Introduction
Among the large number of chronic diseases
worldwide, myocardial infarction is one of the most lethal.
According to statistical data, there were ≥2 million cases of
myocardial infarction in China in 2015, with 500,000 new cases
every year and a falling trend in the age of patients (1). Therefore, the pathophysiology of this
disease has long been the subject of intense scientific research.
According to their pathogenic profiles, acute myocardial infarction
(AMI) can be roughly divided into the following types: i) Type 1
myocardial infarction, which is caused by the interruption of or
insufficient blood supply to the myocardium, mainly due to acute
coronary artery occlusion associated with atherosclerosis or
superimposed thrombosis (2), ii)
type 2 myocardial infarction, which is normally caused by the
metabolic capacity of cardiomyocytes far exceeding blood supply,
where inflammation and fever increase the metabolic demand of
tissues and organs, thereby increasing the heart rate and
ultimately compromising coronary perfusion (3); and iii) myocardial infarctions caused
by cytokine storms, which can inhibit aerobic respiration in the
mitochondria and subsequent acute heart failure (4). In clinical practice, the main cause
of AMI remains to be acute viral or bacterial infections caused by
influenza, pneumonia and acute bronchitis (5). However, the precise molecular
mechanism underlying AMI remain unclear.
An increasing number of studies have reported that
miRNAs can serve important roles in the occurrence of diseases
(6,7). miRNAs belong to a family of
post-transcriptional regulators for gene expression. These small
non-coding RNAs, which are 20-25 nucleotides in length, bind to
target recognition sites (seed sequence) in the 3'-untranslated
regions of their mRNA transcript, leading to mRNA degradation
and/or inhibition of protein translation (8).
A number of studies have demonstrated that the
cardiovascular system exhibits high expression of miRNAs, which are
related to the occurrence and development of heart failure,
atherosclerosis, arrhythmia and coronary heart disease (9,10).
Additionally, a growing number of miRNAs have been reported to
regulate angiogenesis after AMI. Angiogenesis, which is the
formation of new capillaries from pre-existing vessels, that occurs
in the ischemic area after myocardial infarction (MI) promotes
cardiomyocyte survival and is considered to be essential to the
recovery of patients with MI (11). miR-1 was discovered to be a
potential biomarker for AMI diagnosis and prognosis, since it was
released within 3 h after the onset of acute chest pain (12). It has also been demonstrated that
miR-199 and miR-208 exerted similar roles as miR-1 in assisting the
clinical diagnosis of AMI (13).
In addition, it has been reported that increased levels of serum
miR-214 is closely related to the occurrence of cardiomyocyte
apoptosis in elderly patients with AMI (14). Mechanistically, other studies have
demonstrated that manipulating the expression of several miRNAs can
exert direct effects on angiogenesis. Yucel and Sahin (15) reported that silencing of miR-10b
expression could significantly inhibit the angiogenesis of vessel
endothelial cells. In colon cancer, miR-524-5p has been found to
target WNK lysine deficient protein kinase 1, thereby inhibiting
the expression of cell epidermal growth factor to inhibit cell
angiogenesis (16). In addition,
another study revealed that silencing of miR-212/132 expression
could significantly reduce the ability of human vessel endothelial
cells to form blood vessels (17).
In the present study, clinical blood samples of
patients with AMI were analyzed to measure the expression of
miR-124. An in vitro cell model using pcDNA3.1-miR-124
transfected human umbilical vein endothelial cells (HUVECs) was
constructed in the present study. The apoptosis and proliferation
of HUVECs was assessed using flow cytometry, TUNEL and Cell
Counting Kit-8 (CCK-8) assays. Subsequently, the transcriptome of
HUVECs overexpressing miR-124 was sequenced and analyzed. Exploring
the effect of miR-124 overexpression on gene expression in vascular
endothelial cells may provide new avenues for the development of
novel treatment strategies for AMI.
Materials and methods
Patient information
Blood samples were collected from 20 patients (male,
16; female, 4; average age, 60.2±3.7 years) admitted to the Weihai
Central Hospital with myocardial infarction between March 2016 and
June 2017. In addition, blood samples were collected at the Weihai
Central Hospital from 14 healthy volunteers between March 2016 and
June 2017 (male, 11; female, 3; aged >55 years; average age,
60.5±3.1 years; no symptoms or history of AMI). Blood samples were
collected from healthy people on admission and from patients with
myocardial infarction 7 days after the symptoms of myocardial
infarction had been controlled. The study protocol was approved by
the Ethics Committee of Weihai Central Hospital (no. WKY-2019-012).
Informed consent was provided by all patients, and written
permission was obtained before inclusion. Patients were included in
the current study if they exhibited ST segment elevation myocardial
infarction or non-ST segment elevation myocardial infarction.
Healthy individuals were included in the present study if they were
>55 years of age. Patients with AMI were excluded if they
exhibited: Diabetes mellitus, infectious disease and autoimmune
disease. Healthy patients were excluded if they exhibited diabetes
mellitus, myocardial infarction, and infectious and autoimmune
diseases.
Extraction of free RNA from peripheral
blood and reverse transcription-quantitative PCR (RT-qPCR)
identification
Blood was collected in a 5-ml heparin
anticoagulation tube and immediately used to extract free RNA
(GSPureTM Blood RNA Isolation kit, Geneseed; cat. no. E0203). When
extracting miRNA, the ethanol concentration in the cleaning buffer
was increased to 80% to increase the final miRNA concentration. The
RNA was reverse transcribed using a reverse transcription kit at
37˚C for 60 min, followed by termination at 85˚C for 5 min to
inactivate the enzymes (Takara Bio, Inc.; cat. no. 638315). In
total, 25% of the RT product was mixed with a pre-formulated 2X
SYBR Green PCR mix containing the miR-124 RT-qPCR primers, which
were dissolved to 20 µl with ddH2O (Roche Diagnostics;
cat. no. 06402712001). Amplification was performed to collect the
dissolution curve with the following thermocycling conditions: 94˚C
for 5 min; 40 cycles of 94˚C for 2 min, 60˚C for 1 min and 72˚C for
2 min; last step at 72˚C for 2 min. U6 mRNA was used as an internal
reference and the relative expression of miR-124 was calculated
using the 2-ΔΔCq method (18). The primers used for PCR were as
follows: p38 forward, 5'-AGGGCGATGTGACGTTT-3' and reverse,
5'-CTGGCAGGGTGAAGTTGG-3'; AKT forward, 5'-TGGACTACCTGCACTCGGAGAA-3'
and reverse, 5'-GTGCCGCAAAAGGTCTTCATGG-3'; PI3K forward,
5'-GAAGCACCTGAATAGGCAAGTCG-3' and reverse,
5'-GAGCATCCATGAAATCTGGTCGC-3'; β-actin forward,
5'-AAAGACCTGTACGCCAACAC-3' and reverse,
5'-GTCATACTCCTGCTTGCTGAT-3'; miR-124 forward,
5'-TAAGGCACGCGGTGAATGCC-3' and reverse,
5'-CAGGTCCAGTTTTTTTTTTTTTTTVN-3'; U6 forward,
5'-GCTTCGGCAGCACATATACTAAAAT-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'.
Cell culture, miR-124 overexpression
and RT-qPCR
HUVECs were obtained from the American Type Culture
Collection. The cells were cultured in DMEM medium (Hyclone,
Cytiva) containing 10% (v/v) FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin, which were incubated at 37˚C in an
atmosphere with 10% CO2.
To construct the pcDNA3.1-miR-124 vector, cDNA
encoding pri-miR-124 (accession no. MI0000443) was artificially
synthesized and cloned into the pcDNA3.1 vector (Sangon Biotech
Co., Ltd.). Samples were then transfected with 0.5 µg (1 µg/µl)
pcDNA3.1-miR-124 or pcDNA3.1 vector into HUVECs using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol at 37˚C
in the presence of 5% CO2 for 24 h. The empty vector
pcDNA3.1 was similarly transfected into HUVECs as negative control
(NC group). RNA was extracted from HUVECs by cell lysis with the
RNeasy Mini kit (Qiagen GmbH; cat. no. 74104) lysis buffer on ice,
after washing with ice-cold PBS. RNA was extracted with a spin
column Qiagen GmbH). Transcriptor First Strand cDNA Synthesis Kit
(Roche Diagnostics) was used to reverse transcribed the isolated
mRNA into complementary DNA at 65˚C for 10 min.
The One Step miR cDNA Synthesis kit (cat. no. D1801;
Xinhai Gene Testing Co., Ltd.) was used for the reverse
transcription of miR-124 at 37˚C for 60 min.
The expression level of miR-124 was measured by qPCR
performed with SYBR Green Kit (Takara Bio, Inc.). U6 was used as an
internal reference, and the relative expression of miR-124 was
calculated using the aforementioned 2-ΔΔCq method.
HUVEC proliferation and apoptosis
assay
The CCK-8 assay (Bimake; cat. no. B34302) was used
to evaluate the changes in HUVEC proliferation after miR-124
overexpression. Briefly, after transfection with pcDNA3.1-miR-124
or pcDNA3.1, HUVECs were seeded into a 96-well plate at a density
of 5,000 cells/well, before cell viability was analyzed. A total of
20 µl CCK-8 reagent was added into each well and incubated at 37˚C
for 4 h. The optical density at 450 nm was read with a microplate
reader (Thermo Fisher Scientific, Inc.). In total, three
independent experiments were performed.
For the TUNEL assay, HUVECs were fixed with 4%
paraformaldehyde for 20 min at room temperature. Cells were washed
three times with PBS and stained with 50 µl TUNEL reaction mixture
containing FITC-labelled dUTP and TdT for 1 h at 37˚C (In
situ Cell Death Detection kit, Roche Diagnostics; cat. no.
11684795910). Each sample was randomly counted in five fields. The
TUNEL index (%) is the average ratio of the number of
TUNEL-positive cells divided by the total number of cells under
bright-field.
Flow cytometry was used to measure the cell
apoptosis status following miR-124 overexpression in HUVECs. The
apoptosis rate was evaluated using Annexin V-FITC/PI Apoptosis
Detection kit (Tianjin Sungene Biotech Co., Ltd; cat. no.
AO2001-02P-G) according to the manufacturer's protocol. Briefly,
after transfection with pcDNA3.1-miR-124 or pcDNA3.1, the cells
were collected, washed with PBS and resuspended in 500 µl binding
buffer. Subsequently, 100 µl 1x106/ml cells were
incubated at room temperature for 1 h with 5 µl Annexin V-FITC and
5 µl PI in the dark, before analysis by flow cytometry (BD
FACSCanto™; BD Biosciences). Data were analyzed using FlowJo
software (FlowJo LLC; version no. V10).
Western blot assay
Total proteins from HUVECs were extracted by lysis
with ice-cold RIPA buffer (Pierce; Thermo Fisher Scientific, Inc.;
cat. no. 89900) with protease inhibitors (Roche Diagnostics; cat.
no. 04693159001). The protein concentration of the sample was
determined using a BCA assay (Thermo Fisher Scientific, Inc.; cat.
no. 23225). Electrophoresis was conducted with equal amount of
protein samples (40 µg) by 10% SDS-PAGE, before being transferred
onto PVDF membranes. The membranes were incubated in 5% fat-free
milk in PBST (0.1% Tween-20) at room temperature for 30 min. Then
incubated with primary antibodies against PI3K (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 4249), AKT (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 9272), p38 (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 8690) and β-actin as a loading
control (1:1,000; Sangon Biotech Co., Ltd.; cat. no. D191047) at
4˚C overnight. After washing 3 times with PBS-Tween (0.1%
Tween-20), membranes were incubated with HRP-conjugated goat
anti-rabbit IgG secondary antibodies (1:10,000; Cell Signaling
Technology, Inc.; cat. no. 7074S) at room temperature for 2 h.
Protein bands were visualized using the chemiluminescence kit
(Millipore Sigma; cat. no. WBKLS0500).
Sequencing and bioinformatics
analysis
Total RNA was isolated using RNeasy Mini Kit
(Qiagen) according to the manufacturer's protocol. RNA with an RNA
integrity number >8, as assessed using the Agilent Technologies
2100 Bioanalyzer (Agilent Technologies, Inc.), was used to prepare
the c-DNA library using the Hieff NGS® MaxUP II DNA
Library Prep kit for Illumina (Shanghai Yeasen Biotechnology Co.,
Ltd.; cat. no. 12300) following the manufacturer's protocols.
Sequencing was performed by Novogene Co., Ltd. using the HiSeq X
Ten system (v 2.5; Illumina, Inc.) for 2x150 cycles (paired-end
read length of 150 bp). The loading DNA concentration was 300 pM.
Concentration of libraries were determined using the Agilent
Technologies 2100 Bioanalyzer (Agilent Technologies, Inc.). Reads
that were contaminated by adaptors, exhibited a Phred quality score
(19) <5 accounting for >50%
of total bases and reads with an N content >10% were removed
using Fastp software (https://github.com/OpenGene/fastp; version 0.20.0).
Clean reads were mapped to the human genome (hg19) using TopHat
software (v2.1.0) (20). Kallisto
(version v0.43.0; https://pachterlab.github.io/kallisto) (21) was used to quantify the reads
obtained by sequencing and the sleuth package (22) in the R language was used for
differential expression analysis (22). PC analysis was carried out using
the plotPCA (23), whereas the
heatmap was drawn according to gene expression using the pheatmap
package (https://CRAN.R-project.org/package=pheatmap) in the R
language. The ggplot2 package (https://CRAN.R-project.org/package=ggplot2) was used
to draw the volcano plots. Pearson's correlations were calculated
using the R (v3.6) core function. The threshold for differentially
expressed genes was q val<0.1 and abs(log2FC) >0.
The RIdeogram package was used to determine the chromosomal
localization of the differentially expressed genes (24). The CMAP database collects the gene
expression profiles of human cells treated with multiple single
FDA-approved small-molecule drugs. The gseKEGG function in the
clusterprofiler package (25) was
used to perform Gene Set Enrichment Analysis (GSEA) of the Kyoto
Encyclopedia of Genes and Genomes pathway (threshold, q-values
<0.05) and Connectivity Map (CMAP) analysis on the expression
matrix (threshold, P<0.05).
Statistical analysis
Graphpad Prism Software v. 8.0 (GraphPad Software,
Inc.) was used for statistical analysis. All experiments were
performed in triplicates and data is presented as the mean ± SD.
Single comparisons were performed using paired or unpaired
Student's t-test. The statistical significance data involving ≥
three groups and ≥ two treatments was assessed by two-way ANOVA
with Bonferroni post hoc test. P<0.05 was considered to indicate
a statistically significant difference.
Results
Free miR-124 levels in the blood of
patients with AMI is significantly higher compared with those in
healthy individuals
The peripheral blood of patients with AMI and of
healthy controls was collected. When extracting miRNA, the ethanol
concentration in the cleaning buffer was increased to 80% to
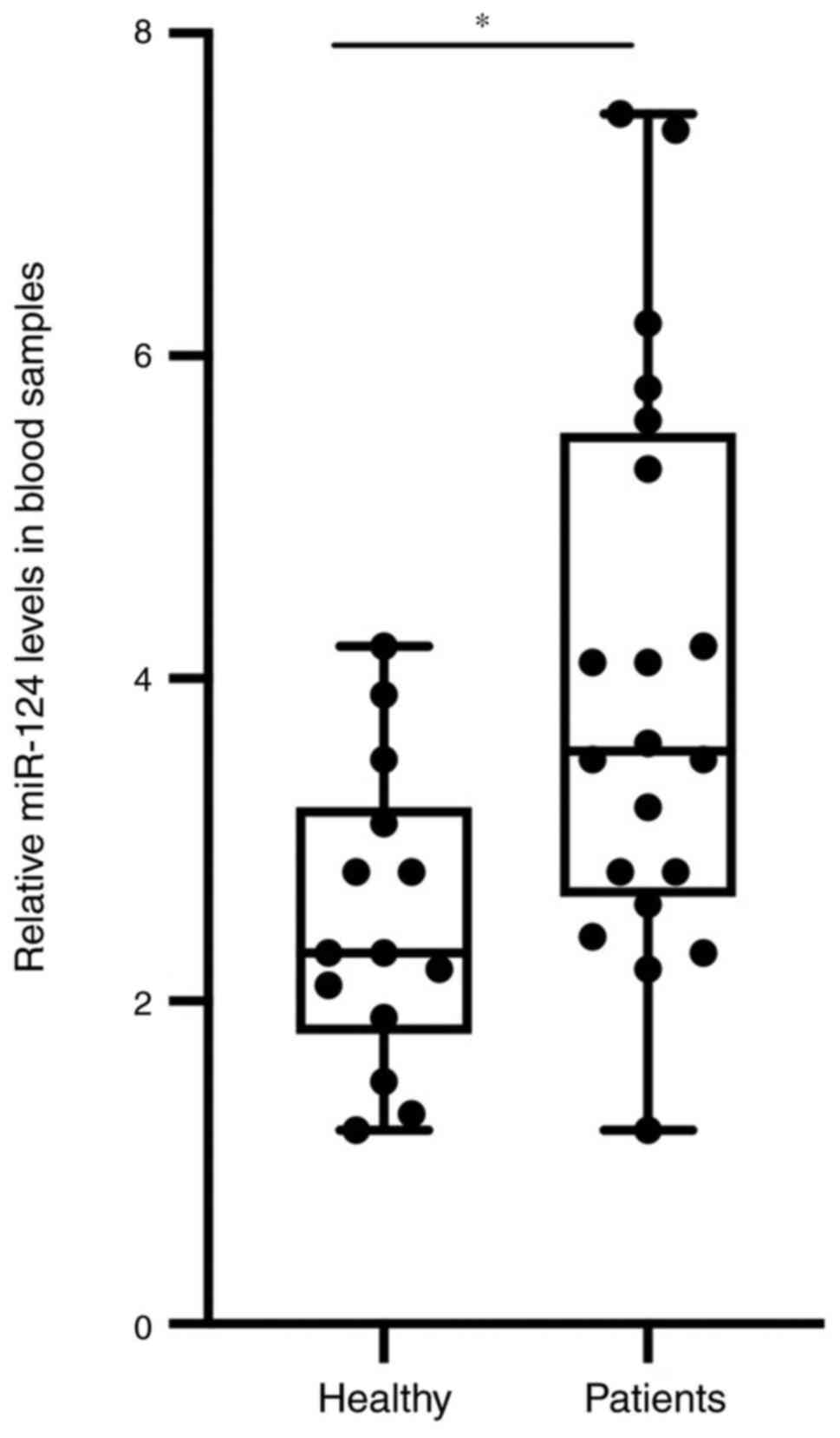
increase the final miRNA concentration. As presented in Fig. 1, the expression level of miR-124 in
the samples of the patients with AMI was significantly higher
compared with that in the healthy control group.
Overexpression of miR-124 can promote
the apoptosis of HUVECs and suppress their proliferation
ability
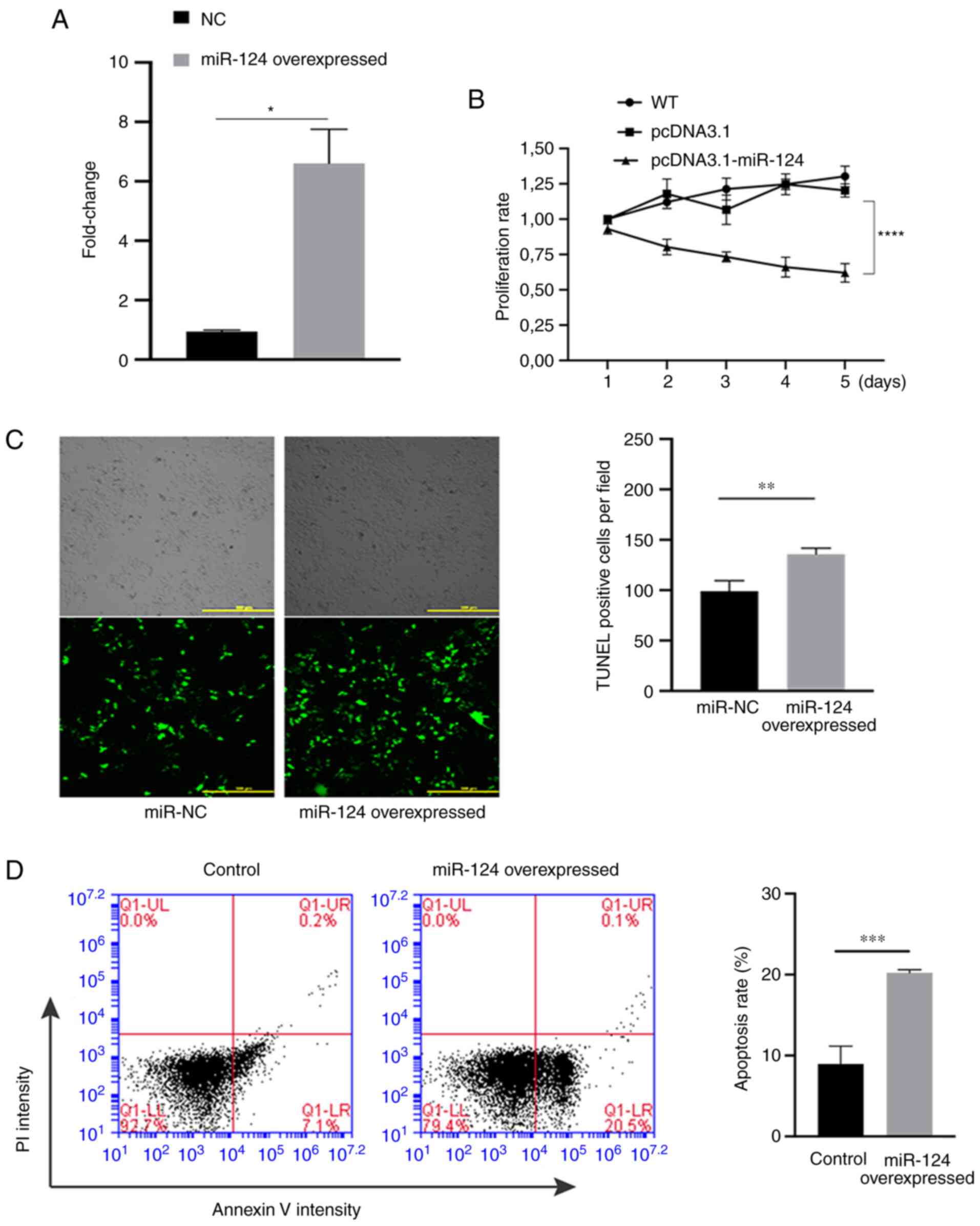
Subsequently, the effects of miR-124 on the
proliferation and apoptosis in HUVECs were evaluated. First,
RT-qPCR was performed on samples from HUVECs that were transfected
with miR-124 overexpression plasmid or empty pcDNA3.1 plasmid. The
experimental results indicated that the expression of miR-124 in
the group transfected with the overexpression plasmid was
significantly higher compared with that in the NC group (Fig. 2A), suggesting that miR-124 was
successfully overexpressed in HUVECs. The proliferation ability of
miR-124-overexpressing HUVECs was also significantly lower compared
with that in the NC group after 5 days (Fig. 2B). TUNEL assay was subsequently
used to detect the effect of miR-124 overexpression on the
apoptotic ability of HUVECs. In terms of apoptosis, the number of
TUNEL-positive cells in the miR-124 overexpression group was
significantly higher compared with that in the empty pcDNA3.1 group
(Fig. 2C), suggesting that
overexpression of miR-124 may promote apoptosis in HUVECs. In
addition, flow cytometry was also performed to measure the
apoptosis, where miR-124 overexpression significantly promoted cell
apoptosis compared with that in the NC group (Fig. 2D).
Overexpression of miR-124 can
significantly change the gene expression profile of HUVECs
To investigate the expression characteristics of
genes related to miR-124, RNA sequencing was performed on the NC
and the miR-124 overexpression groups. Principal component analysis
was then performed to determine the effect of miR-124
overexpression in HUVEC cells. The results indicated that the two
methods of transfections, miR-124 overexpression and NC, formed a
distinct cluster, in which the expression patterns of the two
groups were different (Fig. 3A).
The heatmap was generated using Pearson's correlation analysis,
where differential gene expression was calculated between samples.
The results demonstrated that similarity within
miR-124-overexpressing groups is higher than the similarity between
miR-124-overexpressing group and empty pcDNA3.1 group which means
that they have distinct expression patterns (Fig. 3B). The present results indicated
that the overexpression of miR-124 can significantly alter the gene
expression profile of HUVEC.
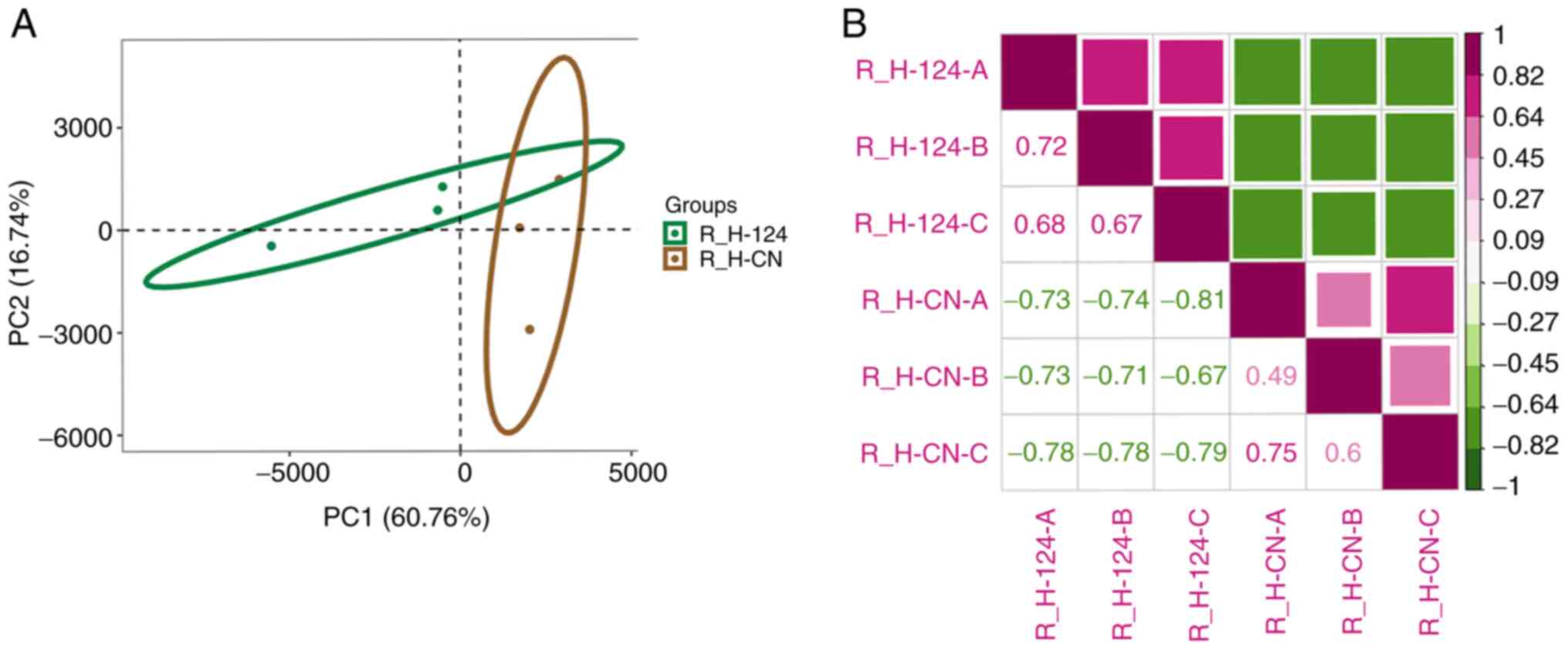 | Figure 3Analysis of gene expression patterns.
(A) plotPCA was used to analyze human umbilical vein endothelial
cell RNA-seq samples that demonstrated tight grouping of biological
replicates. The green oval represents the 95% confidence interval
of principal component distribution in the microRNA
124-overexpression samples. The brown oval represents the 95%
confidence interval of principal component distribution in NC
samples. (B) Pearson's correlation analysis was performed using the
R (v3.6)'s core function. PCA, principal component analysis; NC,
negative control; miR, microRNA; R_H-124-A, R_H-124-B, R_H-124-C
represent microRNA 124-overexpression samples; R_H-CN-A, R_H-CN-B,
R_H-CN-C represent negative control samples. R represents the use
of R language for data analysis, H represents HUVEC cells, and A, B
and C represent three different repeated samples. CN, negative
control. |
Screening of differentially expressed
genes and their chromosomal location analysis
When using the Sleuth package to calculate
differentially expressed genes, q val<0.1 and
abs(log2FC)>0 were used as the screening conditions.
The results revealed a total of 238 differential genes, of which
103 genes were downregulated and 135 genes were upregulated in the
miR-124-overexpressing cells (Fig.
4A). In addition, cluster analysis was performed on these
differentially expressed genes. As shown in Fig. 4B, the differentially expressed
genes presented significantly different expression patterns in the
miR-124-overexpressing and NC groups. For example, the most
downregulated genes were SLC48A1, ALPP and PPL. The most
upregulated genes were IL1A, HMGA2 and SPOCK1. Chromosomal location
analysis using the RIdeogram package was used to determine the
chromosomal localization of the differentially expressed genes
(Fig. 4C). The differentially
expressed genes were found to be scattered on all chromosomes
except the Y chromosome. Among them, chromosome 1 contained the
highest distribution of genes with 29 (accounting for 12.2%),
followed by 22% on chromosome 12 and 16% on chromosome 19. There
were no differentially expressed genes on chromosome Y, which could
mean that miR-124 overexpression did not mediate any effects on
gene expression on the Y chromosome.
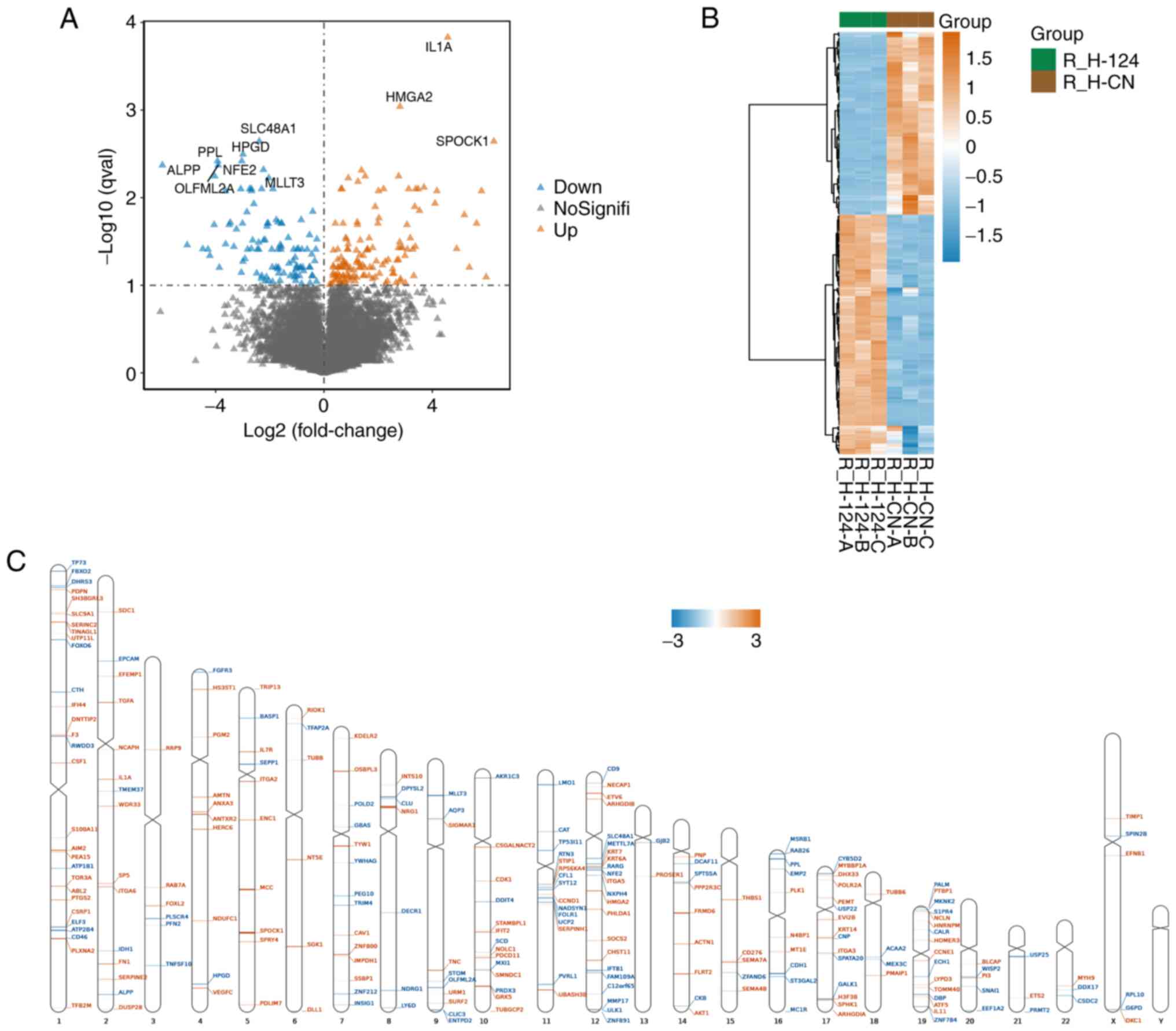 | Figure 4Analysis of differentially expressed
genes (A) Volcano plot of the differentially expressed genes in
human umbilical vein endothelial cells overexpressing miR-124 or a
NC based on their expression fold change [thresholds, q val<0.1
and abs(log2FC)>0]. (B) Heatmap and hierarchical clustering
analysis of the differentially expressed genes. In samples that
were not classified in advance, hierarchical cluster analysis
revealed a different hierarchical clustering algorithm. (C)
Distribution of differentially expressed genes associated with
miR-124 overexpression on each chromosome. Chromosome distribution
indicated that upregulated and downregulated mRNAs were spread on
different chromosomal locations. Brown represents upregulated genes
in microRNA 124-overexpression samples. Blue represents
downregulated genes in microRNA 124-overexpression samples.
R_H-124-A, R_H-124-B, R_H-124-C represent microRNA
124-overexpression samples; R_H-CN-A, R_H-CN-B, R_H-CN-C represent
negative control samples R represents the use of R language for
data analysis, H represents HUVEC cells, and A, B and C represent
three different repeated samples. CN, negative control. |
Enrichment analysis and CMAP analysis
of differentially expressed genes
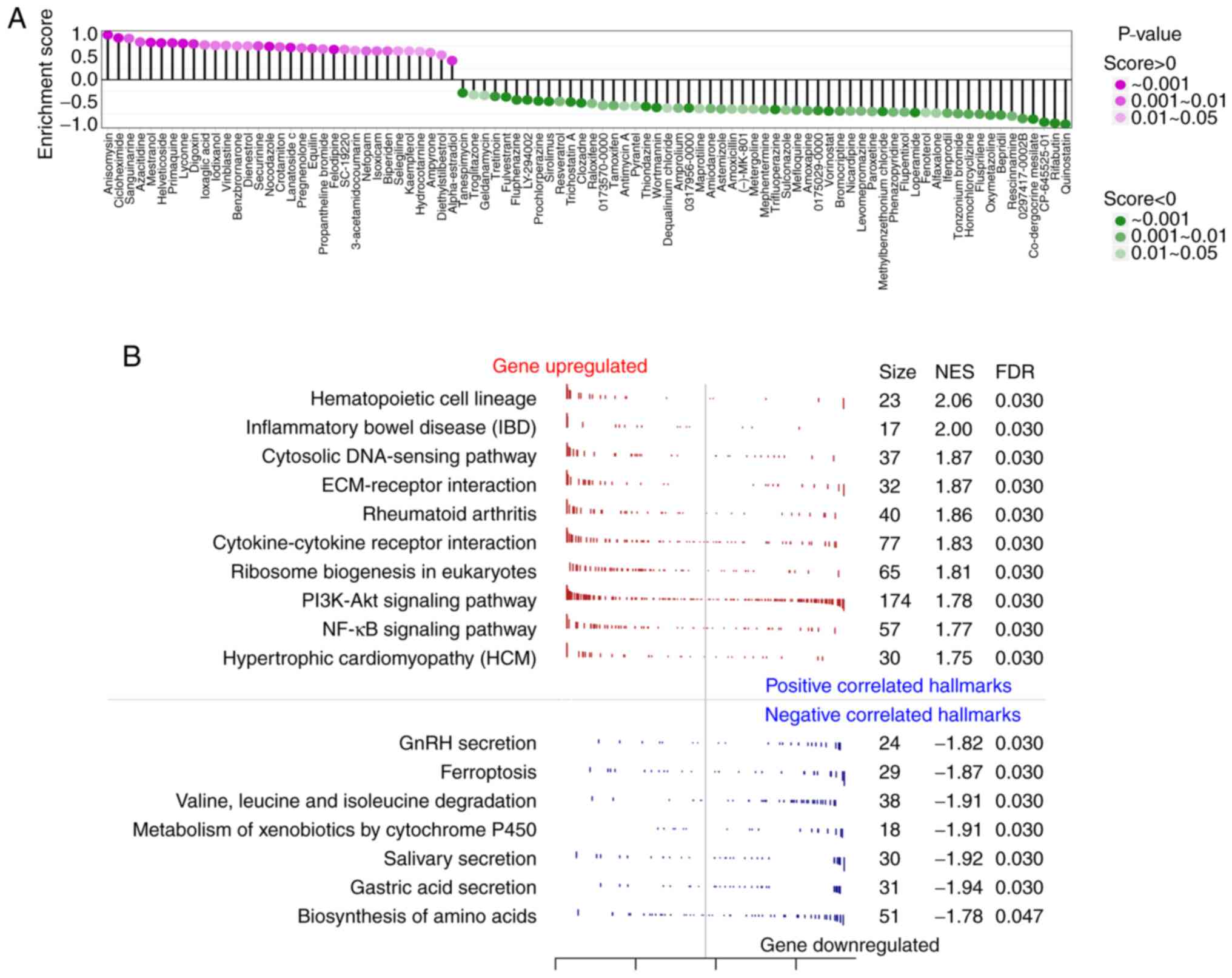
Small-molecule drugs causing gene expression changes
in human cells similar to miR-124 overexpression were researched
using the CMAP database, where total of 90 small-molecule drugs
with statistically significant similarity were found (Fig. 5A). This analysis was performed to
screen for drug candidates that could block miR-124 from causing
vascular endothelial cell damage. Among the three drugs with
enrichment scores >0.9, anisomycin and sanguinarine are reported
to be agonists of the p38/MAPK signaling pathway and could inhibit
angiogenesis, an unfavorable factor for the recurrence of
myocardial infarction (26). This
suggested that the results of CMAP were relatively accurate. The
present analysis also suggested that the three drugs with the
lowest enrichment score, CP-645525-01, rifabutin and quinostatin,
had the potential to promote angiogenesis and to reduce the risk of
myocardial infarction recurrence (Fig.
5A). GSEA was performed to further investigate the inhibitory
effect of miR-124 on angiogenesis. As presented in Fig. 5B, 17 pathways (qvalue <0.05)
were obtained, which were associated with the decrease of HUVEC
proliferation after miR-124 overexpression. Among them, the pathway
that included the most related genes was the PI3K-AKT signaling
pathway. The peak in the GSEA results of the PI3K-AKT signaling
pathway shifted left, with NES=1.78, suggesting that most of the
genes in the PI3K-AKT signaling pathway were downregulated.
Meanwhile, the upregulated genes were mainly enriched in various
metabolism-related signaling pathways, including the metabolism of
xenobiotics via the cytochrome P450 pathway.
miR-124 can influence the p38/MAPK and
PI3K/AKT pathways
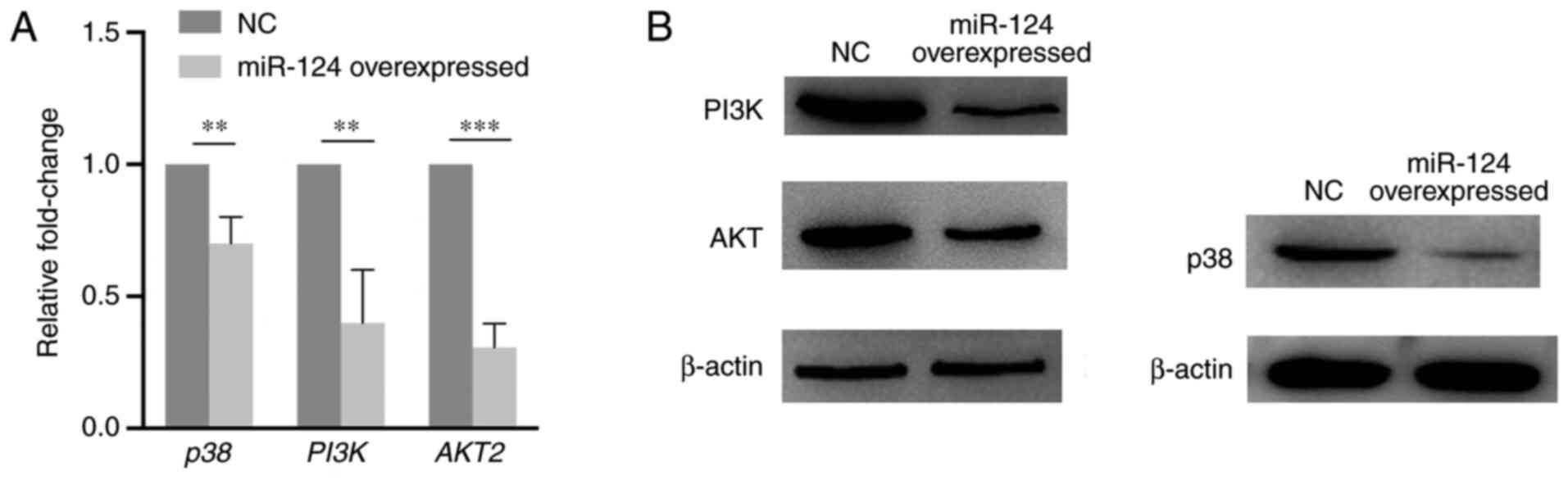
RT-qPCR and western blotting assays were performed
to explore the expression change of key components of the p38/MAPK
and PI3K/AKT pathways caused by miR-124 overexpression in HUVECs.
As presented in Fig. 6, PI3K, AKT
and p38 expression levels significantly decreased on both mRNA and
protein levels compared with that in the NC group. The present
results suggest that miR-124 can influence the p38/MAPK and
PI3K/AKT pathways.
Discussion
In the present study, the effects of miR-124
overexpression on gene expression in HUVECs were explored. Previous
studies on the role of miR-124 suggest that it can regulate
cardiomyocyte apoptosis and myocardial infarction (27). Furthermore, it was shown that
peripheral blood miR-124 can be used to predict AMI (28). In AMI, the regenerative capacity of
blood vessels is an important indicator for evaluating myocardial
recovery (29,30). Previous studies found miR-124 to be
mainly expressed in the human brain, suggesting that its functions
are considered to be related to neural differentiation (31,32).
A previous study demonstrated that miR-124 could target the nuclear
receptor subfamily 3 group C member 2 gene and regulate the
renin-angiotensin-aldosterone system, which is mainly associated
with the regulation of blood pressure (33). In addition, it was demonstrated
that miR-124 expression is associated with impaired cardiac
function (27). For this reason,
clinical samples were obtained to detect the expression of miR-124
in patients with AMI in the present study, which revealed that
miR-124 expression in patients with AMI was significantly higher
compared with that in the healthy control group. Therefore, the
present results strongly indicated that miR-124 function is related
to AMI pathophysiology.
The present study also found that miR-124
overexpression markedly altered gene expression in HUVECs. Compared
with those in the NC group, there were ≥238 differentially
expressed genes; which were relatively enriched in the PI3K/AKT
signaling pathway. AKT serves an important role in regulating
postnatal cardiac development, myocardial angiogenesis and cell
death in cardiac myocytes (34).
In a clinical setting, PI3K/AKT signaling activators are frequently
used as therapeutic options for clinical improvement of cardiac
related diseases (35). By
contrast, an increasing number of biological macromolecules, such
as 17 beta-estradiol (36) and
TO901317(37), have been found to
activate the PI3K/AKT signaling pathway. It was previously reported
that high mobility group box protein 1 could protect the heart from
ischemia-reperfusion injury by the PI3K/AKT pathway-mediated
upregulation of VEGF expression (38). There are a number of other similar
reports in which there is a close correlation between the
expression of PI3K/AKT and the heart-related disease (39-42).
The restoration of myocardial blood supply is an
important therapeutic strategy for AMI recovery (39). Therefore, the extent of
angiogenesis mediated by vascular endothelial cells has become an
important indicator for evaluating the prognosis of AMI (43). The PI3K/AKT signaling pathway is an
important anti-apoptotic signaling pathway, for example, in
colorectal, gastric and breast cancer as well as others (44). Blocking the PI3K/AKT/mTOR signaling
pathway can not only induce cell cycle arrest, apoptosis and
autophagy, but can also inhibit prostate tumor cell angiogenesis
(45). In this regard, a number of
organic and biological agents have been reported to regulate
angiogenesis through the expression of genes in this signaling
pathway. Celastrol can inhibit the angiogenic ability of glioma by
inhibiting the expression and activation of hypoxia inducible
factor-1α, PI3K, AKT and mTOR, thereby inhibiting angiogenesis
(46). Additionally, miR-9 can
regulate the PI3K/AKT signaling pathway by targeting the transient
receptor potential cation channel subfamily M member 7 gene to
promote the angiogenesis of endothelial progenitor cells (47). miR-221 can also regulate the
function of endothelial cells, including HUVECs, through the
PI3K/AKT signaling pathway (48).
Overexpression of miR-221 can significantly improve the angiogenic
ability of HUVECs, whilst the inhibition of miR-221 directly
increased the apoptosis rate of HUVECs (48). Low-level laser therapy has also
been shown to increase the phosphorylation levels of PI3K, AKT and
mTOR proteins, where weakening or blocking the PI3K signaling
pathway could significantly reduce the proliferation, migration and
angiogenesis of HUVECs (49). In
the present study, similar functions of miR-124 on PI3K/AKT
pathways were identified in HUVECs. Therefore, in future studies,
the role of the miR-124/PI3K/AKT axis in HUVEC angiogenesis
warrants further study.
Although the present study provided evidence for the
association between high miR-124 levels in peripheral blood and the
occurrence of AMI, the potential mechanism in which miR-124 can
regulate the PI3K/AKT signaling pathway and affect the
proliferation of vascular endothelial cells remain unclear.
Therefore, present results need to be interpreted with caution.
The current study constructed a connectivity map
database between small molecules and pathways, and analyzed the
topological properties of this map. Anisomycin and sanguinarine had
the highest positive scores, meaning that Anisomycin and
sanguinarine may suppress angiogenesis by inhibiting the PI3K/AKT
signaling pathway, which was consistent with the results of
previous reports indicating that they may be inhibitors of p38 MAPK
activation (26,50). Additionally, rifabutin and
quinostatin had the lowest scores, indicating that quinostatin is
an inhibitor of the PI3K (51),
while rifabutin is a wide spectrum antibiotic that is particularly
effective on atypical and rifampicin-resistant mycobacteria
(52). However, their potential
roles in promoting angiogenesis and reducing the risk of myocardial
infarction recurrence requires further study.
In addition, the present study has some limitations.
A small cohort of patients was included, rendering the power of
confidence insufficient. In future studies, the size of the cohort
will need to be expanded. The present study also did not consider
the blood pressure, a setting of diabetes, drug history or a
history of other related diseases. Therefore, it is not possible to
completely rule out that the observed differential expression of
miR-124 in the blood samples was caused by the confounding factors
aforementioned. Therefore, in future studies, stratified analysis
will be conducted based on these potential confounding factors to
determine the role of the miR-124/PI3K/AKT signaling pathway axis
in angiogenesis to improve the recovery of patients with AMI.
Acknowledgements
No applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus database
repository (accession no. GSE180765), (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180765).
Authors' contributions
WM performed all the experiments. XZ performed
clinic sample collection and preparation. YL performed
bioinformatics analysis. All authors confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the ethics
committees of Weihai Central Hospital (approval no. WKY-2019-012).
Informed consent was provided by all patients, and written
permission was obtained before inclusion.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arora S, Stouffer GA, Kucharska-Newton AM,
Qamar A, Vaduganathan M, Pandey A, Porterfield D, Blankstein R,
Rosamond WD, Bhatt DL and Caughey MC: Twenty year trends and sex
differences in young adults hospitalized with acute myocardial
infarction. Circulation. 139:1047–1056. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Libby P: Mechanisms of acute coronary
syndromes and their implications for therapy. N Engl J Med.
368:2004–2013. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ferro G, Duilio C, Spinelli L, Liucci GA,
Mazza F and Indolfi C: Relation between diastolic perfusion time
and coronary artery stenosis during stress-induced myocardial
ischemia. Circulation. 92:342–347. 1995.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Martín Giménez VM, de Las Heras N, Ferder
L, Lahera V, Reiter RJ and Manucha W: Potential effects of
melatonin and micronutrients on mitochondrial dysfunction during a
cytokine storm typical of oxidative/inflammatory diseases.
Diseases. 9(30)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Musher DM, Abers MS and Corrales-Medina
VF: Acute infection and myocardial infarction. N Engl J Med.
380:171–176. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ardekani AM and Naeini MM: The role of
MicroRNAs in human diseases. Avicenna J Med Biotechnol. 2:161–179.
2010.PubMed/NCBI
|
|
7
|
Condrat CE, Thompson DC, Barbu MG, Bugnar
OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM and Voinea SC: MiRNAs
as biomarkers in disease: Latest findings regarding their role in
diagnosis and prognosis. Cells. 9(276)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen J and Wang DZ: MicroRNAs in
cardiovascular development. J Mol Cell Cardiol. 52:949–957.
2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Silva DCPD, Carneiro FD, Almeida KC and
Fernandes-Santos C: Role of miRNAs on the pathophysiology of
cardiovascular diseases. Arq Bras Cardiol. 111:738–746.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Luo F, Wu P, Chen J, Guo Y, Wang J, Li X
and Fang Z: ANGPTL3 possibly promotes cardiac angiogenesis through
improving proangiogenic ability of endothelial progenitor cells
after myocardial infarction. Lipids Health Dis.
17(184)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Su T and Shao X, Zhang X, Yang C and Shao
X: Value of circulating miRNA-1 detected within 3 h after the onset
of acute chest pain in the diagnosis and prognosis of acute
myocardial infarction. Int J Cardiol. 307:146–151. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pinchi E, Frati P, Aromatario M, Cipolloni
L, Fabbri M, La Russa R, Maiese A, Neri M, Santurro A, Scopetti M,
et al: MiR-1, miR-499 and miR-208 are sensitive markers to diagnose
sudden death due to early acute myocardial infarction. J Cell Mol
Med. 23:6005–6016. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yin Y, Lv L and Wang W: Expression of
miRNA-214 in the sera of elderly patients with acute myocardial
infarction and its effect on cardiomyocyte apoptosis. Exp Ther Med.
17:4657–4662. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yücel EI and Sahin M: Fenretinide reduces
angiogenesis by downregulating CDH5, FOXM1 and eNOS genes and
suppressing microRNA-10b. Mol Biol Rep. 47:1649–1658.
2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li X, Li Z, Zhu Y, Li Z, Yao L, Zhang L,
Yuan L, Shang Y, Liu J and Li C: MiR-524-5p inhibits angiogenesis
through targeting WNK1 in colon cancer cells. Am J Physiol
Gastrointest Liver Physiol. 318:G827–G839. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lei Z, Klasson TD, Brandt MM, van de Hoek
G, Logister I, Cheng C, Doevendans PA, Sluijter JPG and Giles RH:
Control of angiogenesis via a VHL/miR-212/132 axis. Cells.
9(1017)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–410.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Blankenberg D, Gordon A, Von Kuster G,
Coraor N, Taylor J and Nekrutenko A: Galaxy Team. Manipulation of
FASTQ data with galaxy. Bioinformatics. 26:1783–1788.
2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and cufflinks. Nat Protoc. 7:562–578.
2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bray NL, Pimentel H, Melsted P and Pachter
L: Near-optimal probabilistic RNA-seq quantification. Nat
Biotechnol. 34:525–527. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Pimentel H, Bray NL, Puente S, Melsted P
and Pachter L: Differential analysis of RNA-seq incorporating
quantification uncertainty. Nat Methods. 14:687–690.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G
and Chen J: RIdeogram: Drawing SVG graphics to visualize and map
genome-wide data on the idiograms. PeerJ Comput Sci.
6(e251)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chai W, Wu Y, Li G, Cao W, Yang Z and Liu
Z: Activation of p38 mitogen-activated protein kinase abolishes
insulin-mediated myocardial protection against ischemia-reperfusion
injury. Am J Physiol Endocrinol Metab. 294:E183–E189.
2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Han F, Chen Q, Su J, Zheng A, Chen K, Sun
S, Wu H, Jiang L, Xu X, Yang M, et al: MicroRNA-124 regulates
cardiomyocyte apoptosis and myocardial infarction through targeting
Dhcr24. J Mol Cell Cardiol. 132:178–188. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guo ML, Guo LL and Weng YQ: Implication of
peripheral blood miRNA-124 in predicting acute myocardial
infarction. Eur Rev Med Pharmacol Sci. 21:1054–1059.
2017.PubMed/NCBI
|
|
29
|
Lazar E, Benedek T, Korodi S, Rat N, Lo J
and Benedek I: Stem cell-derived exosomes-an emerging tool for
myocardial regeneration. World J Stem Cells. 10:106–115.
2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sondergaard C, Hess DA, Maxwell DJ,
Weinheimer C, Rosová I, Creer MH, Piwnica-Worms D, Kovacs A,
Pedersen L and Nolta JA: Human cord blood progenitors with high
aldehyde dehydrogenase activity improve vascular density in a model
of acute myocardial infarction. J Transl Med. 8(24)2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain-specific alternative Pre-mRNA splicing. Mol
Cell. 27:435–448. 2007.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Angelopoulou E, Paudel YN and Piperi C:
MiR-124 and Parkinson's disease: A biomarker with therapeutic
potential. Pharmacol Res. 150(104515)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sõber S, Laan M and Annilo T: MicroRNAs
miR-124 and miR-135a are potential regulators of the
mineralocorticoid receptor gene (NR3C2) expression. Biochem Biophys
Res Commun. 391:727–732. 2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hao Q, Zhang F, Wang Y, Li Y and Qi X:
Cardiac contractility modulation attenuates chronic heart failure
in a rabbit model via the PI3K/AKT pathway. Biomed Res Int.
2020(1625362)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang X, Chen L, Zhao X, Xiao L, Yi S, Kong
Y, Jiang Y and Zhang J: A cathelicidin-related antimicrobial
peptide suppresses cardiac hypertrophy induced by pressure overload
by regulating IGFR1/PI3K/AKT and TLR9/AMPKα. Cell Death Dis.
11(96)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Guo RX, Wei LH, Tu Z, Sun PM, Wang JL,
Zhao D, Li XP and Tang JM: 17 beta-estradiol activates PI3K/Akt
signaling pathway by estrogen receptor (ER)-dependent and
ER-independent mechanisms in endometrial cancer cells. J Steroid
Biochem Mol Biol. 99:9–18. 2006.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chen J, Zacharek A, Cui X, Shehadah A,
Jiang H, Roberts C, Lu M and Chopp M: Treatment of stroke with a
synthetic liver X receptor agonist, TO901317, promotes synaptic
plasticity and axonal regeneration in mice. J Cereb Blood Flow
Metab. 30:102–109. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhou YH, Han QF, Gao L, Sun Y, Tang ZW,
Wang M, Wang W and Yao HC: HMGB1 protects the heart against
ischemia-reperfusion injury via PI3K/AkT pathway-mediated
upregulation of VEGF expression. Front Physiol.
10(1595)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chen X, Wang R, Chen W, Lai L and Li Z:
Decoy receptor-3 regulates inflammation and apoptosis via PI3K/AKT
signaling pathway in coronary heart disease. Exp Ther Med.
17:2614–2622. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cheng S, Zhang X, Feng Q, Chen J, Shen L,
Yu P, Yang L, Chen D, Zhang H, Sun W and Chen X: Astragaloside IV
exerts angiogenesis and cardioprotection after myocardial
infarction via regulating PTEN/PI3K/Akt signaling pathway. Life
Sci. 227:82–93. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xing X, Guo S, Zhang G, Liu Y, Bi S, Wang
X and Lu Q: MiR-26a-5p protects against myocardial
ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT
signaling pathway. Braz J Med Biol Res. 53(e9106)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Coco D and Leanza S: A review on aorta
mesenteric bypass in surgical management of mesenteric ischemia:
Indications, techniques and outcomes. Maedica (Bucur). 15:381–390.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
van der Laan AM, Piek JJ and van Royen N:
Targeting angiogenesis to restore the microcirculation after
reperfused MI. Nat Rev Cardiol. 6:515–523. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gao N, Zhang Z, Jiang BH and Shi X: Role
of PI3K/AKT/mTOR signaling in the cell cycle progression of human
prostate cancer. Biochem Biophys Res Commun. 310:1124–1132.
2003.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhu Y, Liu X, Zhao P, Zhao H, Gao W and
Wang L: Celastrol suppresses glioma vasculogenic mimicry formation
and angiogenesis by blocking the PI3K/Akt/mTOR signaling pathway.
Front Pharmacol. 11(25)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhou DM, Sun LL, Zhu J, Chen B, Li XQ and
Li WD: MiR-9 promotes angiogenesis of endothelial progenitor cell
to facilitate thrombi recanalization via targeting TRPM7 through
PI3K/Akt/autophagy pathway. J Cell Mol Med. 24:4624–4632.
2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Peng H, Yang H, Xiang X and Li S:
ΜicroRNA-221 participates in cerebral ischemic stroke by modulating
endothelial cell function by regulating the PTEN/PI3K/AKT pathway.
Exp Ther Med. 19:443–450. 2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li Y, Xu Q, Shi M, Gan P, Huang Q, Wang A,
Tan G, Fang Y and Liao H: Low-level laser therapy induces human
umbilical vascular endothelial cell proliferation, migration and
tube formation through activating the PI3K/Akt signaling pathway.
Microvasc Res. 129(103959)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Niu X, Fan T, Li W, Xing W and Huang H:
The anti-inflammatory effects of sanguinarine and its modulation of
inflammatory mediators from peritoneal macrophages. Eur J
Pharmacol. 689:262–269. 2012.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Yang J, Shamji A, Matchacheep S and
Schreiber SL: Identification of a small-molecule inhibitor of class
Ia PI3Ks with cell-based screening. Chem Biol. 14:371–377.
2007.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Brughera M, Scampini G, Newman AJ,
Castellino S, Sammartini U and Mazué G: Overview of toxicological
data on rifabutin. Exp Toxicol Pathol. 47:1–9. 1995.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Tiedemann RE, Schmidt J, Keats JJ, Shi CX,
Zhu YX, Palmer SE, Mao X, Schimmer AD and Stewart AK:
Identification of a potent natural triterpenoid inhibitor of
proteosome chymotrypsin-like activity and NF-kappaB with
antimyeloma activity in vitro and in vivo. Blood. 113:4027–4037.
2009.PubMed/NCBI View Article : Google Scholar
|