matrix deposition by hHSFs.
miR-18a-5p upregulation inhibits
proliferation, induces apoptosis and represses extracellular matrix
deposition in hHSFs
Finally, the effect of miR-18a-5p upregulation on
hHSFs was investigated, and hHSFs were transfected with miR-18a-5p
mimic, mimic control or miR-18a-5p mimic + Smad2-plasmid for 48 h.
In addition, in order to confirm the transfection efficiency of
Smad2-plasmid, hHSFs were also transfected with control-plasmid or
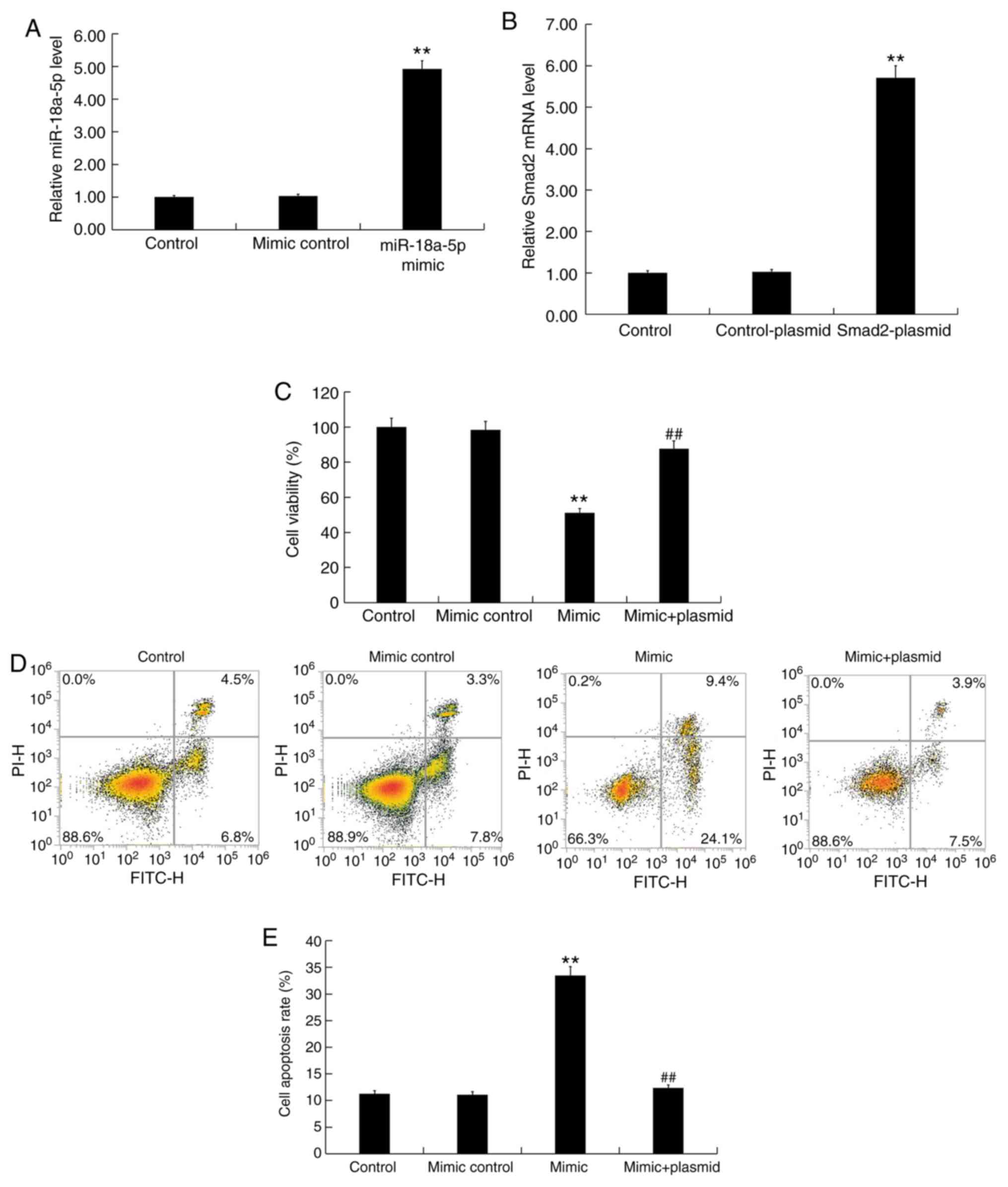
Smad2-plasmid for 48 h. The results confirmed that miR-18a-5p mimic
transfection significantly increased the level of miR-18a-5p in
hHSFs (Fig. 5A), while
Smad2-plasmid transfection significantly enhanced the mRNA level of
Smad2 in hHSFs (Fig. 5B). Further
analysis indicated that compared with the control groups,
miR-18a-5p mimic transfection significantly inhibited the
proliferation (Fig. 5C) and
increased the apoptosis (Fig. 5D
and E) of hHSFs, while it markedly
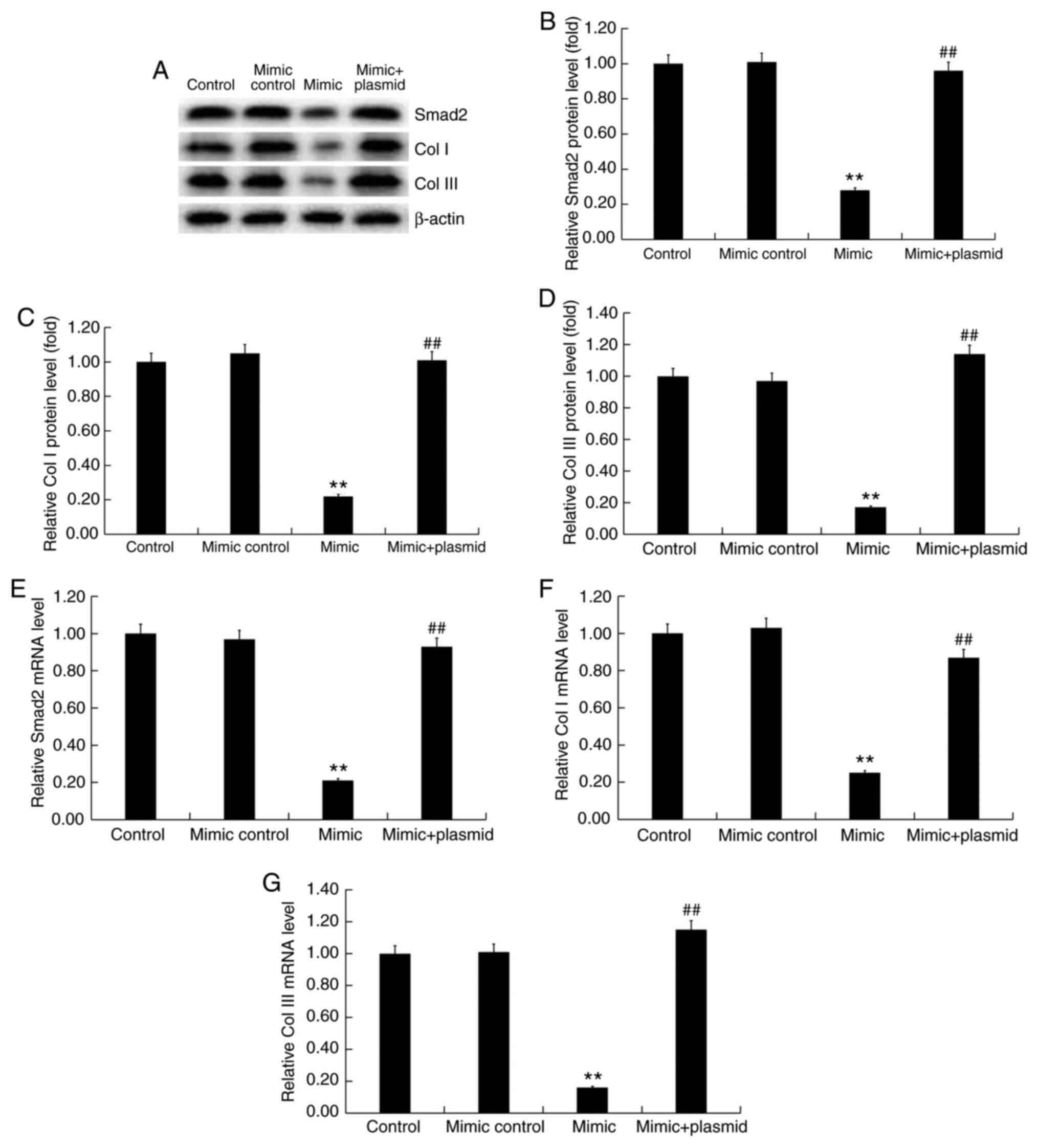
reduced the protein (Fig. 6A-D) and
mRNA (Fig. 6E-G) expression levels
of Smad2 (Fig. 6A, B and E),
Col I (Fig. 6A, C and F),
and Col III (Fig. 6A, D and G) in
hHSFs. Notably, upon co-transfection of cells with miR-18a-5p mimic
and Smad2-plasmid, all the effects of miR-18a-5p mimic on hHSFs
were significantly reversed by the Smad2 overexpression (Figs. 5C-E and 6). These findings suggest that miR-18a-5p
mimic can reduce extracellular matrix deposition by hHSFs.
Discussion
In the present study, miR-18a-5p was demonstrated to
be significantly downregulated in HS tissues and hHSFs. In
addition, the results revealed that Smad2 is a target of
miR-18a-5p, and that it was upregulated in HS tissues and hHSFs.
Further analysis indicated that miR-18a-5p downregulation
significantly promoted cell proliferation and decreased cell
apoptosis, as well as enhanced the expression levels of Smad2, Col
I and Col III in hHSFs. By contrast, miR-18a-5p upregulation
significantly inhibited hHSF proliferation, increased cell
apoptosis, and reduced the expression levels of Smad2, Col I and
Col III in hHSFs. All the effects of miR-18a-5p upregulation on
hHSFs were significantly reversed by Smad2 overexpression. Taken
together, the data suggested that the miR-18a-5p/Smad2 axis may be
a potential therapeutic target for HS treatment.
HS formation does not only cause cosmetic
disfigurement of the human body, but also leads to discomfort, such
as pain and itching, and even serious deformity and loss of
function, affecting the patient's quality of life (29,30,32-34).
Therefore, the formation mechanism and treatment methods of HSs
have received increasing attention, and methods for effectively
eliminating scars have great clinical significance. Current
research has confirmed the important role of miRNAs in the
development of skin wound repair and skin diseases (35,36),
while numerous studies have also reported the important involvement
of miRNAs in HS formation (14-17).
miR-18a-5p, which has been well studied in cancer, serves a
critical role in the regulation of cell growth (18-25).
As fibroblast hyperplasia and extracellular matrix deposition are
the main features of HS formation (28,29),
it was hypothesized in the present study that miR-18a-5p may also
serve an important role in the formation of HS.
Initially, the expression of miR-18a-5p in HS and
normal skin tissues was measured, and the results indicated that
miR-18a-5p was significantly downregulated in HS tissues. However,
it is worth noting that the age of patients who participated in the
current study ranged between 27 and 51 years. Since age affects the
characteristics of scar tissue and the quality of life of patients
(37,38), the wide age range of patients
enrolled in the present study was a limitation of the study. The
level of miR-18a-5p in the human embryonic skin fibroblasts
CCC-ESF-1 and hHSFs was also detected in the current study using
RT-qPCR, and the results indicated that miR-18a-5p was
significantly downregulated in hHSFs. Subsequently, Smad2 was
identified to be a target of miR-18a-5p. Consistent with the
findings of a previous study (39),
it was observed herein that Smad2 was significantly upregulated in
HS tissues. An earlier study reported that silencing of Smad2 gene
expression was able to inhibit the TGF-β signaling pathway and thus
reduce HS formation (40). The
Smad2/3-TGF-β1 signaling pathway serves a key role in the
regulation of target gene transcription, matrix protein production
(including fibronectin and collagen) and extracellular matrix
synthesis and degradation (41,42).
A previous study demonstrated that miR-486-5p
inhibited the proliferation, induced apoptosis and G1/S phase
arrest in hHSFs by targeting Smad2(43). In the present study, the effect of
miR-18a-5p on the proliferation of hHSFs was investigated.
Downregulation of miR-18a-5p in hHSFs significantly promoted the
proliferation and inhibited the apoptosis of hHSFs, whereas
miR-18a-5p upregulation inhibited the proliferation and increased
the apoptosis of hHSFs. All the effects of miR-18a-5p upregulation
on the proliferation and apoptosis of hHSFs were significantly
reversed by Smad2 overexpression. Conversely, previous studies have
indicated that miR-18a-5p promoted the cell proliferation in
certain epithelial cells (18,25),
while another study has reported that Smad2 suppressed the
proliferation of epithelial cells. These contradictory results may
be due to the different functions of miRNAs/genes in different
microenvironments, and may also be caused by the varying
sensitivity of different genes to different signaling pathways
under specific conditions. For instance, miR-18a-5p may inhibit the
endothelial-mesenchymal transition and cardiac fibrosis through
repressing Notch2 pathway (26). In
addition, miR-18a-5p induced intrinsic keratinocyte apoptosis in
patients with toxic epidermal necrolysis through downregulating
BCL2L10 expression (44).
miR-18a-5p was also able to promote MG-63 osteosarcoma cell
invasion and migration by directly regulating the expression of
interferon regulatory factor 2(18). However, the specific mechanisms of
the different roles of miR-18a-5p/Smad2 in epithelial cells and
fibroblasts require further in-depth research.
Collagen deposition caused by collagen metabolism
disorder is one of the main pathological bases of HS. Collagen is
the main component of extracellular matrix and is mainly secreted
by skin fibroblasts. Increased expression of Col I and Col III has
been detected in the skin scar tissues following trauma (45). In the present study, the effect of
miR-18a-5p on extracellular matrix production was also explored in
hHSFs. As expected, the data demonstrated that miR-18a-5p
downregulation significantly enhanced the expression levels of Col
I and Col III in hHSFs, while miR-18a-5p upregulation resulted in
the opposite effect. It is worth mentioning that the effects of
miR-18a-5p upregulation on Col I and Col III expression in hHSFs
were significantly reversed by Smad2 overexpression. However,
extracellular matrix deposition was only investigated by western
blot assay in the present study, whereas it would be more
appropriate to characterize ECM deposition using immunochemistry or
immunofluorescence, which is thus a limitation of the present
study.
In conclusion, to be best of our knowledge, the
present study is the first to reveal that miR-18a-5p is
downregulated in HS, and that its upregulation inhibited hHSF
proliferation, promoted apoptosis, and reduced Col I and Col III
expression levels in hHSFs by targeting Smad2. Therefore,
miR-18a-5p may serve as a novel target for the diagnosis and
treatment of HS. However, the present study is only a preliminary
investigation of the role of miR-18a-5p in HSs. In order to verify
the study conclusions, more in-depth research is necessary. For
example, the sample size examined was small, and thus the
expression of miR-18a-5p in larger HS tissue samples should be
analyzed. Furthermore, the correlation between the expression of
miR-18a-5p and the clinical features of HS patients, as well as the
expression and role of Smad2 in HS, should be further investigated.
Besides, the specific mechanism of the positive regulation of Col I
and Col III expression levels by Smad2 observed in the present
study needs further exploration. Finally, in vivo studies
should be performed to confirm these findings. These issues will be
addressed in future studies.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TSL contributed to the study design, data
collection, statistical analysis, data interpretation and
manuscript preparation. YGW and DDL contributed to data collection
and data interpretation. LDZ contributed to statistical analysis
and literature search. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from each
patient enrolled in the present study, and the study was approved
by Human Ethical Committee of Peking University Shenzhen Hospital
(Guangdong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tyack ZF, Pegg S and Ziviani J: Postburn
dyspigmentation: Its assessment, management, and relationship to
scarring-a review of the literature. J Burn Care Rehabil.
18:435–440. 1997.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Atiyeh BS, El Khatib AM and Dibo SA:
Pressure garment therapy (PGT) of burn scars: Evidence-based
efficacy. Ann Burns Fire Disasters. 26:205–212. 2013.PubMed/NCBI
|
|
3
|
Gauglitz GG, Korting HC, Pavicic T,
Ruzicka T and Jeschke MG: Hypertrophic scarring and keloids:
Pathomechanisms and current and emerging treatment strategies. Mol
Med. 17:113–125. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Younai S, Nichter LS, Wellisz T, Reinisch
J, Nimni ME and Tuan TL: Modulation of collagen synthesis by
transforming growth factor-beta in keloid and hypertrophic scar
fibroblasts. Ann Plast Surg. 33:148–154. 1994.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Aarabi S, Bhatt KA, Shi Y, Paterno J,
Chang EI, Loh SA, Holmes JW, Longaker MT, Yee H and Gurtner GC:
Mechanical load initiates hypertrophic scar formation through
decreased cellular apoptosis. FASEB J. 21:3250–3261.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
van der Veer WM, Bloemen MC, Ulrich MM,
Molema G, van Zuijlen PP, Middelkoop E and Niessen FB: Potential
cellular and molecular causes of hypertrophic scar formation.
Burns. 35:15–29. 2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Atiyeh BS: Nonsurgical management of
hypertrophic scars: Evidence-based therapies, standard practices,
and emerging methods. Aesthetic Plast Surg. 31:468–492, Discussion
493-494. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zuccaro J, Ziolkowski N and Fish J: A
systematic review of the effectiveness of laser therapy for
hypertrophic burn scars. Clin Plast Surg. 44:767–779.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mo YY: MicroRNA regulatory networks and
human disease. Cell Mol Life Sci. 69(3529)2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Laffont B and Rayner KJ: MicroRNAs in the
pathobiology and therapy of atherosclerosis. Can J Cardiol.
33:313–324. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen L and Li J, Li Q, Yan H, Zhou B, Gao
Y and Li J: Non-coding RNAs: The new insight on hypertrophic scar.
J Cell Biochem. 118:1965–1968. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li P, He QY and Luo CQ: Overexpression of
miR-200b inhibits the cell proliferation and promotes apoptosis of
human hypertrophic scar fibroblasts in vitro. J Dermatol.
41:903–911. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang X, Zhang Y, Jiang BH, Zhang Q, Zhou
RP, Zhang L and Wang C: Study on the role of Hsa-miR-31-5p in
hypertrophic scar formation and the mechanism. Exp Cell Res.
361:201–209. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xiao YY, Fan PJ, Lei SR, Qi M and Yang XH:
MiR-138/peroxisome proliferator-activated receptor β signaling
regulates human hypertrophic scar fbroblast proliferation and
movement in vitro. J Dermatol. 42:485–495. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lu C, Peng K, Guo H, Ren X, Hu S, Cai Y,
Han Y, Ma L and Xu P: miR-18a-5p promotes cell invasion and
migration of osteosarcoma by directly targeting IRF2. Oncol Lett.
16:3150–3156. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fei D, Zhang X, Liu J, Tan L, Xing J, Zhao
D and Zhang Y: Long noncoding RNA FER1L4 suppresses tumorigenesis
by regulating the expression of PTEN targeting miR-18a-5p in
osteosarcoma. Cell Physiol Biochem. 51:1364–1375. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu Q, Yu W, Zhu S, Cheng K, Xu H, Lv Y,
Long X, Ma L, Huang J, Sun S and Wang K: Long noncoding RNA GAS5
regulates the proliferation, migration, and invasion of glioma
cells by negatively regulating miR-18a-5p. J Cell Physiol.
234:757–768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang N, Zhang H, Liu Y, Su P, Zhang J,
Wang X, Sun M, Chen B, Zhao W, Wang L, et al: SREBP1, targeted by
miR-18a-5p, modulates epithelial-mesenchymal transition in breast
cancer via forming a co-repressor complex with Snail and HDAC1/2.
Cell Death Differ. 26:843–859. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang G, Han G, Zhang X, Yu Q, Li Z, Li Z
and Li J: Long non-coding RNA FENDRR reduces prostate cancer
malignancy by competitively binding miR-18a-5p with RUNX1.
Biomarkers. 23:435–445. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang Y, Qian W, Feng F, Cao Q, Li Y, Hou
Y, Zhang L and Fan J: Upregulated lncRNA CASC2 may inhibit
malignant melanoma development through regulating miR-18a-5p/RUNX1.
Oncol Res. 27:371–377. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang W, He W, Gao J, Wang Y, Zang W, Dong
Z and Zhao G: RETRACTED: The long noncoding RNA CASC2 inhibits
tumorigenesis through modulating the expression of PTEN by
targeting miR-18a-5p in esophageal carcinoma. Exp Cell Res.
361:30–38. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhou L, Li Z, Pan X, Lai Y, Quan J, Zhao
L, Xu J, Xu W, Guan X, Li H, et al: Identification of miR-18a-5p as
an oncogene and prognostic biomarker in RCC. Am J Transl Res.
10:1874–1886. 2018.PubMed/NCBI
|
|
26
|
Geng H and Guan J: MiR-18a-5p inhibits
endothelial-mesenchymal transition and cardiac fibrosis through the
Notch2 pathway. Biochem Biophys Res Commun. 491:329–336.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang Q, Ye H, Xiang F, Song LJ, Zhou LL,
Cai PC, Zhang JC, Yu F, Shi HZ, Su Y, et al: miR-18a-5p inhibits
sub-pleural pulmonary fibrosis by targeting TGF-β receptor II. Mol
Ther. 25:728–738. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tredget EE, Nedelec B, Scott PG and
Ghahary A: Hypertrophic scars, keloids, and contractures. The
cellular and molecular basis for therapy. Surg Clin North Am.
77:701–730. 1997.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Syed F, Ahmadi E, Iqbal SA, Singh S,
McGrouther DA and Bayat A: Fibroblasts from the growing margin of
keloid scars produce higher levels of collagen I and III compared
with intralesional and extralesional sites: Clinical implications
for lesional site-directed therapy. Br J Dermatol. 164:83–96.
2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lu Q, Guo Z and Qian H: Role of
microRNA-150-5p/SRCIN1 axis in the progression of breast cancer.
Exp Ther Med. 17:2221–2229. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Aarabi S, Longaker MT and Gunner GC:
Hypertrophic scar formation following burns and trauma: New
approaches to treatment. PLoS Med. 4(e234)2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gangemi EN, Gregori D, Berchialla P,
Zingarelli E, Cairo M, Bollero D, Ganem J, Capocelli R, Cuccuru F,
Cassano P, et al: Epidemiology and risk factors for pathologic
scarring after burn wounds. Arch Facial Plast Surg. 10:93–102.
2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Schneider JC, Holavanahalli R, Helm P,
Goldstein R and Kowalske K: Contractures in burn injury: Defining
the problem. J Burn Care Res. 27:508–514. 2006.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Aberdam D, Candi E, Knight RA and Melino
G: miRNAs, ‘sternness’ and skin. Trends Biochem Sci. 33:583–591.
2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Botchkareva NV: MicroRNA/mRNA regulatory
networks in the control of skin development and regeneration. Cell
Cycle. 11:468–474. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Butzelaar L, Ulrich MM, Mink van der Molen
AB, Niessen FB and Beelen RH: Currently known risk factors for
hypertrophic skin scarring: A review. J Plast Reconstr Aesthet
Surg. 69:163–169. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Xiao Y, Sun Y, Zhu B, Wang K, Liang P, Liu
W, Fu J, Zheng S, Xiao S and Xia Z: Risk factors for hypertrophic
burn scar pain, pruritus, and paresthesia development. Wound Repair
Regen. 26:172–181. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Qi J, Liu Y, Hu K, Zhang Y, Wu Y and Zhang
X: MicroRNA-26a inhibits hyperplastic scar formation by targeting
Smad2. Exp Ther Med. 15:4332–4338. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yin L, Zhao X, Ji S, He C, Wang G, Tang C,
Gu S and Yin C: The use of gene activated matrix to mediate
effective Smad2 gene silencing against hypertrophic scar.
Biomaterials. 35:2488–2498. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Cao S, Xiao L, Rao JN, Zou T, Liu L, Zhang
D, Turner DJ, Gorospe M and Wang JY: Inhibition of Smurf2
translation by miR-322/503 modulates TGF-β/Smad2 signaling and
intestinal epithelial homeostasis. Mol Biol Cell. 25:1234–1243.
2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yin K, Yin W, Wang Y, Zhou L, Liu Y, Yang
G, Wang J and Lu J: MiR-206 suppresses epithelial mesenchymal
transition by targeting TGF-β signaling in estrogen receptor
positive breast cancer cells. Oncotarget. 7:24537–24548.
2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shi Y, Wang L, Yu P, Liu Y and Chen W:
MicroRNA-486-5p inhibits the growth of human hypertrophic scar
fibroblasts by regulating Smad2 expression. Mol Med Rep.
19:5203–5210. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ichihara A, Wang Z, Jinnin M, Izuno Y,
Shimozono N, Yamane K, Fujisawa A, Moriya C, Fukushima S, Inoue Y
and Ihn H: Upregulation of miR-18a-5p contributes to epidermal
necrolysis in severe drug eruptions. J Allergy Clin Immunol.
133:1065–1074. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhou R, Zhang Q, Zhang Y, Fu S and Wang C:
Aberrant miR-21and miR-200b expression and its pro-fbrotic
potential in hypertrophic scars. Exp Cell Res. 339:360–366.
2015.PubMed/NCBI View Article : Google Scholar
|