Introduction
Intervertebral disc degeneration (IDD) is a
degenerative disease of the spine originating from the
intervertebral disc, which results in instability of the spine,
disc herniation, spinal stenosis and cervical spondylosis (1). IDD is one of the most significant
causes of musculoskeletal disability and it is a contributor to the
increased motor dysfunction in the population (2). The incidence of lumbar disc
degeneration is associated with age; an increase in age results in
a higher incidence of lumbar disc degeneration (3). In addition, there are multiple other
factors that are associated with IDD, such as obesity, bone
density, smoking, diabetes, occupation and exercise (4-8).
Lumbar disc degeneration significantly and adversely impacts the
quality of life of patients (9).
The combination of basic treatment, biomechanical adjustment and
active exercise rehabilitation is a novel concept and represents a
novel trend in the rehabilitative treatment of degenerative disc
disease (10). Notably, previous
studies have revealed that microRNAs (miRs/miRNAs) influence the
occurrence and development of IDD (11).
At present, increasing evidence has indicated that
miRNAs serve a crucial role in various types of disease (12), such as cancer (13), atherosclerosis (14) and cardiovascular diseases (15). miRNAs are a class of small
non-coding RNAs that are ~20-22 nucleotides in length (in contrast
with mRNA-transcribed proteins) which inhibit the expression of
multiple target genes by binding to their 3'untranslated region
(3'UTR) (16-18).
Previous studies have demonstrated that miR-25-3p regulates the
proliferation and apoptosis of cancer cells in a variety of types
of cancer. In addition, miR-25-3p was reported to be associated
with the degradation of human nucleus pulposus (NP) cells (19); Lv et al (20) revealed that miR-146a may represent a
novel target for IDD treatment; and Liu et al (21) demonstrated that miR-132 accelerated
extracellular matrix (ECM) degradation in human NP cells via the
targeting of the expression levels of growth differentiation factor
5. Moreover, miR-21 has been reported to promote ECM degradation by
inhibiting autophagy via the PTEN/AKT/mTOR signaling pathway in
human degenerated NP cells (22).
Previous research has also revealed that miR-489-3p
serves an important role in secondary spinal cord injury (23). Other studies have also indicated the
important roles of miR-489-3p in cell apoptosis, ECM and
inflammation regulation (23-25).
However, the role of miR-489-3p in IDD remains poorly
understood.
Human NP cells secrete type II collagen, aggrecan
and other components of the ECM, which serve a crucial role in
maintaining intervertebral disc integrity (26,27).
During the progression of IDD, the excessive apoptosis of IVD cells
and excessive degradation of ECM are observed (28). In addition, proinflammatory
cytokines, such as tumor necrosis factor-α (TNF-α), interleukin
(IL)-1β and IL-6, also serve important roles in IDD (29). Currently, lipopolysaccharide
(LPS)-stimulated NP cells are widely used as an in vitro
model for research into disc degeneration (20-22).
It was hypothesized that miR-489-3p may be involved in IDD via
regulating human NP cell apoptosis and ECM deposition. Thus, the
present study aimed to investigate whether miR-489-3p influences
IDD via the regulation of human NP cells.
Materials and methods
Cell culture and treatment
Human NP cells were obtained from Procell Life
Science & Technology Co., Ltd. (cat. no. CP-H097). The cells
were cultured in DMEM/F12 medium (Invitrogen; Thermo Fisher
Scientific, Inc.), supplemented with 10% FBS (Invitrogen; Thermo
Fisher Scientific, Inc.) and 1% (v/v) penicillin-streptomycin
(Gibco; Thermo Fisher Scientific, Inc.), and maintained in a
humidified atmosphere at 5% CO2 and 37˚C. When cells
reached 80% confluency, the cells were subsequently treated with 10
ng/ml LPS (Sigma-Aldrich; Merck KGaA) at 37˚C for 24 h to establish
the IDD cell model in vitro. Cells without any treatment
were used as the control.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from human NP cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. All processes were
carried out on ice. After extracting the RNA, the concentration of
each sample was determined using an ultraviolet spectrophotometer.
Total RNA was reverse transcribed into cDNA using HiScript III 1st
Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd.), according to
the manufacturer's protocol. The reverse transcription reaction
conditions were: 70˚C for 5 min, 37˚C for 5 min and 42˚C for 60
min. qPCR was subsequently performed using the ChamQ SYBR qPCR
Master Mix (Vazyme Biotech Co., Ltd.), according to the
manufacturer's protocol. The following thermocycling conditions
were used for the qPCR: Initial denaturation at 95˚C for 3 min;
followed by 40 cycles of 95˚C for 30 sec, 56˚C for 30 sec and 72˚C
for 30 sec. The following primer sequences were used for the qPCR:
miR-489-3p forward, 5'-GTGACATCACATATACGG-3' and reverse,
5'-GAACATGTCTGCGTATCTC-3'; TLR4 forward,
5'-CCTGACACCAGGAAGCTTGAA-3' and reverse,
5'-TCTGATCCATGCATTGGTAGGT-3'; aggrecan forward,
5'-CTACCAGTGGATCGGCCTGAA-3' and reverse,
5'-CGTGCCAGATCATCACCACA-3'; collagen type II forward,
5'-GGCAATAGCAGGTTCACGTACA-3' and reverse,
5'-CGATAACAGTCTTGCCCCACTT-3'; U6 forward,
5'-GCTTCGGCAGCACATATACTAAAAT-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'; and GAPDH forward,
5'-CTTTGGTATCGTGGAAGGACTC-3' and reverse,
5'-GTAGAGGCAGGGATGATGTTCT-3'. Expression levels were calculated
using the 2-ΔΔCq method (30) and GAPDH or U6 served as the internal
control for normalization.
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Total protein was quantified using a
bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific,
Inc.) and 30 µg protein/lane was separated via 15% SDS-PAGE. The
separated proteins were subsequently transferred onto a PVDF
membrane (EMD Millipore) and blocked at room temperature using 5%
fat-free powdered milk dissolved in TBS-0.1% Tween for 1.5 h. The
membranes were incubated with the following primary antibodies at
4˚C overnight: Anti-TLR4 antibody (cat. no. ab13556; 1:1,000;
Abcam), anti-GAPDH (cat. no. ab181602; 1:1,000; Abcam),
anti-aggrecan (cat. no. ab3778; 1:1,000; Abcam), anti-collagen type
II (cat. no. ab34712; 1:1,000; Abcam), anti-p65 (cat. no. ab16502;
1:1,000; Abcam) and anti-phosphorylated (p)-p65 (cat. no. ab86299;
1:1,000; Abcam). Following the primary antibody incubation, the
membranes were incubated with a horseradish peroxidase-conjugated
secondary antibody (cat. no. ab7090; 1:2,000; Abcam) at room
temperature for 2 h. Protein bands were visualized using an
enhanced chemiluminescence substrate (EMD Millipore) and analyzed
using ImageJ version 2.0 software (National Institutes of Health).
The expression levels were normalized to GAPDH.
Dual-luciferase reporter assay
Bioinformatics analysis using TargetScan 7.2
(http://www.targetscan.org/vert_72/)
was performed to determine the binding sites between miR-489-3p and
TLR4. The wild-type (WT) or mutant (MUT) 3'UTR of TLR4 was cloned
into the pmiRGLO vector (Promega Corporation) and the recombinant
plasmids were acquired using an EndoFree Plasmid Maxi kit (Vazyme
Biotech Co., Ltd.). To point-mutate the miR-489-3p binding domain
in the 3'UTR of TLR4, a QuikChange Site-Directed Mutagenesis kit
(Stratagene; Agilent Technologies, Inc.) was used according to the
manufacturer's instructions. Cells were seeded into 24-well plates
at a density of 5x104 cells/well and co-transfected with
a miR-489-3p mimic or mimic control and the MUT or WT 3'UTR of TLR4
using Fugene transfection reagent (Promega Corporation) according
to the manufacturer's protocol, together with the Renilla
luciferase pRL-TK vector (Promega Corporation) as a control.
Following transfection at 37˚C for 48 h, firefly and Renilla
luciferase activities were determined using a Dual-Luciferase
Reporter assay system (Promega Corporation) according to the
manufacturer's protocol. Firefly luciferase activity was normalized
to Renilla luciferase activity.
Cell transfection
Human NP cells (5x104 cells per well)
were transiently transfected with a 100 nM mimic control
(5'-UUGUCCGAACGUGUCACGUTT-3'; Suzhou GenePharma Co., Ltd.), 100 nM
miR-489-3p mimic (5'-GUGACAUCACAUAUACGGCAGC-3'; Suzhou GenePharma
Co., Ltd.), 1 µg Control CRISPR Activation Plasmid (cat. no.
sc-437275; Santa Cruz Biotechnology, Inc.), 1 µg TLR4 CRISPR
Activation Plasmid (cat. no. sc-400068-ACT; Santa Cruz
Biotechnology, Inc.), 100 nM miR-489-3p mimic + 1 µg
control-plasmid or 100 nM miR-489-3p mimic + 1 µg TLR4-plasmid at
37˚C for 24 h using Fugene transfection reagent (Promega
Corporation) according to the manufacturer's protocol. The
transfection efficiency was detected using a RT-qPCR assay.
Following 24 h of cell transfection, the cells were treated with 10
ng/ml LPS at 37˚C for 24 h and then the cells were subjected to
subsequent experiments (Fig.
S1).
MTT assay
Cell viability was determined via an MTT assay.
Briefly, transfected human NP cells were treated with 10 ng/ml LPS
at 37˚C for 24 h and then cells were plated in 96-well plates at a
density of 5x103 cells/well. Following 48 h of
incubation at 37˚C, 20 µl MTT reagent (Beyotime Institute of
Biotechnology) was added into each well and incubated for 4 h at
37˚C. Subsequently, 150 µl DMSO was added into each well and the
solution was agitated at 37˚C for 15 min. The optical density
values were measured at 570 nm using a microplate reader.
Flow cytometric analysis of
apoptosis
Cell apoptosis was analyzed using an Annexin V-FITC
Apoptosis Detection kit (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. Briefly, the cells
(1x106) were washed with 1X PBS three times, centrifuged
at 4˚C for 5 min at 1,000 x g, and trypsinized into single cell
suspensions with 500 µl buffer (Beyotime). The cells were stained
with 5 µl Annexin V-FITC and 5 µl propidium iodide at room
temperature for 15 min. Apoptotic cells were subsequently analyzed
using BD FACSCalibur flow cytometer (BD Biosciences) and analyzed
using FlowJo 7.6.1 software (FlowJo LLC).
ELISAs
Following 24 h of cell transfection, NP cells were
treated with 10 ng/ml LPS, and the supernatants were subsequently
harvested through centrifugation at 500 x g at 4˚C for 5 min.
Subsequently, specific ELISA kits (Beyotime Institute of
Biotechnology) were used, according to the manufacturers' protocol,
to determine the concentrations of TNF-α (cat. no. PT518), IL-1β
(cat. no. PI305) and IL-6 (cat. no. PI330) in the cell culture
supernatant from the different groups.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6.0 software (GraphPad Software, Inc.); each experiment was
performed in triplicate and all data are presented as the mean ±
SD. Statistical differences between two groups were determined
using a unpaired Student's t-test, whereas an one-way ANOVA
followed by Tukey's post hoc test was used to analyze the
statistical differences between multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
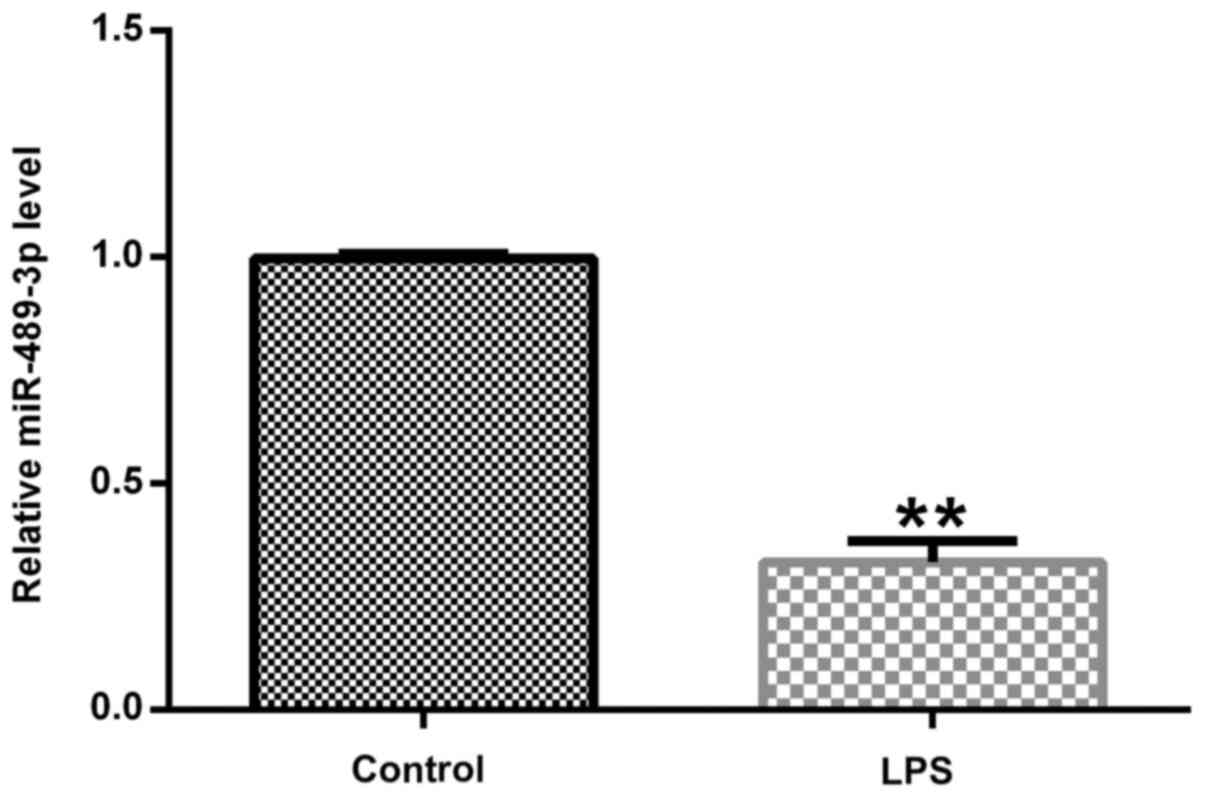
Expression levels of miR-489-3p in
human NP cells
To investigate the effects of miR-489-3p on an IDD
model in vitro, RT-qPCR was performed to detect the
expression levels of miR-489-3p. The expression levels of
miR-489-3p were significantly downregulated in the LPS-treated
group compared with the control group (Fig. 1).
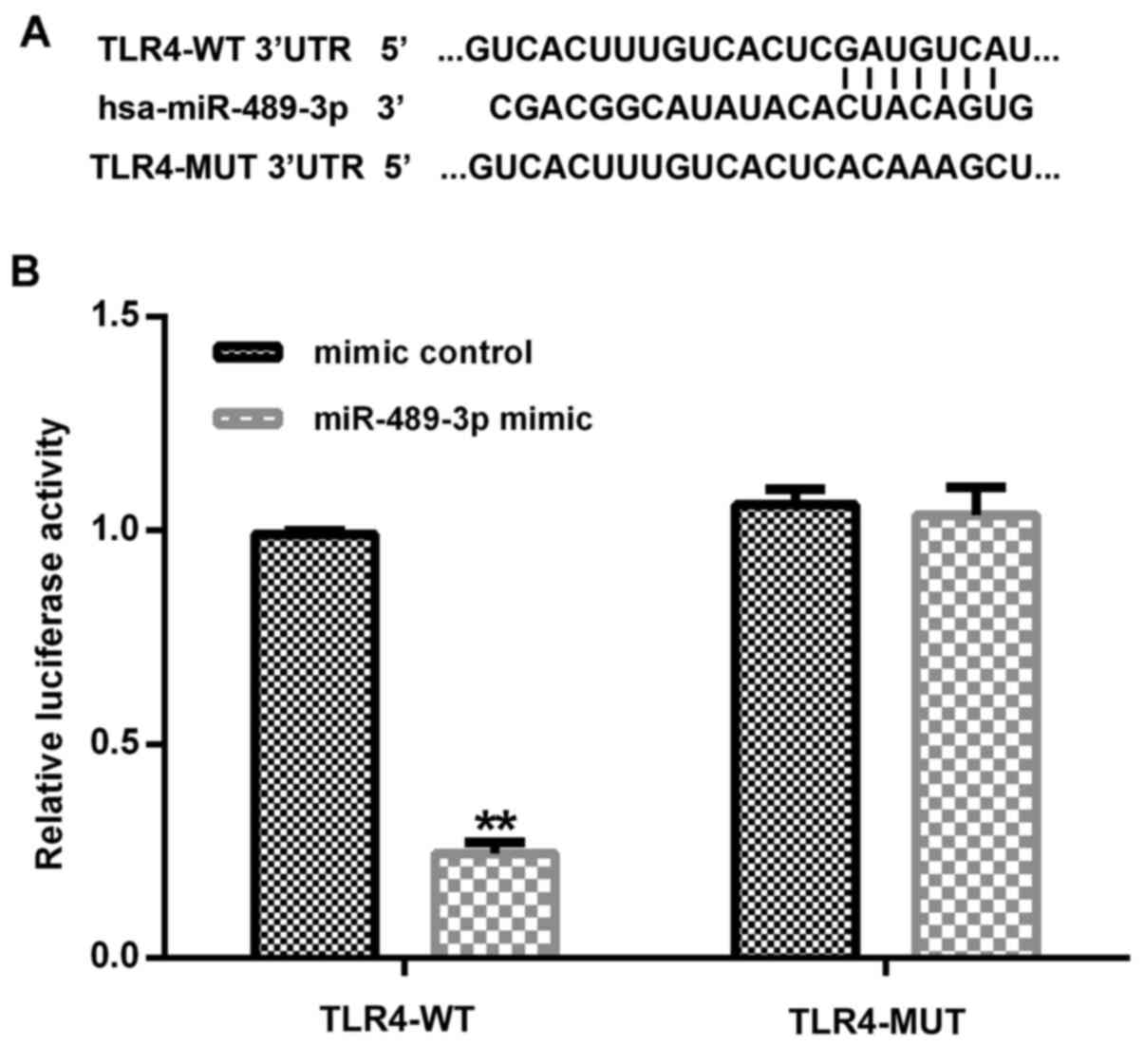
TLR4 is a direct target gene of
miR-489-3p
Subsequently, to determine the interaction between
miRNA and its target genes, the TargetScan tool was used to predict
the target genes of miR-489-3p; it was revealed that the TLR4 3'UTR
contained a putative site that was partially complementary to
miR-489-3p (Fig. 2A). Furthermore,
the dual-luciferase reporter assay was used to determine whether
miR-489-3p interacted directly with the target gene TLR4. The
reporter containing the TLR4-WT 3'UTR exhibited significantly
decreased relative luciferase activity in the human NP cells
co-transfected with the miR-489-3p mimic compared with the mimic
control (Fig. 2B). Taken together,
the current results indicated that TLR4 may be a direct target gene
of miR-489-3p.
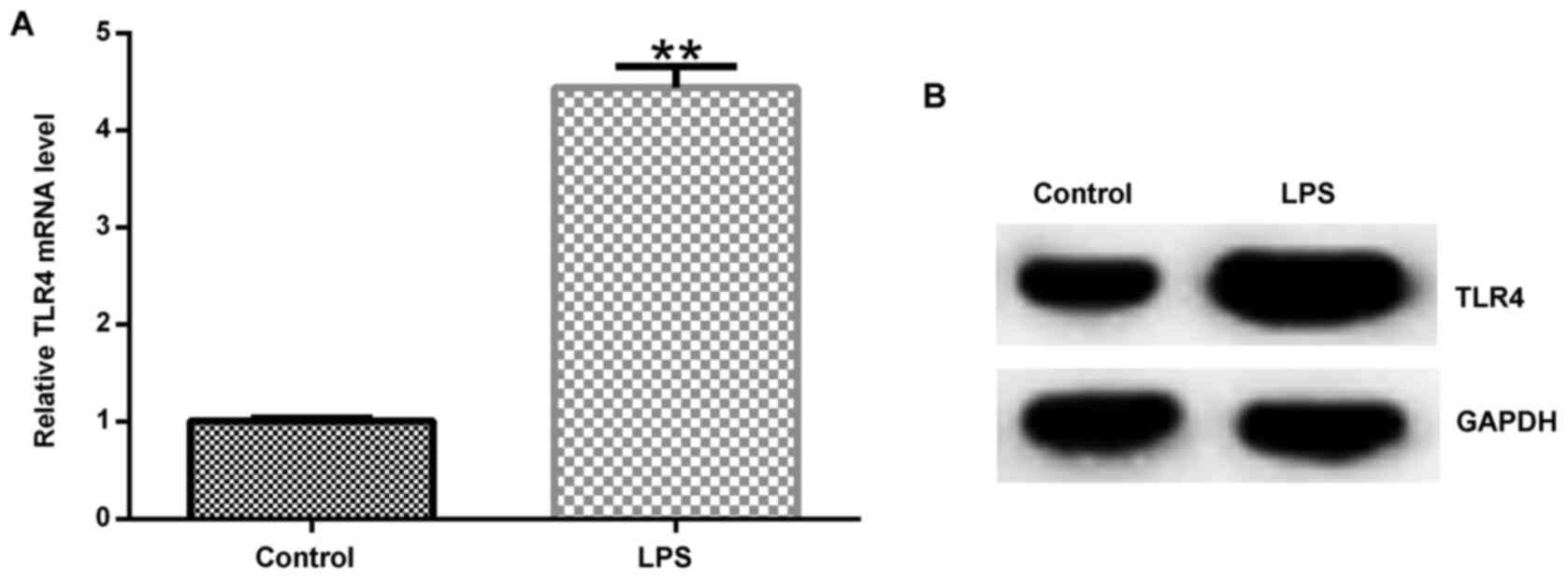
Subsequently, RT-qPCR and western blotting assays
were used to determine the expression levels of TLR4. The results
of these assays revealed that TLR4 expression levels were
upregulated in the LPS-treated group compared with control group at
both the mRNA and protein level (Fig.
3A and B).
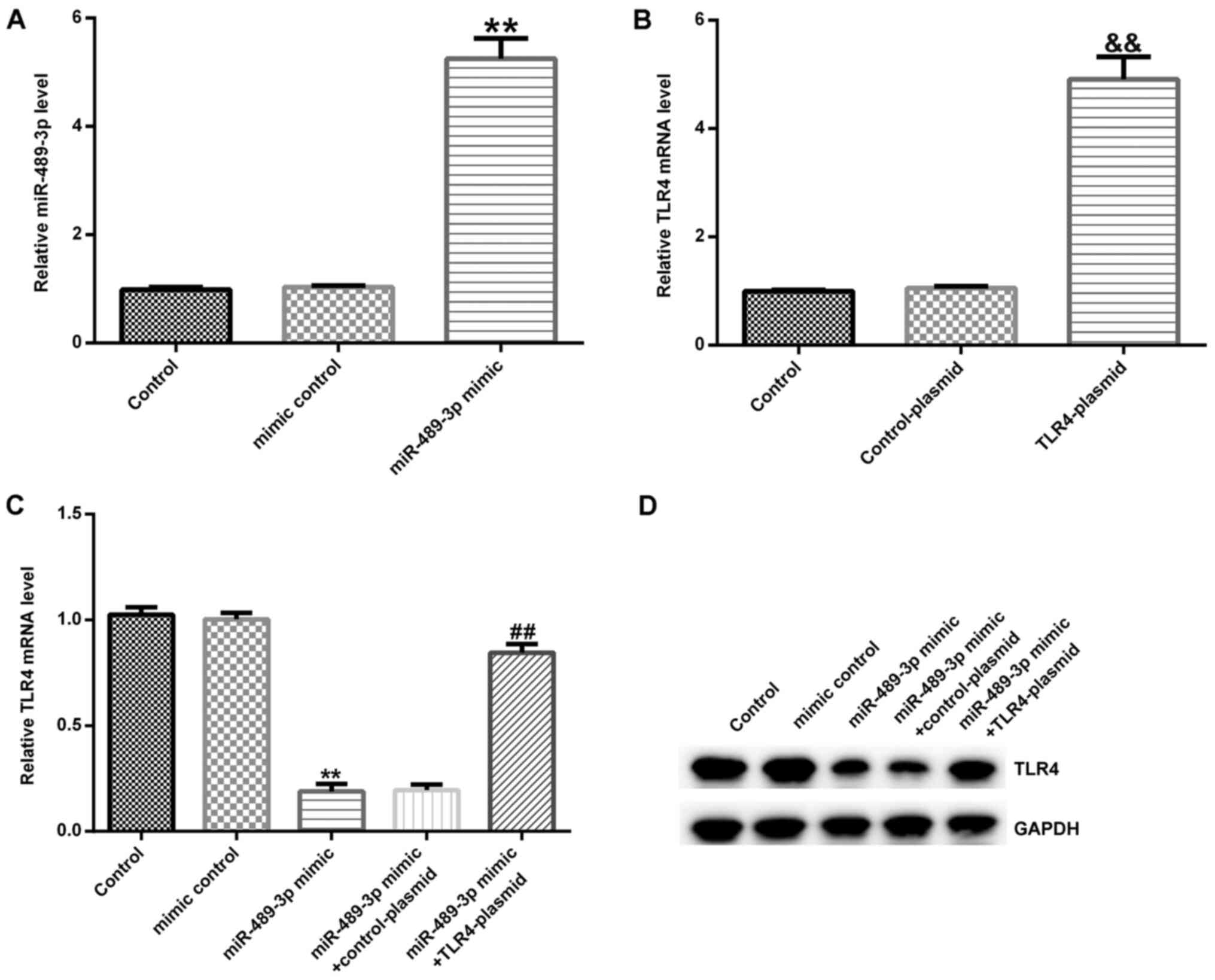
TLR4 is negatively regulated by
miR-489-3p in human NP cells
Human NP cells were transfected with either a mimic
control or miR-489-3p mimic. RT-qPCR revealed that compared with
the mimic control group, the miR-489-3p mimic significantly
increased the expression levels of miR-489-3p in human NP cells
(Fig. 4A). Subsequently, the cells
were transfected with either the control- or TLR4-plasmid; the
results indicated that compared with the control-plasmid group,
TLR4 mRNA expression levels were significantly upregulated in the
TLR4-plasmid group (Fig. 4B). Then,
the effect of miR-489-3p on TLR4 expression levels was examined.
Human NP cells were transfected with either a miR-489-3p mimic +
control-plasmid or miR-489-3p mimic + TLR4-plasmid. It was
demonstrated that compared with the mimic control group, the
miR-489-3p mimic significantly decreased the expression levels of
TLR4 in human NP cells, which was partially restored following the
co-transfection with the TLR4-plasmid (Fig. 4C and D).
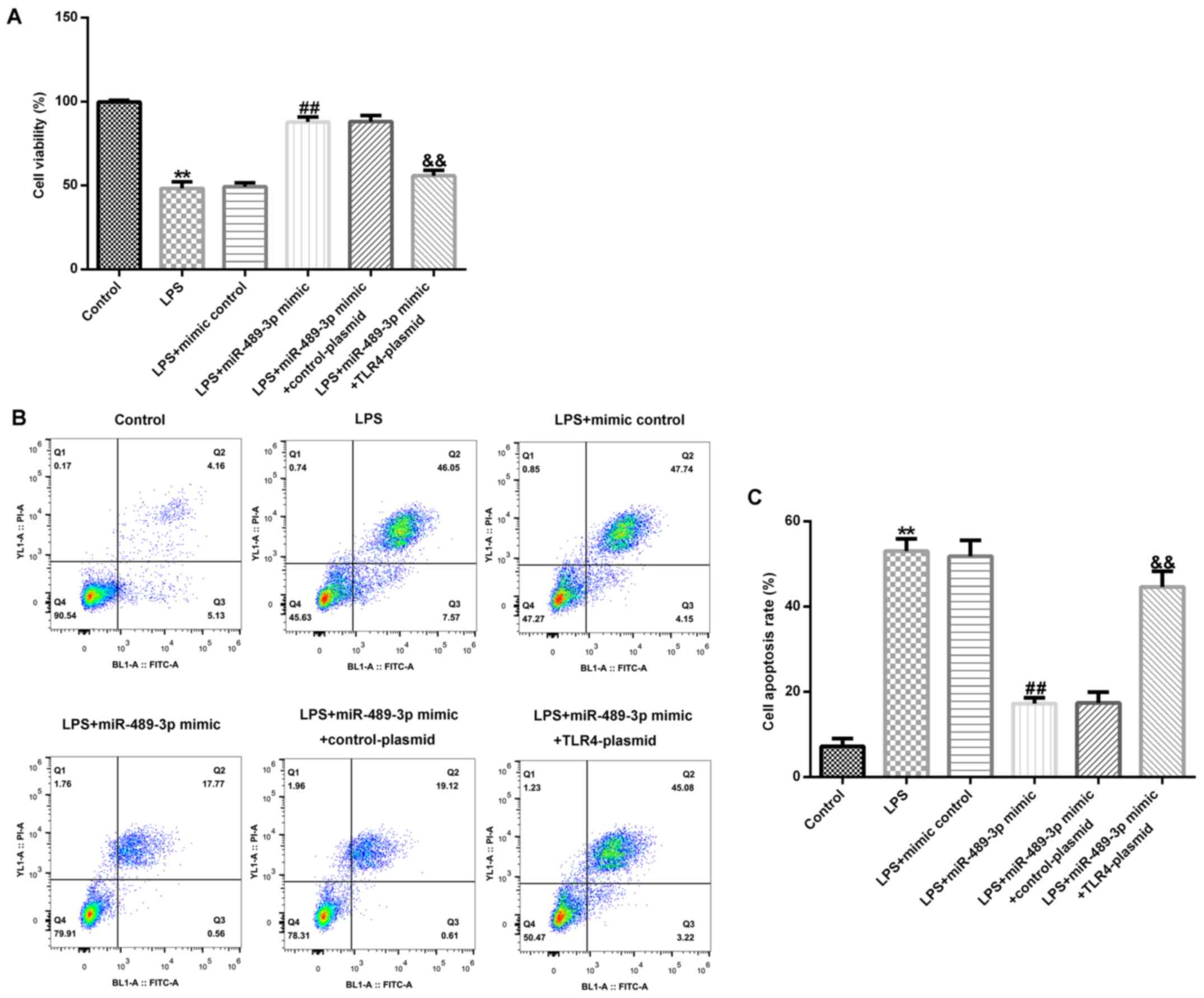
miR-489-3p promotes cell viability and
inhibits apoptosis in LPS-induced human NP cells
The effect of miR-489-3p on cell viability and
apoptosis in LPS-induced human NP cells was examined. Compared with
the control group, LPS treatment significantly inhibited NP cell
viability and induced apoptosis (Fig.
5A-C). However, compared with the LPS-treated group, the
miR-489-3p mimic group exhibited significantly increased NP cell
viability (Fig. 5A) and decreased
cell apoptosis (Fig. 5B and
C), which was partially reversed
following the co-transfection with the TLR4-plasmid.
miR-489-3p inhibits the levels of
inflammatory cytokines in LPS-induced human NP cells
To determine the effect of miR-489-3p on the levels
of inflammatory factors in LPS-induced human NP cells, ELISAs were
performed to analyze the expression levels of TNF-α, IL-1β and
IL-6. The results indicated that compared with the control group,
LPS treatment significantly increased the secretion of TNF-α, IL-1β
and IL-6 in human NP cells (Fig.
6A-C). Notably, the transfection with the miR-489-3p mimic
significantly reduced these LPS-induced increases in the expression
levels of TNF-α, IL-1β and IL-6 (Fig.
6A-C), which were significantly restored following transfection
with the TLR4-plasmid.
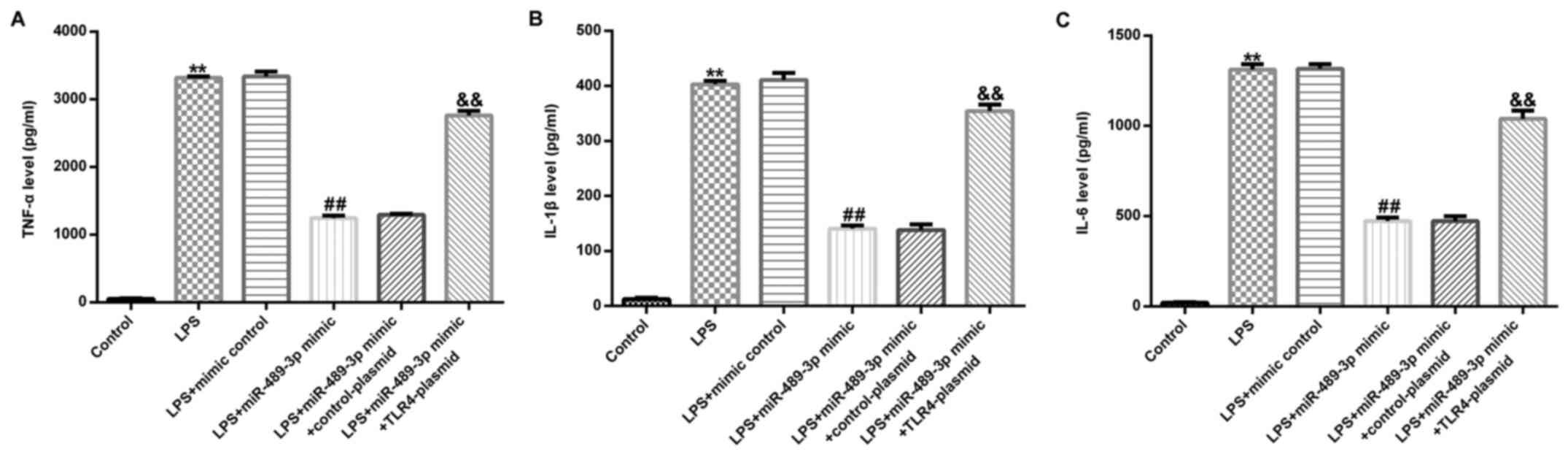 | Figure 6miR-489-3p suppresses the secretion
of TNF-α, IL-1β and IL-6 in LPS-induced human NP cells. Human NP
cells were transfected with the mimic control, miR-489-3p mimic,
miR-489-3p mimic + control-plasmid or miR-489-3p mimic +
TLR4-plasmid for 24 h and treated with 10 ng/ml LPS for 24 h.
ELISAs were used to analyze the expression levels of (A) TNF-α, (B)
IL-1β and (C) IL-6 in human NP cells. TLR4, Toll-like receptor 4;
miR, microRNA; NP, nucleus pulposus; LPS, lipopolysaccharide; IL,
interleukin; TNF, tumor necrosis factor. **P<0.01 vs.
Control; ##P<0.01 vs. LPS+mimic control;
&&P<0.01 vs. LPS+miR-489-3p
mimic+control-plasmid. |
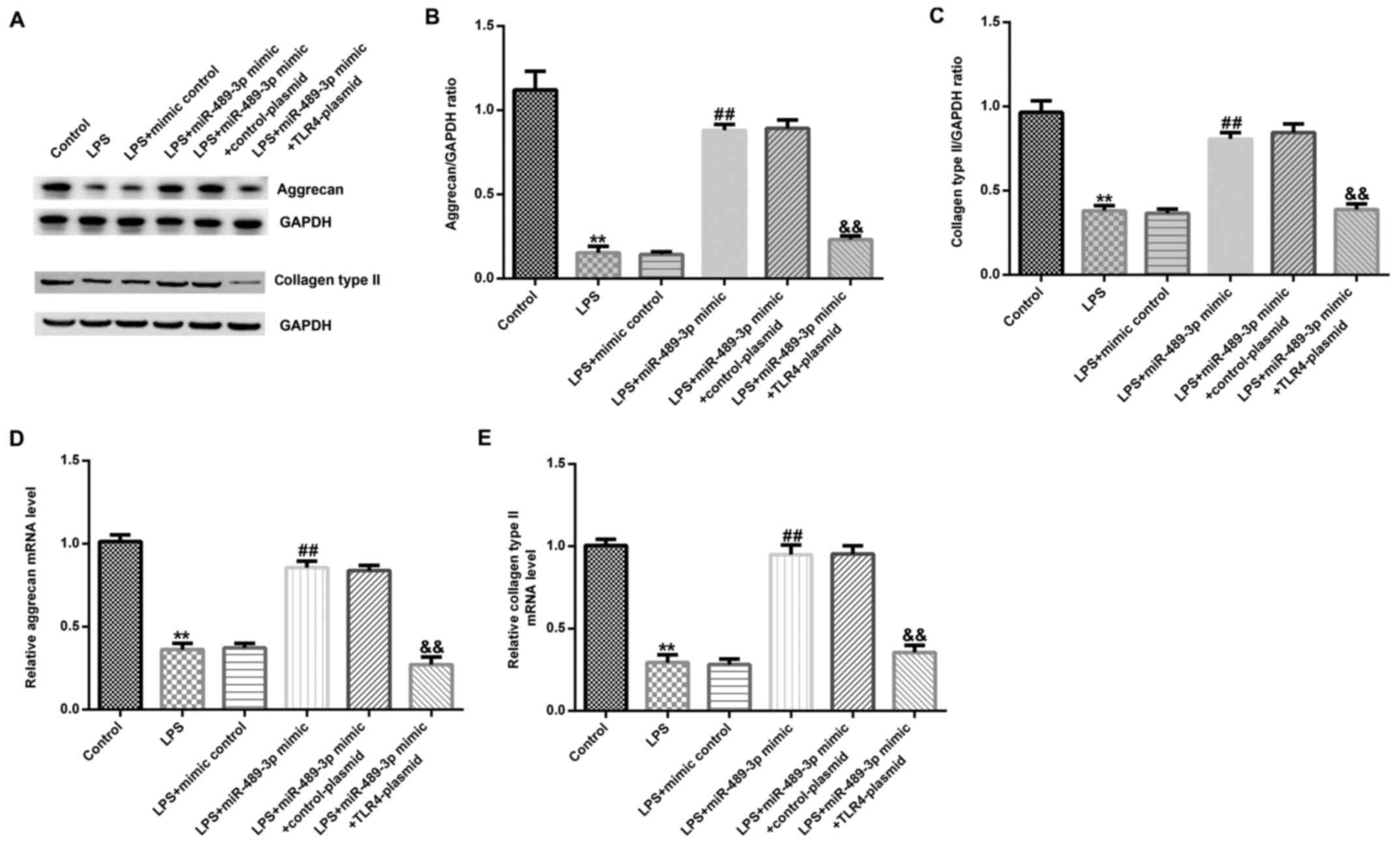
miR-489-3p inhibits ECM breakdown in
LPS-induced human NP cells
RT-qPCR and western blot assays were conducted to
analyze the expression levels of ECM-associated proteins. Compared
with the control group, LPS treatment significantly reduced the
expression levels of aggrecan and collagen type II in human NP
cells at the protein (Fig. 7A-C)
and mRNA level (Fig. 7D and
E). Moreover, compared with the
LPS-treated group, the miR-489-3p mimic significantly increased the
expression levels of aggrecan and collagen type II in human NP
cells, and this effect was partially reversed following the
co-transfection with the TLR4-plasmid.
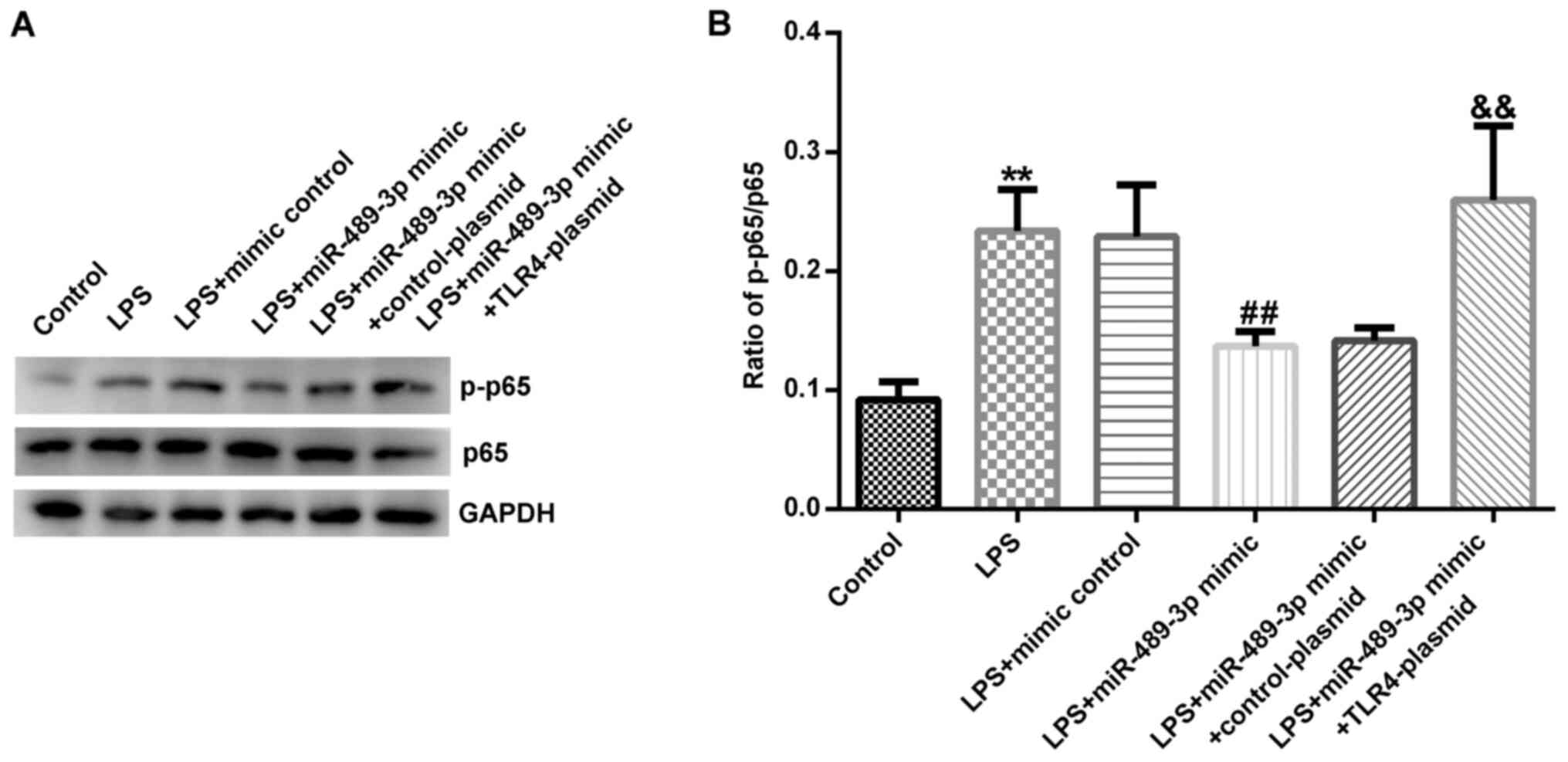
miR-489-3p inhibits the activation of
the NF-κB signaling pathway in LPS-induced human NP cells
Finally, the specific mechanism underlying the
influence of miR-489-3p in IDD was investigated. Western blotting
revealed that LPS treatment significantly increased the protein
expression levels of p-p65 and the ratio of p-p65/p65 in human NP
cells compared with the control group (Fig. 8A and B). However, the miR-489-3p mimic decreased
the protein expression levels of p-p65 (Fig. 8A) and the ratio of p-p65/p65
(Fig. 8B); this effect was
significantly restored following co-transfection with the
TLR4-plasmid.
Discussion
In the present study, human NP cells were treated
with LPS to establish an IDD model in vitro and it was
revealed that TLR4 is a direct target gene of miR-489-3p. Notably,
miR-489-3p expression levels were discovered to be downregulated in
LPS-treated human NP cells and TLR4 expression levels were observed
to be negatively associated with miR-489-3p expression levels.
Subsequently, NP cells were transfected with a miR-489-3p mimic and
TLR4 overexpression plasmid to study the effect of miR-489-3p on
human NP cells. The results indicated that miR-489-3p suppressed
the LPS-induced decreases in cell viability and increases in cell
apoptosis in human NP cells. Moreover, miR-489-3p decreased the
expression levels of the inflammatory cytokines, TNF-α, IL-1β and
IL-6, and inhibited the degradation of the ECM (evidenced by
increased expression levels of the ECM proteins), in LPS-induced
human NP cells. Finally, it was also revealed that miR-489-3p
decreased the expression levels of p-p65, which is associated with
the NF-κB signaling pathway (31).
IDD is the primary cause of lower back pain and it
is a medical condition that places a heavy burden on the global
medical system, resulting in significant socioeconomic consequences
(32-34).
At present, due to both professional and personal factors, the
incidence of IDD is increasing, particularly in China (35). Currently, rehabilitation is
important following lumbar spine fusion surgery for degenerative
diseases and it is critical for the healing of degenerative
diseases (36).
Previous studies have indicated that miRNAs are
regulators of gene expression and serve important roles in the
prevention and treatment of IDD (37,38).
It has been reported that several miRNAs were found to be
dysregulated in IDD, including miR-21, -10b, -155 and -27 (37,39-41),
whereas miR-200c was discovered to be upregulated in degenerative
NP tissues (42). In the present
study, miR-489-3p was demonstrated to be downregulated in
LPS-treated human NP cells. miR-489 expression levels were
decreased in numerous types of cancer tissue, such as gastric
cancer (43) and breast cancer
(44). In addition, miR-489 was
revealed to influence multiple pathological processes, such as
proliferation, apoptosis, invasiveness and the metastasis of tumor
cells (45). It has also been
reported that the miR-489-3p sequence shares high homology to the
miR-489 sequence.
The majority of miRNAs serve their biological
function by complementarily binding their target gene (46). In the current study, it was
discovered that TLR4 was a target gene of miR-489-3p. TLR4 is an
innate and adaptive immune cell receptor (47), which serves a vital role in the
inflammatory response (48).
Notably, Yang et al (47)
also indicated that TLR4 may be a target gene of miR-760. In
addition, TLR4 has been reported to be regulated by several miRNAs,
including miR-708-5p (49),
miR-106a (50) and miR-20a
(51). Wu et al (52) reported that the primary pathological
changes in IDD were NP apoptosis and the significant degradation of
the ECM. miRNAs have been identified to regulate cell apoptosis and
ECM protein expression (53-56).
Notably, several studies have demonstrated that miRNAs influence
the occurrence and development of IDD by regulating the apoptosis
and ECM deposition of NP cells (20-22,57,58).
For example, miR-132 was revealed to promote matrix degradation in
IDD via increased ECM catabolic factors (matrix metalloproteinase
13 and A disintegrin and metalloproteinase with thrombospondin
motifs 4), and decreased anabolic proteins (type II collagen and
aggrecan) in NP cells (21). In
other previous studies, miR-21 was also demonstrated to contribute
to type II collagen and aggrecan catabolism in human NP cells
(22); miR-145 suppressed apoptosis
and promoted ECM synthesis in NP cells (57); and miR-499a-5p was revealed to
suppress the apoptosis of human NP cells and inhibit the
degradation of the ECM via targeting the transcription factor
SOX-4(58). In the present study it
was determined that miR-489-3p suppressed the LPS-induced decreases
in cell viability and the increase in cell apoptosis in human NP
cells. However, the cell viability was analyzed following the
treatment of human NP cells with 10 ng/ml LPS for 24 h; this was a
limitation of the present study and the study protocol could be
improved by subjecting the cells to longer term viability assays.
In addition, the results indicated that miR-489-3p may inhibit the
LPS-induced degradation of the ECM in human NP cells.
Altogether, the findings of the present study
suggest that miR-489-3p may regulate the LPS-induced NP cell
inflammatory response, apoptosis and ECM-related protein
expression; however, their interaction and relationship with each
other requires further investigation in the future. Furthermore,
the present study did not use a miR-489-3p mimic control +
TLR4-plasmid + LPS group, which is also represents a limitation.
Thus, the targeting of miR-489-3p may represent a promising
therapeutic strategy and the current study may have identified
novel therapeutic targets for the development of treatments for
IDD.
In conclusion, miR-489-3p was discovered to inhibit
the LPS-induced decreases in cell viability and increases in
apoptosis, the inflammatory response and ECM degradation in human
NP cells via the suppression of the NF-κB signaling pathway via the
targeting of TLR4. Therefore, miR-489-3p may represent a potential
target for IDD treatment. However, the current study is only a
preliminary study of the role of miR-489-3p in IDD. To clarify the
mechanism underlying the role of miR-489-3p in IDD, numerous
additional in-depth experiments are required. For example, the role
of TLR4-plasmid in LPS-treated NP cells should be studied and
investigations into the role of the miR-489-3p/TLR4 axis in IDD
in vivo should be performed. In addition, the expression
levels of miR-489-3p/TLR4 in patients with IDD and the association
between miR-489-3p and TLR4 expression levels and the
clinicopathological characteristics of patients with IDD should be
investigated; our future studies aim to cover some of these
investigations.
Supplementary Material
Flow chart of the methods used in the
present study. NP, nucleus pulposus; LPS, lipopolysaccharide; miR,
microRNA; TLR4, toll-like receptor 4; RT-qPCR, reverse
transcription-quantitative PCR; WB, western blotting.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LD contributed to the design of the study, data
collection, statistical analysis, data interpretation and
manuscript preparation. BD contributed to the data collection,
statistical analysis and manuscript preparation. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kadow T, Sowa G, Vo N and Kang JD:
Molecular basis of intervertebral disc degeneration and
herniations: What are the important translational questions? Clin
Orthop Relat Res. 473:1903–1912. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tang P, Gu JM, Xie ZA, Gu Y, Jie ZW, Huang
KM, Wang JY, Fan SW, Jiang XS and Hu ZJ: Honokiol alleviates the
degeneration of intervertebral disc via suppressing the activation
of TXNIP-NLRP3 inflammasome signal pathway. Free Radic Biol Med.
120:368–379. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Powell MC, Wilson M, Szypryt P, Symonds EM
and Worthington BS: Prevalence of lumbar disc degeneration observed
by magnetic resonance in symptomless women. Lancet. 2:1366–1367.
1986.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hangai M, Kaneoka K, Kuno S, Hinotsu S,
Sakane M, Mamizuka N, Sakai S and Ochiai N: Factors associated with
lumbar intervertebral disc degeneration in the elderly. Spine J.
8:732–740. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liuke M, Solovieva S, Lamminen A, Luoma K,
Leino-Arjas P, Luukkonen R and Riihimäki H: Disc degeneration of
the lumbar spine in relation to overweight. Int J Obes (Lond).
29:903–908. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Oda H, Matsuzaki H, Tokuhashi Y,
Wakabayashi K, Uematsu Y and Iwahashi M: Degeneration of
intervertebral discs due to smoking: Experimental assessment in a
rat-smoking model. J Orthop Sci. 9:135–141. 2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jhawar BS, Fuchs CS, Colditz GA and
Stampfer MJ: Cardiovascular risk factors for physician-diagnosed
lumbar disc herniation. Spine J. 6:684–691. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kauppila LI: Atherosclerosis and disc
degeneration/low-back pain-a systematic review. Eur J Vasc Endovasc
Surg. 37:661–670. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Enercan M, Kahraman S, Cobanoglu M, Yilar
S, Gokcen BH, Karadereler S, Mutlu A, Ulusoy LO, Ozturk C, Erturer
E, et al: Selective thoracic fusion provides similar health-related
quality of life but can cause more lumbar disc and facet joint
degeneration: A comparison of adolescent idiopathic scoliosis
patients with normal population 10 years after surgery. Spine
Deform. 3:469–475. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Canbulat N, Oktenoglu T, Ataker Y, Sasani
M, Ercelen O, Cerezci O, Suzer T and Ozer AF: A rehabilitation
protocol for patients with lumbar degenerative disc disease treated
with posterior transpedicular dynamic stabilization. Turk
Neurosurg. 27:426–435. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ji ML, Jiang H, Zhang XJ, Shi PL, Li C, Wu
H, Wu XT, Wang YT, Wang C and Lu J: Preclinical development of a
microRNA-based therapy for intervertebral disc degeneration. Nat
Commun. 9(5051)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lv K: Expression profiles of miRNAs in
polarized macrophages. Int J Mol Med. 31:797–802. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen X, Slack FJ and Zhao H: Joint
analysis of expression profiles from multiple cancers improves the
identification of microRNA-gene interactions. Bioinformatics.
29:2137–2145. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wei Y, Nazari-Jahantigh M, Chan L, Zhu M,
Heyll K, Corbalán-Campos J, Hartmann P, Thiemann A, Weber C and
Schober A: The microRNA-342-5p fosters inflammatory macrophage
activation through an akt1- and microRNA-155-depend-ent pathway
during atherosclerosis. Circulation. 127:1609–1619. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ono K, Kuwabara Y and Han J: MicroRNAs and
cardiovascular diseases. FEBS J. 278:1619–1633. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ro S, Park C, Young D, Sanders KM and Yan
W: Tissue-dependent paired expression of miRNAs. Nucleic Acids Res.
35:5944–5953. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Mallory AC and Vaucheret H: MicroRNAs:
Something important between the genes. Curr Opin Plant Biol.
7:120–125. 2004.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu JY, Yang LL, Ma C, Huang YL, Zhu GX and
Chen QL: MiR-25-3p attenuates the proliferation of tongue squamous
cell carcinoma cell line Tca8113. Asian Pac J Trop Med. 6:743–747.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lv F, Huang Y, Lv W, Yang L, Li F, Fan J
and Sun J: MicroRNA-146a ameliorates inflammation via TRAF6/NF-κB
pathway in intervertebral disc cells. Med Sci Monit. 23:659–664.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu W, Xia P, Feng J, Kang L, Huang M,
Wang K, Song Y, Li S, Wu X, Yang S and Yang C: MicroRNA-132
upregulation promotes matrix degradation in intervertebral disc
degeneration. Exp Cell Res. 359:39–49. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang WJ, Yang W, Ouyang ZH, Xue JB, Li XL,
Zhang J, He WS, Chen WK, Yan YG and Wang C: MiR-21 promotes ECM
degradation through inhibiting autophagy via the PTEN/akt/mTOR
signaling pathway in human degenerated NP cells. Biomed
Pharmacother. 99:725–734. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jiang R, Zhang C, Gu R and Wu H:
MicroRNA-489-3p inhibits neurite growth by regulating PI3K/AKT
pathway in spinal cord injury. Pharmazie. 72:272–278.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu Q, Han L, Yan W, Ji X, Han R, Yang J,
Yuan J and Ni C: miR-489 inhibits silica-induced pulmonary fibrosis
by targeting MyD88 and Smad3 and is negatively regulated by lncRNA
CHRF. Sci Rep. 6(30921)2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wiese CB, Zhong JY, Xu ZQ, Zhang Y,
Ramirez Solano MA, Zhu W, Linton MF, Sheng Q, Kon V and Vickers KC:
Dual inhibition of endothelial miR-92a-3p and miR-489-3p reduces
renal injury-associated atherosclerosis. Atherosclerosis.
282:121–131. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G
and Liu J: The role of leptin on the organization and expression of
cytoskeleton elements in nucleus pulposus cells. J Orthop Res.
31:847–857. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li Z, Liang J, Wu WK, Yu X, Yu J, Weng X
and Shen J: Leptin activates RhoA/ROCK pathway to induce
cytoskeleton remodeling in nucleus pulposus cells. Int J Mol Sci.
15:1176–1188. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ding F, Shao ZW and Xiong LM: Cell death
in intervertebral disc degeneration. Apoptosis. 18:777–785.
2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cildir G, Low KC and Tergaonkar V:
Noncanonical NF-κB signaling in health and disease. Trends Mol Med.
22:414–429. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Juniper M, Le TK and Mladsi D: The
epidemiology, economic burden, and pharmacological treatment of
chronic low back pain in France, Germany, Italy, Spain and the UK:
A literature-based review. Expert Opin Pharmacother. 10:2581–2592.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Phillips C, Main C, Buck R, Aylward M,
Wynne-Jones G and Farr A: Prioritising pain in policy making: The
need for a whole systems perspective. Health Policy. 88:166–175.
2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Waddell G: Low back pain: A twentieth
century health care enigma. Spine (Phila Pa 1976).
21(2820)1996.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang YG, Sun Z, Zhang Z, Liu J and Guo X:
Risk factors for lumbar intervertebral disc herniation in Chinese
population: A case-control study. Spine (Phila Pa 1976).
34:918–922. 2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Madera M, Brady J, Deily S, McGinty T,
Moroz L, Singh D, Tipton G and Truumees E: for the Seton Spine
Rehabilitation Study Group. The role of physical therapy and
rehabilitation after lumbar fusion surgery for degenerative
disease: A systematic review. J Neurosurg Spine. 26:694–704.
2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis
D, Jia LT, Wu SX, Huang J, Chen J and Luo ZJ: Deregulated mir-155
promotes Fas-mediated apoptosis in human intervertebral disc
degeneration by targeting FAdd and caspase-3. J Pathol.
225:232–242. 2011.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ji ML, Lu J, Shi PL, Zhang XJ, Wang SZ,
Chang Q, Chen H and Wang C: Dysregulated mir-98 contributes to
extracellular matrix degradation by targeting IL-6/StAt3 signaling
pathway in human intervertebral disc degeneration. J Bone Miner
Res. 31:900–909. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X
and Qiu G: MicroRNA-10b promotes nucleus pulposus cell
proliferation through RhoC-Akt pathway by targeting HOXD10 in
intervetebral disc degeneration. PLoS One. 8(e83080)2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liu H, Huang X, Liu X, Xiao S, Zhang Y,
Xiang T, Shen X, Wang G and Sheng B: miR-21 promotes human nucleus
pulposus cell proliferation through PTEN/AKT signaling. Int J Mol
Sci. 15:4007–4018. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu G, Cao P, Chen H, Yuan W, Wang J and
Tang X: MiR-27a regulates apoptosis in nucleus pulposus cells by
targeting PI3K. PLoS One. 8(e75251)2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang H, Li L, Yuan C, Wang C, Gao T and
Zheng Z: MiR-489 inhibited the development of gastric cancer via
regulating HDAC7 and PI3K/AKT pathway. World J Surg Oncol.
18(73)2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li F: Expression of miR-221 and miR-489 in
breast cancer patients and their relationship with prognosis. Oncol
Lett. 19:1523–1529. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sun D, Yu M, Huang ZH, et al: Research
progress in the action mechanism of miR-489 in tumors. Chem Life.
37:329–335. 2017.
|
|
46
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yang YZ, Zhang YF, Yang L, Xu J, Mo XM and
Peng W: MiR-760 mediates hypoxia-induced proliferation and
apoptosis of human pulmonary artery smooth muscle cells via
targeting TLR4. Int J Mol Med. 42:2437–2446. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wang YC, Lin S and Yang QW: Toll-like
receptors in cerebral ischemic inflammatory injury. J
Neuroinflammation. 8(134)2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li WT and Zhang Q: MicroRNA-708-5p
regulates mycobacterial vitality and the secretion of inflammatory
factors in Mycobacterium tuberculosis-infected macrophages by
targeting TLR4. Eur Rev Med Pharmacol Sci. 23:8028–8038.
2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yang J, Chen Y, Jiang K, Yang Y, Zhao G,
Guo S and Deng G: MicroRNA-106a provides negative feedback
regulation in lipopolysaccharide-induced inflammation by targeting
TLR4. Int J Biol Sci. 15:2308–2319. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Gong XY and Zhang Y: Protective effect of
miR-20a against hypoxia/reoxygenation treatment on cardiomyocytes
cell viability and cell apoptosis by targeting TLR4 and inhibiting
p38 MAPK/JNK signaling. In Vitro Cell Dev Biol Anim. 55:793–800.
2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wu D, Zheng C, Wu J, Huang R, Chen X,
Zhang T and Zhang L: Molecular biological effects of weightlessness
and hypergravity on intervertebral disc degeneration. Aerosp Med
Hum Perform. 88(1123)2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wang J, Liew OJ, Richards AM and Chen YT:
Overview of MicroRNAs in cardiac hypertrophy, fibrosis, and
apoptosis. Int J Mol Sci. 17(749)2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Moro A, Driscoll TP, Boraas LC, Armero W,
Kasper DM, Baeyens N, Jouy C, Mallikarjun V, Swift J, Ahn SJ, et
al: MicroRNA-dependent regulation of biomechanical genes
establishes tissue stiffness homeostasis. Nat Cell Biol.
21:348–358. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Toyono T, Usui T, Villarreal G Jr, Kallay
L, Matthaei M, Vianna LM, Zhu AY, Kuroda M, Amano S and Jun AS:
MicroRNA-29b overexpression decreases extracellular matrix mRNA and
protein production in human corneal endothelial cells. Cornea.
35:1466–1470. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhou J, Sun J, Markova DZ, Li S, Kepler
CK, Hong J, Huang Y, Chen W, Xu K, Wei F and Ye W: MicroRNA-145
overexpression attenuates apoptosis and increases matrix synthesis
in nucleus pulposus cells. Life Sci. 221:274–283. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Sun JC, Zheng B, Sun RX, Meng YK, Wang SM,
Yang HS, Chen Y, Shi JG and Guo YF: MiR-499a-5p suppresses
apoptosis of human nucleus pulposus cells and degradation of their
extracellular matrix by targeting SOX4. Biomed Pharmacother.
113(108652)2019.PubMed/NCBI View Article : Google Scholar
|