Introduction
Oral squamous cell carcinoma (OSCC) accounts for 90%
of oral cancer cases and has become a major global public health
problem, as the eighth most common type of cancer worldwide
(1). Due to its highly recurrent
and heterogeneous nature, OSCC commonly occurs in the oral
epithelium and is characterized by an insidious onset, a difficult
diagnosis and rapid progression (2). In total, 300,000 new cases of OSCC are
diagnosed worldwide each year, and the prevalence has significantly
increased in recent years, particularly amongst the younger
population (3). Risk factors for
OSCC are usually lifestyle-associated and include smoking,
excessive alcohol consumption and betel nut chewing (4). Although OSCC is superficial, due to
the fact that individuals do not show indicative symptoms at the
early stages, the diagnosis of the disease largely occurs at an
extremely late stage (stage III or IV), resulting in a poor
prognosis for patients, ineffective treatment and an increased
social burden (5). A previous study
has reported a higher rate of pathogenic tumor node metastasis in
stage IV patients with OSCC, which was accompanied with an overall
survival rate of 41.2%, compared with in patients with stage I and
II disease (6). In addition, the
5-year survival rate of patients with OSCC was found to be
substantially decreased (7).
Another study has revealed that the 5-year survival rate of
patients with OSCC may be up to 90% if the disease is diagnosed
early and treated appropriately (8). The high morbidity and mortality rates
of OSCC highlight the urgency of identifying effective biomarkers
for the early detection of OSCC.
Analysis of the Gene Expression Omnibus (GEO)
dataset, GSE138206, has demonstrated that the expression levels of
protein tyrosine kinase 7 (PTK7) are markedly upregulated in OSCC
tissues compared with in normal tissues (9). PTK7, which is also known as colon
cancer kinase 4, belongs to the receptor tyrosine kinase family and
is an evolutionary, highly conserved cell surface planar cell
polarity receptor (10,11). Previous studies have demonstrated
that PTK7 serves an extensive role in tissue development and
homeostasis in vivo, affecting various aspects of
communication and migration between cells (12,13).
Furthermore, PTK7 can control tissue morphogenesis and pattern
formation by affecting cell polarity, migration and tissue
regeneration and wound healing (14-16).
In numerous previous studies, the dysregulation of PTK7 expression
has been found to be closely associated with cancer development and
with a number of cellular processes, including cell proliferation,
migration and angiogenesis (17-19).
Existing preclinical data have demonstrated that antibody-drug
complexes targeting PTK7 may exert anti-angiogenic and immune
cell-stimulating antitumor effects, thereby improving the long-term
survival of patients with numerous types of cancer (20). Although the role of PTK7 in numerous
human malignancies, including thyroid (21), cervical (22), adrenocortical (23) and colorectal cancer (24), has been reported, the role of PTK7
in OSCC has not yet been determined.
Dishevelled segment polarity protein (DVL)3 is a
member of the dishevelled protein family, and acts as a cytoplasmic
scaffold protein that links receptors to their downstream targets
(25). A previous study has
reported that DVL3-knockdown suppresses breast cancer cell
proliferation by mediating Wnt/β-catenin signaling (26). In esophageal squamous cell
carcinoma, DVL3 downregulation can inhibit the proliferation and
promote apoptosis of tumor cells (27). A meta-analysis has revealed that the
DVL3 gene, which is involved in the Notch signaling pathway, is
upregulated in OSCC samples, and the inhibition of Notch signaling
by γ-secretase inhibitors markedly decrease the proliferation of
OSCC cells in vitro (28).
However, to the best of our knowledge, the biological function and
clinical significance of DVL3 in OSCC has not been reported.
Therefore, the present study aimed to investigate the biological
role of PTK7 in OSCC and to determine its underlying mechanisms of
action.
Materials and methods
Cell lines and culture
Healthy human oral keratinocytes (HOKs) were bought
from ScienCell Research Laboratories, Inc. (cat. no. 2610) and OSCC
cell lines (CAL-27, HSC-4 and SCC-9) were purchased from The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences.
Cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Invitrogen; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin in a humidified
incubator at 37˚C with 5% CO2.
Cell transfection
Short hairpin (sh) RNAs targeting PTK7 (sh-PTK7-1
and sh-PTK7-2) were used to silence PTK7 expression, and a
scrambled shRNA was used as a negative control (NC) shRNA.
Overexpression plasmids for DVL3 (pcDNA 3.1-DVL3) were synthesized
to overexpress DVL3, and pcDNA 3.1 empty vector was used as a NC.
sh-PTK7-1 (1 µg), sh-PTK7-2 (1 µg), NC shRNA (1 µg), pcDNA-DVL3 (50
nM) and pcDNA (50 nM) were all designed by Shanghai GenePharma Co.,
Ltd. SCC-9 cells were seeded into 6-well plates
(1x106/well), and then transfection was performed for 8
h at 37˚C using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Cells were maintained under normal culture conditions and were
harvested for analysis 72 h after transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from OSCC cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse-transcribed into cDNA using a
PrimeScript™ RT Master Mix kit (Takara Bio, Inc.) according to the
manufacturer's protocol. qPCR was subsequently performed using a
TaqMan assay (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol to analyze the mRNA expression levels of
PTK7 and DVL3. The following thermocycling conditions were used:
Initial denaturation at 95˚C for 10 min; followed by 35 cycles of
denaturation at 95˚C for 15 sec, annealing at 60˚C for 1 min and
extension of 10 min at 65˚C. The following primers were used for
qPCR: PTK7 forward, 5'-CAGTTCCTGAGGATTTCCAAGAG-3' and reverse,
5'-TGCATAGGGCCACCTTC-3'; DVL3 forward, 5'-CGC AAGTATGCCAGCAAC-3'
and reverse, 5'-GCAGAGGTC ACCGAAGAT-3'; and GAPDH forward,
5'-GGAGCGAGA TCCCTCCAAAAT-3' and reverse, 5'-GGCTGTTGTCAT
ACTTCTCATGG-3'. The expression levels were quantified using the
2-ΔΔCq method (29) and
normalized to GAPDH.
Western blotting
OSCC cells were collected and lysed in RIPA lysis
buffer (Beyotime Institute of Biotechnology) on ice to obtain the
precipitate. Protein concentration was determined using a BCA
protein assay kit (EMD Millipore) and equal amounts of protein (35
µg/lane) were separated via 12% SDS-PAGE. The separated proteins
were subsequently transferred onto PVDF membranes (EMD Millipore)
and blocked at room temperature with 5% skimmed milk for 1 h. The
membranes were then incubated with the following primary antibodies
diluted in TBS-0.1% Tween-20 (TBST) overnight at 4˚C: Anti-PTK7
(1:200; sc-100304), anti-DVL3 (1:100; sc-271295), anti-Ki67 (1:200;
sc-23900), anti-proliferating cell nuclear antigen (PCNA; 1:200;
sc-56), anti-MMP2 (1:200; sc-13594), anti-MMP9 (1:100; sc-393859)
and anti-GAPDH (1:200; sc-32233) (all Santa Cruz Biotechnology,
Inc.). Following the primary antibody incubation, the membranes
were washed thrice with TBST for 10 min each and incubated with
HRP-conjugated goat anti-rabbit IgG (1:5,000; cat. no. SA00001-9)
or goat anti-mouse IgG secondary antibodies (1:5,000; cat. no.
SA00001-8) (both from ProteinTech Group, Inc.) at room temperature
for 2 h. Protein bands were visualized using a luminol reagent
(Santa Cruz Biotechnology, Inc.) and analyzed using ImageJ software
(version 1.48; National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
SCC-9 cells transfected with NC shRNA, sh-PTK7,
pcDNA or pcDNA-DVL3 were cultured in 96-well plates at 37˚C in 5%
CO2. Following 24 h of incubation, 10 µl CCK-8 solution
(Dojindo Molecular Technologies, Inc.) was added to each well and
further incubated for 1 h at 37˚C with 5% CO2. The
absorbance was measured at a wavelength of 450 nm using a
microplate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
Cell proliferation was determined using a colony
formation assay. Briefly, SCC-9 cells transfected with NC shRNA,
sh-PTK7, pcDNA or pcDNA-DVL3 were plated into 6-well plates (with 3
replicates per condition). Following 14 days of incubation, the
cells were washed with PBS, fixed with methanol for 10 min at room
temperature and stained with 0.1% crystal violet solution for 15
min at room temperature (Sigma-Aldrich; Merck KGaA). The number of
positively stained cells was counted.
Wound healing assay
Cell migration was analyzed using a wound healing
assay. Briefly, transfected cells were seeded into 6-well plates
and serum-free medium replaced normal medium. The artificial wounds
were created in the cell monolayer by a single scratch with a
100-µl pipette tip (0 h). Following 24 h of incubation, the wound
closure area was visualized using a light microscope (Olympus
Corporation) (magnification, x100) and the cell migration rate was
calculated using ImageJ (1.52r; National Institutes of Health).
Transwell invasion assay
A Transwell assay was used to determine cell
invasion. The upper chambers of Transwell plates (Corning, Inc.)
were precoated with Matrigel (BD Biosciences) overnight at 37˚C.
Both transfected and non-transfected cells were seeded into the
upper chamber of each well in serum-free medium (Thermo Fisher
Scientific, Inc.), while 600 µl medium supplemented with 10% FBS
was plated into the lower chambers. Following incubation at 37˚C
with 5% CO2 for 24 h, non-invasive cells in the upper
chamber were removed with a cotton swab, while invasive cells in
the lower chambers were fixed with 4% paraformaldehyde for 30 min
at room temperature and stained with 0.1% crystal violet solution
for 20 min at room temperature. The invasive cells were visualized
using a light microscope (Olympus Corporation; magnification,
x100).
Bioinformatics analysis
Search Tool for Recurring Instances of Neighboring
Genes (STRING) database 11.5 (https://www.string-db.org/) was used to analyze the
connection between PTK7 and DVL3.
Co-immunoprecipitation (Co-IP)
assay
To determine protein interactions in SCC-9 cells,
cells were collected and lysed in lysis buffer [50 mM Tris-HCl (pH
7.5), 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 10% glycerol, 1 mM
protease inhibitor PMSF] (#20-188; MilliporeSigma). The cell
lysates (100 µl per antibody) were incubated with an anti-PTK7 (1
µg per 50 µg total protein; #sc-100304; Santa Cruz Biotechnology,
Inc.), anti-DVL3 (1 µg per 50 µg total protein, #sc-271295; Santa
Cruz Biotechnology, Inc.) or anti-IgG negative control antibody and
40 µl protein A/G magnetic beads (#LSKMAGAG; MilliporeSigma). The
beads were subsequently washed thrice with lysis buffer and
centrifuged at 500 x g for 5 min at 4˚C. The precipitated proteins
were eluted in 1X SDS-PAGE loading buffer and boiled for 10 min.
Western blotting was performed to analyze the precipitated proteins
and cell lysate, as aforementioned.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 (GraphPad Software, Inc.). All experiments were repeated
in triplicate and the results are presented as the mean ± SD.
Statistical differences between groups were determined using an
unpaired Student's t-test or one-way ANOVA followed by Tukey's
post-hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
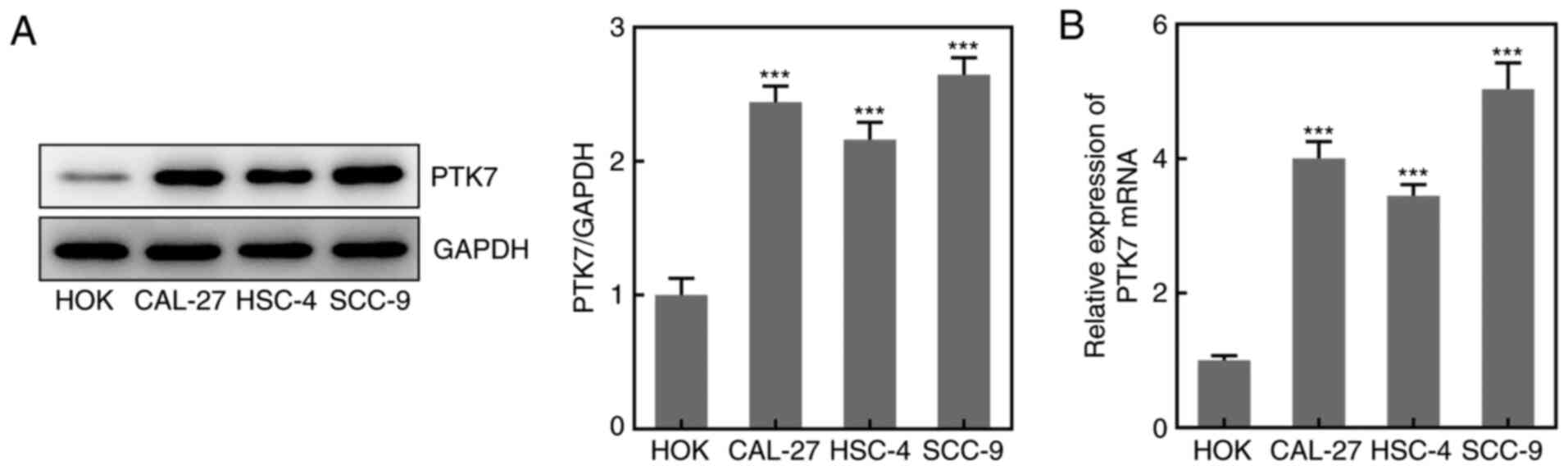
Expression levels of PTK7 in OSCC cell
lines
To analyze the expression levels of PTK7 in OSCC
cell lines, RT-qPCR and western blotting were performed. The
results revealed that the protein and mRNA expression levels of
PTK7 were significantly upregulated in CAL-27, HSC-4 and SCC-9
cells compared with in HOKs, with the highest expression observed
in SCC-9 cells (Fig. 1A and
B); therefore, SCC-9 cells were
selected for use in subsequent experiments. These results suggested
that the expression levels of PTK7 were upregulated in OSCC cell
lines.
Effect of PTK7 on OSCC cell viability
and proliferation
To investigate the effect of PTK7 on OSCC cell
viability and proliferation, sh-PTK7 was used to knock down PTK7
expression. As shown in Fig. 2A,
both sh-PTK7-1 and sh-PTK7-2 significantly downregulated the
protein expression levels of PTK7; however, sh-PTK7-1 downregulated
the expression to a greater extent than sh-PTK7-2. The results
obtained from the RT-qPCR analysis were similar (Fig. 2B); thus, sh-PTK7-1 was selected to
knock down PTK7 expression in subsequent experiments. Subsequently,
CCK-8 assays were conducted to analyze cell viability. Compared
with the control and shRNA groups, the optical density value
(measured at a wavelength of 450 nm) was significantly decreased in
the sh-PTK7 group at 24, 48 and 72 h (Fig. 2C). The results of the colony
formation assay revealed that the number of colonies formed was
markedly decreased following PTK7-knockdown (Fig. 2D). In addition, the expression
levels of the proliferation-associated proteins, Ki67 and PCNA,
were analyzed and were found to be significantly downregulated in
SCC-9 cells following the transfection with sh-PTK7 compared with
the control group (Fig. 2E). These
data suggested that the knockdown of PTK7 expression inhibited OSCC
cell proliferation.
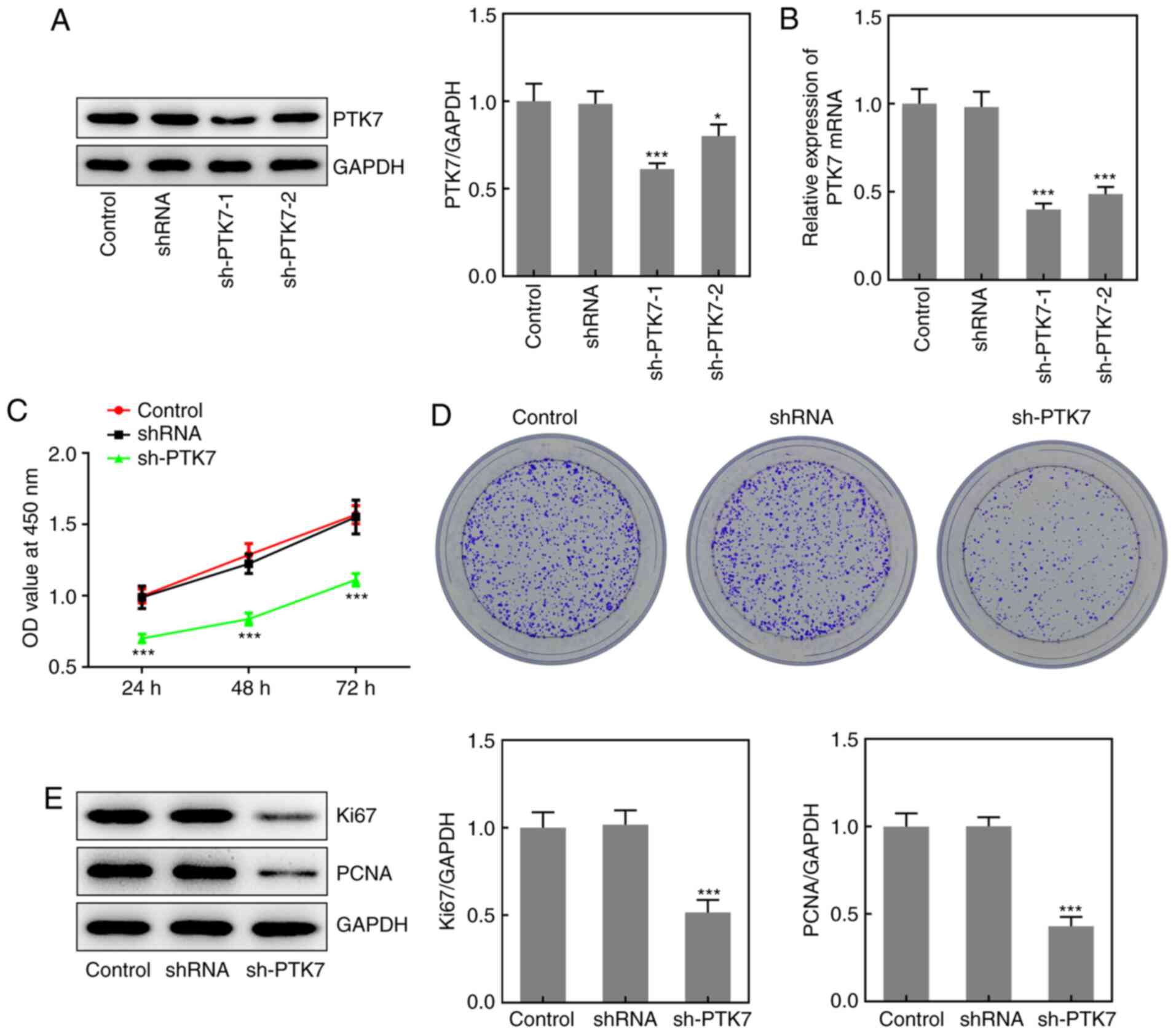 | Figure 2PTK7-knockdown inhibits SCC-9 cell
viability and proliferation. PTK7 expression was measured in SCC-9
cells following transfection with sh-PTK7-1, sh-PTK7-2 or shRNA
using (A) western blotting and (B) reverse
transcription-quantitative PCR. (C) SCC-9 cells were transfected
with sh-PTK7-1, sh-PTK7-2 or shRNA for 24, 48 or 72 h, then cell
viability was determined using a Cell Counting Kit-8 assay. (D)
Proliferation of SCC-9 cells transfected with sh-PTK7-1, sh-PTK7-2
or shRNA was analyzed using a colony formation assay. (E) Ki67 and
PCNA expression in SCC-9 cells following transfection with
sh-PTK7-1, sh-PTK7-2 or shRNA was analyzed using western blotting.
*P<0.05 and ***P<0.001 vs. shRNA. PTK7,
protein tyrosine kinase 7; sh/shRNA, short hairpin RNA; PCNA,
proliferating cell nuclear antigen; OD, optical density. |
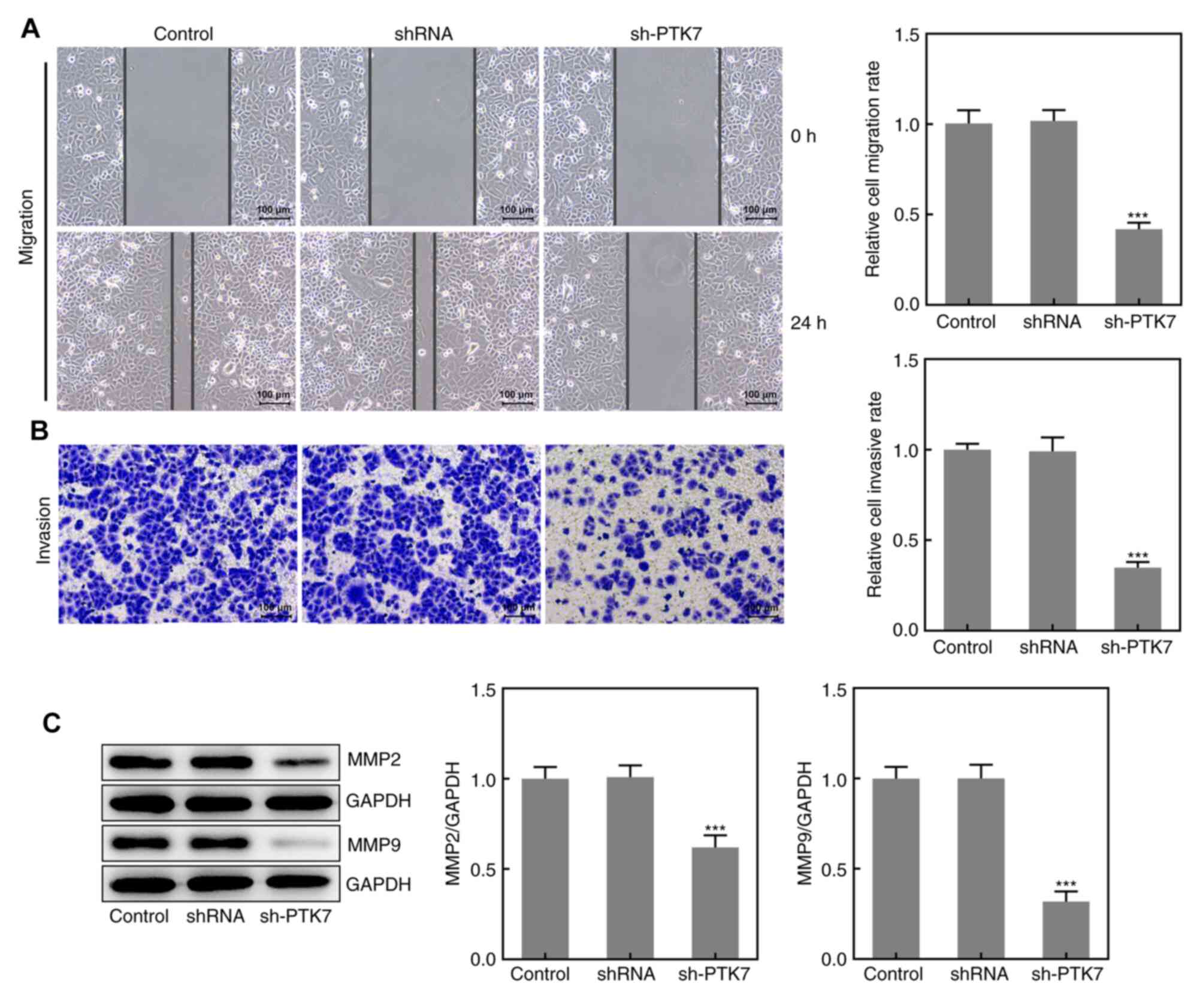
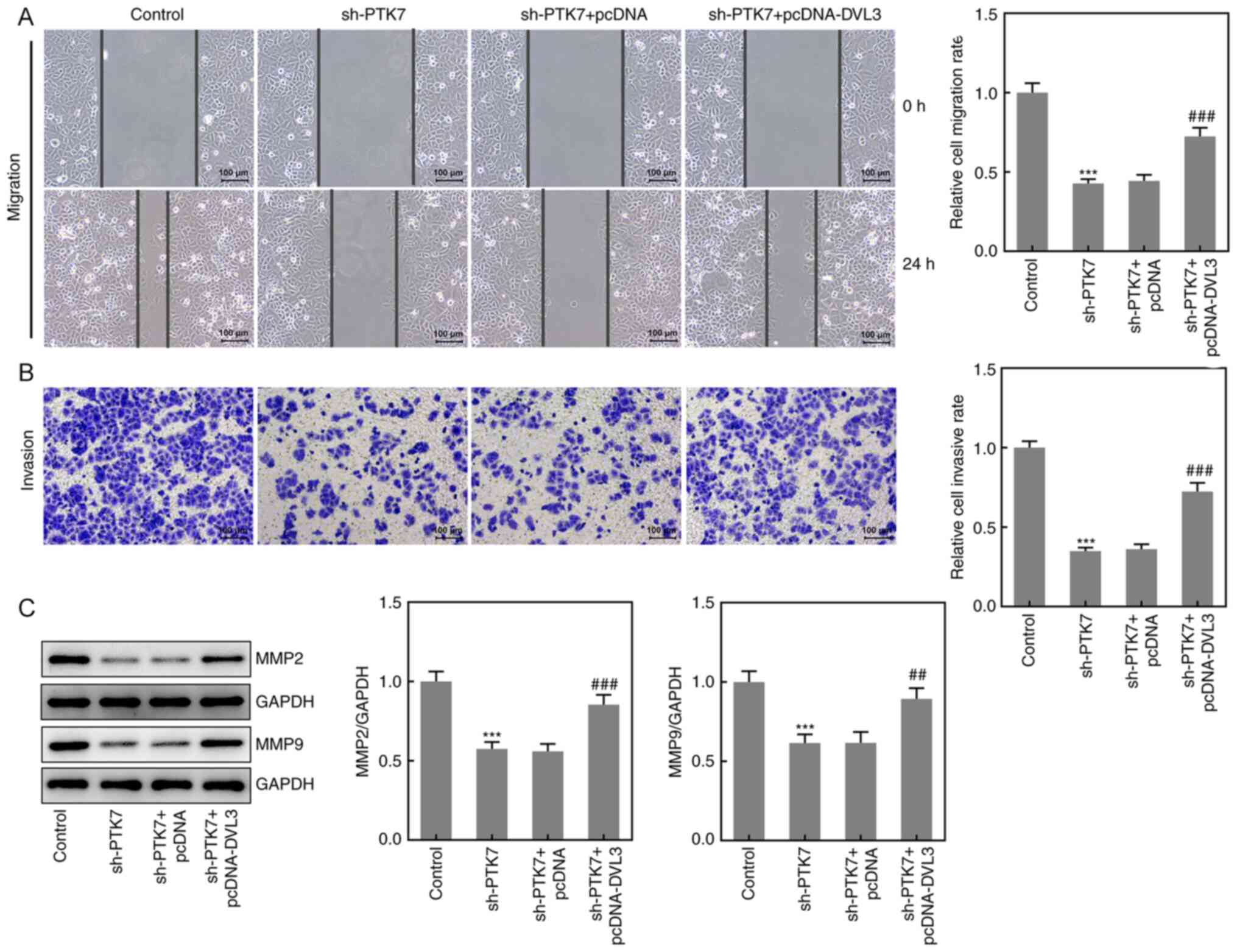
Effect of PTK7 on OSCC cell migration
and invasion
The migration and invasion of SCC-9 cells were
analyzed using wound healing and Transwell assays, respectively. As
presented in Fig. 3A and B, the area of the wound was larger at 24 h
and the number of invasive cells was decreased in the sh-PTK7 group
compared with the NC shRNA group, which suggested that the
knockdown of PTK7 expression may inhibit SCC-9 cell migration and
invasion. Moreover, the expression levels of MMP2 and MMP9 were
significantly downregulated following PTK7-knockdown, which further
indicated the inhibitory effect of PTK-knockdown on OSCC cell
migration and invasion (Fig. 3C).
These results suggested that the knockdown of PTK7 expression
inhibited the migration and invasion of SCC-9 cells.
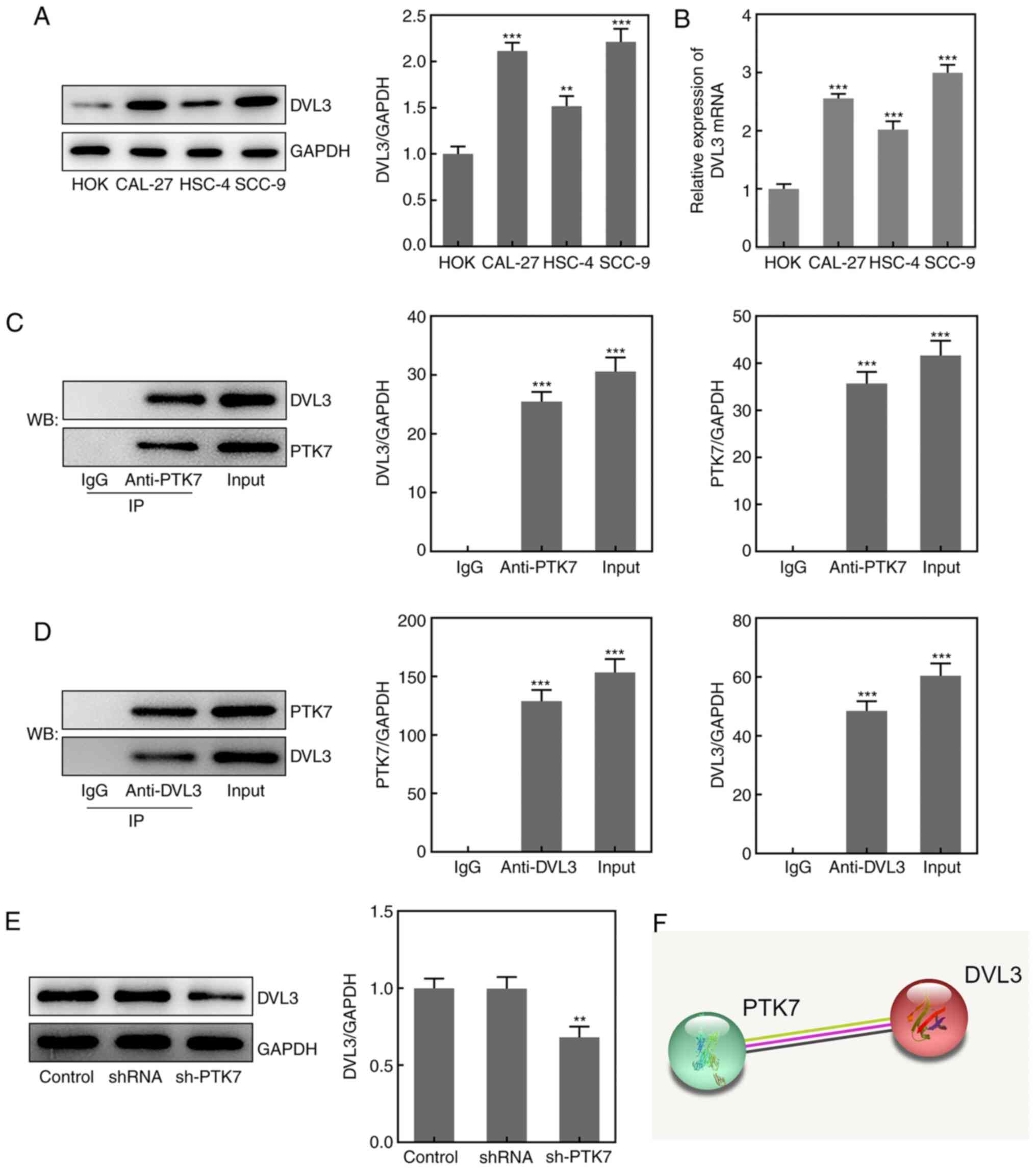
Association between PTK7 and DVL3 in
OSCC
To determine the potential mechanism underlying the
function of PTK7 in OSCC, the Search Tool for the Retrieval of
Interacting Genes/Proteins (STRING) database was used to predict
that PTK7 could bind to and regulate DVL3 expression (Fig. 4F). RT-qPCR analysis and western
blotting revealed that the expression levels of DVL3 were
significantly upregulated in OSCC cell lines compared with in HOKs,
with the highest expression levels observed in SCC-9 cells
(Fig. 4A and B). A Co-IP assay was performed to verify
the binding between PTK7 and DVL3; the results revealed that DVL3
was enriched by the anti-PTK7 antibody, while PTK7 was enriched by
the anti-DVL3 antibody (Fig. 4C and
D). These findings indicated that
PTK7 may bind to and interact with DVL3. In addition, the
expression levels of DVL3 in SCC-9 cells were significantly
downregulated following transfection with sh-PTK7 (Fig. 4E). These results suggested that PTK7
expression may be positively associated with DVL3 expression in
OSCC.
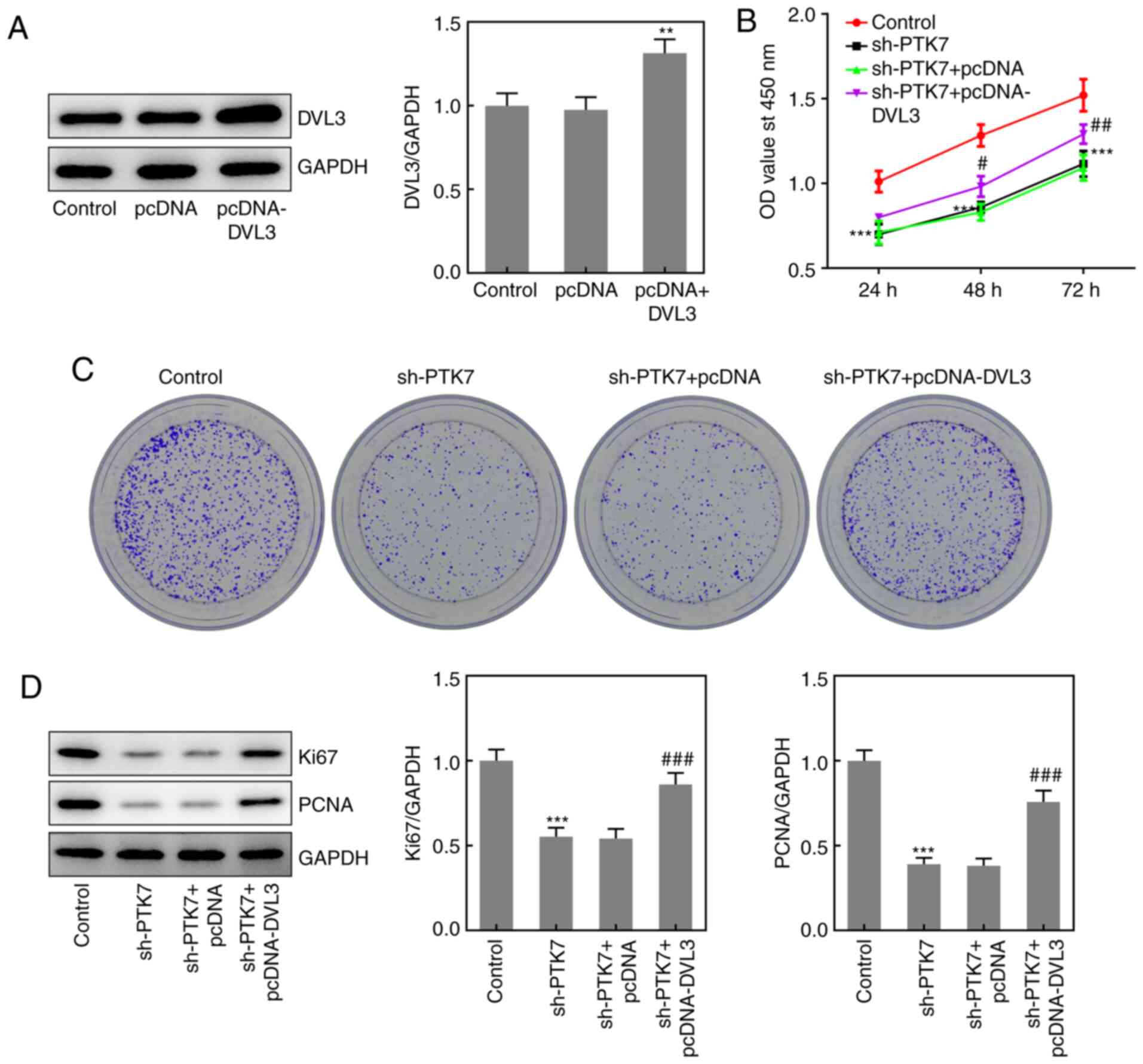
Effect of DVL3 on OSCC cell viability
and proliferation
To further determine the role of the PTK7/DVL3
interaction in regulating OSCC cell viability and proliferation,
pcDNA-DVL3 was transfected into OSCC cells. The transfection
efficiency of pcDNA-DVL3 was verified using western blotting
(Fig. 5A). Subsequently, CCK-8 and
colony formation assays were performed to determine cell viability
and proliferation, respectively. The results revealed that the
overexpression of DVL3 significantly alleviated the inhibitory
effects of PTK7-knockdown on cell viability and colony formation
(Fig. 5B and C). In addition, the expression levels of
Ki67 and PCNA were significantly upregulated in the sh-PTK7 +
pcDNA-DVL3 group compared with the sh-PTK7 + pcDNA group, while no
significant differences were observed in the expression levels of
either protein between the sh-PTK7 + pcDNA and sh-PTK7 groups
(Fig. 5D). These results suggested
that the overexpression of DVL3 may reverse the
PTK7-knockdown-induced inhibitory effects on the proliferation and
viability of OSCC cells.
Effect of DVL3 on OSCC cell migration
and invasion
Wound healing and Transwell assays were performed to
determine the effects of the overexpression of DVL3 on the
migration and invasion of SCC-9 cells, respectively. The results
demonstrated that cell migration and invasion were significantly
increased following the co-transfection of sh-PTK7 and pcDNA-DVL3
compared with sh-PTK7 alone, indicating that the overexpression of
DVL3 may ameliorate the inhibitory effects of sh-PTK7 (Fig. 6A and B). The protein expression levels of MMP2
and MMP9 were also significantly upregulated in the sh-PTK7 +
pcDNA-DVL3 group compared with the sh-PTK7 + pcDNA group (Fig. 6C). These results suggested that the
overexpression of DVL3 may reverse the sh-PTK7-induced inhibitory
effects on the migration and invasion of OSCC cells.
Discussion
The present study aimed to investigate the
tumor-promoting effect of PTK7 in OSCC. The results revealed that
the expression levels of PTK7 were upregulated in OSCC cell lines
and that PTK7-knockdown inhibited the viability, proliferation,
migration and invasion of OSCC cells. In addition, DVL3 was
identified to be positively associated with PTK7, and the
overexpression of DVL3 ameliorated the inhibitory effects of
sh-PTK7 on these biological processes.
Numerous previous studies have reported the role of
PTK7 in several types of cancer. For example, PTK7 has been
identified as a potential target for cervical cancer treatment,
since knocking down PTK7 expression inhibits the proliferative,
migratory and invasive abilities of cervical cancer cells both
in vivo and in vitro (22). Another previous study has reported
that microRNA-205-5p inhibits the migration and invasion of colon
cancer cells by downregulating PTK7 expression (24). PTK7 expression has been also found
to be upregulated in OSCC cells compared with in normal squamous
cells, and high PTK7 expression has been positively associated with
TNM stage, tumor differentiation and lymph node metastasis. In
addition, patients with higher PTK7 expression exhibit a poorer
overall survival (30). These data
indicate that PTK7 is associated with OSCC prognosis and may
represent a potential therapeutic target (30). The present study first analyzed the
expression levels of PTK7 in OSCC cell lines, and the results
obtained were consistent with the observed expression levels of
PTK7 in OSCC tissues from the GEO database (9), indicating that PTK7 expression may be
upregulated in OSCC. Next, the potential biological role of PTK7 in
OSCC cells was investigated using sh-PTK7. Cancer cell
proliferation determines cancer progression, and metastasis,
characterized by cancer cell migration and invasion, represents a
significant challenge in the clinical treatment of various types of
cancer (31). Therefore, the
current study sought to determine the effect of PTK7 on OSCC cell
proliferation, migration and invasion. The results demonstrated
that the knockdown of PTK7 expression suppressed the viability,
proliferation, migration and invasion of OSCC cells.
The potential mechanisms underlying the biological
effects of PTK7 were investigated. The STRING database was used to
predict that PTK7 could bind to and regulate DVL3 expression.
Notably, DVL3, as a gene of the Notch signaling pathway, has been
previously reported to be upregulated in OSCC samples (28). In addition, the knockdown of DVL3
expression has been found to control the progression of esophageal
squamous cell carcinoma, inhibit cell proliferation and promote the
apoptosis of tumor cells (27).
Moreover, a recent study has revealed that testis-specific
transcript, Y-linked 15 downregulates DVL3 expression in colorectal
cancer tissues and promotes the proliferation, migration and
invasion of colorectal cancer cells (32). The findings of the present study
revealed that the expression levels of DVL3 were upregulated in
OSCC cell lines, and a Co-IP assay confirmed the interaction
between PTK7 and DVL3. Subsequently, pcDNA-DVL3 was co-transfected
with sh-PTK7 into OSCC cells, and the results demonstrated that the
overexpression of DVL3 reversed the inhibitory effects of sh-PTK7
on OSCC cells. Notably, it was found that PTK7 positively
associated with DVL3 expression and that inhibition of PTK7
expression inhibited the proliferation, migration and invasion of
OSCC cells. A previous study has also indicated that PTK7 interacts
with AMIGO2 and is able to act as a survival factor in melanoma
(33). Therefore, it is possible
that PTK7 and DVL3 also interact with each other in OSCC, and this
should be further explored in future studies.
In conclusion, the findings of the current study
revealed that the expression levels of PTK7 and DVL3 were
upregulated in OSCC. PTK7-knockdown inhibited OSCC cell viability,
proliferation, migration and invasion by downregulating DVL3
expression. These results may provide novel insight into the
potential role of PTK7 and DVL3 in the clinical prognosis and
treatment of OSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XJ and WT conceived and designed the experiments.
TH, CM and JD performed the experiments. RL and WZ analyzed the
data. XJ and TH wrote the manuscript. All authors read and approved
the final manuscript and confirm the authenticity of the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chai AWY, Lim KP and Cheong SC:
Translational genomics and recent advances in oral squamous cell
carcinoma. Semin Cancer Biol. 61:71–83. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hübbers CU and Akgül B: HPV and cancer of
the oral cavity. Virulence. 6:244–248. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sasahira T and Kirita T: Hallmarks of
cancer related newly prognostic factors of oral squamous cell
carcinoma. Int J Mol Sci. 19(2413)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Huang SH and O'Sullivan B: Overview of the
8th Edition TNM Classification for Head and Neck Cancer. Curr Treat
Options Oncol. 18(40)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Roi A, Roi CI, Negrutiu ML, Rivis M,
Sinescu C and Rusu LC: The Challenges of OSCC diagnosis: Salivary
cytokines as potential biomarkers. J Clin Med.
9(2866)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Campisi G, Calvino F, Carinci F, Matranga
D, Carella M, Mazzotta M, Rubini C, Panzarella V, Santarelli A,
Fedele S, et al: Peri-tumoral inflammatory cell infiltration in
OSCC: A reliable marker of local recurrence and prognosis? An
investigation using artificial neural networks. Int J Immunopathol
Pharmacol. 24:113–120. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kreppel M, Nazarli P, Grandoch A, Safi AF,
Zirk M, Nickenig HJ, Scheer M, Rothamel D, Hellmich M and Zöller
JE: Clinical and histopathological staging in oral squamous cell
carcinoma - comparison of the prognostic significance. Oral Oncol.
60:68–73. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cristaldi M, Mauceri R, Di Fede O,
Giuliana G, Campisi G and Panzarella V: Salivary biomarkers for
oral squamous cell carcinoma diagnosis and follow-up: Current
Status and Perspectives. Front Physiol. 10(1476)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Guo ZC, Jumatai S, Jing SL, Hu LL, Jia XY
and Gong ZC: Bioinformatics and immunohistochemistry analyses of
expression levels and clinical significance of CXCL2 and TANs in an
oral squamous cell carcinoma tumor microenvironment of
Prophyromonas gingivalis infection. Oncol Lett.
21(189)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Berger H, Wodarz A and Borchers A: PTK7
faces the Wnt in development and disease. Front Cell Dev Biol.
5(31)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mossie K, Jallal B, Alves F, Sures I,
Plowman GD and Ullrich A: Colon carcinoma kinase-4 defines a new
subclass of the receptor tyrosine kinase family. Oncogene.
11:2179–2184. 1995.PubMed/NCBI
|
|
12
|
Golubkov VS, Prigozhina NL, Zhang Y,
Stoletov K, Lewis JD, Schwartz PE, Hoffman RM and Strongin AY:
Protein-tyrosine pseudokinase 7 (PTK7) directs cancer cell motility
and metastasis. J Biol Chem. 289:24238–24249. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
González P, González-Fernández C,
Campos-Martín Y, Mollejo M, Carballosa-Gautam M, Marcillo A,
Norenberg M, García-Ovejero D and Rodríguez FJ: Spatio-temporal and
Cellular Expression Patterns of PTK7 in the Healthy and
Traumatically Injured Rat and Human Spinal Cord. Cell Mol
Neurobiol. 40:1087–1103. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bie J, Hu X, Yang M, Shi X, Zhang X and
Wang Z: PTK7 promotes the malignant properties of cancer stem-like
cells in esophageal squamous cell lines. Hum Cell. 33:356–365.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Peradziryi H, Tolwinski NS and Borchers A:
The many roles of PTK7: A versatile regulator of cell-cell
communication. Arch Biochem Biophys. 524:71–76. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Katoh M: Canonical and non-canonical WNT
signaling in cancer stem cells and their niches: Cellular
heterogeneity, omics reprogramming, targeted therapy and tumor
plasticity (Review). Int J Oncol. 51:1357–1369. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tian X, Yan L, Zhang D, Guan X, Dong B,
Zhao M and Hao C: PTK7 overexpression in colorectal tumors:
Clinicopathological correlation and prognosis relevance. Oncol Rep.
36:1829–1836. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kadioglu O, Saeed MEM, Mahmoud N, Hussein
Azawi SS, Rincic M, Liehr T and Efferth T: Identification of
metastasis-related genes by genomic and transcriptomic studies in
murine melanoma. Life Sci. 267(118922)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nweke EE, Naicker P, Aron S, Stoychev S,
Devar J, Tabb DL, Omoshoro-Jones J, Smith M and Candy G: SWATH-MS
based proteomic profiling of pancreatic ductal adenocarcinoma
tumours reveals the interplay between the extracellular matrix and
related intracellular pathways. PLoS One.
15(e0240453)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Damelin M, Bankovich A, Bernstein J, Lucas
J, Chen L, Williams S, Park A, Aguilar J, Ernstoff E, Charati M, et
al: A PTK7 targeted antibody drug conjugate reduces tumor
initiating cells and induces sustained tumor regressions. Sci
Transl Med. 9(eaag2611)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Duan F, Tang J, Kong FL, Zou HW, Ni BL and
Yu JC: Identification of PTK7 as a promising therapeutic target for
thyroid cancer. Eur Rev Med Pharmacol Sci. 24:6809–6817.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sun JJ, Li HL, Guo SJ, Ma H, Liu SJ, Liu D
and Xue FX: The increased PTK7 expression is a malignant factor in
cervical cancer. Dis Markers. 2019(5380197)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bie J, Liu K, Song G, Hu X, Xiong R, Zhang
X, Shi X and Wang Z: ENST00000489707.5 is a preferred alternative
splicing variant of PTK7 in adrenocortical cancer and shows
potential prognostic value. Med Sci Monit. 25:8326–8334.
2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen S, Wang Y, Su Y, Zhang L, Zhang M, Li
X, Wang J and Zhang X: miR 205 5p/PTK7 axis is involved in the
proliferation, migration and invasion of colorectal cancer cells.
Mol Med Rep. 17:6253–6260. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sharma M, Castro-Piedras I, Simmons GE Jr
and Pruitt K: Dishevelled: A masterful conductor of complex Wnt
signals. Cell Signal. 47:52–64. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zou YF, Xie CW, Yang SX and Xiong JP: AMPK
activators suppress breast cancer cell growth by inhibiting
DVL3-facilitated Wnt/β-catenin signaling pathway activity. Mol Med
Rep. 15:899–907. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chen XQ, Jiang J, Wang XT, Zhang CL, Ji AY
and Chen XJ: Role and mechanism of Dvl3 in the esophageal squamous
cell carcinoma. Eur Rev Med Pharmacol Sci. 22:7716–7725.
2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Osathanon T, Nowwarote N and Pavasant P:
Expression and influence of Notch signaling in oral squamous cell
carcinoma. J Oral Sci. 58:283–294. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dong Y, Chen X, Li H, Ni Y, Han W and Wang
J: PTK7 is a molecular marker for metastasis, TNM stage, and
prognosis in oral tongue squamous cell carcinoma. Pol J Pathol.
68:49–54. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zuo K, Qi Y, Yuan C, Jiang L, Xu P, Hu J,
Huang M and Li J: Specifically targeting cancer proliferation and
metastasis processes: The development of matriptase inhibitors.
Cancer Metastasis Rev. 38:507–524. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zheng XY, Cao MZ, Ba Y, Li YF and Ye JL:
LncRNA testis specific transcript, Y linked 15 (TTTY15) promotes
proliferation, migration and invasion of colorectal cancer cells
via regulating miR 29a 3p/DVL3 axis. Cancer Biomark. 31:1–11.
2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fontanals-Cirera B, Hasson D, Vardabasso
C, Di Micco R, Agrawal P, Chowdhury A, Gantz M, de
Pablos-Aragoneses A, Morgenstern A, Wu P, et al: Harnessing BET
inhibitor sensitivity reveals AMIGO2 as a melanoma survival gene.
Mol Cell. 68:731–744.e9. 2017.PubMed/NCBI View Article : Google Scholar
|