Introduction
Low density lipoprotein cholesterol (LDL-C) has been
considered a key predictor for cardiovascular disease, which is at
present the leading cause of mortality and morbidity worldwide
(1-3).
Circulating LDL is mainly cleared by the LDL receptor (LDLR) on the
cell membrane of hepatocytes. The mature LDLR is a transmembrane
glycoprotein of 839 amino acids, with five major structural
domains: the ligand-binding domain (LBD); an EGF precursor homology
domain, which includes the EGF-A domain, the EGF-B domain, a
6-bladed β-propeller and the EGF-C domain; an O-linked sugar
domain; a transmembrane domain; and a cytoplasmic tail (4,5). The
LDL particles are recognized and bound by the LBD of LDLR and then
internalized by endocytosis. At the low pH found in endosomes, the
LDLR/LDL complex dissociates and LDLR returns to the cell surface,
while the released LDL proceeds to lysosomes and is degraded
(6-8).
Therefore, elevation of LDLR protein has been the main strategy to
reduce LDL-C (9).
The expression of LDLR protein is tightly regulated
at the transcriptional, post-transcriptional and post-translational
levels. In hepatocytes, the transcription of LDLR is
negatively regulated by cellular cholesterol through the sterol
responsive element 1 (SRE1) motif located in the promoter region of
the LDLR gene (10). When
the cellular cholesterol level is low, SRE1 binding proteins, which
are located in the endoplasmic reticulum (ER) membrane, are
activated by two-step proteolytic cleavage and released into the
nucleus to bind with the SRE1 motif, resulting in the activation of
LDLR transcription (11).
However, the newly synthesized LDLR mRNA is short-lived
because of the three adenosine and uridine-rich elements, AU-rich
elements (AREs) in the 3' untranslated region (UTR) of LDLR
mRNA (12,13). The half-life of LDLR mRNA is
~30 min (12). The activation of
ERK protein has been found to be involved in LDLR mRNA
stabilization at the post-transcriptional level (4,14-16).
Proprotein convertase subtilisin/kexin type 9
(PCSK9) protein, a secretory protein mainly expressed and secreted
by the liver tissues, functions at the post-translational level by
promoting the degradation of LDLR protein (17-19).
PCSK9 is a single peptide and includes a prodomain, a catalytic
domain and a C-terminal domain (CTD) (20). Following autocleavage between
Gln-152 and Ser-153, the prodomain remains noncovalently attached
to the catalytic domain (5). PCSK9
serves as a natural eliminator of LDLR by binding to the EGF-A
extracellular domain of LDLR protein, triggering its degradation in
lysosomes and thus increasing the level of circulating LDL-C
(21). The first humanized PCSK9
antibodies, alirocumab and evolocumab, were approved by the US FDA
in 2015 and introduced a significant reduction in cardiovascular
death (22,23). However, the daunting cost and the
troublesome subcutaneous administration are evident drawbacks for
the two antibodies. Therefore, small, orally available compounds
that act as PCSK9 regulators and have excellent efficacy and low
cost are desired.
Our previous study identified euphornin L, a
jatrophane macrocyclic diterpenoid extracted from Euphorbia
helioscopia, as an active compound for promoting LDL uptake and
improving LDLR protein levels in HepG2 cells. An in vivo
study showed that treatment with euphornin L in high-fat diet-fed
Golden Syrian hamsters significantly reduced serum LDL-C by 38.9%
(24). The current study
investigated the mechanisms underlying the lipid-lowering effects
of euphornin L, which is designated C21 in this article, in HepG2
cells.
Materials and methods
Materials and reagents
Dulbecco's modified Eagle's medium (DMEM; cat. no.
SH30243.01) was purchased from HyClone (Cytiva). Fetal bovine serum
(FBS; cat. no. 10091-148) and Lipofectamine® 3000 (cat.
no. L3000-015) were obtained from Thermo Fisher Scientific, Inc.
Atorvastatin calcium salt trihydrate (Atorvastatin; cat. no.
PZ0001; ≥98%-HPLC), Phorbol 12-myristate 13-acetate (PMA; cat. no.
P1585; ≥99%-HPLC) and DMSO (cat. no. D2650; ≥99.7%-HPLC) were
purchased from Sigma-Aldrich; Merck KGaA.
Rabbit anti-human LDLR monoclonal antibody (cat.
no.; ab52818; 1:1,000) and rabbit anti-human PCSK9 monoclonal
antibody (cat. no.; ab181142; 1:2,000) were purchased from Abcam.
Rabbit anti-human/hamster p-ERK polyclonal antibody (cat. no. 9101;
1:2,000), rabbit anti-human/hamster ERK monoclonal antibody (cat.
no. 4695; 1:2,000) and mouse anti-human/hamster β-actin monoclonal
antibody (cat. no. 4967, 1:3,000) were purchased from Cell
Signaling Technology, Inc. Rabbit anti-hamster LDLR polyclonal
antibody (cat. no. 3839; 1:2,000) was purchased from BioVision,
Inc. (cat. no. 3839; 1:2,000). Rabbit anti-hamster PCSK9 polyclonal
antibody was a kind gift from laboratory of Professor Jingwen Liu
(Department of Veterans' Affairs, Palo Alto Health Care
System).
Hamster serum triglyceride (cat. no. CH0101151;
Maccura Biotechnology Co., Ltd.), total cholesterol (cat. no. L873;
Shino-Test Corporation), high density lipoprotein cholesterol (cat.
no. AQ986; FujiFilm Wako Pure Chemical Corporation), LDL-C (cat.
no. AG251; FujiFilm Wako Pure Chemical Corporation) and the
activities of aspartate aminotransferase (AST; cat. no. A871;
Shino-Test Corporation) and alanine aminotransferase (ALT; cat. no.
B871; Shino-Test Corporation) were measured with commercial kits
and the Hitachi 7020 autoanalyzer (Hitachi, Ltd.).
Compound extraction and isolation
Euphorbia helioscopia was harvested in March
2015 from Hebei (China) and identified by Professor Heming Yang
(Shanghai Institute of Materia Medica, China). A voucher specimen
(no. SIMM 238) has been deposited in the Herbarium of Shanghai
Institute of Materia Medica, Chinese Academy of Sciences.
The air-dried, powdered E. helioscopia (10
kg) was extracted three times with 95% aqueous ethanol (EtOAc) at
room temperature. A crude extract (934 g) was obtained after
concentration in vacuo, which was dissolved in water (2l)
and then extracted with EtOAc to get the EtOAc-soluble fraction
(377 g). The latter was subjected to a silica gel column and eluted
with petroleum ether-acetone (100:0-0:100) to obtain eight
fractions A-H. Fractions D1-D9 were yield from fraction D after
eluting with petroleum ether (PE)/EtOAc (20:1 to 1:1). Sephadex
LH-20 gel column was then used to get two fractions (D9A-D9B) from
fraction D9. Then fractions D9B1-D9B3 were acquired by eluting with
PE/EtOAc (8:1-3:1) from fraction D9B. Fraction D9B2 was then put
into a RP-C18 column methanol (MeOH)/H2O, 75:25 to 95:5)
to obtain fractions D9B2A-D9B2D, among which C21 (812.5 mg) was
harvested from fraction D9B2C by preparative HPLC
(MeOH/H2O, 85:15). C21 was identified as Euphornin L by
comparison of the 13C NMR spectra with the data reported
previously (25).
Euphornin L (C21): 13C NMR (100 MHz,
CDCl3) δ: 15.8, 17.1, 20.3, 20.6, 21.2, 21.3,
21.4, 22.6, 22.8, 32.4, 36.6, 39.7, 39.8, 41.9, 43.6, 73.2, 74.0,
74.0, 83.0, 92.2, 121.3, 128.4 (x2), 129.4, 129.4 (x2), 130.5,
132.7, 133.0, 136.0, 166.2, 169.4, 169.6, 170.0, 170.5. HR-ESI-MS:
m/z 565.2770 (M + Na)+ (calculated for
C31H42O8Na, 565.2772). HPLC
analysis: MeOH-H2O (0.1% formic acid; 90:10), eluted at
3.52 min, 97.77% purity.
Cell culture
The liver cancer cell line HepG2 was obtained from
the American Type Culture Collection (cat. no. HB-8065) and
maintained in DMEM with 10% FBS (v/v) and incubated under a
humidified atmosphere of 95% O2 and 5% CO2 at
37˚C. Cells were sub-cultured once every two days. Cells within 4
to 11 passages were used for experiments.
Cell viability analysis
HepG2 cells were seeded in 96-well plates
(1.0x104 cells/well) for 24 h prior to treatment with
indicated concentrations of C21. After another 24 h, cell viability
was detected with Enhanced Cell Counting Kit-8 (cat. no. C0043;
Beyotime Institute of Biotechnology) according to the
manufacturer's instructions.
Lipoprotein isolation and DiI-LDL
preparation
Human plasma (200 ml) was obtained from Shanghai
Xuhui Central Hospital, China (healthy donors, n=4, male, 20-40
years old), after informed consent and approval by the Ethics
Committee of Shanghai Xuhui Central Hospital (approval no.
2018-038). The procedures conformed to the principles outlined in
the Declaration of Helsinki (26).
LDL and lipoprotein-deficient serum (LPDS) were separated from the
pooled plasma by ultracentrifugation. Briefly, density of the
pooled plasma was adjusted to 1.019 g/ml with NaBr before
ultracentrifugation at 4˚C, 250,000 x g for 4 h. After removing the
first layer of chylomicron and VLDL, the density of the remaining
part was adjusted to 1.063 g/ml with NaBr before
ultracentrifugation at 4˚C, 250,000 x g for 24 h. The resulting
first layer was LDL. After removing LDL, density of the remaining
part was adjusted to 1.21 g/ml with NaBr before ultracentrifugation
at 4˚C, 250,000 x g for 48 h; the bottom layer was LPDS. LDL and
LPDS were dialyzed in dialysis buffer and PBS for 24 h and 48 h,
respectively. LDL was then labeled with the fluorescent probe 1,
1'-dioctadecyl-3, 3, 3', 3'-tetramethylindocarbocyanine perchlorate
(DiI; Biotium) as previously described, with some modifications
(27). Briefly, DiI was dissolved
in DMSO (15 mg/ml) and added into the LDL/LPDS mixture (1:2 v/v) to
a final concentration of 300 mg DiI/mg LDL protein and incubated
for 18 h at 37˚C. Density of the mixture was adjusted to 1.063 g/ml
with NaBr before ultracentrifugation at 10˚C, 250,000 x g for 24 h
to obtain the first layer of DiI-labeled LDL (DiI-LDL), followed by
dialyzing in dialysis buffer and PBS for 24 and 48 h, respectively.
The obtained DiI-LDL was sterilized using 0.45 µm filters (EMD
Millipore).
DiI-LDL uptake assay
DiI-LDL uptake assays were conducted as described
previously (27), with minor
modifications. Briefly, HepG2 cells were seeded in 24-well plates.
At 12 h later, the culture medium was changed to 2% LPDS DMEM
(v/v). After incubation for 24 h, the drugs were added into the
medium and incubated with cells for another 24 h. Then the culture
medium was changed to DiI-LDL DMEM (20 µg/ml) and incubated for 3 h
at 37˚C in the dark. Subsequently, the cells were rinsed twice with
ice-cold PBS buffer containing 0.4% albumin (Sigma-Aldrich; Merck
KGaA) and washed twice with ice-cold PBS buffer. Then, 500 µl of
isopropanol was added into each well followed by a 20-min
incubation in the dark at room temperature with constant shaking.
Aliquots (200 µl) were used for fluorescence detection with
SpectraMax M2e Microplate Reader at 520-578 nm (Molecular Devices,
LLC).
Western blot analysis of proteins in
hamster liver tissues and in HepG2 cells
Western blotting with HepG2 cell lysates was
described previously (24). HepG2
cells were cultured in 6-well plates. After 12 h, the culture
medium was changed to 2% LPDS DMEM (v/v). Following incubation for
24 h, the drugs were added into the medium and incubated with cells
for another 24 h. The cells were washed three times with ice-cold
PBS buffer and total cellular proteins were extracted using 100
µl/well lysis buffer containing protease and phosphatase inhibitor
cocktail (cat. no. 539134; Calbiochem; Merck KGaA) and centrifuged
at 12,000 x g, 4˚C for 10 min.
Proteins in liver tissues were extracted using 200
µl lysis buffer containing protease and phosphatase inhibitor
cocktail from 20-40 mg frozen liver tissues.
Protein concentrations were determined with BCA
Protein Assay kit (cat. no. P0010; Beyotime Institute of
Biotechnology). A total of 30 µg protein was loaded per lane in 8%
SDS-PAGE and then transferred onto PVDF membranes (cat. no.
162-0177; Bio-Rad Laboratories, Inc.), blocked with 5% skimmed milk
(cat. no. 232100; Becton, Dickinson and Company) for 2 h at room
temperature and incubated with primary antibodies overnight at 4˚C.
Membranes were then washed three times in TBST solution followed by
incubation with secondary antibodies (cat. no. 1706515 or 1706516;
Bio-Rad Laboratories, Inc.) for 2 h and visualized using Clarity
Western ECL Blotting Substrates (cat. no. 170-5061; Bio-Rad
Laboratories, Inc.). Values were normalized to the housekeeping
protein β-actin. Quantity One (v4.6.2, Bio-Rad Laboratories, Inc.)
was used for densitometry.
RNA interference
Small interfering RNA (siRNA; 1,500 ng/well) was
transfected into HepG2 cells in 12-well plates using Lipofectamine
3000 reagent according to the manufacturer's instructions
(Invitrogen; Thermo Fisher Scientific, Inc.). At 24 h post
transfection, cell culture medium was discarded and replaced with
fresh DMEM with 10% FBS (v/v). At 24 h later, cells were used for
subsequent experiments. siRNAs against human LDLR (forward:
5'-CGGCUUAAGAACAUCAACAdTdT-3', reverse:
5'-UGUUGAUGUUCUUAAGCCGdTdT-3') and scrambled siRNA were synthesized
by Shanghai GenePharma Co., Ltd.
RNA isolation and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from HepG2 cells after
treatment with drugs for 6 h, or from hamster liver tissues, using
TRIzol® Reagent (cat. no. 15596-018; Thermo Fisher
Scientific, Inc.) and reverse transcribed into cDNA (cat. no.
RR036A; Takara Biotechnology Co., Ltd.). RT-qPCR was performed
using SYBR Green PCR Supermix (cat. no. 172-5125; Bio-Rad
Laboratories, Inc.) with specific primers as listed in Table I. The relative signal intensity was
measured by CFX Real-time PCR Detection System (Bio-Rad
Laboratories, Inc.). The following thermocycling conditions were
used: Initial denaturation at 95˚C for 2 min; followed by 40 cycles
of 95˚C for 15 sec, 58˚C for 25 sec and 72˚C for 25 sec. Values
were normalized to the housekeeping gene GAPDH and then analyzed in
Bio-Rad CFX manager (v3.0) using the mode for normalized expression
(2-∆∆Cq) (28). Results
were presented as relative fold changes to the control group.
 | Table IReal-time QPCR primers. |
Table I
Real-time QPCR primers.
| Gene name | Accession no. | Primer (5'-3') |
|---|
| Human | | |
|
LDLR | NM 000527.4 |
ctgaaatcgccgtgttactg |
| | |
gccaatcccttgtgacatct |
|
PCSK9 | NM 174936.3 |
ccaagcctcttcttacttcacc |
| | |
gcatcgttctgccatcact |
|
GAPDH | NM 002046.3 |
aagaaggtggtgaagcagg |
| | |
aggtggaggagtgggtgtcg |
| Hamster | | |
|
LDLR | NM 010700 |
ttgggttgattccaaactcc |
| | |
gattggcactgaaaatggct |
|
PCSK9 | NM 153565 |
tgctccagaggtcatcacag |
| | |
gtcccactctgtgacatgaag |
|
GAPDH | DQ 403055 |
aactttggcattgtggaagg |
| | |
ggatgcagggatgatgttct |
Transient transfections of luciferase
reporter constructs
Wild-type or mutant LDLR (gifts from
Professor Lene Holland, University of Missouri School of Medicine,
Columbia, MO, USA) and PCSK9 (gifts from Professor Sahng Wook Park,
Yonsei University College of Medicine, Seoul, Republic of Korea)
promoter reporter constructs were co-transfected with the
Renilla luciferase vector (cat. no. E2231; Promega
Corporation) respectively, which was constitutively expressed as a
transfection control, into HepG2 cells using Lipofectamine 3000
reagent (Thermo Fisher Scientific, Inc.). At 24 h post
transfection, cell culture medium was discarded and changed to new
DMEM with 10% FBS (v/v). At 24 h later, cells were used for
subsequent experiments. Luciferase activity was evaluated by
dual-luciferase reporter assay system according to the
manufacturer's instructions (cat. no. E1910; Promega Corporation).
Data were normalized to Renilla luciferase activity.
mRNA stability assay
HepG2 cells (1x105 cells/well) were
cultured at 37˚C in 24-well plates for 12 h before the culture
medium was then changed to 2% LPDS DMEM (v/v). Following incubation
for 24 h, C21 (5 and 20 µM) or DMSO were added into the medium and
incubated with cells at 37˚C for another 24 h. Actinomycin D (cat.
no. SBR00013; Sigma-Aldrich; Merck KGaA) was added into medium to
inhibit RNA transcription in cells, as previously described
(29). After incubated for
predetermined time (30-180 min), cells were lysed to collect total
RNA and remaining LDLR and PCSK9 mRNA in cells was
determined by RT-qPCR.
Animals and drug treatment
Our previous study evaluated the lipid-lowering
effects of C21 (intragastric administration, i.g., 30 mg/kg/day) in
high-fat diet induced hyperlipidemia Golden Syrian hamster model
(24). A total of 12 male LVG
golden Syrian hamsters (strain code: 501), with body weight of
80-90 g (age, 6-9 weeks), were obtained from the Beijing Vital
River Laboratory Animal Technology Co., Ltd. and housed in
ventilated cages under 20-26˚C, humidity of 40-70%, and a 12-h
light/dark cycle with free access to water. After feeding with a
high-fat (HF) diet, which contained 0.5% cholesterol and 10% lard
oil, the animals were separated into two groups: HF Group (n=6) and
C21 Group (n=6), according to the serum lipid profiles. During the
three weeks' treatment (D1-D21), the body weight of hamsters and
lipid profiles in the serum were monitored at following time
points: The day before switching food to high-fat diet, one week
after switching, on the tenth day (D10) of treatment with C21 and
at the end of the treatment (D21). To collect the blood, hamsters
were anaesthetized with 2% isoflurane (30) and 300 µl blood was collected via the
jugular vein. At the end of the experiment (D21), hamsters were
sacrificed with CO2 euthanasia followed by bilateral
thoracotomy, the flow rate of CO2 used in the present
study was 25% displacement of the chamber volume per minute
following which the liver tissue samples were collected. All animal
procedures were in compliance with PREPARE guidelines (31) and were approved by the Institutional
Ethics Committee of Shanghai Institute of Materia Medica, Chinese
Academy of Sciences (approval no. 2018-08-WYP-30).
In order to confirm the effects of C21, tissue
protein lysates and total RNA were extracted from liver tissues and
subjected to western blot and RT-qPCR assays respectively to assess
protein and mRNA levels. The serum PCSK9 protein level and the
serum ALT and AST activities were assessed with commercial
kits.
ELISA
The PCSK9 protein level in serum was measured using
Quantikine® ELISA kit according to the manufacturer's
instructions (cat. no. MPC900; R&D Systems, Inc.) (32,33).
Statistical analysis
For each experiment, the values were expressed as
the mean ± standard error of the mean. All data were obtained from
at least three independent experiments. Statistical analysis was
performed using GraphPad Prism (v7.0, GraphPad Software Inc.).
Student's t-test was used to determine if two sets of data were
significantly different from each other. One-way ANOVA was used to
compare the statistical differences between at least three groups
of data and Dunnett's multiple comparison test was used to compare
all groups with the control group. P<0.05 was considered to
indicate a statistically significant difference.
Results
C21 improves LDL uptake by increasing
LDLR protein levels in HepG2 cells
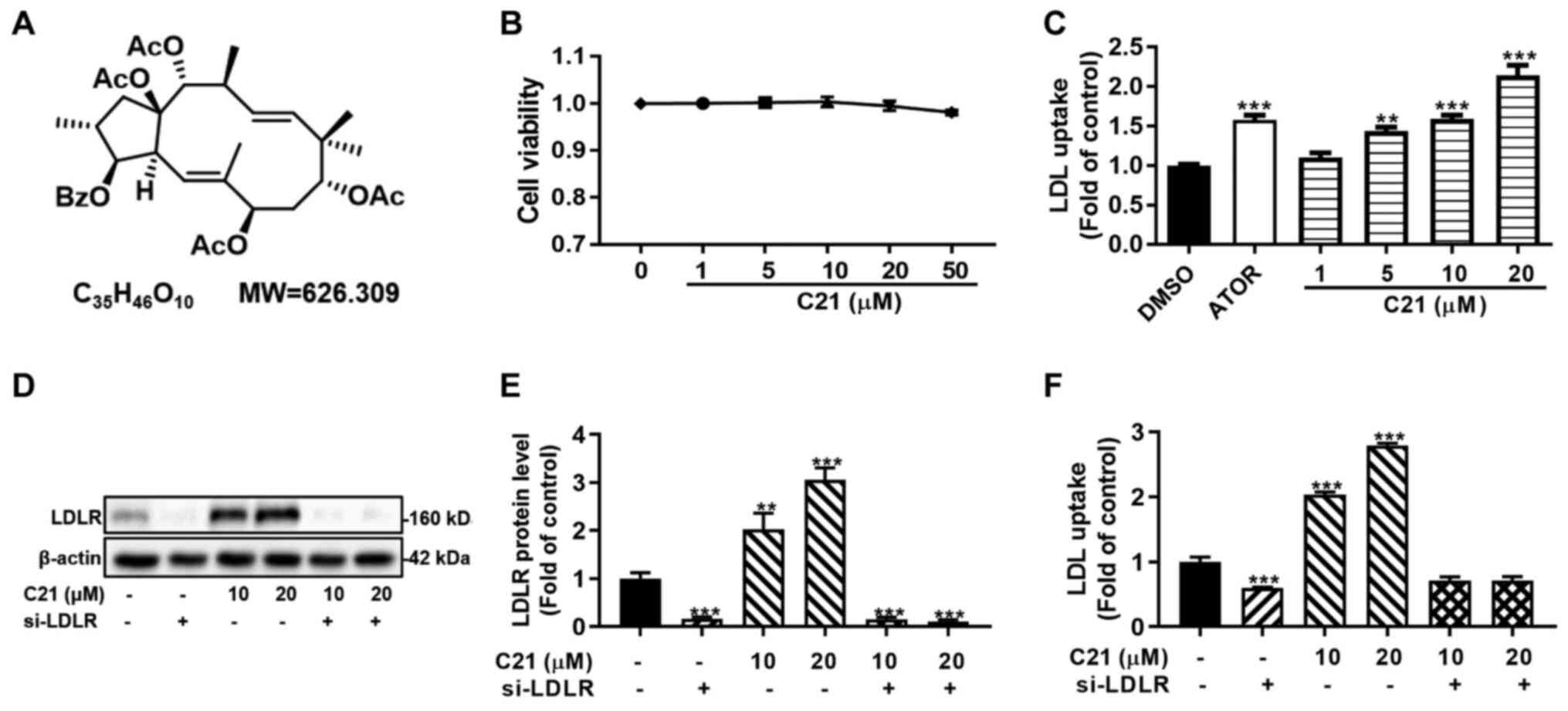
Since C21 (Fig. 1A)
had little effect on the viability of HepG2 cells at experimental
concentrations in this study (1-50 µM; Fig. 1B), DiI-LDL uptake assays and western
blot assays were conducted to verify the effect of C21 on LDL
uptake and LDLR protein. In line with our previous study (24), C21 dose-dependently promoted DiI-LDL
uptake and LDLR protein abundance in HepG2 cells (Fig. 1C-E). In addition, knocking down LDLR
with a specific siRNA abolished the C21-induced increase in DiI-LDL
uptake (Fig. 1D-F). These results
suggested that the promotion of DiI-LDL uptake by C21 was
LDLR-dependent and that C21 improved LDL uptake by increasing LDLR
protein levels in HepG2 cells.
C21 has little effect on the promoter
activities of LDLR and PCSK9 genes
As mentioned above, PCSK9 serves an important role
in the post-translational regulation of LDLR protein. The present
study further assessed whether PCSK9 was involved in C21-induced
LDLR elevation in HepG2 cells using western blot assay. As
expected, LDLR protein levels increased dose-dependently. Notably,
PCSK9 protein levels were also dose-dependently increased in HepG2
cells after treatment with C21 (Fig.
2A and B). To further
investigate the effects of C21 on mRNA levels and promoter
activities of both LDLR and PCSK9, RT-qPCR assays and
dual luciferase reporter assays were conducted. It was found that
C21 dose-dependently increased LDLR mRNA levels but had
little effect on PCSK9 mRNA levels (Fig. 2C). Notably, C21 induced no
significant difference in the promoter activities of both
LDLR and PCSK9 genes (Fig. 2D). These results indicated that
post-transcriptional regulation of LDLR and PCSK9 might be induced
by C21 to elevate LDLR and PCSK9 protein levels in HepG2 cells.
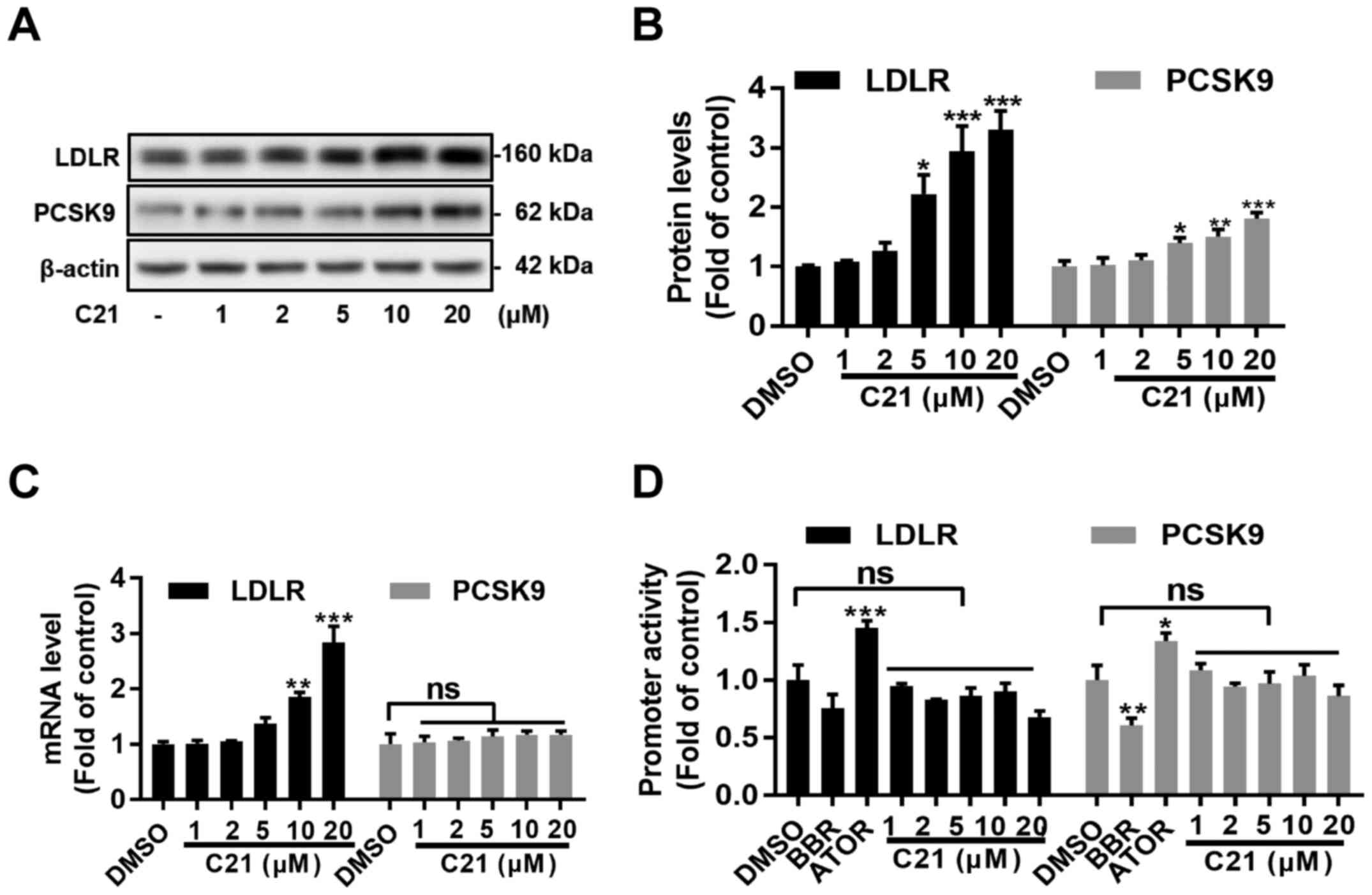 | Figure 2C21 has little effect on the promoter
activities of LDLR and PCSK9. (A and B) C21 elevated
LDLR and PCSK9 protein levels in HepG2 cells after treatment for 24
h. (C) C21 boosted LDLR mRNA levels dose-dependently after
treatment for 6 h but had little effect on PCSK9 mRNA levels. (D)
C21 induced no significant effects on LDLR and PCSK9
promoter activities. Results are presented as the means ± standard
error of the mean, n≥5. *P<0.05,
**P<0.01, ***P<0.001 vs. vehicle
control by one-way ANOVA (with Dunnett's multiple comparisons
test). C21, euphornin L; LDL, Low density lipoprotein; LDLR, LDL
receptor; PCSK9, Proprotein convertase subtilisin/kexin type 9;
BBR, berberine; ATOR, atorvastatin; ns, no significance. |
C21 increases the half-life of LDLR
mRNA and reduces the PCSK9 protein abundance in cell culture
medium
Given the instability of LDLR mRNA, the
half-life of LDLR mRNA in HepG2 cells was evaluated after
treatment with C21. C21 (20 µM) increased the half-life of
LDLR mRNA by almost 3-fold (Fig.
3A, DMSO group, T1/2=31.49 min; C21-20 µM group,
T1/2=92.45 min).
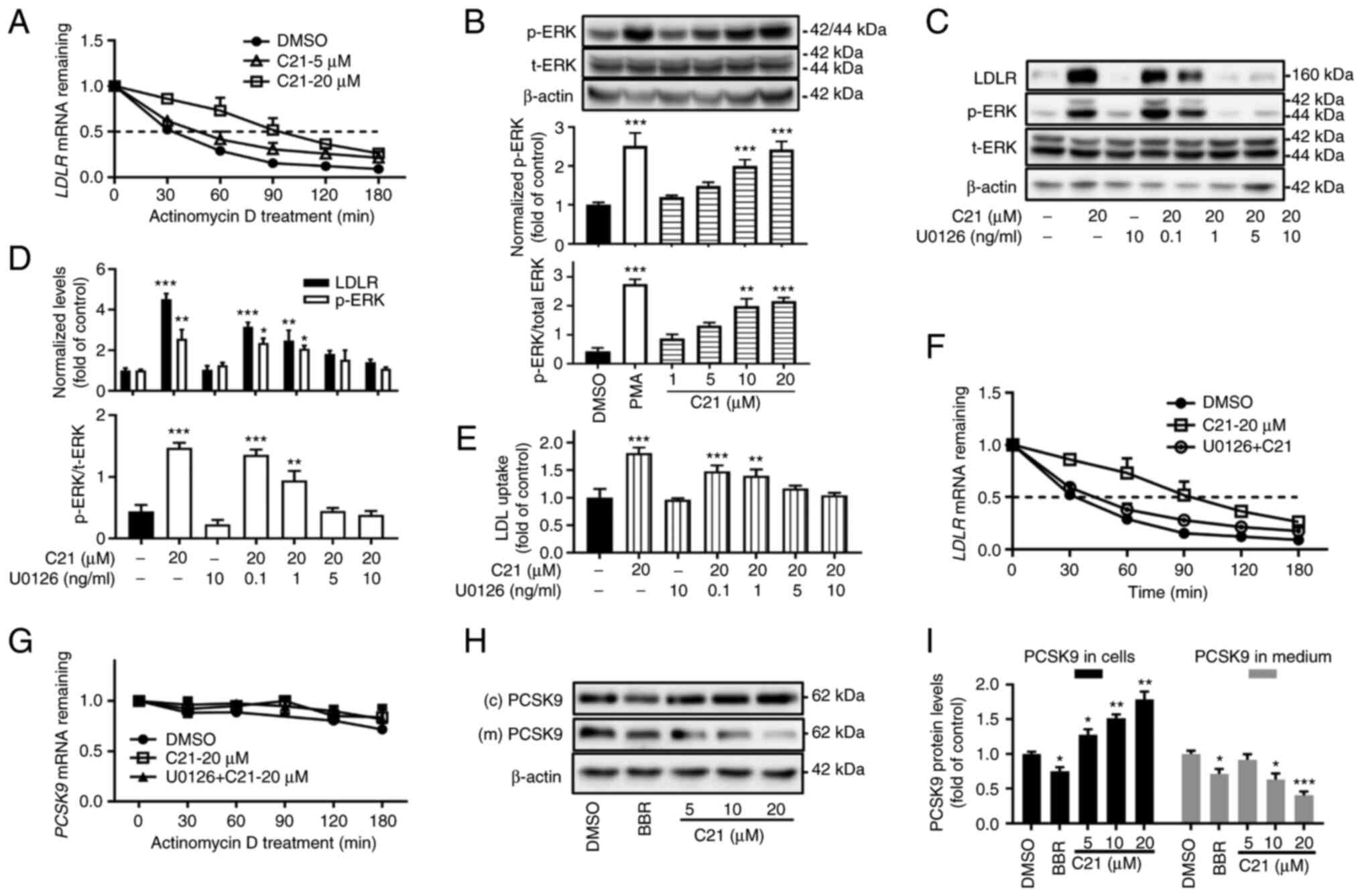 | Figure 3C21 increases the half-life of
LDLR mRNA and reduced the secretion of PCSK9 protein. (A)
C21 increased the half-life of LDLR mRNA. (B) After
treatment for 2 h, C21 dose-dependently promoted the
phosphorylation of ERK protein. PMA (5 ng/ml) was used as a
positive control. (C-F) The ERK activation inhibitor U0126
counteracted the effects of C21 on LDLR protein, LDL uptake and
half-life of LDLR mRNA in HepG2 cells. (G) C21 had little
effect on the half-life of PCSK9 mRNA. (H and I) C21
decreased the secretion of PCSK9 protein into cell culture medium.
BBR (40 µM) was used as a positive control. Results are presented
as the means ± standard error of the mean, n≥5.
*P<0.05, **P<0.01,
***P<0.001 vs. vehicle control by one-way ANOVA
(Dunnett's multiple comparisons test). C21, euphornin L; LDL, Low
density lipoprotein; LDLR, LDL receptor; PMA, phorbol 12-myristate
13-acetate; p-, phosphorylated; t-, total; PCSK9, Proprotein
convertase subtilisin/kexin type 9; (c) PCSK9, PCSK9 protein in
whole cell lysates; (m) PCSK9, PCSK9 protein in cell culture
medium; BBR, berberine. |
Since the activation of ERK protein has been found
to be involved in LDLR mRNA stabilization (4,14-16),
the effect of C21 on ERK was assessed. The results showed that
treatment with C21 for 2 h dose-dependently elevated the
phosphorylation of ERK (Fig. 3B).
Furthermore, when cells were pretreated with U0126, a specific
inhibitor of mitogen-activated protein kinase (MAPKK or MEK), the
effects of C21 on LDLR protein (Fig.
3C and D), LDL uptake (Fig. 3E) and LDLR mRNA half-life
(Fig. 3F) were abolished, which
demonstrated that C21 boosted LDLR expression by stabilizing
LDLR mRNA and the activation of ERK was essential in this
post-transcriptional regulation.
C21 had little effect on the half-life of
PCSK9 mRNA (Fig. 3G).
However, unexpectedly, following treatment with C21 (5-20 µM),
PCSK9 protein levels in cell culture medium were decreased even
though PCSK9 protein levels in whole cell lysates were increased.
The plant-derived compound berberine, which has been reported to
transcriptionally suppress PCSK9 expression in HepG2 cells
(4), was used as a positive
control. As C21 evoked little effect on the promoter activity and
mRNA level of PCSK9 in HepG2 cells, it was hypothesized that
C21 might inhibit the secretion of PCSK9 protein.
C21 upregulates LDLR and PCSK9 protein
levels in hamster liver tissues and downregulates PCSK9 protein
levels in hamster serum
Consistent with the results obtained from HepG2
cells, LDLR, PCSK9 and p-ERK protein levels as well as LDLR
mRNA levels in hamster liver tissues were significantly elevated
(P<0.05 or P<0.001, Fig.
4A-C, E and F). In addition, compared with the HF
group, the C21 group showed a significant reduction in PCSK9
protein abundance in serum (P<0.05, Fig. 4D) but little change in PCSK9
mRNA levels in liver tissues (Fig.
4C).
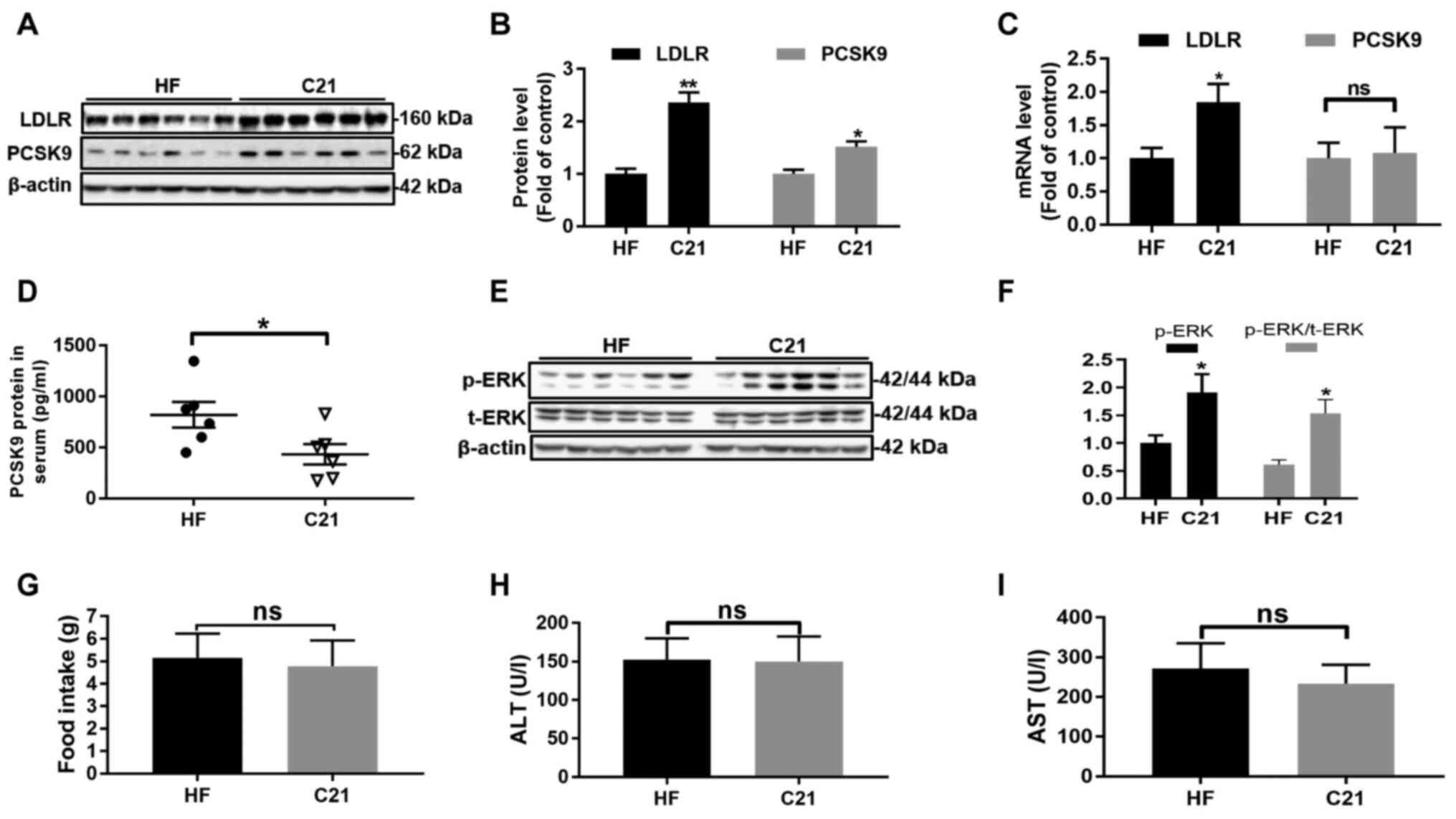 | Figure 4C21 upregulated LDLR and PCSK9
protein levels in hamster liver tissues and downregulated PCSK9
protein levels in hamster serum. (A-C) mRNA and protein levels of
LDLR and PCSK9 in hamster liver tissues (n=6/group). (D) PCSK9
protein levels in hamster serum were significantly reduced after 3
weeks of treatment with C21 (30 mg/kg/d). (E and F) C21 promoted
the phosphorylation of ERK protein in hamster liver tissues. (G)
The food intake of hamsters in the two groups was not significantly
different. (H and I) The activities of ALT and AST in hamster serum
were not significantly different between the two groups. Results
are presented as the means ± standard error of the mean, n=6.
*P<0.05, **P<0.01 vs. vehicle control
by Student's t-test. C21, euphornin L; LDL, Low density
lipoprotein; LDLR, LDL receptor; PCSK9, Proprotein convertase
subtilisin/kexin type 9; ALT, alanine aminotransferase; AST,
aspartate aminotransferase; HF, high-fat diet; p-, phosphorylated;
t-, total; ns, no significance. |
In addition, treatment with C21 caused no obvious
damage to hamster liver tissues since no significant differences in
serum ALT and AST activities, nor in the food intake of hamsters,
were observed between the two groups (Fig. 4G-I).
Discussion
The current study explored the mechanisms underlying
the lipid-lowering effect of C21 in HepG2 cells. It demonstrated
that C21 increased LDLR protein levels by stabilizing LDLR
mRNA and inhibiting the secretion of PCSK9 protein.
Following treatment with C21 for 6 h, the
LDLR mRNA in HepG2 cells was dose-dependently increased
(Fig. 2C). However, in our previous
study, HepG2 cells were treated for 24 h with C21 (1-10 µM) and the
LDLR mRNA levels were not significantly changed (24). It is hypothesized that 24 h may not
be an appropriate time to assess the effects of C21 on LDLR
mRNA since the LDLR mRNA half-life was 92.45 min following
treatment with 20 µM C21.
The half-life of ARE-containing mRNAs is mainly
regulated by ARE-BPs (ARE binding proteins) (34). Some ARE-BPs, such as TTP, ZFP36,
AUF1/hnRNP D and KSRP proteins, have been shown to promote the
degradation of mRNA by binding with AREs (35,36)
and other ARE-BPs, such as ELAVL1/HuR protein, prolong the
half-life of mRNA (34,37). The activation of ERK protein has
been found to be involved in LDLR mRNA stabilization
(4,14-16).
The mechanism remains to be elucidated, but the phosphorylation of
ERK may influence the binding of specific ARE-BPs, such as ZFP36L1
and ZFP36L2(36) as well as HuR
protein (37), to the SRE1 motif in
the LDLR promoter. The present study found that the
activation of ERK protein was essential in the stabilization of
LDLR mRNA induced by C21. It is planned to determine whether
and which ARE-BPs are involved downstream of ERK activation in
future studies.
Following treatment with C21, PCSK9 protein in HepG2
cells increased while PCSK9 protein levels in cell culture medium
decreased without a significant influence on the promoter activity
and mRNA level of PCSK9. Therefore, it was hypothesized that
the secretion of PCSK9 might be inhibited by C21. The inhibition
was confirmed when the serum PCSK9 level was decreased while the
hepatocellular PCSK9 protein level was increased in hamsters from
the C21 group. During the cellular transportation of secretory
proteins, coat protein complex II, consisting of SAR1 gene homolog
B (SAR1B) GTPase, SEC23/SEC24 (heterodimers) and SEC13/SEC31
(heterotetramers), is responsible in the early secretion phase from
the ER to the Golgi bodies (38).
It has been reported that genetic deficiency of SEC24A in mice
results in decreased secretion of PCSK9 protein (39). However, SEC24A cannot interact
directly with PCSK9 since the former is located outside of the ER
membrane while the latter is limited to the luminal side (40). A study published in 2018 proposed
that SURF4 protein, an ER cargo receptor, might serve as a primary
mediator of PCSK9 in the ER, as knocking down SURF4 results in an
accumulation of PCSK9 in the ER (40). Sortilin 1 (SORT1), a trans-membrane
trafficking receptor localized to the trans-Golgi network and early
endosomes (41), has been
demonstrated to be significantly associated with coronary artery
disease, LDL-C levels and abdominal aortic aneurysm (41-43).
In 2014, Gustafsen et al (44) reported that SORT1 promotes the
secretion of PCSK9. Furthermore, a SORT1 inhibitor, AF38469,
decreases plasma cholesterol levels in Western diet-fed mice,
suggesting that SORT1 may serve as a potential target for
hyperlipidemia treatment (45). The
current study found that PCSK9 protein levels in cell culture
medium or hamster serum decreased after treatment with C21, without
any significant changes in PCSK9 promoter activity or
PCSK9 mRNA levels. Thus, the secretion process of PCSK9
might be inhibited by C21. However, how C21 is involved in this
process remains to be elucidated, which will be attempted in future
studies.
In conclusion, the present study demonstrated that
euphornin L increased LDLR protein levels in HepG2 cells by dual
regulation of LDLR mRNA and PCSK9 and represented an active
compound for lipid-lowering drug development.
Acknowledgements
The authors thank Professor Jing-wen Liu (Department
of Veterans' Affairs, Palo Alto Health Care System) for kindly
providing anti-hamster PCSK9 antibody and Professor Lene Holland
(University of Missouri School of Medicine, Columbia, MO, USA) and
Professor Sahng Wook Park (Yonsei University College of Medicine,
Seoul, Republic of Korea) for kindly providing LDLR and
PCSK9 promoter reporter constructs as gifts. The authors
would also like to express our sincere gratitude and deep
condolence to our mentor, Professor Yi-ping Wang, who passed away
on April 11, 2018, for his enlightening mentoring and strong
support throughout.
Funding
The authors gratefully acknowledge grants from the Drug
Innovation Major Project of China (grant no.
2018ZX09735001-002-005), the Personalized Medicines-Molecular
Signature-based Drug Discovery and Development Strategic Priority
Research Program of the Chinese Academy of Sciences (grant no.
XDA12040335), the National Natural Science Foundation of China
(grant no. 81773863) and the Youth Innovation Promotion Association
(grant no. 2019284).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LX and HL designed the study. HL, JuL and XZ
performed the experiments and drafted the manuscript. JiL, CX, WW
and YL were involved in data analysis and interpretation, All
authors confirm the authenticity of all data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Shanghai Institute of Materia Medica (approval no. 2018-08-WYP-30)
and the Ethics Committee of Shanghai Xuhui Central Hospital
(approval no. 2018-038). Informed consents were signed by the
healthy volunteers.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wadhera RK, Steen DL, Khan I, Giugliano RP
and Foody JM: A review of low-density lipoprotein cholesterol,
treatment strategies, and its impact on cardiovascular disease
morbidity and mortality. J Clin Lipidol. 10:472–489.
2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Stone NJ, Robinson JG, Lichtenstein AH,
Bairey Merz CN, Blum CB, Eckel RH, Goldberg AC, Gordon D, Levy D,
Lloyd-Jones DM, et al: American College of Cardiology/American
Heart Association Task Force on Practice Guidelines: 2013 ACC/AHA
guideline on the treatment of blood cholesterol to reduce
atherosclerotic cardiovascular risk in adults: A report of the
American College of Cardiology/American Heart Association Task
Force on Practice Guidelines. Circulation. 129 (Suppl 2):S1–S45.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Grundy SM, Arai H, Barter P, Bersot TP,
Betteridge DJ, Carmena R, Cuevas A, Davidson MH, Genest J,
Kesäniemi YA, et al: Expert Dyslipidemia Panel of the International
Atherosclerosis Society Panel members: An International
Atherosclerosis Society Position Paper: Global recommendations for
the management of dyslipidemia - full report. J Clin Lipidol.
8:29–60. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Li H, Dong B, Park SW, Lee HS, Chen W and
Liu J: Hepatocyte nuclear factor 1alpha plays a critical role in
PCSK9 gene transcription and regulation by the natural
hypocholesterolemic compound berberine. J Biol Chem.
284:28885–28895. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang DW, Garuti R, Tang WJ, Cohen JC and
Hobbs HH: Structural requirements for PCSK9-mediated degradation of
the low-density lipoprotein receptor. Proc Natl Acad Sci USA.
105:13045–13050. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kesaniemi YA, Witztum JL and Steinbrecher
UP: Receptor-mediated catabolism of low density lipoprotein in man.
Quantitation using glucosylated low density lipoprotein. J Clin
Invest. 71:950–959. 1983.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Brown MS and Goldstein JL: A
receptor-mediated pathway for cholesterol homeostasis. Science.
232:34–47. 1986.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lo Surdo P, Bottomley MJ, Calzetta A,
Settembre EC, Cirillo A, Pandit S, Ni YG, Hubbard B, Sitlani A and
Carfí A: Mechanistic implications for LDL receptor degradation from
the PCSK9/LDLR structure at neutral pH. EMBO Rep. 12:1300–1305.
2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Goldstein JL and Brown MS: A century of
cholesterol and coronaries: From plaques to genes to statins. Cell.
161:161–172. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Südhof TC, Van der Westhuyzen DR,
Goldstein JL, Brown MS and Russell DW: Three direct repeats and a
TATA-like sequence are required for regulated expression of the
human low density lipoprotein receptor gene. J Biol Chem.
262:10773–10779. 1987.PubMed/NCBI
|
|
11
|
Miserez AR, Muller PY, Barella L, Barella
S, Staehelin HB, Leitersdorf E, Kark JD and Friedlander Y:
Sterol-regulatory element-binding protein (SREBP)-2 contributes to
polygenic hypercholesterolaemia. Atherosclerosis. 164:15–26.
2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wilson GM, Vasa MZ and Deeley RG:
Stabilization and cytoskeletal-association of LDL receptor mRNA are
mediated by distinct domains in its 3' untranslated region. J Lipid
Res. 39:1025–1032. 1998.PubMed/NCBI
|
|
13
|
Zhang T, Kruys V, Huez G and Gueydan C:
AU-rich element-mediated translational control: complexity and
multiple activities of trans-activating factors. Biochem Soc Trans.
30:952–958. 2002.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kong W, Wei J, Abidi P, Lin M, Inaba S, Li
C, Wang Y, Wang Z, Si S, Pan H, et al: Berberine is a novel
cholesterol-lowering drug working through a unique mechanism
distinct from statins. Nat Med. 10:1344–1351. 2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yashiro T, Yokoi Y, Shimizu M, Inoue J and
Sato R: Chenodeoxycholic acid stabilization of LDL receptor mRNA
depends on 3'-untranslated region and AU-rich element-binding
protein. Biochem Biophys Res Commun. 409:155–159. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li Z, Jiang JD and Kong WJ: Berberine
up-regulates hepatic low-density lipoprotein receptor through
Ras-independent but AMP-activated protein kinase-dependent Raf-1
activation. Biol Pharm Bull. 37:1766–1775. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang CP, Tian Y, Zhang M, Tuo QH, Chen JX
and Liao DF: IDOL, inducible degrader of low-density lipoprotein
receptor, serves as a potential therapeutic target for
dyslipidemia. Med Hypotheses. 86:138–142. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tavori H, Fan D, Blakemore JL, Yancey PG,
Ding L, Linton MF and Fazio S: Serum proprotein convertase
subtilisin/kexin type 9 and cell surface low-density lipoprotein
receptor: Evidence for a reciprocal regulation. Circulation.
127:2403–2413. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Abifadel M, Varret M, Rabès JP, Allard D,
Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich
D, et al: Mutations in PCSK9 cause autosomal dominant
hypercholesterolemia. Nat Genet. 34:154–156. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Horton JD, Cohen JC and Hobbs HH: PCSK9: A
convertase that coordinates LDL catabolism. J Lipid Res. 50
(Suppl):S172–S177. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
McNutt MC, Kwon HJ, Chen C, Chen JR,
Horton JD and Lagace TA: Antagonism of secreted PCSK9 increases low
density lipoprotein receptor expression in HepG2 cells. J Biol
Chem. 284:10561–10570. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Raal FJ, Stein EA, Dufour R, Turner T,
Civeira F, Burgess L, Langslet G, Scott R, Olsson AG, Sullivan D,
et al: RUTHERFORD-2 Investigators: PCSK9 inhibition with evolocumab
(AMG 145) in heterozygous familial hypercholesterolaemia
(RUTHERFORD-2): A randomised, double-blind, placebo-controlled
trial. Lancet. 385:331–340. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Robinson JG, Farnier M, Krempf M, Bergeron
J, Luc G, Averna M, Stroes ES, Langslet G, Raal FJ, El Shahawy M,
et al: ODYSSEY LONG TERM Investigators: Efficacy and safety of
alirocumab in reducing lipids and cardiovascular events. N Engl J
Med. 372:1489–1499. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li J, Li HH, Wang WQ, Song WB, Wang YP and
Xuan LJ: Jatrophane diterpenoids from Euphorbia helioscopia and
their lipid-lowering activities. Fitoterapia. 128:102–111.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yamamura S, Shizuri Y, Kosemura S, Ohtsuka
J, Tayama T, Ohba S, Ito M, Saito Y and Terada Y: Diterpenes from
Euphorbia helioscopia. Phytochemistry. 28:3421–3436. 1989.
|
|
26
|
World Med A; World Medical Association.
World Medical Association Declaration of Helsinki: Ethical
principles for medical research involving human subjects. JAMA.
310:2191–2194. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Stephan ZF and Yurachek EC: Rapid
fluorometric assay of LDL receptor activity by DiI-labeled LDL. J
Lipid Res. 34:325–330. 1993.PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sobell HM: Actinomycin and DNA
transcription. Proc Natl Acad Sci USA. 82:5328–5331.
1985.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Greenfield EA: Administering anesthesia to
mice, rats, and hamsters. Cold Spring Harb Protoc: Jun 3, 2019.
doi: 10.1101/pdb.prot100198.
|
|
31
|
Smith AJ, Clutton RE, Lilley E, Hansen KEA
and Brattelid T: PREPARE: Guidelines for planning animal research
and testing. Lab Anim. 52:135–141. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Dong B, Young M, Liu X, Singh AB and Liu
J: Regulation of lipid metabolism by obeticholic acid in
hyperlipidemic hamsters. J Lipid Res. 58:350–363. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Dong B, Li H, Singh AB, Cao A and Liu J:
Inhibition of PCSK9 transcription by berberine involves
down-regulation of hepatic HNF1α protein expression through the
ubiquitin-proteasome degradation pathway. J Biol Chem.
290:4047–4058. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Barreau C, Paillard L and Osborne HB:
AU-rich elements and associated factors: Are there unifying
principles? Nucleic Acids Res. 33:7138–7150. 2006.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wilusz CJ and Wilusz J: Bringing the role
of mRNA decay in the control of gene expression into focus. Trends
Genet. 20:491–497. 2004.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Adachi S, Homoto M, Tanaka R, Hioki Y,
Murakami H, Suga H, Matsumoto M, Nakayama KI, Hatta T, Iemura S, et
al: ZFP36L1 and ZFP36L2 control LDLR mRNA stability via the ERK-RSK
pathway. Nucleic Acids Res. 42:10037–10049. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yashiro T, Nanmoku M, Shimizu M, Inoue J
and Sato R: 5-Aminoimidazole-4-carboxamide ribonucleoside
stabilizes low density lipoprotein receptor mRNA in hepatocytes via
ERK-dependent HuR binding to an AU-rich element. Atherosclerosis.
226:95–101. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang Q, Meng F, Chen S, Plouffe SW, Wu S,
Liu S, Li X, Zhou R, Wang J, Zhao B, et al: Hippo signalling
governs cytosolic nucleic acid sensing through YAP/TAZ-mediated
TBK1 blockade. Nat Cell Biol. 19:362–374. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Miller E, Antonny B, Hamamoto S and
Schekman R: Cargo selection into COPII vesicles is driven by the
Sec24p subunit. EMBO J. 21:6105–6113. 2002.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Emmer BT, Hesketh GG, Kotnik E, Tang VT,
Lascuna PJ, Xiang J, Gingras AC, Chen XW and Ginsburg D: The cargo
receptor SURF4 promotes the efficient cellular secretion of PCSK9.
eLife. 7(7)2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Willnow TE, Petersen CM and Nykjaer A:
VPS10P-domain receptors - regulators of neuronal viability and
function. Nat Rev Neurosci. 9:899–909. 2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kathiresan S, Melander O, Guiducci C,
Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF,
Havulinna AS, et al: Six new loci associated with blood low-density
lipoprotein cholesterol, high-density lipoprotein cholesterol or
triglycerides in humans. Nat Genet. 40:189–197. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Linsel-Nitschke P, Heeren J, Aherrahrou Z,
Bruse P, Gieger C, Illig T, Prokisch H, Heim K, Doering A, Peters
A, et al: Genetic variation at chromosome 1p13.3 affects sortilin
mRNA expression, cellular LDL-uptake and serum LDL levels which
translates to the risk of coronary artery disease. Atherosclerosis.
208:183–189. 2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Gustafsen C, Kjolby M, Nyegaard M,
Mattheisen M, Lundhede J, Buttenschøn H, Mors O, Bentzon JF, Madsen
P, Nykjaer A, et al: The hypercholesterolemia-risk gene SORT1
facilitates PCSK9 secretion. Cell Metab. 19:310–318.
2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen C, Li J, Matye DJ, Wang Y and Li T:
Hepatocyte sortilin 1 knockout and treatment with a sortilin 1
inhibitor reduced plasma cholesterol in Western diet-fed mice. J
Lipid Res. 60:539–549. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Barile E, Borriello M, Di Pietro A, Doreau
A, Fattorusso C, Fattorusso E and Lanzotti V: Discovery of a new
series of jatrophane and lathyrane diterpenes as potent and
specific P-glycoprotein modulators. Org Biomol Chem. 6:1756–1762.
2008.PubMed/NCBI View Article : Google Scholar
|