Introduction
Ulcerative colitis (UC), which is a type of
inflammatory bowel disease (IBD), is a chronic intestinal disorder
of multifactorial etiology (1,2). The
principal outcome of UC is the development of colorectal cancer
(3,4).
The pathogenesis of IBD is not fully understood. It
is believed that chronic inflammation and immune response disorders
are the key pathological features in UC, whereas an imbalance in
gut microbiota is a key factor leading to inflammation and abnormal
immune response, thereby contributing to disease initiation and
progression (5). Under a normal
physiological state, the intestinal flora remains stable and
interacts with the host, serving an important role in nutrient
absorption, the prevention of pathogen invasion and the maintenance
of normal immune function (6). As
the microbiota is an important factor that is associated with the
maturation of the early immune system, it effectively establishes
an interaction with the host and is the principal factor leading to
chronic inflammatory disorders, such as UC (7). Previous observations from clinical and
experimental studies have indicated that an intestinal bacteria
imbalance is associated with disease initiation and progression in
UC (8,9). The alterations in the characteristics
of the intestinal flora of patients with IBD include variations in
the total number of mucosal bacteria and diversities in the
composition of the flora early in the disease (10).
A previous study reported that the amount of total
bacteria is decreased in patients with active UC, especially the
balance of Staphylococci/Bacilli, which is characterized by
an increased abundance of Staphylococci and a decreased
abundance of Bacilli. Subsequently, UC was attributed to the
alterations in the abundance of several bacterial species (11). In our previous study (12), the abundance of different types of
bacteria was examined using PCR, and it was revealed that UC was
attributed to the alterations in the abundance of several bacterial
species.
A recent study revealed that the manifestation of
dysbiosis in patients with UC is characterized by a decrease in
bacterial diversity, and this study concluded that an alteration in
the diversity and composition of the gut microbiome, rather than
the presence of specific pathogens, is likely to serve a critical
role in disease progression (11).
Furthermore, the abundance of invasive bacteria types was indicated
to increase, and that of protective bacteria types was revealed to
decrease in patients with UC (13).
For example, it was demonstrated that the increased abundance of
Enterobacteriaceae, Pasteurellaceae,
Veillonellaceae and Fusobacteriaceae, and the
decreased abundance of Erysipelotrichales,
Bacteroidales and Clostridiales, were associated with
UC disease status (14). Another
study has indicated that in patients with both Crohn's disease (CD)
and UC, a decreased biodiversity, a reduced proportion of
Firmicutes and an increased abundance of
Gammaproteobacteria were observed when compared with healthy
controls (15). In addition, it is
widely believed that the gut microbiota of patients with active UC
is different from that of healthy controls, whereas to the best of
our knowledge, no evidence for a difference between remission and
control groups exists. Hence, the current study aimed to assess
whether gut microbiota may be a potential target for controlling UC
progression.
Currently, there is little evidence to support the
alterations in intestinal aerobes and common anaerobes in patients
with UC, owing to the differences in the disease course, stage,
treatment, analysis method and microbiota complexity. Additionally,
their specific role in the occurrence of the host disease has not
yet been fully elucidated. Furthermore, to the best of our
knowledge, a relationship between microbiota alterations and
mucosal damage has not yet been demonstrated. The majority of the
studies on intestinal inflammation have focused on the association
between interleukins and the immune system, which demonstrated that
interleukin-17 (IL-17) and interleukin-23 (IL-23) serve a critical
role in intestinal inflammation and exhibit a typical positive
correlation with UC (16-18).
In the present study, the differences in intestinal flora between
patients with UC and healthy controls were compared, and the
relationship between microbiota alterations and inflammation was
explored, which provided novel insights that may be used for the
diagnosis and treatment of UC.
Materials and methods
Patient recruitment
A total of 16 patients with UC (age range, 40-60
years, median age, 53 years; 7 male and 9 female) were recruited at
Huaihe Hospital of Henan University (Kaifeng, China) from July 2016
to June 2017. Patients with different degrees of abdominal pain,
diarrhea and mucopurulent bloody stool, who were diagnosed with UC
by fibro-colonoscopy and routine pathological examination [meeting
the diagnostic criteria published in the Consensus on the Standards
for the Diagnosis and Cure of Inflammatory Bowel Diseases in China
(19)], were included in the
current study according to the Montreal standard for the evaluation
of clinical performance (20). A
total of 10 healthy control volunteers (age range, 35-60 years old,
median age, 46 years old; 4 male and 6 female) who had not received
antibiotics in the previous three months, were recruited at Huaihe
Hospital of Henan University (Kaifeng, China) from July 2016 to
June 2017. The healthy controls displayed no evidence of active
pathology. Written informed consent was obtained from all patients
prior to the procedure. Ethical approval was obtained from the
Medical Ethics Committee of Huaihe Hospital of Henan University
(Kaifeng, China). The patients were excluded from the present study
if at least one of the following criteria were met: i) Received
antibiotics or probiotics within 4 weeks before specimen
collection; ii) diagnosed with infective enteritis, such as
bacterial dysentery, intestinal tuberculosis, schistosomiasis,
Crohn's disease, ischaemic enteritis, radiation enteritis,
irritable bowel syndrome or colon carcinoma, via fibro-colonoscopy;
iii) suffered from coronary heart disease, hypertensive disease,
diabetes, active pulmonary tuberculosis or peptic ulcers.
Histological evaluation
Colon tissues were fixed in 4% paraformaldehyde at
room temperature overnight. Paraffin-embedded colon tissues were
cut into 5-µm-thick sections for hematoxylin and eosin (H&E)
staining, the slices were stained at room temperature for 5 min
with hematoxylin and 1min with eosin. The histological index was
estimated based on the histological severity of colitis. The
pathological sections, which were stained with H&E, were
observed under a light microscope (magnification, x200) and were
scored independently by two pathologists via the blind method.
According to the specific scoring criteria (21), inflammatory cell infiltration (0
point, sporadic inflammatory cells in lamina propria; 1 point,
number of inflammatory cells in lamina propria increased; 2 points,
number of inflammatory cells extended to the submucosa; and 3
points, infiltration of inflammatory cells into the intestinal
wall) and tissue damage (0 point, no mucosal injury; 1 point,
lymphoepithelial injury; 2 points, surface mucosal erosion or focal
ulcer; and 3 points, extensive mucosal injury and structure
extending to deeper intestinal wall) were combined. The details are
presented in Table I. The two
sub-scores were added and the combined histological score ranged
from 0 (no alterations) to 6 (highest score with extensive cell
infiltration and tissue damage).
 | Table IScores of histology activity. |
Table I
Scores of histology activity.
| Property | Score |
|---|
| Inflammatory cell
infiltration | |
| Sporadic
inflammatory cells in lamina propria | 0 |
| Number of
inflammatory cells in lamina propria increased | 1 |
| Number of
inflammatory cells extended to the submucosa | 2 |
| Infiltration of
inflammatory cells into the intestinal wall | 3 |
| Tissue damage | |
| No mucosal
injury | 0 |
| Lymphoepithelial
injury | 1 |
| Surface mucosal
erosion or focal ulcer | 2 |
| Extensive mucosal
injury and structure extending to deeper intestinal wall | 3 |
Immunohistochemistry for IL-17 and
IL-23 protein
Colon tissues were fixed in 4% paraformaldehyde at
room temperature overnight. Paraffin-embedded colon tissues were
sliced into sections (5-µm thick), and were subjected to dewaxing
(60˚C for 2 h), followed by soaking in dimethylbenzene twice for 15
min, hydration (ethyl alcohol was used twice, 95% alcohol and 80%
alcohol once, all for 1 min respectively; 70% alcohol once for 2
min. All alcohols used were analytically pure), antigen retrieval
and washing with PBS. 3% H2O2 was used to
block the endogenous peroxidase for 10 min at room temperature, and
the slides were incubated with anti-IL-17 (1:50; cat. no. ab79056;
Abcam) or anti-IL-23 (1:200; cat. no. ab45420; Abcam) antibody
dissolved in blocking solution (QuickBlock™; Beyotime
Institute of Biotechnology) at 4˚C overnight. After incubation, the
slides were washed with PBS and incubated with an HRP-labelled
polymer system (used neat; cat. no. SP-0023; Beijing Boaosen
Biotechnology Co., Ltd.) at 37˚C for 15 min, followed by an
incubation with 3,3-diaminobenzidine detection reagent at room
temperature for 5 min, and finally observed under light microscope
with a magnification of x200. The semi-quantitative expression of
each protein was analyzed by Image-Pro Plus software v.6.0 (Media
Cybernetics, Inc.).
DNA extraction
Fresh stool specimen from the control and UC groups
were collected in a sterile container, and then stored in the
refrigerator at -80˚C within 30 min. A total of 26 fecal bacterial
DNA samples were extracted using a fecal nucleic acid extraction
kit (cat. no. DP328-02; Tiangen Biotech Co., Ltd.), with additional
proteinase K treatment at 70˚C for 10 min to ensure adequate
bacterial cell lysis. The DNA extraction for all included
biospecimens was performed at a single center by the same person
using an identical protocol and the same extraction kit for all
samples.
Amplicon sequencing and bioinformatics
analysis
The MetaVx™ library preparation and
Illumina MiSeq next generation sequencing (NGS) were performed at
Genewiz, Inc.. The DNA samples were quantified using a Qubit 2.0
Fluorometer (Thermo Fisher Scientific, Inc.). In total, 30-50 ng
DNA was used to generate amplicons using a MetaVx™
Library Preparation kit (Genewiz, Inc.). The V3 and V4
hypervariable regions of prokaryotic 16S rRNA were selected for the
generation of amplicons and subsequent taxonomic analysis. Genewiz,
Inc. designed a panel of proprietary primers targeting relatively
conserved regions bordering the V3 and V4 hypervariable regions of
the 16S rRNA of bacteria and archaea. The V3 and V4 regions were
amplified using forward primers containing the sequence
5'-CCTACGGRRBGCASCAGKVRVGAAT-3' and reverse primers containing the
sequence 5'-GGACTACNVGGGTWTCTAATCC-3'. The first-round PCR products
were used as templates for the second-round amplicon enrichment
PCR. Indexed adapters were added to the ends of the 16S rRNA
amplicons to generate indexed libraries ready for the downstream
NGS Illumina MiSeq sequencing. The DNA libraries were validated by
an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) and
quantified by a Qubit 2.0 Fluorometer. The DNA libraries were
multiplexed and loaded on an Illumina MiSeq instrument according to
the manufacturer's instructions (Illumina, Inc.). Sequencing was
performed using a 2x300 paired-end configuration; image analysis
and base calling were performed using the MiSeq Control Software
v.2.5.0.5 (Illumina, Inc.) on the aforementioned MiSeq
instrument.
Data analysis
The QIIME data analysis packagev.1.8.0 (http://bio.cug.edu.cn/qiime/) was used for the 16S
rRNA data analysis. The forward and reverse reads were combined and
assigned to samples based on the barcode. Raw reads were trimmed
using QIIME. Quality filtering on the combined sequences was
performed, and sequences that did not fulfil the following
criterion were discarded: Sequence length <20 nucleotides.
Subsequently, the sequences were compared with those of the
reference ribosomal database project (RDP) Gold database v.2.2
(http://rdp.cme.msu.edu/classifier/classifier.jsp;jsessionid=D5D6C78C6C197C015E237D0FD7A85246.10.0.0.9)
using the UCHIME (http://www.drive5.com/uchime/uchime_download.html)
algorithm to detect chimeric sequences, and the chimeric sequences
were removed. The remaining sequences were used in the final
analysis. The sequences were grouped into operational taxonomic
units (OTUs) using the clustering program VSEARCH (v.1.9.6;
https://github.com/torognes/vsearch)
against the SILVA 119 database (https://www.arb-silva.de/search/), pre-clustered at
97% sequence identity. The RDP classifier was used to assign
taxonomic categories to all OTUs at a confidence threshold of 0.8.
The RDP classifier used the SILVA 123 database (https://www.arb-silva.de/search/), which had
taxonomic category predictions down to the species level. The
sequences were rarefied prior to the calculation of α- and
β-diversity statistics. α-diversity indexes were calculated in
QIIME from rarefied samples using the Shannon index for diversity
(22) and the Chao1 index (23) for richness. Rarefaction curve was
used to judge whether the sample size was sufficient and to
estimate the species richness. β-diversity was calculated using
weighted and unweighted UniFrac distances, and a principal
coordinate analysis (PCoA) and a principal component analysis (PCA)
were performed. The unweighted pair group method with arithmetic
mean (UPGMA) was used to build a tree from the β-diversity distance
matrix. Nonmetric Mutidimensional Scaling (NMDS) was used to
reflect the differences between the samples by the distance between
points. Analysis of similarities (ANOSIM) (24) was confirmed according to
Bray-Curtis.
Statistical analysis
The data are presented as the mean ± standard
deviation. To determine the statistical significance, all the data
were analyzed with independent-samples t-tests for normally
distributed data or Mann-Whitney tests for non-normally distributed
data, with analyses performed using SPSS version 13.0 software
(SPSS, Inc.). The associations between the average optical density
of IL-17/IL-23 and the histopathological scores in the intestinal
mucosa were analyzed using Spearman's correlation test, and the
associations between the average optical density of IL-17/IL-23 and
the abundance of Enterococcus, Lactobacillus, Bacteroides,
Bifidobacterium and Escherichia-Shigella, were analyzed
using Pearson's correlation test. P<0.05 was considered to
indicate a statistically significant difference.
Results
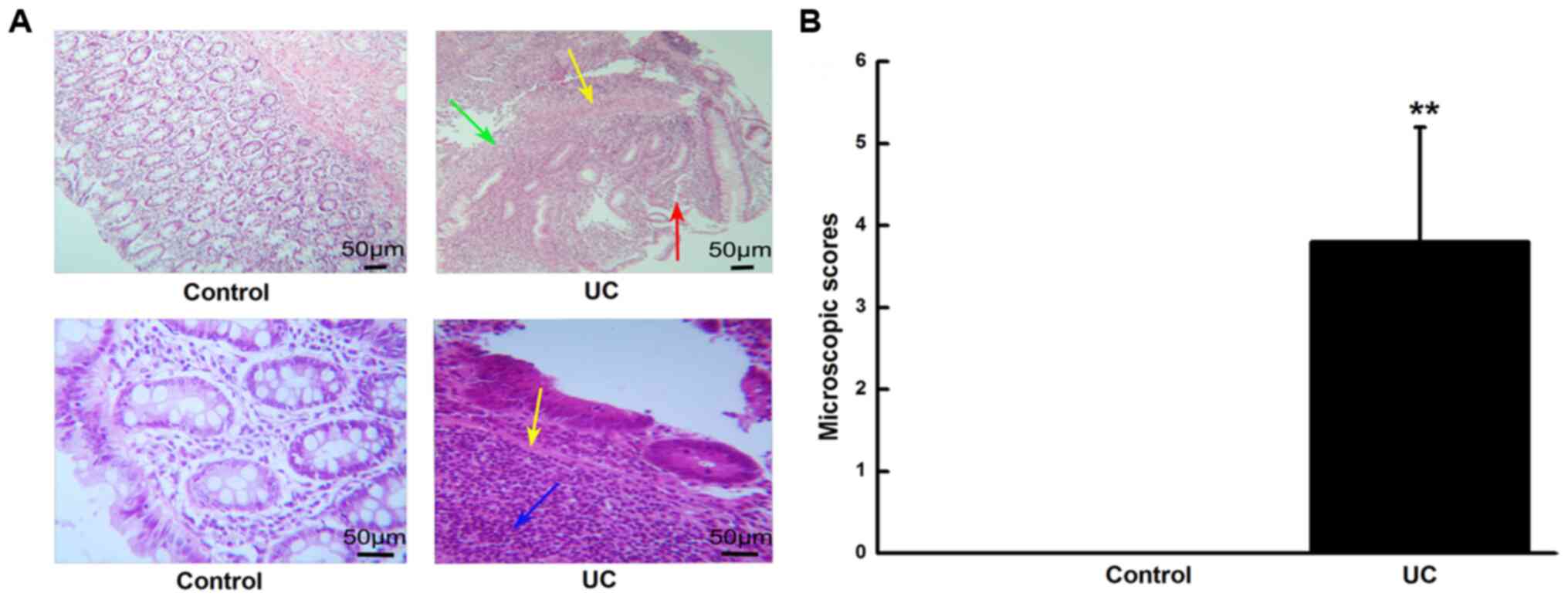
Histological examination
A histological examination of the colon was
performed to validate the degree of inflammation. As illustrated in
Fig. 1, the H&E staining showed
that no signs of inflammation were observed in the control group.
The epithelial cells of the normal colonic mucosa were intact, and
the glands were arranged neatly in proximity. The goblet cells were
abundant, with a few neutrophils and lymphocytes being scattered in
the lamina propria, and the capillaries did not display a
compressed or enlarged appearance. In the UC group, the colonic
mucosa was damaged and eroded, the epithelial cells and the lacunae
were destroyed, the glands were arranged in a distorted manner, the
goblet cells were absent, and a large number of neutrophils and
lymphocytes had infiltrated the lamina propria and the myometrium.
Capillary hyperemia and dilatation, as well as a formation of
multiple ulcers, were observed. In total, an apparent colitis with
characteristic ulcers, multiple erosive lesions, a loss of entire
crypts in the colon, as well as a marked inflammatory cell
infiltration into the colonic submucosa, were observed; thus, the
histological score was higher compared with the control group
(P<0.01).
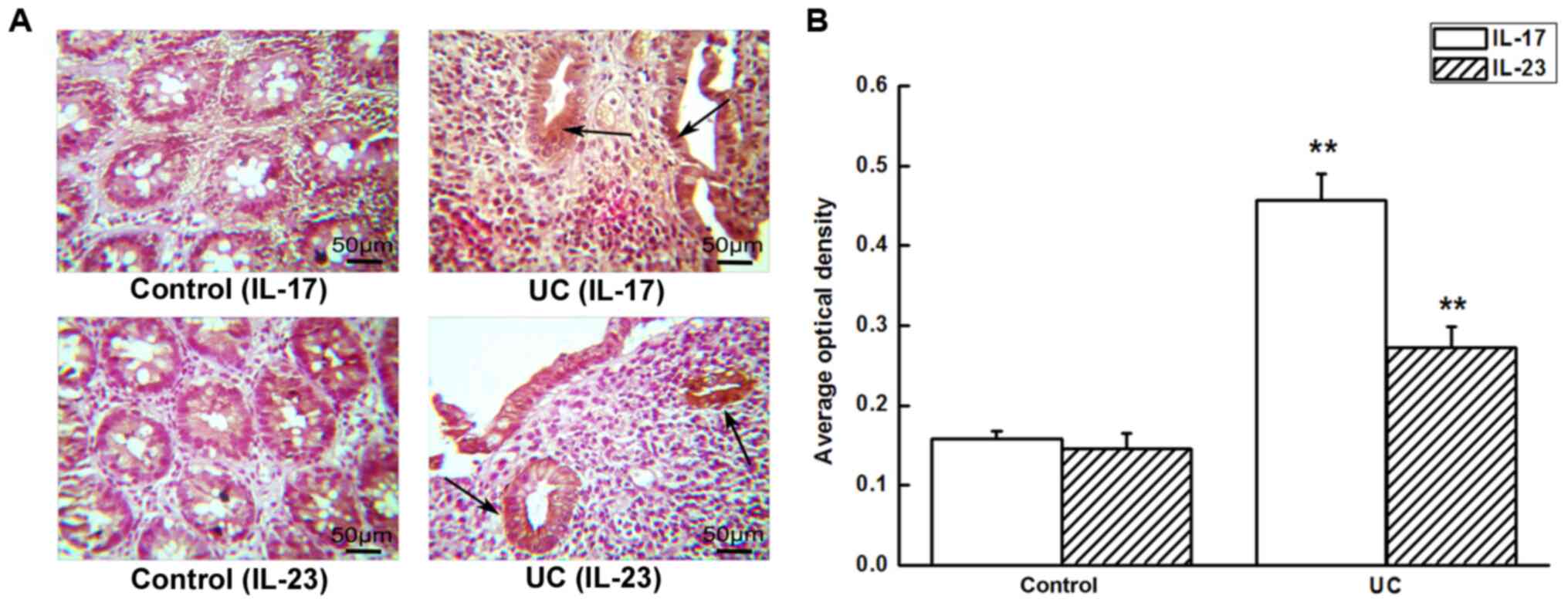
IL-17 and IL-23 expression in colonic
tissues
Immunohistochemical analysis showed that IL-17 and
IL-23 proteins were weakly expressed in normal colonic mucosal
tissues, while they were increased in UC tissues, and were
primarily localized in mucosal epithelial cells and lamina propria.
The cytoplasm of the mononuclear cells in UC tissues was stained in
a brown-yellow color, which indicated positive IL-17 and IL-23
staining, and the average optical density of IL-17 and IL-23
expression was increased in UC compared with the control groups
(P<0.01; Fig. 2).
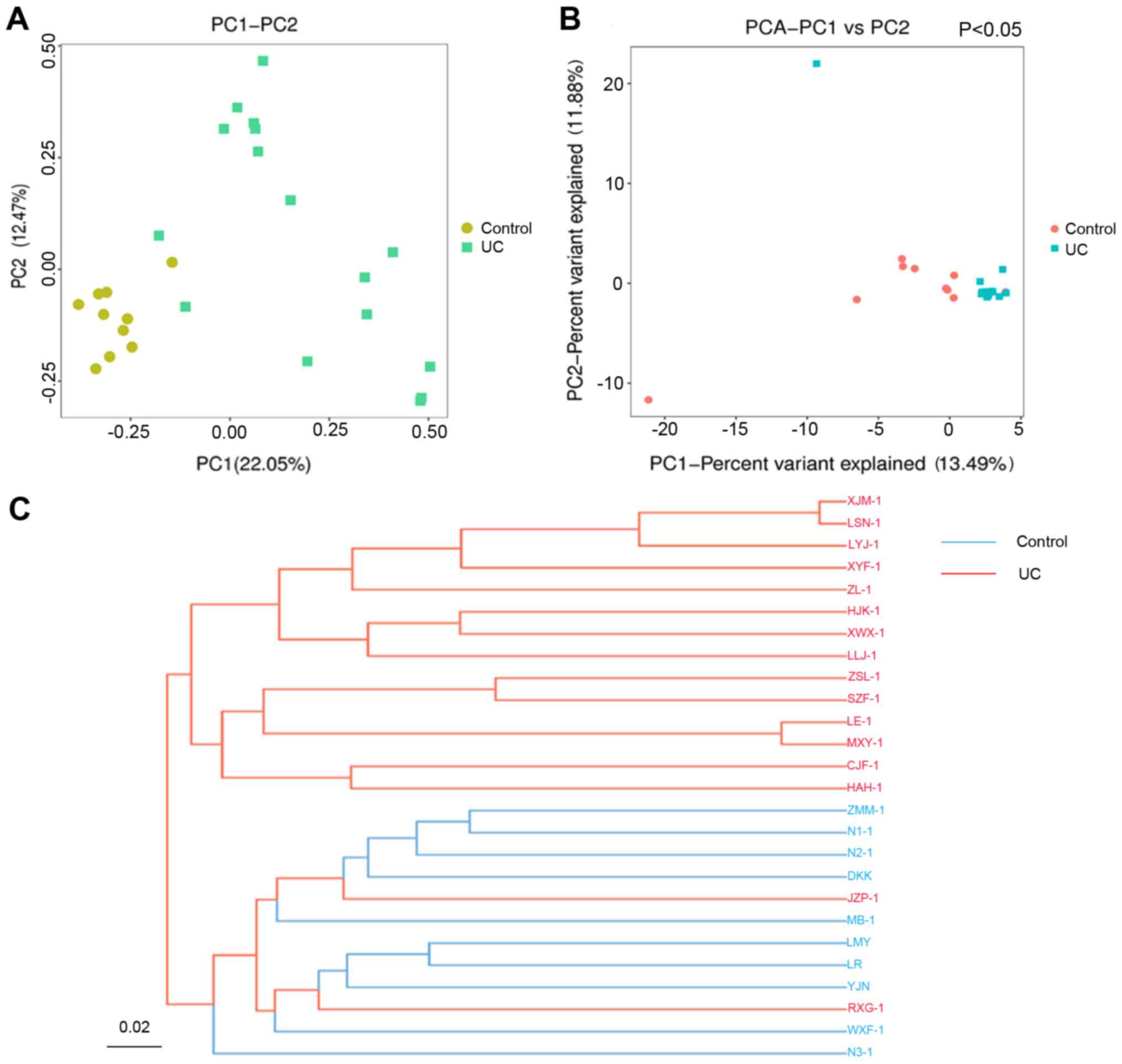
Gut microbiota variation PCoA, PCA and
UPGMA clustering analysis
After the unqualified sequences were removed, a
total of 3,706,470 raw reads and an average of 142,557 reads per
sample were obtained. Following quality filtering of the reads, a
total of 1,853,235 high-quality reads were generated, and each
fecal sample produced an average of 71,278 high-quality reads. The
samples with a low number of high-quality reads (<3,000) were
not analyzed.
According to the UniFrac-based PCoA (Fig. 3A), the gut microbiota in both the
control and UC groups was divided into two groups. The gut
microbiota composition exhibited a high level of variation between
the two groups. The principal coordinates 1 and 2 explained 22.05
and 12.47% of the total composition variation, respectively. The
multivariate analysis of variance of the PCoA matrix scores
indicated a statistically significant difference between the
microbiota of the control and that of the UC group. The UC group
was statistically different from the control group, according to
the PCA analysis (P<0.05; Fig.
3B). The results of the control group clustering analysis
suggested that the samples were similar. The distribution in the UC
group was fragmented but was distinct from that of the control
group. The intestinal flora composition exhibited multiple cluster
directions but was statistically different from that of the control
group in total.
According to the UPGMA clustering analysis (Fig. 3C), these two groups were separated
into two different tree clusters, and the samples of each group
were clustered well together and were separated into two distinct
subgroups.
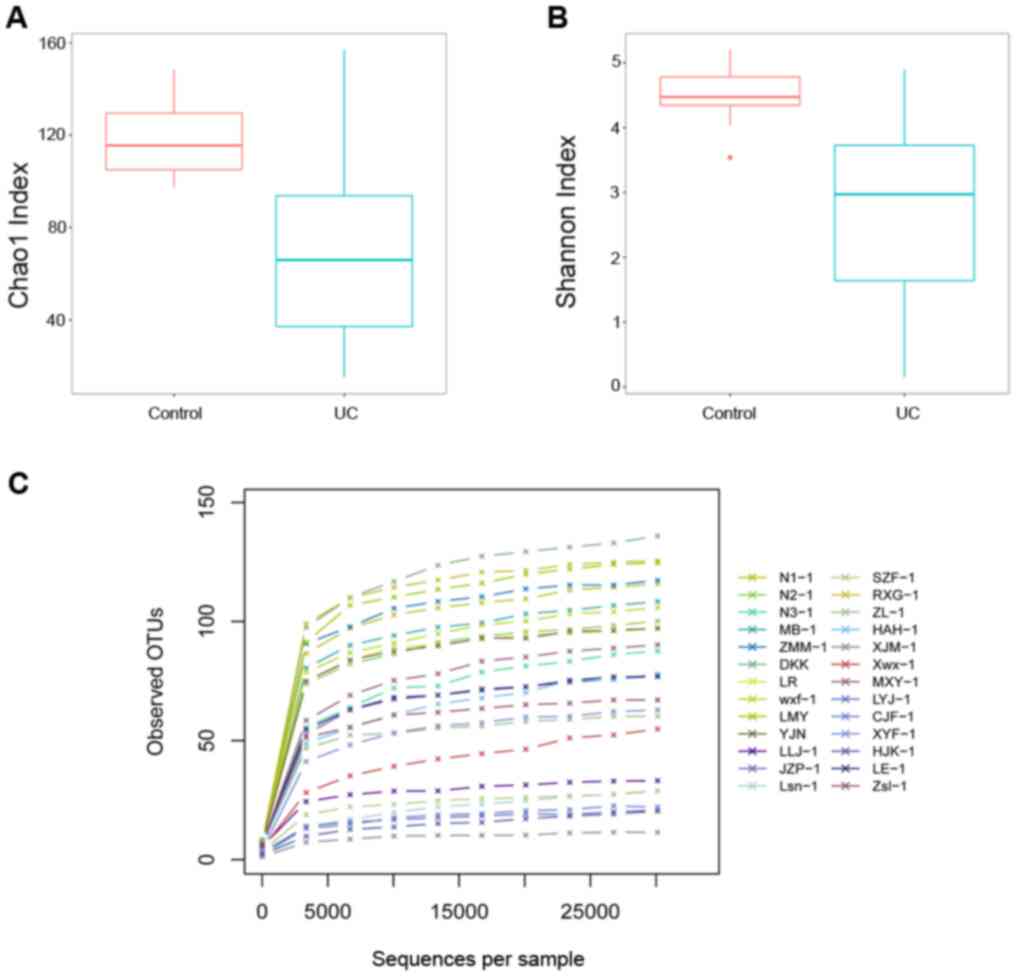
α-diversity and rarefaction
analysis
Fig. 4A and B revealed that microbial α-diversity,
which was assessed by the Chao 1 and Shannon indexes, was decreased
in the UC group compared with the healthy control group. A
rarefaction analysis was performed to cluster all OTUs, which were
present in the data. As shown in Fig.
4C, all the rarefaction curves reached a plateau, suggesting
that the sequencing depth for all samples captured the bacterial
diversity in these communities.
Variations in bacterial community
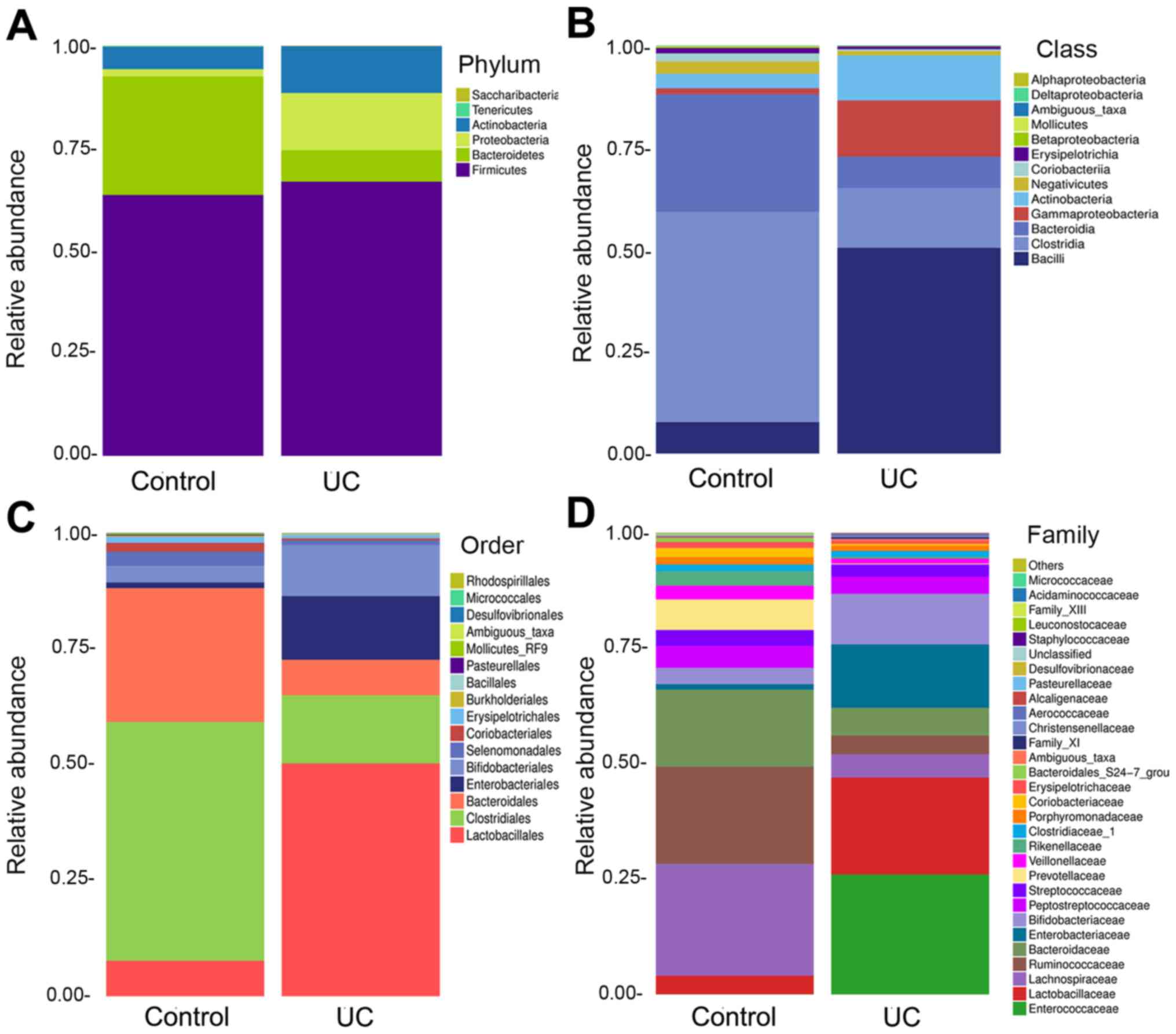
diversity Bacterial community composition at the phylum level
Compared with that in the control group, the
abundance of Bacteroidetes was decreased, the abundance of
Proteobacteria and Actinobacteria was increased, and
the abundance of Firmicutes was slightly increased in the UC
group (Fig. 5A).
Bacterial community composition at the
class/order level
At the class level, the abundance of Bacilli,
Gammaproteobacteria and Actinobacteria in the UC
group was increased, the abundance of Clostridia and
Bacteroidia was decreased, and that of Negativicutes
and Coriobacteriia exhibited a slight decrease (Fig. 5B).
At the order level, the abundance of
Lactobacillales and Enterobacteriales was increased
in the UC group compared with that in the control group, the
abundance of Bifidobacteriales and Coriobacteriales
was increased to a lesser extent, and that of Clostridiales
and Bacteroidales was reduced (Fig. 5C).
Bacterial community composition at the
family level
At the family level, there were prominent
differences between the UC and the control group. The abundance of
Enterococcaceae in the UC group was higher than that in the
control group. The abundance of Lactobacillaceae and
Enterobacteriaceae was also increased in the UC group, and
that of Bifidobacteriaceae was slightly increased; however,
the abundance of Lachnospiraceae and Bacteroidaceae was decreased
(Fig. 5D).
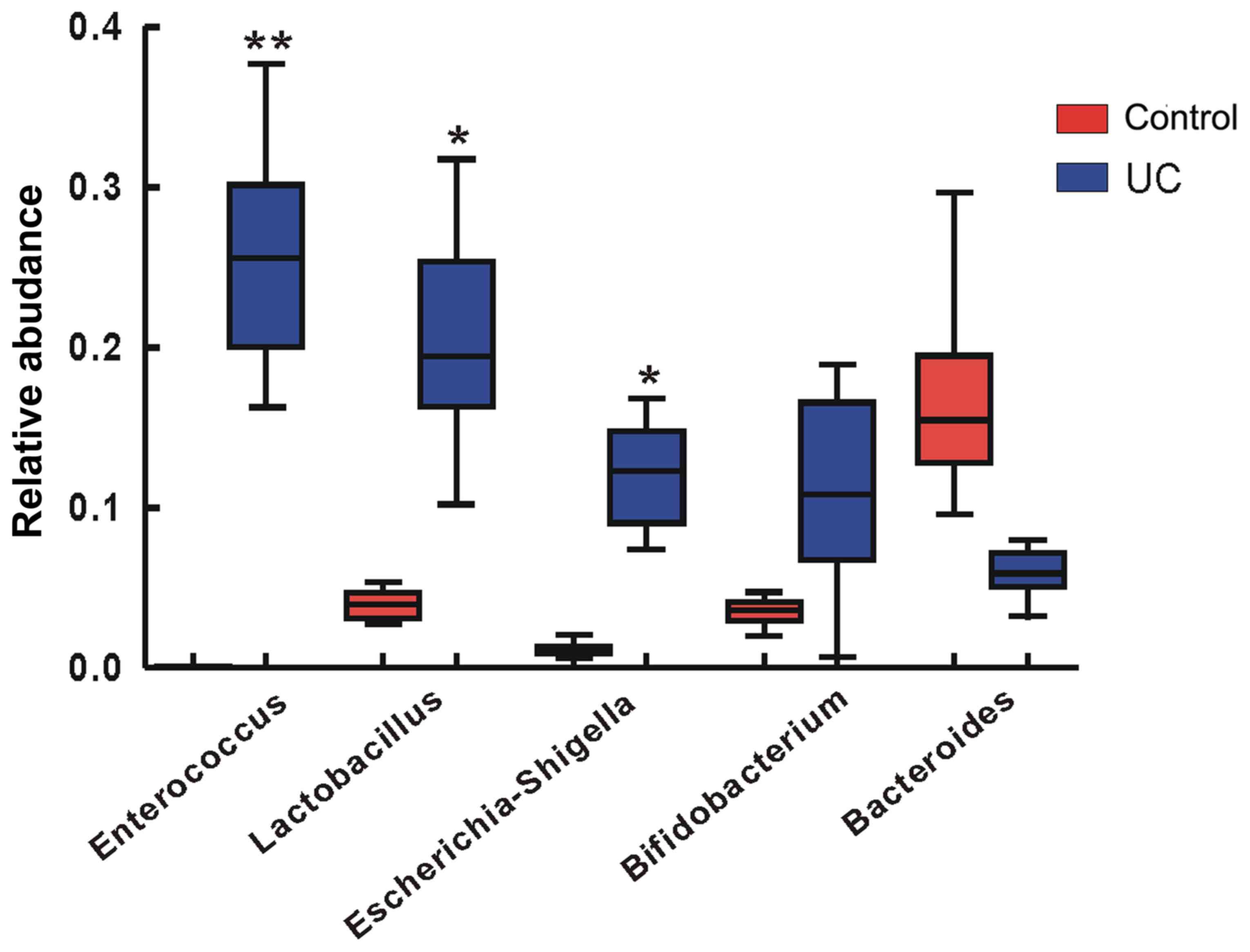
Top five abundant bacteria at the
genus level
In the analysis of the species diversity differences
between the disease group and the control group by Metastats
(http://metastats.cbcb.umd.edu/), the top
five species with the greatest abundance differences were
Enterococcus (P<0.01), Lactobacillus (P<0.05),
Escherichia-Shigella (P<0.05), Bifidobacterium and
Bacteroides (Fig. 6).
Enterococcus exhibited the highest abundance among the gut
bacteria of the disease group.
Key OTU analysis of the gut
microbiota
The key OTUs were identified using partial least
square discriminate analysis (PLS-DA). The comparison of the
control with the UC group revealed that each group had increased
numbers of OTUs belonging to different species (data not shown). In
the Venn diagram, there were 176 OTUs shared between the two
groups. There were 30 types of unique OTUs in the healthy control
group and 14 types in the UC group; the two groups displayed a
small number of differences in their OTU types (data not
shown).
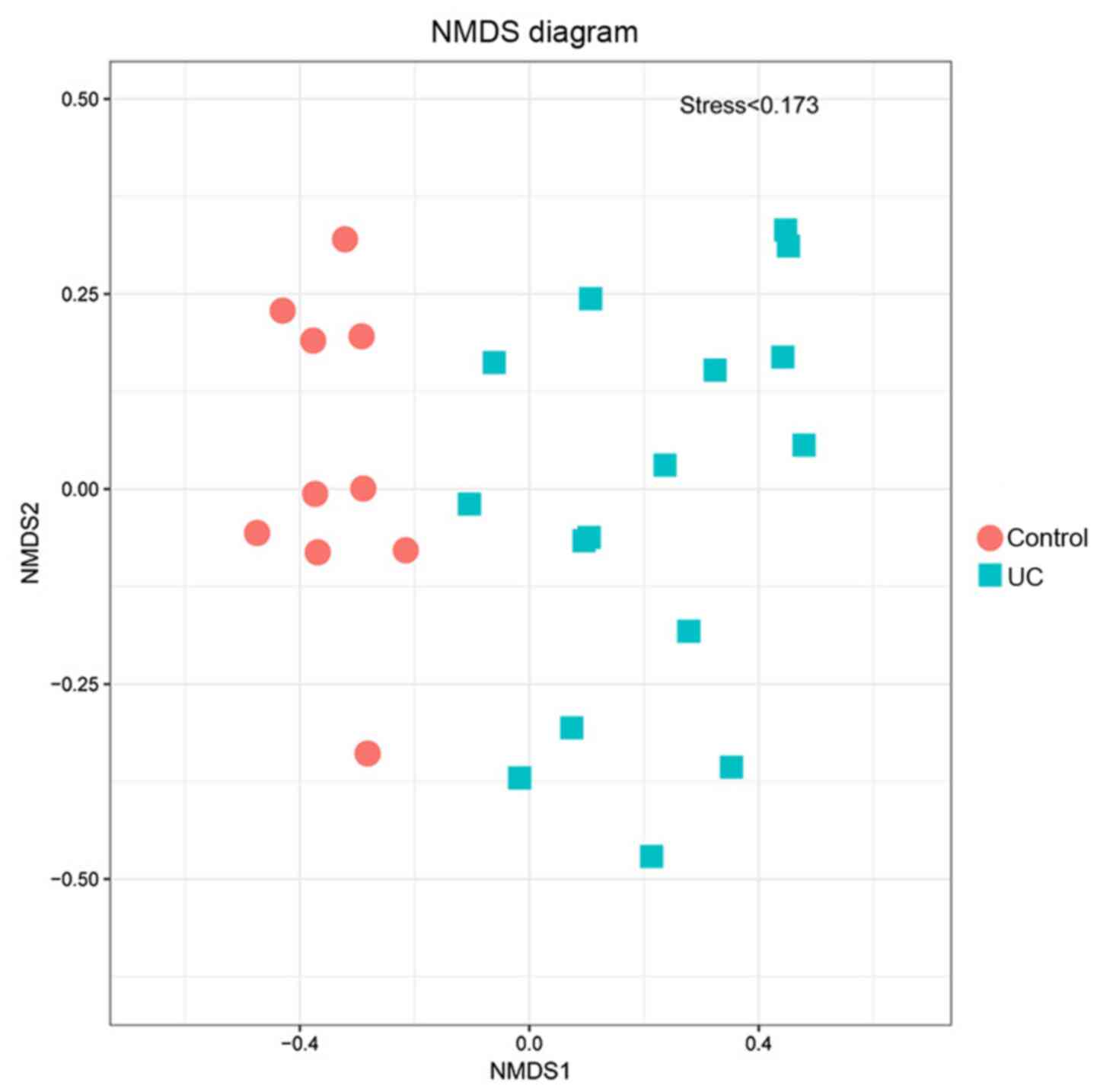
Non-metric multidimensional scaling
(NMDS) analysis
The UC and the control group were distributed on
both sides of the coordinate axis, they were relatively
concentrated in distance, and the stress value was <0.173
(Fig. 7). This analysis accurately
reflected the differences between the two groups.
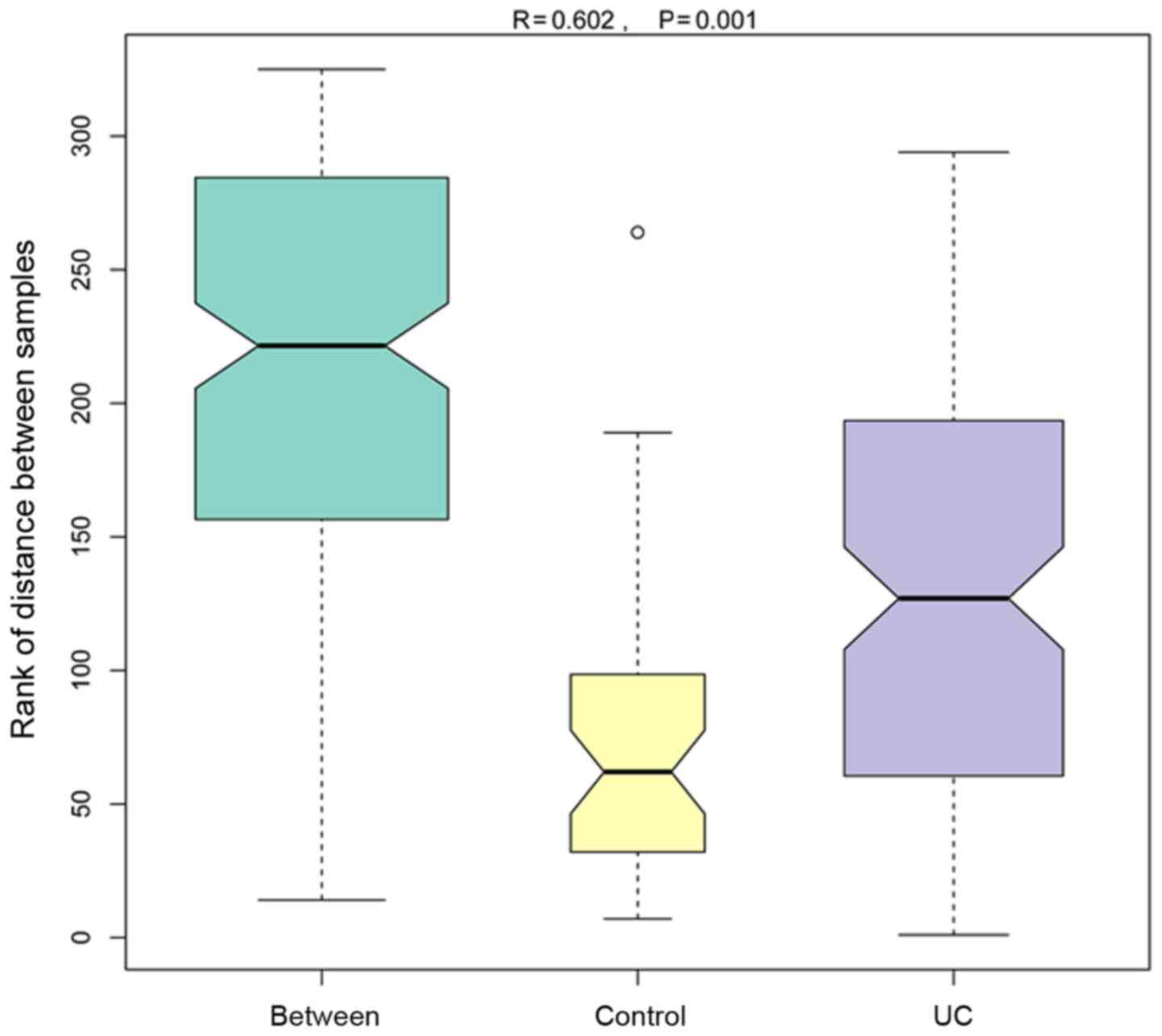
Analysis of similarities (ANOSIM)
ANOSIM is a non-parametric test, which examines
whether the differences between groups (≥2 groups) are higher than
those within the group, ensuring that the grouping is accurate
(Fig. 8). The R-value was obtained
via the analysis of the distance between the sample matrices;
usually, the R-value range is -1 to 1, and the current result was
between 0 and 1. An R-value close to zero demonstrates that no
significant difference between the groups exists. The R value was
close to 1 in the present comparison, indicating that the
between-group difference was higher than the within-group
difference. The P-value represents the reliability of the
statistical analysis, and P<0.05 indicated a statistical
significance. In the current two groups of samples, the R-value was
0.602, and the P-value was 0.001, which indicated that a
significant difference between the two groups was observed, and the
analysis was credible.
Correlation between the expression of
IL-17/IL-23 and the histopathological score
The average optical density values of IL-17
(ρ=0.669; P=0.035; Table II) and
IL-23 (ρ=0.733; P=0.016; Table II)
were positively correlated with the histological score.
 | Table IICorrelation of IL-17/IL-23 with the
histological score. |
Table II
Correlation of IL-17/IL-23 with the
histological score.
| Inflammatory
cytokines | ρ | P-value |
|---|
| IL-17 | 0.669 | 0.035 |
| IL-23 | 0.733 | 0.019 |
Correlation between the expression of
IL-17/IL-23 and intestinal bacteria
Pearson's correlation analysis revealed that the
average optical density of IL-17 was positively correlated with the
abundance of Enterococcus (r=0.843; P<0.001),
Lactobacillus (r=0.737; P=0.001), Bifidobacterium
(r=0.773; P<0.001) and Escherichia-Shigella (r=0.663;
P=0.005). The average optical density of IL-23 was positively
correlated with the abundance of Enterococcus (r=0.771;
P<0.001), Lactobacillus (r=0.566; P=0.022),
Bifidobacterium (r=0.517; P=0.041) and
Escherichia-Shigella (r=0.613; P=0.012). The results are
presented in Table III.
 | Table IIICorrelation of IL-17/IL-23 with top
five differentially abundant bacteria. |
Table III
Correlation of IL-17/IL-23 with top
five differentially abundant bacteria.
| Bacteria | r (IL-17) | P-value
(IL-17) | r (IL-23) | P-value
(IL-23) |
|---|
|
Enterococcus | 0.843 | <0.001 | 0.771 | <0.001 |
|
Lactobacillus | 0.737 | 0.001 | 0.566 | 0.022 |
|
Bacteroides | 0.454 | 0.077 | 0.475 | 0.063 |
|
Bifidobacterium | 0.773 | <0.001 | 0.517 | 0.041 |
|
Escherichia-Shigella | 0.663 | 0.005 | 0.613 | 0.012 |
Discussion
The human body is estimated to be composed of
3x1013 eukaryotic cells and 3.9x1013
colonizing microorganisms, indicating that host cells and
microbiota are represented in similar numbers within an individual
(25). The gut microbiota
constitutes a complex intestinal microecosystem and serves an
important role in the host intestinal immune system, the synthesis
and absorption of nutrients and the inhibition of colonization by
pathogenic bacteria (26,27). An alteration in the bacterial
numbers and proportions results in an imbalance in the intestinal
mucosa, as well as in the dysfunction of the intestinal epithelial
cells and the dysregulation of the immune response (28,29).
Several lines of evidence support the hypothesis
that gut microbiota dysbiosis contributes to the pathogenesis of
IBD (30,31). Certain gene knockout or
gene-deficient mice did not develop UC when they were placed in a
sterile environment, but developed UC following an intestinal flora
enema (32), which introduced the
concept of ‘sterility without inflammation’. It has also been
suggested that the composition of the intestinal flora is an
important factor that is associated with the pathogenesis of
enteritis. Gradel et al (33) demonstrated that patients infected
with Campylobacter or Salmonella were susceptible to
IBD. Li et al (34) observed
fecal bacteria in 16 healthy controls and 41 patients with UC, and
revealed that the microbial diversity of the healthy control feces
was higher than that of patients with UC. Compared with that in the
normal, static and mild active stages, the abundance of the
principal types of fecal bacteria were prominently altered in the
moderate and severe active stages, and the proportion of the
principal bacteria types was negatively correlated with disease
activity. Additionally, the number of Clostridium
perfringens was increased, and the number of
Enterobacteriaceae and rectal bacteria was decreased in
patients with UC. The results from Rajilić-Stojanović et al
(35) revealed that the composition
of fecal bacteria in patients with UC was different from that in
healthy controls. In the active disease period, the primary markers
of bacterial dysbiosis were as follows: The bacterial diversity of
the Clostridium IV cluster was decreased, the abundance of
bacteria involved in the metabolism of butyric and propionic acid,
including Rumen, Enterococcus, Roche and
Akkermansia, was decreased, however, the abundance of the
conditionally pathogenic bacteria Clostridium,
Streptococcus, Helicobacter, Campylobacter and
Clostridium difficile was increased. Other studies have also
indicated that microbiota alterations, namely dysbiosis, are common
in IBD (36-39).
These alterations include an increased number of
Enterobacteriaceae and Bacteroides-Prevotella, an
increased or decreased abundance of Bifidobacteria and a
decrease in the numbers of Firmicutes (especially
Clostridium coccoides, Eubacterium rectale and
Faecalibacterium prausnitzii). Therefore, recent
technological advances have provided evidence that gut dysbiosis is
one of the triggers and/or mediators of the progression of
intestinal inflammation in IBD.
In the current study, a prominent decrease in the
abundance of Bacteroides in the UC group was observed,
accompanied by an increased abundance of Proteus and
Actinomycetes, as well as a slightly increased abundance of
Firmicutes. These results are not consistent with the
results of other reports (40-42).
Several studies (43-46)
have indicated that the reduced diversity of the gut microbiota in
patients with IBD is primarily associated with the reduced
abundance of the phylum Firmicutes, particularly the
Clostridium cluster IV of anaerobic bacteria. The reason for
this difference may be accounted for by the samples being collected
from different research centers, therefore additional supporting
evidence is required.
The increased appreciation of the role of the
microbiota in host physiology and disease has resulted in a
vigorous effort to understand the precise mechanisms underlying the
involvement of the microbiota in the pathogenesis of IBD (47). The alterations in bacterial numbers
and proportions result in an ecological imbalance in the intestinal
mucosa, a decreased production of metabolites, butyrate and
anti-inflammatory factors, and an increased production of
pro-inflammatory factors (48).
This is followed by a dysfunction of intestinal epithelial cell
responses to the gut flora signals, and a dysregulation of the
signal transfer and the immune responses (49). Dysbacteriosis also leads to a lack
of necessary micronutrients, such as short chain fatty acids, and
redox potential in the intestinal mucosa, thereby resulting in
increased permeability and damage of the intestinal mucosa
(28,29).
Another important manifestation of dysbacteriosis is
the alteration in the microbiota composition. Several studies have
indicated that the bacterial diversity was reduced in patients with
UC (50-52),
however, no differences were observed in the gut microbiota
diversity between patients with CD and those with UC, but prominent
differences existed between the microbiota of patients with IBD and
healthy controls. In addition, α-diversity was demonstrated to be
lower in patients with UC and CD compared with non-IBD controls
(53). In the present study,
microbial α-diversity was substantially decreased in the UC
compared with the healthy control group, as assessed by the Chao1
and Shannon indexes, suggesting that the composition of the
microbiome may be different between the two groups. The hypothesis
of decreased α-diversity contends that various intestinal bacteria
constitute a normal barrier (54).
In response to various stresses, such as inflammation and
antibiotics, the microbial diversity decreases, the normal
bacterial functions cannot be performed and anecological imbalance
occurs, which indicates that the gut microbiota diversity is
required to maintain balance (13,46,55,56).
The β-diversity, which reflects the diversity of
constituents in different samples, was also dissimilar between the
two groups. PCoA is a method of research data visualization to
analyze similarities or differences, and involves ranking the
samples through a series of eigenvalues and eigenvectors, and
subsequently selecting the most prominent of several main
characteristic values to calculate the coordinates of the main
distance matrix (57). In the
present study, the principal coordinate sample difference
contribution rate was 22.05%, the second was 12.47%, and the two
groups were distinctly separated based on the distinguished
coordinates. Similarly, the PCA and NMDS results also supported
this conclusion. In patients with IBD, differences in the
abundances of pro-inflammatory and non-inflammatory microbiota have
been identified based on β-diversity (48). The exploration of inflamed and
non-inflamed sites per patient also resulted in similar conclusions
(58).
At the class level, the Bacillus class was
the most abundant in the UC group, and the abundance of
Gammaproteobacteria also appeared to increase, compared with
that in the control group. By contrast, the abundance of
Clostridia and Bacteroidia was reduced.
Bacilli, as a taxonomic class of bacteria, include two
orders, Bacillales and Lactobacillales.
Lactobacilli have been identified as probiotics that can
alter the intestinal environment and cure or alleviate certain
intestinal diseases (59).
Lactobacillus bulgaricus OLL1181 has been reported to
activate the aryl hydrocarbon receptor pathway to suppress
inflammation (60). Animal
experiments have indicated that Lactobacillus plantarum K68
improved the dextran sulfate sodium-induced UC in BALB/c mice via
anti-inflammatory and immunomodulatory activities (61). Furthermore, Lactobacillales
BR11 were demonstrated to reduce the severity of experimental IBD
due to their probiotic properties, possibly via the production of
thiol using a unique cysteine/cystine-transport system (62). Contrary to these previous studies,
in the current study, the UC group had the highest levels of
Lactobacillus, and at the genus level, Lactobacillus
also displayed high abundance. It was hypothesized that β-diversity
differences existed between the UC group and healthy controls, but
this was not observed in certain species of bacteria, such as
Lactobacillus and Bifidobacterium (63,64).
Therefore, the clinical function of probiotics for the treatment of
intestinal diseases was questioned. Indeed, the therapeutic effects
of some probiotics were not satisfactory (65). In a double-blind, randomized,
placebo-controlled study, VSL#3®, which is a
high-potency probiotic mixture that has been efficiently used in
the treatment of pouchitis, was used to treat relapsing patients
with UC, who were also being treated with 5-aminosalicylic acid
and/or immunosuppressants at stable doses (27). Although VSL#3 appeared to exert an
alleviative effect, no significant differences were observed in the
measured parameters. The association of Lactobacillus with
UC should be the focus of future research.
Although the gut microbiota has important
physiological functions, it may also pose a considerable threat to
the immune homeostasis of the host. Bacteria may induce an immune
response if they are recognized by mucosal immune cells as foreign
species (66). In the present
study, it was also demonstrated that Enterococcus was the
most abundant genus in the UC group, which was clearly distinct
from the control group. Furthermore, in the bacterial abundance
diagram, the difference in the abundance of Enterococcus was
higher compared with other bacteria. These results are consistent
with those of previous studies, which aimed at elucidating the
association between Enterococcus faecalis and chronic
intestinal inflammation. Steck et al (67) reported that Enterococcus
resulted in chronic intestinal inflammation via producing
gelatinase GelE, which is a type of metalloproteinase that can
damage the intestinal mucosal barrier. Another study also
demonstrated that Enterococcus faecalis was enriched in
patients with IBD, and an increased Enterococcus faecalis
abundance was associated with clinically active CD (68). However, the mechanisms underlying
the pathogenesis of Enterococcus remain to be further
elucidated.
Previous studies investigating IL-17 and IL-23
demonstrated that these two factors were associated with intestinal
inflammation. Xie et al (69) revealed that the occurrence of human
colon cancer was associated with an increased inflammatory response
driven by IL-17. Another study, which investigated the lamina
propria T cells of the intestinal mucosa, revealed that an
increased IL-17 expression and a reduced suppressor activity were
observed in patients with UC compared with the control group
(70). In a meta-analysis, the
level of IL-17 was indicated to increase in association with the
increase of the degree of UC (17).
Furthermore, drugs inhibiting the IL-17/IL-23 axis were
demonstrated to exhibit a therapeutic effect on UC (18,71).
In summary, IL-17 and IL-23 were associated with intestinal
inflammation. Previous studies have comprehensively described the
intestinal flora of patients with IBD, suggested that the
alterations of the IBD microbiome are closely associated with the
development of inflammation, identified the function of certain
cells and mechanisms which regulate the interaction between the
host and the microorganisms, or explored the flora of patients with
UC with/without primary biliary cirrhosis (46,72-76).
In the current study, the altered intestinal flora and IL-17/IL-23
expression were discussed, and it was demonstrated that IL-17 and
IL-23 expression was correlated with histological score. The
association of IL-17/IL-23 with intestinal flora was also analyzed,
and it was revealed that IL-17/IL-23 expression was correlated with
the abundance of Enterococcus, Lactobacillus,
Bifidobacterium and Escherichia-Shigella.
However, certain limitations are still present in
this field. For example, the challenges of the microbiota studies,
which are associated with the remarkable variation in the
microbiome, owing to a variety of factors, including body site,
age, location, lifestyle, diet and host genetics, still exist, and
the importance of isolation standards and sample processing in
these studies requires improvement (75,77).
In addition, as demonstrated in early genetic association studies
(75,77,78),
the current studies investigating the microbiota may not be
adequate to identify the microbiota-phenotype associations, and
future studies are likely to require an increased numbers of
samples. Whether a more general intestinal microbial profile of
fecal samples or a specific localized profile of endoscopic tissue
samples is optimal for the study of microbiota in UC remains to be
elucidated. The mucosa-associated organisms, however, are not
limited to a particular intestinal location and are observed in
fecal and tissue samples, albeit at different abundances (14). Until a consensus is reached on the
optimal biospecimen type for the characterization of the intestinal
microbiota, future studies may consider collecting both biospecimen
types in their design. Next generation sequencing technologies
continue to develop, with their cost progressively decreasing,
which results in an increase in the number of sequences that are
generated per sample (78). As a
consequence, more in-depth sequencing will be feasible in future
studies, resulting in low-abundance organisms being
characterized.
Despite the results of the present study, it is
possible that the presence of the pathogens examined in fecal
samples may not accurately reflect the microbiome dynamics in the
gut. Adherent bacteria are in direct contact with the host
epithelium, and may exert greater effects on gene expression in the
colonic mucosal cells than transient bacteria, which are removed in
fecal samples (79). Therefore, the
lack of mucosal microbiota is a potential limitation of the current
study. As the bacterial abundance in the colonic tissue is low, and
the small size of the collected tissues was not adequate for the
detection of microbiota, fecal samples were selected, which reflect
the colonization of flora to a certain extent. In addition, the
number of patients in the current study was small. However, more
patients will be included in order to collect adequate samples and
deeply sequence each sample.
In conclusion, the present study indicated that the
intestinal microbiota of patients with UC was different from that
of healthy controls at multiple taxonomic levels, and an altered
intestinal microflora was closely associated with the degree of
inflammation. The IL-23/IL-17 axis, as a key factor in the
development of UC, maybe associated with the alterations of
intestinal microflora. The interaction between intestinal
microflora and the IL-23/IL-17 axis may serve an important role in
the pathogenesis of UC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the National Natural
Science Foundation of China (grant nos. 81500430 and U1304802 to Dr
Xu Hong Lin), Henan Medical Science and Technology Tackling Project
(grant no. 201702136 to Mr. Zhi Feng Dai; grant no. 2018020320 to
Mr. Hui Chao Wang), the Key Project of Science and Technology
Research of the Education Department of Henan (grant no. 17A320019
to Mr. Hui Chao Wang), Research of the Shanghai Municipal
Commission of Health and Family Planning (grant no. 201440313 to Dr
Kun Li) and the Science and Technology Planning Project of Henan
Province (grant nos. 192102310045 and 182102310544 to Dr Xu Hong
Lin; grant no. 182102310566 to Mr. Hui Chao Wang; grant no.
182102310567to Mrs. Rui Lin Yang; grant no. 172102310150 to Mr. Yao
Xu).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
XHL designed the research protocols and revised the
manuscript. ZFD, XYM and RLY performed the experiments and wrote
the manuscript. HCW, DDX, XBG, SSM, RX and JNY acquired and
analyzed the data and performed the experiments. YXL, YX and KL
helped to prepare the figures and tables. DDX, XBG, YXL, KL and YX
interpretated and confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from each
patient prior to the procedure. Ethical approval was obtained from
the medical Ethics Committee of Huaihe Hospital of Henan University
(Kaifeng, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sheehan D, Moran C and Shanahan F: The
microbiota in inflammatory bowel disease. J Gastroenterol.
50:495–507. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sun M, Wu W, Liu Z and Cong Y: Microbiota
metabolite short chain fatty acids, GPCR, and inflammatory bowel
diseases. J Gastroenterol. 52:1–8. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rogler G: Chronic ulcerative colitis and
colorectal cancer. Cancer Lett. 345:235–241. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Adams SM and Bornemann PH: Ulcerative
colitis. Am Fam Physician. 87:699–705. 2013.PubMed/NCBI
|
|
5
|
Al Bander Z, Nitert MD, Mousa A and
Naderpoor N: The gut microbiota and inflammation: An overview. Int
J Environ Res Public Health. 17(7618)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yap YA and Mariño E: An insight into the
intestinal web of mucosal immunity, microbiota, and diet in
inflammation. Front Immunol. 9(2617)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Shen ZH, Zhu CX, Quan YS, Yang ZY, Wu S,
Luo WW, Tan B and Wang XY: Relationship between intestinal
microbiota and ulcerative colitis: Mechanisms and clinical
application of probiotics and fecal microbiota transplantation.
World J Gastroenterol. 24:5–14. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jostins L, Ripke S, Weersma RK, Duerr RH,
McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et
al: Host-microbe interactions have shaped the genetic architecture
of inflammatory bowel disease. Nature. 491:119–124. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Macdonald TT and Monteleone G: Immunity,
inflammation, and allergy in the gut. Science. 307:1920–1925.
2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kostic AD, Xavier RJ and Gevers D: The
microbiome in inflammatory bowel disease: Current status and the
future ahead. Gastroenterology. 146:1489–1499. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nishikawa J, Kudo T, Sakata S, Benno Y and
Sugiyama T: Diversity of mucosa-associated microbiota in active and
inactive ulcerative colitis. Scand J Gastroentero. l44:180–186.
2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ma XY, Dai ZF, Wang HC, Yang JN, Tang XY,
Kang YH, Ding CS, Li YX, Yang RL and Lin XH: Change of intestinal
flora and its relationship with IL-23 /IL-17 axis in patients with
ulcerative colitis. Chinese Journal of Pathophysiology. 34:884–892.
2008.(In Chinese).
|
|
13
|
Sartor RB and Wu GD: Roles for intestinal
bacteria, viruses, and fungi in pathogenesis of inflammatory bowel
diseases and therapeutic approaches. Gastroenterology.
152:327–339.e4. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gevers D, Kugathasan S, Denson LA,
Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song
SJ, Yassour M, et al: The treatment-naive microbiome in new-onset
Crohn's disease. Cell Host Microbe. 15:382–392. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sokol H and Seksik P: The intestinal
microbiota in inflammatory bowel diseases: Time to connect with the
host. Curr Opin Gastroenterol. 26:327–331. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yu P, Shen F, Zhang X, Cao R, Zhao X, Liu
P, Tu H, Yang X, Shi R and Zhang H: Association of single
nucleotide polymorphisms of IL23R and IL17 with ulcerative colitis
risk in a Chinese Han population. PLoS One.
7(e44380)2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li J, Tian H, Jiang HJ and Han B:
Interleukin-17 SNPs and serum levels increase ulcerative colitis
risk: A meta-analysis. World J Gastroenterol. 20:15899–15909.
2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Larussa T, Oliverio M, Suraci E, Greco M,
Placida R, Gervasi S, Marasco R, Imeneo M, Paolino D, Tucci L, et
al: Oleuropein decreases cyclooxygenase-2 and interleukin-17
expression and attenuates inflammatory damage in colonic samples
from ulcerative colitis patients. Nutrients. 9(391)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Inflammatory bowel disease collaboration
group of Chinese Society of Gastroenterology. Consensus on the
diagnosis and treatment of inflammatory bowel disease in China.
Chin J Intern Med. 47:73–79. 2008.(In Chinese).
|
|
20
|
Liang J, Zhou L, Sha SM, Lei SN, Luo GH
and Wu KC: Consensus on Diagnosis and Management of Inflammatory
Bowel Disease (Guangzhou, 2012), an interpretation on the diagnosis
of UC. Chin J Gastroenterol. 17:712–720. 2012.(In Chinese).
|
|
21
|
Siegmund B, Lehr HA, Fantuzzi G and
Dinarello CA: IL-1β-converting enzyme (caspase-1) in intestinal
inflammation. Proc Natl Acad Sci USA. 98:13249–13254.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kim BR, Shin J, Guevarra R, Lee JH, Kim
DW, Seol KH, Lee JH, Kim HB and Isaacson R: Deciphering diversity
indices for a better understanding of microbial communities. J
Microbiol Biotechnol. 27:2089–2093. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ren G, Yu M, Li K, Hu Y, Wang Y, Xu X and
Qu J: Seleno-lentinan prevents chronic pancreatitis development and
modulates gut microbiota in mice. J Funct Foods. 22:177–188.
2016.
|
|
24
|
Schmidt M, Unterer S, Suchodolski JS,
Honneffer JB, Guard BC, Lidbury JA, Steiner JM, Fritz J and Kölle
P: The fecal microbiome and metabolome differs between dogs fed
bones and raw food (BARF) diets and dogs fed commercial diets. PLoS
One. 13(e201279)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sender R, Fuchs S and Milo R: Are we
really vastly outnumbered? Revisiting the ratio of bacterial to
host cells in humans. Cell. 164:337–340. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tomasello G, Bellavia M, Palumbo VD,
Gioviale MC, Damiani P and Lo Monte AI: From gut microflora
imbalance to mycobacteria infection: Is there a relationship with
chronic intestinal inflammatory diseases? Ann Ital Chir.
82:361–368. 2011.PubMed/NCBI
|
|
27
|
Shen J, Zuo ZX and Mao AP: Effect of
probiotics on inducing remission and maintaining therapy in
ulcerative colitis, Crohn's disease, and pouchitis: Meta-analysis
of randomized controlled trials. Inflamm Bowel Dis. 20:21–35.
2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Verma R, Verma AK, Ahuja V and Paul J:
Real-time analysis of mucosal flora in patients with inflammatory
bowel disease in India. J Clin Microbiol. 48:4279–4282.
2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Marteau P: Bacterial flora in inflammatory
bowel disease. Dig Dis. 27 (Suppl 1):S99–S103. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Mosca A, Leclerc M and Hugot JP: Gut
microbiota diversity and human diseases: Should we reintroduce key
predators in our ecosystem? Front Microbiol. 7(455)2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Manichanh C, Rigottier-Gois L, Bonnaud E,
Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P,
Marteau P, et al: Reduced diversity of faecal microbiota in Crohn's
disease revealed by a metagenomic approach. Gut. 55:205–211.
2006.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Balish E and Warner T: Enterococcus
faecalis induces inflammatory bowel disease in interleukin-10
knockout mice. Am J Pathol. 160:2253–2257. 2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gradel KO, Nielsen HL, Schonheyder HC,
Ejlertsen T, Kristensen B and Nielsen H: Increased short- and
long-term risk of inflammatory bowel disease after salmonella or
campylobacter gastroenteritis. Gastroenterology. 137:495–501.
2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li KY, Wang JL, Wei JP, Gao SY, Zhang YY,
Wang LT and Liu G: Fecal microbiota in pouchitis and ulcerative
colitis. World J Gastroenterol. 22:8929–8939. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Rajilić-Stojanović M, Shanahan F, Guarner
F and de Vos WM: Phylogenetic analysis of dysbiosis in ulcerative
colitis during remission. Inflamm Bowel Dis. 19:481–488.
2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cucchiara S, Iebba V, Conte MP and Schippa
S: The microbiota in inflammatory bowel disease in different age
groups. Dig Dis. 27:252–258. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Swidsinski A, Loening-Baucke V,
Vaneechoutte M and Doerffel Y: Active Crohn's disease and
ulcerative colitis can be specifically diagnosed and monitored
based on the biostructure of the fecal flora. Inflamm Bowel Dis.
14:147–161. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sokol H, Seksik P, Furet JP, Firmesse O,
Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P and
Doré J: Low counts of Faecalibacterium prausnitzii in
colitis microbiota. Inflamm Bowel Dis. 15:1183–1189.
2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Schwiertz A, Jacobi M, Frick JS, Richter
M, Rusch K and Köhler H: Microbiota in pediatric inflammatory bowel
disease. J Pediatr. 157:240–244.e1. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kabeerdoss J, Jayakanthan P, Pugazhendhi S
and Ramakrishna BS: Alterations of mucosal microbiota in the colon
of patients with inflammatory bowel disease revealed by real time
polymerase chain reaction amplification of 16S ribosomal
ribonucleic acid. Indian J Med Res. 142:23–32. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Dziarski R, Park SY, Kashyap DR, Dowd SE
and Gupta D: Pglyrp-regulated gut microflora prevotella falsenii,
parabacteroides distasonis and Bacteroides eggerthii enhance
and alistipes finegoldii attenuates colitis in mice. PLoS One.
11(e146162)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Swidsinski A, Weber J, Loening-Baucke V,
Hale LP and Lochs H: Spatial organization and composition of the
mucosal flora in patients with inflammatory bowel disease. J Clin
Microbiol. 43:3380–3389. 2005.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bamola VD, Ghosh A, Kapardar RK, Lal B,
Cheema S, Sarma P and Chaudhry R: Gut microbial diversity in health
and disease: Experience of healthy Indian subjects, and colon
carcinoma and inflammatory bowel disease patients. Microb Ecol
Health Dis. 28(1322447)2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Frank DN, St Amand AL, Feldman RA,
Boedeker EC, Harpaz N and Pace NR: Molecular-phylogenetic
characterization of microbial community imbalances in human
inflammatory bowel diseases. Proc Natl Acad Sci USA.
104:13780–13785. 2007.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Dong LN, Wang M, Guo J and Wang JP: Role
of intestinal microbiota and metabolites in inflammatory bowel
disease. Chin Med J (Engl). 132:1610–1614. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Becker C, Neurath MF and Wirtz S: The
intestinal microbiota in inflammatory Bowel Disease. ILAR J.
56:192–204. 2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Nagao-Kitamoto H and Kamada N:
Host-microbial cross-talk in inflammatory bowel disease. Immune
Netw. 17:1–12. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Neurath MF: Cytokines in inflammatory
bowel disease. Nat Rev Immunol. 14:329–342. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
McDermott AJ and Huffnagle GB: The
microbiome and regulation of mucosal immunity. Immunology.
142:24–31. 2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ott SJ, Musfeldt M, Wenderoth DF, Hampe J,
Brant O, Fölsch UR, Timmis KN and Schreiber S: Reduction in
diversity of the colonic mucosa associated bacterial microflora in
patients with active inflammatory bowel disease. Gut. 53:685–693.
2004.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Kabeerdoss J, Sankaran V, Pugazhendhi S
and Ramakrishna BS: Clostridium leptum group bacteria
abundance and diversity in the fecal microbiota of patients with
inflammatory bowel disease: A case-control study in India. BMC
Gastroenterol. 13(20)2013.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Imhann F, Vich Vila A, Bonder MJ, Fu J,
Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van
Dullemen HM, et al: Interplay of host genetics and gut microbiota
underlying the onset and clinical presentation of inflammatory
bowel disease. Gut. 67:108–119. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Nishino K, Nishida A, Inoue R, Kawada Y,
Ohno M, Sakai S, Inatomi O, Bamba S, Sugimoto M, Kawahara M, et al:
Analysis of endoscopic brush samples identified mucosa-associated
dysbiosis in inflammatory bowel disease. J Gastroenterol.
53:95–106. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Danilova NA, Abdulkhakov SR, Grigoryeva
TV, Markelova MI, Vasilyev IY, Boulygina EA, Ardatskaya MD,
Pavlenko AV, Tyakht AV, Odintsova AK and Abdulkhakov RA: Markers of
dysbiosis in patients with ulcerative colitis and Crohn's disease.
Ter Arkh. 91:17–24. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Huttenhower C, Knight R, Brown CT,
Caporaso JG, Clemente JC, Gevers D, Franzosa EA, Kelley ST, Knights
D, Ley RE, et al: Advancing the microbiome research community.
Cell. 159:227–230. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lynch SV and Pedersen O: The human
intestinal microbiome in health and disease. N Engl J Med.
375:2369–2379. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Daniels L, Budding AE, de Korte N, Eck A,
Bogaards JA, Stockmann HB, Consten EC, Savelkoul PH and Boermeester
MA: Fecal microbiome analysis as a diagnostic test for
diverticulitis. Eur J Clin Microbiol Infect Dis. 33:1927–1936.
2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hirano A, Umeno J, Okamoto Y, Shibata H,
Ogura Y, Moriyama T, Torisu T, Fujioka S, Fuyuno Y, Kawarabayasi Y,
et al: Comparison of the microbial community structure between
inflamed and non-inflamed sites in patients with ulcerative
colitis. J Gastroenterol Hepatol: Feb 20, 2018 (Epub ahead of
print).
|
|
59
|
Pace F, Pace M and Quartarone G:
Probiotics in digestive diseases: Focus on Lactobacillus GG.
Minerva Gastroenterol Dietol. 61:273–292. 2015.PubMed/NCBI
|
|
60
|
Takamura T, Harama D, Fukumoto S, Nakamura
Y, Shimokawa N, Ishimaru K, Ikegami S, Makino S, Kitamura M and
Nakao A: Lactobacillus bulgaricus OLL1181 activates the aryl
hydrocarbon receptor pathway and inhibits colitis. Immunol Cell
Biol. 89:817–822. 2011.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Liu YW, Su YW, Ong WK, Cheng TH and Tsai
YC: Oral administration of Lactobacillus plantarum K68
ameliorates DSS-induced ulcerative colitis in BALB/c mice via the
anti-inflammatory and immunomodulatory activities. Int
Immunopharmacol. 11:2159–2166. 2011.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Atkins HL, Geier MS, Prisciandaro LD,
Pattanaik AK, Forder RE, Turner MS and Howarth GS: Effects of a
Lactobacillus reuteri BR11 mutant deficient in the
cystine-transport system in a rat model of inflammatory bowel
disease. Dig Dis Sci. 57:713–719. 2012.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Clarke G, Sandhu KV, Griffin BT, Dinan TG,
Cryan JF and Hyland NP: Gut reactions: Breaking down
xenobiotic-microbiome interactions. Pharmacol Rev. 71:198–224.
2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Suez J, Zmora N, Segal E and Elinav E: The
pros, cons, and many unknowns of probiotics. Nat Med. 25:716–729.
2019.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Meijer BJ and Dieleman LA: Probiotics in
the treatment of human inflammatory bowel diseases: Update 2011. J
Clin Gastroenterol. 45 (Suppl 1):S139–S144. 2011.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Shi N, Li N, Duan X and Niu H: Interaction
between the gut microbiome and mucosal immune system. Mil Med Res.
4(14)2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Steck N, Hoffmann M, Sava IG, Kim SC,
Hahne H, Tonkonogy SL, Mair K, Krueger D, Pruteanu M, Shanahan F,
et al: Enterococcus faecalis metalloprotease compromises epithelial
barrier and contributes to intestinal inflammation.
Gastroenterology. 141:959–971. 2011.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zhou Y, Chen H, He H, Du Y, Hu J, Li Y, Li
Y, Zhou Y, Wang H, Chen Y and Nie Y: Increased Enterococcus
faecalis infection is associated with clinically active Crohn
disease. Medicine (Baltimore). 95(e5019)2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Xie Z, Qu Y, Leng Y, Sun W, Ma S, Wei J,
Hu J and Zhang X: Human colon carcinogenesis is associated with
increased interleukin-17-driven inflammatory responses. Drug Des
Devel Ther. 9:1679–1689. 2015.PubMed/NCBI View Article : Google Scholar
|
|
70
|
D'Ambrosio A, Cossu A, Amendola A, Zandri
A, Butera A, Sanchez M, Biffoni M, Pronio A, Montesani C, Kohn A,
et al: Lamina propria CD4+LAP+ regulatory T
cells are increased in active ulcerative colitis but show increased
IL-17 expression and reduced suppressor activity. J Crohns Colitis.
10:346–353. 2016.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Zhu Q, Zheng P, Chen X, Zhou F, He Q and
Yang Y: Andrographolide presents therapeutic effect on ulcerative
colitis through the inhibition of IL-23/IL-17 axis. Am J Transl
Res. 10:465–473. 2018.PubMed/NCBI
|
|
72
|
Kevans D, Tyler AD, Holm K, Jørgensen KK,
Vatn MH, Karlsen TH, Kaplan GG, Eksteen B, Gevers D, Hov JR and
Silverberg MS: Characterization of intestinal microbiota in
ulcerative colitis patients with and without primary sclerosing
cholangitis. J Crohns Colitis. 10:330–337. 2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Bajer L, Kverka M, Kostovcik M, Macinga P,
Dvorak J, Stehlikova Z, Brezina J, Wohl P, Spicak J and Drastich P:
Distinct gut microbiota profiles in patients with primary
sclerosing cholangitis and ulcerative colitis. World J
Gastroenterol. 23:4548–4558. 2017.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Rubio CA, Langner C and Schmidt PT:
Partial to complete abrogation of the subepithelial macrophage
barrier against the gut microbiota in patients with ulcerative
colitis and Crohn's colitis. Histopathology. 72:580–587.
2018.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Kiely CJ, Pavli P and O'Brien CL: The role
of inflammation in temporal shifts in the inflammatory bowel
disease mucosal microbiome. Gut Microbes. 9:477–485.
2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Yang Y and Jobin C: Novel insights into
microbiome in colitis and colorectal cancer. Curr Opin
Gastroenterol. 33:422–427. 2017.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Huttenhower C, Kostic AD and Xavier RJ:
Inflammatory bowel disease as a model for translating the
microbiome. Immunity. 40:843–854. 2014.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Slatko BE, Gardner AF and Ausubel FM:
Overview of next-generation sequencing technologies. Curr Protoc
Mol Biol. 122(e59)2018.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Eckburg PB, Bik EM, Bernstein CN, Purdom
E, Dethlefsen L, Sargent M, Gill SR, Nelson KE and Relman DA:
Diversity of the human intestinal microbial flora. Science.
308:1635–1638. 2005.PubMed/NCBI View Article : Google Scholar
|