Introduction
Acute otitis media (AOM) is one of the most common
infectious diseases in childhood caused by bacteria (1), such as Streptococcus
pneumoniae, non-typeable Haemophilus influenzae (H.
influenzae) and Moraxella catarrhalis (2). AOM is characterized by the presence of
middle ear effusion together with an acute onset of signs and
symptoms caused by middle ear inflammation. The pathophysiology,
pathogenesis and progression of AOM are associated with multiple
factors, such as pathogenic bacteria, reactive oxygen species and
inflammatory cytokines (3).
Currently, due to the extensive use of antibiotics, which may cause
resistance of AOM to treatment, 10-20% of children may experience
continuous recurrences of otitis media and long-term deafness
(4). Therefore, identifying
effective therapies is crucial for the prevention and treatment of
AOM.
Long non-coding (lnc)RNAs are a group of
heterogeneous non-coding RNAs with a length of >200 nucleotides,
which are widely distributed in the genome (5). Previous research has shown that
lncRNAs play an important role in transcription regulation,
maintenance of genomic integrity, alternative splicing, gene
chromatin rearrangement (6,7) and various pathophysiological processes
(8,9). Several studies have revealed that
lncRNAs are implicated in immune defense against bacterial
infections, involving Salmonella, Mycobacterium bovis
and Mycobacterium tuberculosis (10-12).
The lncRNA nuclear-enriched abundant transcript 1 (NEAT1) has two
isoforms (short isoform NEAT1-1 and long isoform NEAT1-2), which
are associated with the prognosis of human tumors (13). In addition, NEAT1 has been reported
to be involved in a number of inflammatory diseases. It has been
reported that overexpression of NEAT1 protected cardiomyocytes from
As2O3-induced injury (14), whereas other studies re-ported that
downregulating NEAT1 inhibited inflammation, fibrosis and apoptosis
(15-17).
In addition, NEAT1 was found to be upregulated by
lipopolysaccharide (LPS) (15,18).
However, the regulatory mechanism of NEAT1 in AOM remains unclear
and must be further investigated. Therefore, the present study
focused on the biological function of NEAT1 in AOM.
MicroRNAs (miRNAs/miRs) are non-coding, endogenous
single-stranded small RNAs. Studies have shown that miRNAs may
serve as nodes of signaling networks, participating in the
regulation of basic cell activities, such as cell inflammation
(19). miR-301b-3p was found to be
significantly reduced in LPS-induced human middle ear epithelial
cells (HMEECs) (20), but the
biological function of miR-301b-3p in AOM has not been reported to
date. Furthermore, miRNAs can negatively regulate protein
expression by targeting the 3'-untranslated region (UTR) of their
target mRNAs (21). Based on
bioinformatics prediction, the toll-like receptor 4 gene
(TLR4) was shown to be a target of miR-301b-3p in the
present study. TLR4 has been proven to play an essential role in
triggering the inflammatory response, and TLR4 signal activation
promotes an increase in the production and upregulation of
pro-inflammatory factors (22,23).
Furthermore, the TLR4 protein was found to be upregulated by
non-typeable H. influenzae infection and LPS induction in
animal models and patients with AOM (24-27).
Hence, it was hypothesized that miR-301b-3p and TLR4 may be
involved in NEAT1-mediated inflammation in AOM. The aim of the
present study was to investigate the effect of NEAT1 on
inflammatory response in LPS-treated HMEECs and explore the
potential interactions among NEAT1, miR-301b-3p and TLR4.
Materials and methods
Cell culture and LPS treatment
HMEECs were purchased from American Type Culture
Collection; human renal epithelial cells (HK2) and human liver
epithelial cells (THLE-3) were purchased from Beijing Beina
Chuanglian Institute of Biotechnology. HMEECs and THLE-3 cells were
routinely cultured in RPMI-1640 medium supplemented with 10% FBS
(both from Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin. HK2 cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 100 U/ml
penicillin and 100 µg/ml streptomycin. The cells were kept in a
humidified atmosphere with 5% CO2 and 95% humidity at
37˚C in an incubator. HMEECs were stimulated with LPS
(Sigma-Aldrich; Merck KGaA) at different concentrations (0, 0.1, 1,
3, 5 and 10 µg/ml) for 24 h, and then cultured for 12, 24 and 48 h
to determine cell viability. The cells were incubated with 5 µg/ml
LPS for 6, 12, 24 and 48 h to determine the optimal action time.
Finally, 5 µg/ml LPS was used for 24 h to establish an inflammatory
cell model.
Cell transfection
Transfections were performed using 1.0 µg small
interfering (si)RNA of NEAT1 (si-NEAT1; 5'-GUGAGA
AGUUGCUUAGAAACUUUCC-3'), 1.0 µg si-TLR4 (5'-TTGC
TAAATGCTGCCGTTTTATC-3'), 1.0 µg si-RNA-negative control (si-NC;
5'-UACUGUCUAGUCGCCGUAC-3'), 100 nM miR-301b-3p mimic
(5'-CAGUGCAAUGAUAUUGUCAA AGC-3'), 100 nM NC mimic
(5'-UUGUACUACACAAAAGU ACUG-3'), 100 nM miR-301b-3p inhibitor
(5'-GCUUUGACAA UAUCAUUGCACUG-3') and 100 nM NC inhibitor (5'-CAG
UACUUUUGUGUAGUACAA-3'), which were obtained from Shanghai
GenePharma Co., Ltd.. To overexpress NEAT1 or TLR4, the
full-length sequence was inserted into the pcDNA3.1 vector (2 µg;
Shanghai GenePharma Co., Ltd.), and an empty vector (2 µg) was used
as a control. HMEECs cells (1x105) were transfected with
si-RNA, miR-mimic, miR-inhbitor, overexpression vectors and
corresponding NCs using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37˚C. After transfection for 48
h, the efficiency of transfection was detected by reverse
transcription-quantitative PCR (RT-qPCR) and western blot
analysis.
RNA extraction and RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The first cDNA of
total RNA was synthesized according to the instructions of
PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.). qPCR
was subsequently performed using the SYBR Green qPCR kit (Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. The following primer sequences were used for RT-qPCR:
Taurine upregulated gene 1, forward 5'-AAATCTGGCTCTGCTGACCC-3' and
reverse 5'-CAGAAAGGCCAGCCATACCA-3'; zinc finger antisense 1,
forward 5'-AACCATTAGCTAGCTGG GGC-3' and reverse
5'-CAAGTTAACCCCGGAGGGAC-3'; plasmacytoma variant translocation 1,
forward 5'-CCTGTGA CCTGTGGAGACAC-3' and reverse 5'-GTCCGTCCAGAGT
GCTGAAA-3'; zinc finger E-box binding homeobox 1 antisense 1,
forward 5'-CCGTGGGCAC-TGCTGAAT-3' and reverse
5'-CTGCTGGCAAGCGGAACT-3'; X-inactive-specific transcript, forward
5'-CTTGGATGGGTTGCCAGCTA-3' and reverse 5'-TCATGCCCCATCTCCACCTA-3';
NEAT1, forward 5'-CAGTTAGTTTATCAG-TTCTCCCATCCA-3' and reverse
5'-GTTGTTGTCGTCACCTTTCAACTCT-3'; miR-301b-3p, forward
5'-TGCTGCTAACGAATGCTCTGA-3' and reverse
5'-CTCTGCTTTCAGATGCTTTGAC-3'; GAPDH, forward
5'-AGAAGGCTGGGGCTCATTTG-3' and reverse 5'-AGGG GCCATCCACAGTCTTC-3';
and U6, forward 5'-CTCGCTTC GGCAGCACATA-3' and reverse
5'-AACGATTCACGAATT TGCGT-3'. RT-qPCR experiments were performed
with the 7900HT Fast Real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The following thermocy-cling
conditions were used for qPCR: Initial denaturation at 95˚C for 7
min, followed by 40 cycles of denaturation at 95˚C for 10 sec,
annealing at 60˚C for 20 sec and elongation at 72˚C for 20 sec,
with final extension at 72˚C for 10 min. Relative expression levels
were calculated using the 2-∆∆Cq method (28) and normalized to the internal
reference genes GAPDH (mRNA) and U6 (miRNA).
Cell viability assay
Cell viability was assessed using the Cell Counting
Kit-8 proliferation de-tection kit (Beyotime Institute of
Biotechnology). Briefly, cells were seeded in 96-well plates at a
density of 104 cells/well; 10 µl CCK-8 solution was
added to each well and cultured for 4 h. The OD value at 450 nm was
measured with a microplate reader (BioTek Instruments, Inc.).
Detection of cell apoptosis
Apoptosis was detected using an Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) cell
apoptosis detection kit (Beijing Solarbio Science & Technology
Co., Ltd.). In brief, HMEECs were collected using cold PBS and then
cultured with 5 µl Annexin V-FITC reagent and 5 µl PI in the dark
for 15 min. The cell apoptosis rate was analyzed by flow cytometry
(BD Biosciences) and FlowJo software (version 10; FlowJo LLC).
ELISA
Cell-free culture supernatant was collected after
treatment. The secretory levels of IL-6 (cat. no. 900-T16), TNF-α
(cat. no. 900-M25) and IL-1β (cat. no. 900-M95) were analyzed using
their corresponding ELISA kits (Neobioscience Technology Co., Ltd.)
according to the manufacturers' instructions. The concentration of
inflammatory cytokines was measured at 450 nm using a microplate
spectrophotometer (BioTek Instruments, Inc.).
Western blot analysis
Cells were lysed with RIPA buffer [50 mM Tris-HCl
(pH 8.0), 50 mM NaCl and 0.5% NP-40]. The total protein (20
µg/well) in the supernatant was separated by 10% SDS-PAGE and then
transferred to PVDF membranes. After blocking with 5% skimmed milk
at room temperature for 2 h, the membrane was incubated with
primary antibodies against TLR4 (dilution 1:1,000; cat. no.
ab13556; Abcam) or GAPDH (dilution 1:1,000; cat. no. ab8245; Abcam)
at 4˚C overnight, and then incubated with horseradish
peroxidase-conjugated secondary antibody (dilution 1:4,000; cat.
no. sc-2357; Santa Cruz Biotechnology, Inc.) for 2 h at room
temperature. The immunoreactive bands were visualized using an
enhanced chemilumi-nescence reagent (Beyotime Institute of
Biotechnology). The blots were semiquantified by ImageJ software,
version 1.47 (National Institutes of Health).
Bioinformatics and luciferase reporter
analysis
The starBase (version 2.0: star-base.sysu.edu.cn) online software was used to
predict targeting binding sites of lncRNA to miRNA and miRNA to
mRNA. Therefore, the targeting associations between miR-301b-3p and
NEAT1 or TLR4 were predicted. Reporter vectors were
constructed using pmirGLO dual-luciferase expression vector
(Promega Corporation), including wild-type (WT) and mutant (MUT)
binding sequences of NEAT1 and TLR4, named as WT-NEAT1,
MUT-NEAT1, WT-TLR4 and MUT-TLR4, and were
co-transfected with miR-301b-3p mimics or negative control
(sequences as aforementioned) into 293 cells (The Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences, Shanghai,
China) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37˚C. After 48 h of transfection,
luciferase activities were measured with a dual-luciferase reporter
assay system (Promega Corporation). Firefly luciferase activity was
normalized to Renilla luciferase activity and all
experiments were performed in triplicate.
Statistical analysis
All experiments were repeated three times. GraphPad
Prism 8 (GraphPad Software, Inc.) was used for statistical
analysis, and data are presented as mean ± standard deviation.
Differences between the two groups were analyzed using unpaired
Student's t-tests and differences between multiple groups data were
analyzed by one-way ANOVA followed by Tukey's post hoc tests.
P<0.05 was considered to indicate statistically significant
differences.
Results
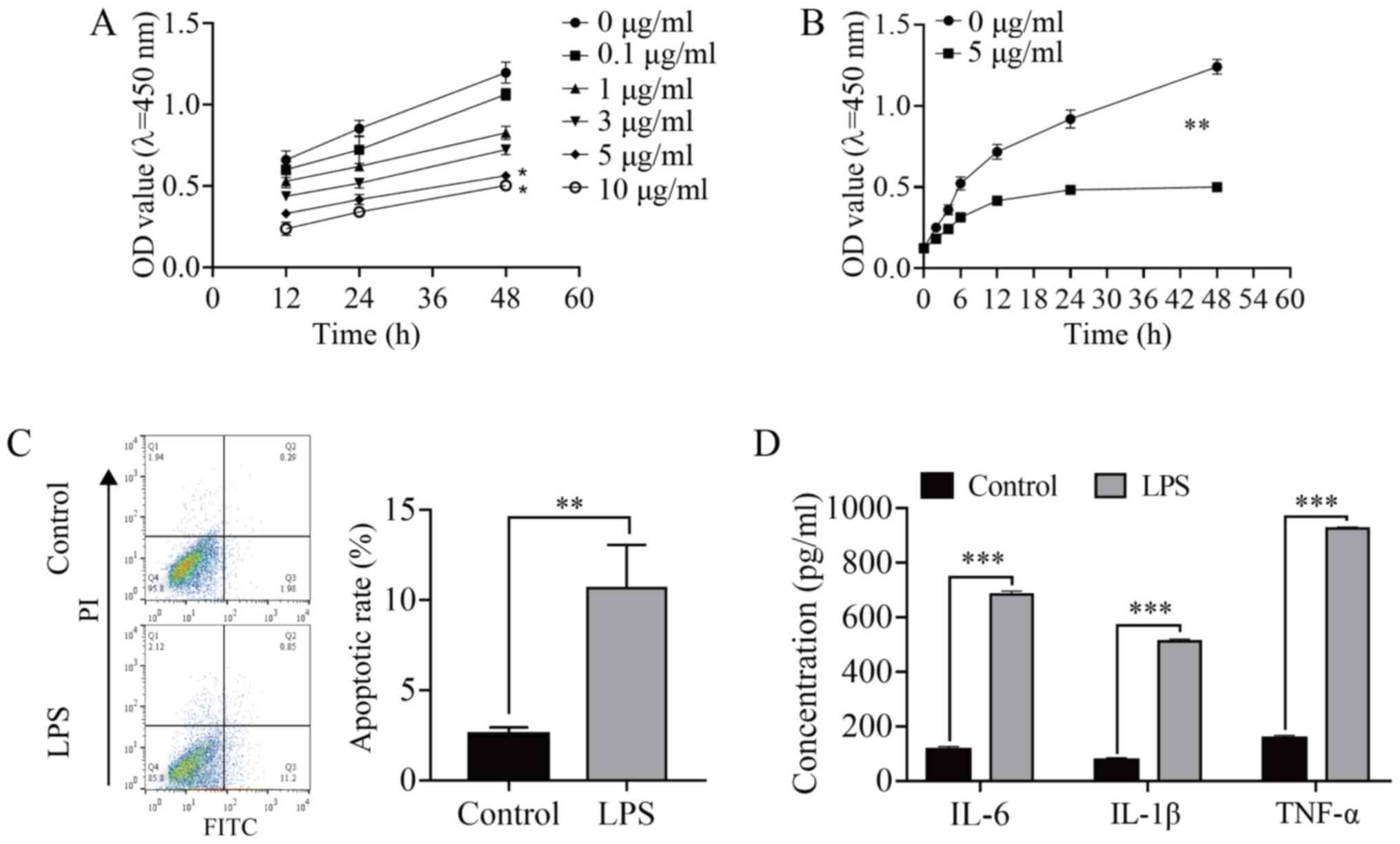
LPS induces apoptosis and inflammatory
response in HMEECs and inhibits cell viability
To investigate whether LPS causes HMEEC injury,
different concentrations (0, 0.1, 1, 3, 5 and 10 µg/ml) of LPS were
used to treat HMEECs for 12, 24 and 48 h. The CCK-8 assay
demonstrated that the viability of HMEECs was inhibited after LPS
induction in a time- and concentration-dependent manner (Fig. 1A). Following treatment with 3 and 5
µg/ml for 48 h, the cell viability decreased by ~50% (P<0.05),
and it was significantly decreased in the 5 µg/ml group compared
with that in the 0 µg/ml (control) group (P<0.01; Fig. 1B). Therefore, HMEECs were treated
with 5 µg/ml for 24 h in the subsequent experiments. LPS-induced
HMEEC apoptosis was significantly increased (P<0.01; Fig. 1C), and the concentration of
inflammatory factors (IL-6, IL-1β and TNF-α) was also significantly
increased (P<0.001; Fig. 1D),
indicating that LPS induced an inflammatory response in HMEECs.
NEAT1 knockdown alleviates cell
apoptosis and inflammatory response in LPS-induced HMEECs
RT-qPCR was conducted to explore the possible
involvement of lncRNAs in the regulation of AOM-related mechanisms.
As shown in Fig. 2A, all six
inflammation-related lncRNAs were found to be upregulated in
LPS-stimulated HMEECs, among which NEAT1 exhibited the most
prominent increase (P<0.001). Subsequently, si-NEAT1 was
transfected into HMEECs to inhibit the expression of NEAT1. The
results demonstrated that NEAT1 was successfully silenced (Fig. 2B; P<0.01). CCK-8 assay, flow
cytometry and ELISA were conducted to elucidate the effect of NEAT1
knockdown on the proliferation, apoptosis and inflammation of
LPS-stimulated HMEECs (Fig. 2C-E).
The results revealed that NEAT1 knockdown relieved the
proliferation inhibition of LPS-induced HMEECs (P<0.01), and
markedly alleviated cell apoptosis and the levels of the
inflammatory factors IL-6, IL-1β and TNF-α (P<0.05 and
P<0.001, respectively). Taken together, these results suggested
that the downregulation of NEAT1 inhibited LPS-induced HMEEC
inflammatory injury.
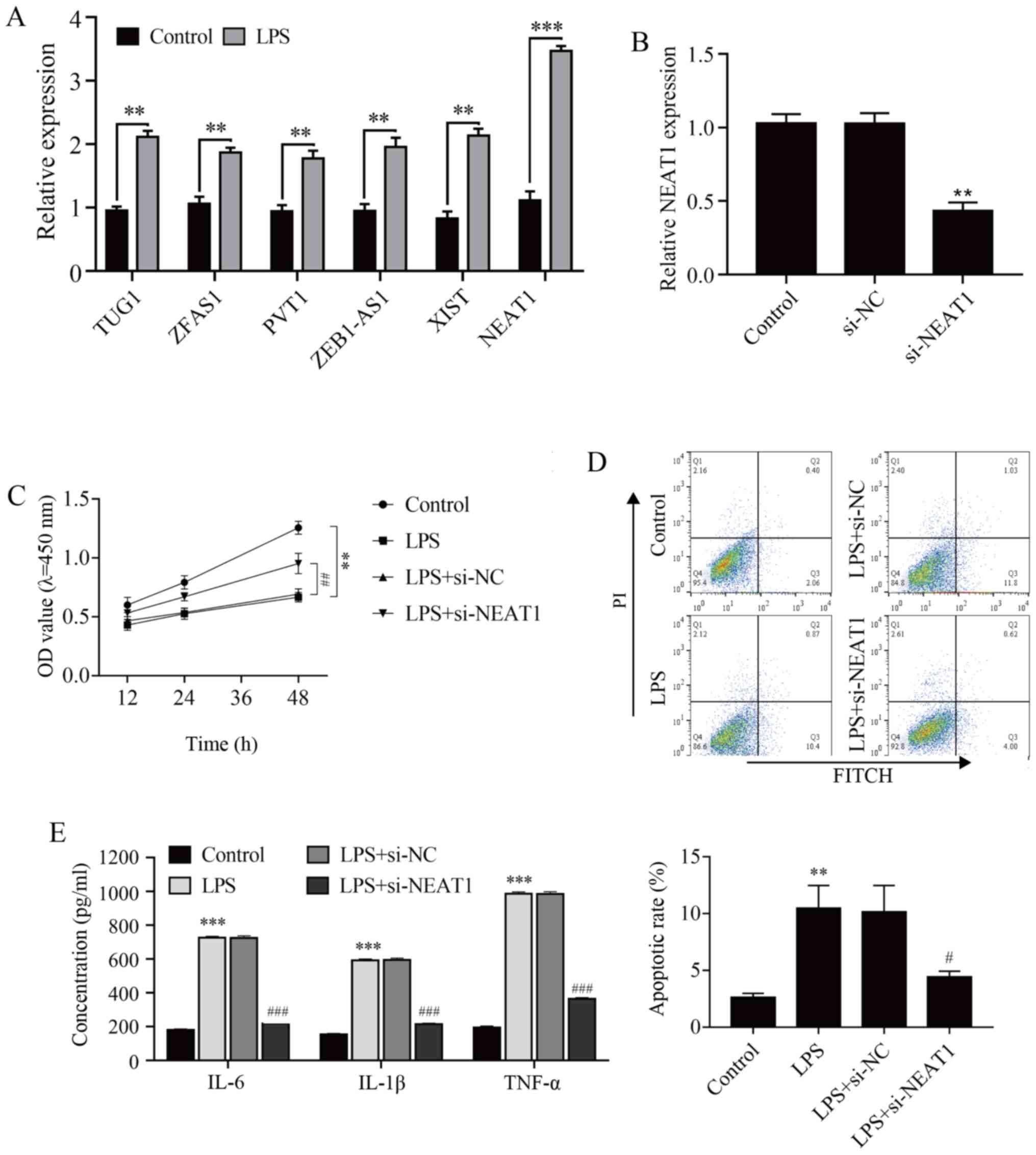 | Figure 2NEAT1 knockdown alleviates cell
apoptosis and inflammatory response in LPS-induced human middle ear
epithelial cells. (A) The expression of different long non-coding
RNAs and (B) transfection efficiency of si-NEAT1 were analyzed by
reverse transcription-quantitative PCR. (C) Cell viability was
analyzed by Cell Counting Kit-8 assay. (D) Cell apoptosis was
evaluated by flow cytometry. (E) IL-6, IL-1β and TNF-α expression
was measured by ELISA. **P<0.01 and
***P<0.001 vs. control; #P<0.05,
##P<0.01 and ###P<0.001 vs. LPS+si-NC.
LPS, LPS-stimulated; NEAT1, nuclear enriched abundant transcript 1;
TUG1, taurine upregulated gene 1; ZFAS1, zinc finger antisense 1;
PVT1, plasmacytoma variant translocation 1; ZEB1-AS1, zinc finger
E-box binding homeobox 1 antisense 1; XIST, X-inactive-specific
transcript; LPS, lipopolysaccharide; NC, negative control; si,
small interfering; OD, optical density; FITC, fluorescein
isothiocyanate; PI, propidium iodide. |
miR-301b-3p targets NEAT1 and TLR4 in
HMEECs
Epithelial cells are protected from injury due to
different stressors through inflammation response regulation. To
investigate the abnor-mal expression of miR-301b-3p and TLR4 in
LPS-induced epithelial cells, as described by Ma et al
(29) in HMEECs, HK-2 and THLE-3
cells were selected, and then exposed to LPS for 48 h. miR-301b-3p
expression in HMEECs was markedly downregulated following LPS
treat-ment (Fig. 3A; P<0.01),
while TLR4 expression was significantly upregulated (Fig. 3B; P<0.05). Moreover, miR-301b-3p
and TLR4 levels in HK-2 and THLE-3 cells, as well as in HMEECs,
were detected by RT-qPCR and western blot assays. miR-301b-3p and
TLR4 levels were down- and upregulated, respectively, following LPS
induction (Fig. 3A and B; P<0.05). These results suggested that
LPS down- and upregulated miR-301b-3p and TLR4 levels,
re-spectively, in HMEECs. A search through the bioinformatics
database starBase revealed a putative interaction between NEAT1 and
miR-301b-3p (the target binding sequence is shown in Fig. 3C). Dual-luciferase reporter assay
demonstrated that NEAT1-WT and miR-301b-3p mimics cotransfection
significantly decreased luciferase activity in HMEECs, while
NEAT1-MUT and miR-301b-3p mimics cotransfection failed to affect
luciferase activity in HMEECs (Fig.
3D; P<0.01). Furthermore, NEAT1 silencing significantly
increased miR-301b-3p expression (Fig.
3E; P<0.01). These results indicated that miR-301b-3p was
targeted by NEAT1. Subsequently, based on the biological
information database starBase, the target gene of miR-301b-3p was
predicted, and it was found that TLR4, which is involved in
inflammation, and miR-301b-3p had a targeted binding sequence
(Fig. 3F). Dual-luciferase reporter
assay demonstrated that the luciferase activity of TLR4-WT was
decreased by miR-301b-3p mimics in HMEECs, while TLR4-MUT and
miR-301b-3p mimics cotransfection failed to affect luciferase
activity (Fig. 3G; P<0.01).
Subsequently, pc-NEAT1 vector and miR-301b-3p mimic were
transfected into HMEECs to upregulate the expression of NEAT1 and
miR-301b-3p, and the results revealed that NEAT1and miR-301b-3p
expression was strongly enhanced (Fig.
3H and 3I; P<0.01). As shown
in Fig. 3J, overexpression of
miR-301b-3p inhibited TLR4 expression, which was reversed by
overexpressing TLR4 (Fig. 3J;
P<0.05). Taken together, these results proved that miR-301b-3p
directly targets both NEAT1 and TLR4.
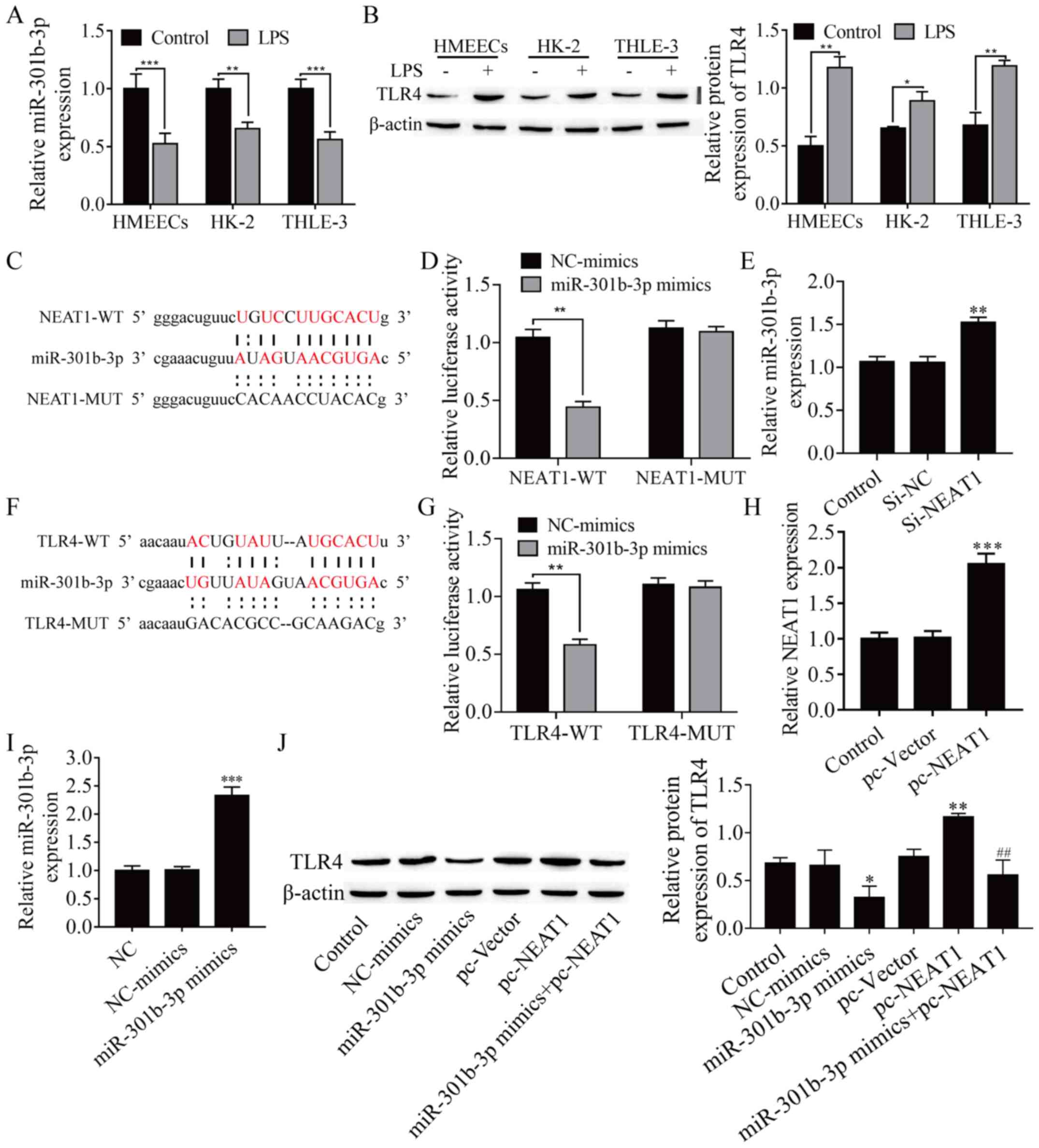 | Figure 3miR-301b-3p directly targets both
NEAT1 and TLR4. (A) miR-301b-3p expression was detected by
RT-qPCR. (B) TLR4 protein level was analyzed with western blotting.
(C) Binding site of miR-301b-3p and NEAT1. (D) Dual-luciferase
reporter assay was used to prove NEAT1 can target miR-301b-3p. (E)
NEAT1 silencing significantly increased miR-301b-3p expression in
HMEECs. (F) Binding site of miR-301b-3p and TLR4. (G)
Dual-luciferase reporter assay was used to prove that miR-301b-3p
can target TLR4. (H) NEAT1 expression was determined by
RT-qPCR. (I) miR-301b-3p expression was detected by RT-qPCR. (J)
TLR4 protein expression was measured by western blotting.
*P<0.05 vs. control or NC-mimics;
**P<0.01 vs. control, NC-mimics or si-NC; and
***P<0.001 vs. control or NC-mimics;
##P<0.01 compared with pc-vector. HMEECs, human
middle ear epithelial cells; NEAT1, nuclear enriched abundant
transcript 1; TLR4, toll-like receptor 4; LPS, lipopolysaccharide;
miR, microRNA; NC, negative control; si, small interfering; WT,
wild-type; MUT, mutant; RT-qPCR, reverse transcription-quantitative
PCR. |
NEAT1 regulates LPS-induced
proliferation, apoptosis and inflammation of HMEECs via the
miR-301-3p/TLR4 axis
To explore how NEAT1 regulates LPS-induced HMEEC
proliferation, inflammation and apoptosis, miR-301b-3p inhibitors,
si-NEAT1, si-TLR4 and pc-TLR4 were transfected into
HMEECs to detect their regulatory effects on inflammation.
Silencing miR-301b-3p significantly decreased miR-301b-3p
expression compared with the NC group (Fig. 4A; P<0.01), and then transfection
of si-TLR4 and pc-TLR4 resulted in effective decrease
and increase in the expression of TLR4, respectively (Fig. 4B and C; P<0.01). The CCK-8 assay revealed
that LPS inhibited cell proliferation, which was recovered with
si-NEAT1 and si-TLR4, but proliferation was finally
suppressed by miR-301b-3p inhibitors or overexpression TLR4
(Fig. 4D). Similarly, the
inflammation and apoptosis of HMEECs was analyzed. Flow cytometry
and ELISA revealed an opposite trend than the CCK-8 assay (Fig. 4E and F). Next, western blotting was performed to
verify whether NEAT1 regulated the miR-301-3p/TLR4 axis and protein
expression of TLR4. As shown in Fig.
4G, miR-301b-3p inhibitors or overexpression of TLR4 abolished
the inhibitory effect of NEAT1 silencing on the protein expression
of TLR4. In addition, miR-301b-3p inhibitors reversed the
suppression of the protein expression of TLR4 by TLR4 silencing.
Taken together, these results indicated that NEAT1 attenuated
LPS-induced inflammation in HMEECs via modulating the
miR-301b-3p/TLR4 axis.
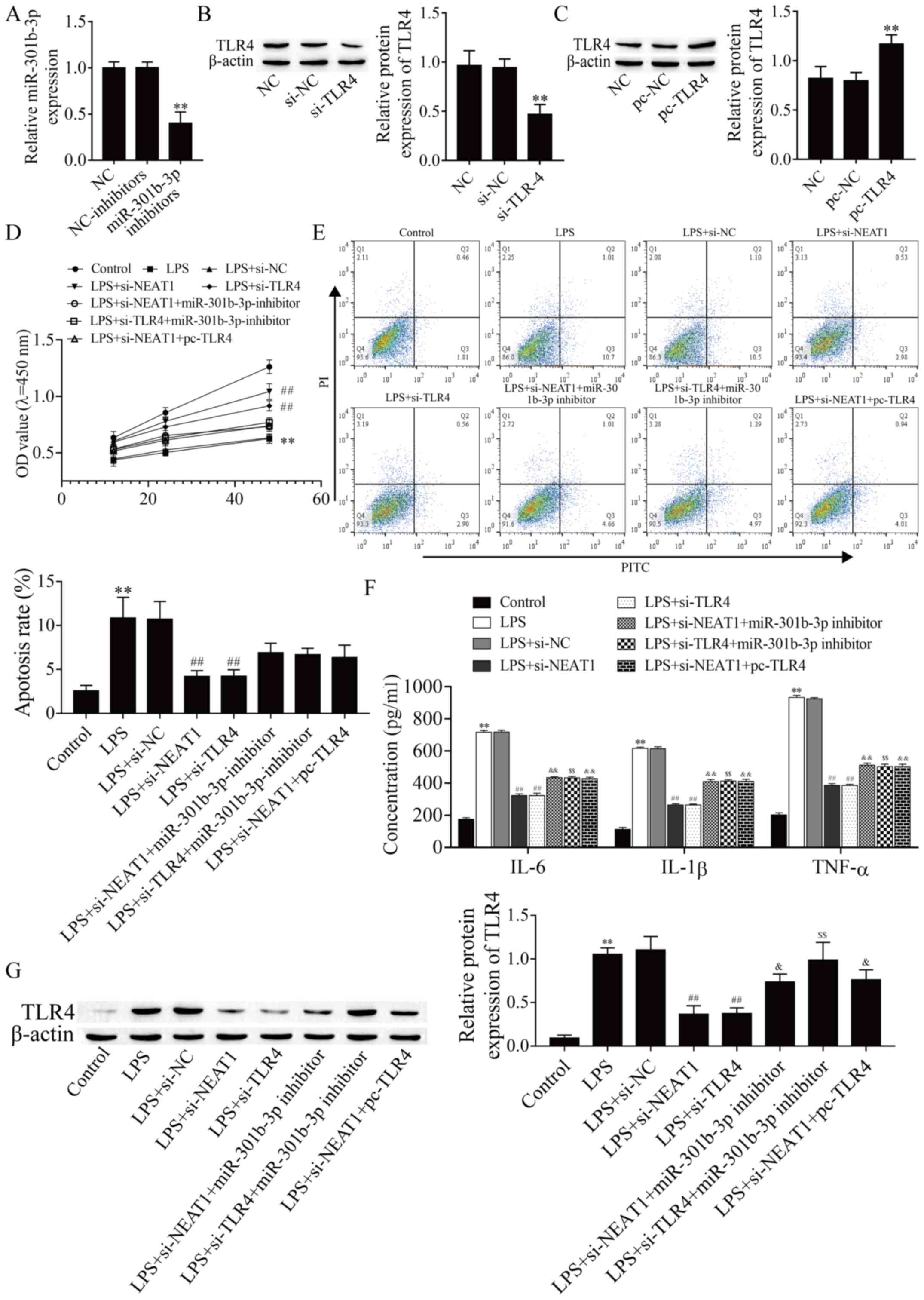 | Figure 4NEAT1 regulates LPS-induced cell
viability, apoptosis and inflammation of human middle ear
epithelial cells via the miR-370-3p/TLR4 axis. (A) miR-301b-3p
expression was determined by reverse transcription-quantitative
PCR. TLR4 protein expression was detected by western blot assay
after transfection with (B) si-TLR4 and (C) pc-TLR4.
(D) Cell viability was analyzed by Cell Counting Kit-8 assay. (E)
Cell apoptosis was evaluated by flow cytometry. (F) IL-6, IL-1β and
TNF-α expression levels were detected by ELISA. (G) TLR4 protein
expression in different groups was detected by western blot assay.
**P<0.01 vs. control, si-NC or pc-NC;
##P<0.01 vs. control and LPS+si-NC;
&P<0.05 vs. LPS+si-NEAT1;
&&P<0.01 vs. LPS+si-NEAT1;
$$P<0.01 vs. LPS+si-TLR4. LPS,
lipopolysaccharide; miR, microRNA; NC, negative control; si, small
interfering; NEAT1, nuclear enriched abundant transcript 1; TLR4,
toll-like receptor 4; OD, optical density; FITC, fluorescein
isothiocyanate; PI, propidium iodide. |
Discussion
AOM is one of the most common infectious diseases in
children. Its pathological characteristic is inflammation, with
induced secretion of cytokines TNF-α, IL-6 and IL-8(30). AOM may develop into recurrent or
persistent otitis media, which causes progressive hearing loss and
severely affects the quality of life of the patients (1), even causing language disability and
intellectual impairment (31). The
specific molecular mechanism of AOM is poorly understood and
finding effective therapeutic strategies is urgently required.
Therefore, the present study aimed to investigate the
NEAT1/miR-301b-3p/TLR4 regulatory network, in the hope of revealing
a new theoretical basis for the application of NEAT1 in the
treatment of AOM. In the present study, LPS was used to establish
the cell model of AOM and investigate the potential mechanism
(26,32). According to previous reports, NEAT1
is induced in multiple immune system diseases, such as pneumonia
(15), hepatitis (17) and osteoarthritis (33). Thus, it was hypothesized that NEAT1
may be induced in LPS-stimulated HMEECs. In the present study,
NEAT1 was found to be significantly increased in LPS-induced
HMEECs, and knockdown of NEAT1 significantly improved cell
proliferation and inhibited cell apoptosis and inflammation. These
data suggested that NEAT1 knockdown attenuated LPS-induced
inflammation and apoptosis.
NEAT1 has been previously found to be induced in
various tumors (13). For example,
NEAT1 was shown to increase cell survival and facilitate
gallbladder cancer progression by sponging miR-335(34). Furthermore, NEAT1 is not only
im-plicated in cancer, but may also affect the inflammatory
response. Li et al (35)
reported that NEAT1 may promote LPS-induced inflammatory injury in
macrophages by regulating miR-17-5p/TLR4. In addition,
downregulation of NEAT1 promoted cell proliferation and inhibited
the apoptosis of rat cardiac muscle cells (36). However, to the best of our
knowledge, the function and mechanism of action of NEAT1 in AOM
have not been reported in the literature to date. The present study
demonstrated that NEAT1 expression was higher in LPS-stimulated
HMEECs compared with other pro-inflammatory lncRNAs. Subsequently,
loss-of-function experiments revealed that NEAT1 silencing
inhibited LPS-stimulated HMEEC inflammation and apoptosis, while it
promoted cell proliferation. Thus, these data suggested that NEAT1
may regulate LPS-induced inflammation of HMEECs.
It has been previously demonstrated that lncRNAs may
serve as miRNA competitive endogenous RNAs (ceRNAs) in several
diseases (37). For example, NEAT1
may serve as a ceRNA, regulating LPS-induced apoptosis and
inflammatory injury of WI-38 cells by spong-ing miR-193a-3p to
target TLR4 (15). In the
present study, miR-301b-3p was downregulated and determined to be a
target of and negatively regulated by NEAT1, thereby inhibiting
LPS-induced apoptosis and inflammation of HMEECs. According to
previous reports, miR-301b-3p was shown to contribute to tumor
growth in human hepatocellular carcinoma and gastric cancer
(38,39). In addition, the expression of
miR-301b-3p was significantly upregulated by Pseudomonas
aeruginosa and Candida albicans (22,40,41),
but miR-301b-3p was downregulated in LPS-induced HMEECs (20). miR-301b-3p and TLR4 expression was
also investigated in the HK2 and THLE-3 cells. The results
demonstrated that the expression of miR-301b-3p was reduced in
LPS-induced HK2 and THLE-3 cells, whereas TLR4 expression was
increased. The present study found that miR-301b-3p was a target of
NEAT1, and miR-301b-3p inhibitors reversed the inhibitory effect of
NEAT1 silencing on the inflammatory response of HMEECs. In this
context, our data suggested that NEAT1 may regulate LPS-induced
inflammation of HMEECs by sponging miR-301b-3p.
Previous studies have reported that TLR4 is highly
expressed in necrotizing enterocolitis (42), oral cancer and brain injury
(43). Of note, TLR4 is also highly
expressed in AOM (31,44). Consistent with the aforementioned
findings, it was observed herein that the enrichment of TLR4
increased in LPS-stimulated HMEECs compared with normal HMEECs. In
addition, miRNAs can regulate gene expression through binding to
the 3'-UTR of their target mRNAs (45). For example, miR-301b-3p targets the
3'-UTR of PTEN and promotes cell migration and invasion in bladder
cancer (46). Previous studies
reported that miR-301b-3p was transcriptionally upregulated by the
TLR4/NF-κB axis in human bladder cancer and Candida
albicans-induced lung injury (40,47).
Notably, in patients with AOM, the expression of TLR4 was
upregulated, and H. influenzae inoculation and LPS-induced
inflammation also upregulated the TLR4 level in animal models with
AOM (24-27),
but LPS induction significantly reduced miR-301b-3p expression in
HMEECs (20). It was also observed
that LPS induction significantly increased TLR4 and decreased
miR-301b-3p expression in HMEECs. Notably, TLR4 was found to
be the target gene of miR-301b-3p, and miR-301b-3p inhibited TLR4
expression in LPS-induced HMEECs. Li et al (35) reported that NEAT1 may promote
LPS-induced inflammatory injury in macrophages by regulating
miR-17-5p/TLR4. Consistently, the present study found that
miR-301b-3p inhibitors and TLR4 overexpression upregulated the
inhibitory effect of NEAT1 silencing on TLR4, indicating that NEAT1
affects TLR4 expression by modulating miR-301b-3p, thereby
promoting LPS-induced apoptosis and inflammation of HMEECs.
However, the present study only investigated the potential role and
mechanism of the NEAT1/miR-301b-3p/TLR4 axis in LPS-induced HMEECs,
and several limitations should also be considered. Preclinical data
are expected to improve our understanding of the potential
mechanism by using mouse models in vivo or primary cells
in vitro in future studies.
In summary, it was herein demonstrated that
interference with lncRNA NEAT1 expression may protect HMEECs from
LPS-induced inflammation and apoptosis via the miR-301b-3p/TLR4
axis, which contributes to improving the pathological status of
AOM. The findings of the present study also indicated that the
NEAT1/miR-301b-3p/TLR4 axis may constitute a potential therapeutic
target for AOM.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81960187).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL and RL conceived and designed the research. TL,
SL, FZ, JY, SD and BR performed the experiments and analyzed the
data. ZL, TL and SL wrote the manuscript. ZL and RL interpreted the
data. RL, BR and RL confirm the authenticity of all the raw data.
All the authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Morris LM, DeGagne JM, Kempton JB, Hausman
F and Trune DR: Mouse middle ear ion homeostasis channels and
intercellular junctions. PLoS One. 7(e39004)2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Block SL: Causative pathogens, antibiotic
resistance and therapeutic considerations in acute otitis media.
Pediatr Infect Dis J. 16:449–456. 1997.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Post JC: Direct evidence of bacterial
biofilms in otitis media 2001. Laryngoscope. 125:2003–2014.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Topcuoglu N, Keskin F, Ciftci S, Paltura
C, Kulekci M, Ustek D and Kulekci G: Relationship between oral
anaerobic bacteria and otitis media with effusion. Int J Med Sci.
9:256–261. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Curtale G and Citarella F: Dynamic nature
of noncoding RNA regulation of adaptive immune response. Int J Mol
Sci. 14:17347–17377. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang J, Lucas BA and Maquat LE: New gene
expression pipelines gush lncRNAs. Genome Biol.
14(117)2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang Y, Zhu Y, Gao G and Zhou Z:
Knockdown XIST alleviates LPS-induced WI-38 cell apoptosis and
inflammation injury via targeting miR-370-3p/TLR4 in acute
pneumonia. Cell Biochem Funct. 37:348–358. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Liang WJ, Zeng XY, Jiang SL, Tan HY, Yan
MY and Yang HZ: Long non-coding RNA MALAT1 sponges miR-149 to
promote inflammatory responses of LPS-induced acute lung in-jury by
targeting MyD88. Cell Biol Int. 44:317–326. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gomez JA, Wapinski OL, Yang YW, Bureau JF,
Gopinath S, Monack DM, Chang HY, Brahic M and Kirkegaard K: The
NeST long ncRNA controls microbial susceptibility and epigenetic
activation of the interferon-γ locus. Cell. 152:743–754.
2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pawar K, Hanisch C, Palma Vera SE,
Einspanier R and Sharbati S: Down regulated lncRNA MEG3 eliminates
mycobacteria in macrophages via autophagy. Sci Rep.
6(19416)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang Y, Zhong H, Xie X, Chen CY, Huang D,
Shen L, Zhang H, Chen ZW and Zeng G: Long noncoding RNA derived
from CD244 signaling epigenetically controls CD8+ T-cell
immune responses in tuberculosis infection. Proc Natl Acad Sci USA.
112:E3883–E3892. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Shen X, Zhao W, Zhang Y and Liang B: Long
non-coding RNA-NEAT1 promotes cell migration and invasion via
regulating miR-124/NF-κB pathway in cervical cancer. OncoTargets
Ther. 13:3265–3276. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen XX, Jiang YJ, Zeng T and Li JJ:
Overexpression of the long noncoding RNA NEAT1 pro-tects against
As2O3-induced injury of cardiomyocyte by
inhibiting the miR-124/NF-κB signaling pathway. Eur Rev Med
Pharmacol Sci. 24:1378–1390. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Nong W: Long non-coding RNA
NEAT1/miR-193a-3p regulates LPS-induced apoptosis and inflammatory
injury in WI-38 cells through TLR4/NF-κB signaling. Am J Transl
Res. 11:5944–5955. 2019.PubMed/NCBI
|
|
16
|
Ma J, Zhao N, Du L and Wang Y:
Downregulation of lncRNA NEAT1 inhibits mouse mesangial cell
proliferation, fibrosis, and inflammation but promotes apoptosis in
diabetic nephropathy. Int J Clin Exp Pathol. 12:1174–1183.
2019.PubMed/NCBI
|
|
17
|
Jin SS, Lin XF, Zheng JZ, Wang Q and Guan
HQ: lncRNA NEAT1 regulates fibrosis and inflammatory response
induced by nonalcoholic fatty liver by regulating miR-506/GLI3. Eur
Cytokine Netw. 30:98–106. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang W and Guo Z: Downregulation of lncRNA
NEAT1 ameliorates LPS-induced inflammatory responses by promoting
macrophage M2 polarization via miR-125a-5p/TRAF6/TAK1 axis.
Inflammation. 43:1548–1560. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou X, Jeker LT, Fife BT, Zhu S, Anderson
MS, McManus MT and Bluestone JA: Selective miRNA disruption in T
reg cells leads to uncontrolled autoimmunity. J Exp Med.
205:1983–1991. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Song JJ, Kwon SK, Cho CG, Park SW and Chae
SW: Microarray analysis of microRNA expression in LPS induced
inflammation of human middle ear epithelial cells (HMEECs). Int J
Pediatr Otorhinolaryngol. 75:648–651. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
(Suppl):R40–R44. 2007.PubMed/NCBI
|
|
22
|
Wree A, McGeough MD, Inzaugarat ME, Eguchi
A, Schuster S, Johnson CD, Peña CA, Geis-ler LJ, Papouchado BG,
Hoffman HM, et al: NLRP3 inflammasome driven liver injury and
fibrosis: Roles of IL-17 and TNF in mice. Hepatology. 67:736–749.
2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang YC, Wang PF, Fang H, Chen J, Xiong XY
and Yang QW: Toll-like receptor 4 antagonist attenuates
intracerebral hemorrhage-induced brain injury. Stroke.
44:2545–2552. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hirano T, Kodama S, Fujita K, Maeda K and
Suzuki M: Role of Toll-like receptor 4 in innate immune responses
in a mouse model of acute otitis media. FEMS Immunol Med Microbiol.
49:75–83. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Leichtle A, Hernandez M, Pak K, Yamasaki
K, Cheng CF, Webster NJ, Ryan AF and Wasser-man SI: TLR4-mediated
induction of TLR2 signaling is critical in the pathogenesis and
resolution of otitis media. Innate Immun. 15:205–215.
2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li P, Chen D and Huang Y: Fisetin
administration improves LPS-induced acute otitis media in mouse
in vivo. Int J Mol Med. 42:237–247. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Guo H, Li M and Xu LJ: Apigetrin treatment
attenuates LPS-induced acute otitis media though suppressing
inflammation and oxidative stress. Biomed Pharmacother.
109:1978–1987. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ma YK, Chen YB and Li P: Quercetin
inhibits NTHi-triggered CXCR4 activation through suppressing
IKKα/NF-κB and MAPK signaling pathways in otitis media. Int J Mol
Med. 42:248–258. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li GD, Li TL, Liu H and Sun L: Correlation
between recovery time of extended high-frequency audiometry and
duration of inflammation in patients with acute otitis media. Eur
Arch Otorhinolaryngol. 277:2447–2453. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang J, Xu M, Zheng Q, Zhang Y, Ma W and
Zhang Z: Blocking macrophage migration inhibitory factor activity
alleviates mouse acute otitis media in vivo. Immunol Lett.
162A:101–108. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tuoheti A, Gu X, Cheng X and Zhang H:
Silencing Nrf2 attenuates chronic suppurative otitis media by
inhibiting pro-inflammatory cytokine secretion through
up-regulating TLR4. Innate Immun. 27:70–80. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang Z, Hao J and Chen D: Long noncoding
RNA nuclear enriched abundant transcript 1(NEAT1) regulates
proliferation, apoptosis, and inflammation of chondrocytes via the
miR-181a/glycerol-3-phosphate dehydrogenase 1-like (GPD1L) axis.
Med Sci Monit. 25:8084–8094. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yang F, Tang Z, Duan A, Yi B, Shen N, Bo
Z, Yin L, Zhu B, Qiu Y and Li J: Long noncoding RNA NEAT1
upregulates survivin and facilitates gallbladder cancer progression
by sponging microRNA-335. OncoTargets Ther. 13:2357–2367.
2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li Y, Guo W and Cai Y: NEAT1 Promotes
LPS-induced inflammatory injury in macrophages by regulating
miR-17-5p/TLR4. Open Med (Wars). 15:38–49. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ren L, Chen S, Liu W, Hou P, Sun W and Yan
H: Downregulation of long non-coding RNA nuclear enriched abundant
transcript 1promotes cell proliferation and inhibits cell apoptosis
by targeting miR-193a in myocardial ischemia/reperfusion injury.
BMC Cardiovasc Disord. 19(192)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fan H, Jin X, Liao C, Qiao L and Zhao W:
MicroRNA-301b-3p accelerates the growth of gastric cancer cells by
targeting zinc finger and BTB domain containing 4. Pathol Res
Pract. 215(152667)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Guo Y, Yao B, Zhu Q, Xiao Z, Hu L, Liu X,
Li L, Wang J, Xu Q, Yang L, et al: MicroRNA-301b-3p contributes to
tumour growth of human hepatocellular carcinoma by repressing
vestigial like family member 4. J Cell Mol Med. 23:5037–5047.
2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wu CX, Cheng J, Wang YY, Wang JJ, Guo H
and Sun H: Microrna expression profiling of macrophage line
RAW264.7 infected by Candida albicans. Shock:. 47:520–530.
2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Li X, He S, Li R, Zhou X, Zhang S, Yu M,
Ye Y, Wang Y, Huang C and Wu M: Pseudomonas aeruginosa
infection augments inflammation through miR-301b repression of
c-Myb-mediated immune activation and infiltration. Nat Microbiol.
1(16132)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen Z, Zhang Y, Lin R, Meng X, Zhao W,
Shen W and Fan H: Cronobacter sakazakiiinduces necrotizing
enterocolitis by regulating NLRP3 inflammasome expression via TLR4.
J Med Microbiol. 69:748–758. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhou K, Cui S, Duan W, Zhang J, Huang J,
Wang L, Gong Z and Zhou Y: Cold-inducible RNA-binding protein
contributes to intracerebral hemorrhage-induced brain injury via
TLR4 signaling. Brain Behav. 10(e01618)2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kaur R, Casey J and Pichichero M:
Cytokine, chemokine, and Toll-like receptor expression in middle
ear fluids of children with acute otitis media. Laryngoscope.
125:E39–E44. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Egawa H, Jingushi K, Hirono T, Ueda Y,
Kitae K, Nakata W, Fujita K, Uemura M, Nonomura N and Tsujikawa K:
The miR-130 family promotes cell migration and invasion in bladder
cancer through FAK and Akt phosphorylation by regulating PTEN. Sci
Rep. 6(20574)2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Man X, Piao C, Lin X, Kong C, Cui X and
Jiang Y: USP13 functions as a tumor suppressor by blocking the
NF-κB-mediated PTEN downregulation in human bladder cancer. J Exp
Clin Cancer Res. 38(259)2019.PubMed/NCBI View Article : Google Scholar
|