Introduction
Pancreatic cancer is a common malignancy of the
digestive system and usually presents as ductal adenocarcinoma
originating from the pancreatic duct epithelium (1). The incidence of pancreatic cancer has
been on the rise due to the lifestyle changes in modern society
(2). The current understanding of
pancreatic carcinogenesis is limited, but habits such as cigarette
smoking, alcohol intake, high-fat diet and excessive consumption of
caffeine have been identified as some of the contributing factors
(3). The 5-year survival rate of
pancreatic cancer after diagnosis is reportedly as low as ~9%,
rendering it one of the malignancies with the poorest prognosis
(4,5). In addition to the strong propensity
of pancreatic cancer for metastasis, patients with early-stage
disease are often asymptomatic; both these factors contribute to
the high mortality rate of this malignancy and, in numerous cases,
delay diagnosis until the disease is at an advanced stage, when
effective treatment options are limited (6-8).
In recent years, gene therapy based on gene signal transduction
suppression for pancreatic cancer has entered the clinical trial
stage as an innovative treatment (9). The screening of core genes in
pancreatic cancer in combination with the development of
conventional chemotherapy and radiotherapy is essential for
accurate targeting of the oncogenes in gene therapy (10,11).
Bone marrow stromal cell antigen 2 (BST2), also
known as CD317/tetherin, is a type II transmembrane protein that is
widely expressed by bone marrow stromal cells, B cells, T cells and
natural killer cells (12).
Notably, it has been established that certain immunocytes and
malignant cells, such as B-cell chronic lymphoid leukemia cells and
pulmonary cancer cells, also exhibit differential expression of
BST2 (13,14). More importantly, the bioinformatics
database Gene Expression Profiling Interactive Analysis (GEPIA;
http://gepia2.cancer-pku.cn) has
demonstrated BST2 upregulation in pancreatic cancer and a distinct
association between high BST2 expression and lower overall
survival. In addition, the transcription factor specificity protein
1 (SP1) has been shown to play a key role in pancreatic cancer, and
high SP1 expression has been reported to be a key tumorigenic
factor by several studies on pancreatic tumorigenesis (15-17).
Thus, the present study was undertaken to investigate the
association of BST2 with pancreatic cancer occurrence and
development, and determine whether transcriptional regulation by
SP1 is involved in this process.
Materials and methods
Cell culture and treatment
A human pancreatic duct epithelial cell line
(HPDE6-C7) and pancreatic cancer cell lines (SW1990, BxPC3, PANC1
and PSN-1) were purchased from EK-Biosciences GmbH. HPDE6-C7 cells
were cultured in 89% DMEM supplemented with 10% FBS and 1%
penicillin-streptomycin solution (P/S); SW1990 cells were cultured
in Leibovitz L15 medium supplemented with 10% FBS and 1% P/S (in an
environment free of CO2 at 37˚C); BxPC3 cells were
cultured in RPMI-1640 medium supplemented with 10% FBS and 1% P/S;
and PANC1 and PSN-1 cells were cultured in RPMI-1640 medium
supplemented with 15% FBS, 0.01 mg/ml insulin and 1% P/S. All the
aforementioned reagents were purchased from Thermo Fisher
Scientific, Inc.
Cell transfection
Small interfering RNA (siRNA) plasmids specific for
BST2 (siBST2-1: 5'-GGCATCTACTTG TATGACTATT-3'; siBST2-2:
5'-TCCTTTGGATGGCCT AGTACTAG-3'), empty siRNA vector negative
control (siNC: 5'-GTAAGCTTCTGCTGGGGATAGG-3'), wild-type BST2
(BST2-WT) (5'-CAGGCCCCGCCCCCA-3') and mutant BST2 (BST2-MUT)
(5'-CAAGCCGGAGGUUUG-3') promoter, siRNA plasmids targeting SP1
(siSP1-1: 5'-CCGAAACC TTCTGACTACTAACC-3'; siSP1-2: 5'-ATGCCTAATAT
TC AGTATCAAGT-3'), pcDNA 3.1(+)/SP1 [overexpression (OE)-SP1)] and
empty pcDNA 3.1(+) vectors were constructed by Guangzhou RiboBio
Co., Ltd. PANC-1 cell transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. Cells were
harvested following 48 h of transfection at 37˚C and the
transfection efficiency was determined using RT-qPCR. The
subsequent experimentation was conducted within 48 h.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from transfected cells by
means of the spin column-based method using the MolPure®
Cell RNA kit (Shanghai Yeasen Biotechnology Co., Ltd.).
Subsequently, cDNA was synthesized at 42˚C for 30 min using a
PrimeScript™ RT Reagent kit (Takara Bio, Inc.). The PCR
system was set in accordance with the instructions of
BeyoFast™ Probe qPCR Mix (Beyotime Institute of
Biotechnology). After pre-denaturation and amplification of the
template (initial denaturation at 95˚C for 10 min; followed by 40
cycles of denaturation at 95˚C for 15 sec and annealing at 60˚C for
1 min; and a final extension of 10 min at 72˚C), the results were
analyzed using software provided by a fluorescent quantitative PCR
instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The 2-ΔΔCq method was used to compare relative
expression levels (18). The
following primer pairs were used: BST2 forward,
5'-AGCGACTGAGAAGAGAAA ACCA-3' and reverse,
5'-TGTTCAAGCGAAAAGCCGAG-3'; and U6 forward,
5'-AAAGCAAATCATCGGACGACC-3' and reverse:
5'-GTACAACACATTGTTTCCTCGGA-3'.
Western blotting
Total proteins were extracted from transfected cells
using the RIPA lysis buffer (Shanghai Yeasen Biotechnology Co.,
Ltd.) followed by protein quantification using a BCA kit (Shanghai
Enzyme-linked Biotechnology Co., Ltd.) and protein separation was
performed on 10% gels using SDS-PAGE. PVDF membranes containing the
proteins were blocked using 5% skimmed milk for 1 h at room
temperature and incubated with anti-BST2 (1:1,000; cat no.
ab243230), anti-SP1 (1:1,000; cat no. ab227383), anti-Ki67
(1:1,000; cat no. ab16667), anti-proliferating cell nuclear antigen
(anti-PCNA; 1:1,000; cat no. ab18197), anti-MMP2 (1:1,000; cat no.
ab92536) and anti-MMP9 (1:1,000; cat no. ab38898) primary
antibodies (all from Abcam) at 4˚C overnight. Mouse anti-rabbit IgG
HRP-conjugated secondary antibody (1:1,000; cat no. sc-2357; Santa
Cruz Biotechnology, Inc.) was used for subsequent incubation with 2
h at room temperature. Protein bands were visualized using
Immobilon™ Chemiluminescent HRP substrate
(MilliporeSigma).
Analysis of cell proliferation
To evaluate cell colony formation ability,
transfected cells were plated into a 6-well plate (1x104
cells/well) and the cells were allowed to grow for 10 days at 37˚C.
The colonies were observed after 3.7% paraformaldehyde fixation for
10 min at room temperature and 0.2% crystal violet staining
(Shanghai Aladdin Biochemical Technology Co., Ltd.) for 5 min at
room temperature.
A Cell Counting Kit-8 (CCK-8) assay (Beijing
Solarbio Science & Technology Co., Ltd.) was used to assess
cell proliferation. Briefly, transfected cells were incubated with
CCK-8 solution for another 4 h at 37˚C before the reading of
OD450 using a microplate reader (Bio-Rad Laboratories,
Inc.).
Analysis of cell migration
Transfected cells (3x105 cells/well) were
cultured in 6-well culture plates to achieve 80-90% confluence. The
cells were then incubated overnight at 37˚C with serum-free
RPMI-1640 medium. A linear scratch was created in the cell
monolayer using the tip of a 200-µl pipette. At 0 and 24 h of wound
healing, cell migration was observed using a light microscope
(magnification, x100).
Analysis of cell invasion
Transfected cells were seeded into the upper chamber
of a Transwell insert pre-coated (at 37˚C overnight) with Matrigel
(Corning, Inc.) in serum-free RPMI-1640 medium. Complete medium
containing 10% FBS was added to the lower chamber. Following a 12-h
incubation at 37˚C, the invading cells in lower chamber were fixed
with 4% paraformaldehyde for 15 min at 37˚C, followed by staining
with 0.1% crystal violet solution for 10 min at 37˚C. Cell invasion
was evaluated by counting the cells under a light microscope
(magnification, x100).
Bioinformatics analysis
The expression of BST2 in pancreatic cancer was
analyzed using the GEPIA database (http://gepia2.cancer-pku.cn), which is a newly
developed interactive web server for analyzing the 9,736 tumors and
8,587 normal samples from The Cancer Genome Atlas and the
Genotype-Tissue Expression project, using a standard processing
pipeline. The JASPAR database 2020 (http://jaspar.genereg.net/) predicted sequence
matching between the transcription factor SP1 and BST2.
Verification of SP1-BST2 binding
For chromatin immunoprecipitation (ChIP), 1%
formaldehyde was added to the culture medium to fix PANC1 cells for
12 min at room temperature, and PBS-washed cells were harvested at
4˚C with 300 x g for 5 min and resuspended in hypotonic buffer
supplemented with 10 mM ethylene diamine tetraacetic acid (EDTA).
After collection of the nuclei using centrifugation at 4˚C with
15,000 x g for 1 min and resuspension in the dilution buffer, 60 µl
Protein A/G (PrimeGene; Bio-Techne) was added to the DNA solution
for antibody incubation. Following elution of the Protein A/G
beads, DNA sequences binding to IgG or SP1 were detected via
RT-qPCR.
For the luciferase reporter assay, PANC1 cells were
co-transfected with siSP1-2 (50 nM; Guangzhou RiboBio Co., Ltd.) or
si-NC (50 nM; Guangzhou RiboBio Co., Ltd.) and 50 ng BST2-WT or
BST2-MUT. Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) was used for transfection. After 48 h of transfection at 37˚C
with the luciferase reporter vector (Promega Corporation), a Dual
Luciferase Reporter Assay kit (Promega Corporation) was used to
evaluate the relative luciferase signals. Relative luciferase
activity was expressed as the ratio of the firefly luciferase
activity to that of the Renilla luciferase activity.
Statistical analysis
Experiments in this study were repeated in
triplicate. The data are expressed as the mean ± SD and were
analyzed using one-way ANOVA followed by Dunnett's and Tukey's post
hoc tests, as appropriate, or using an unpaired Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
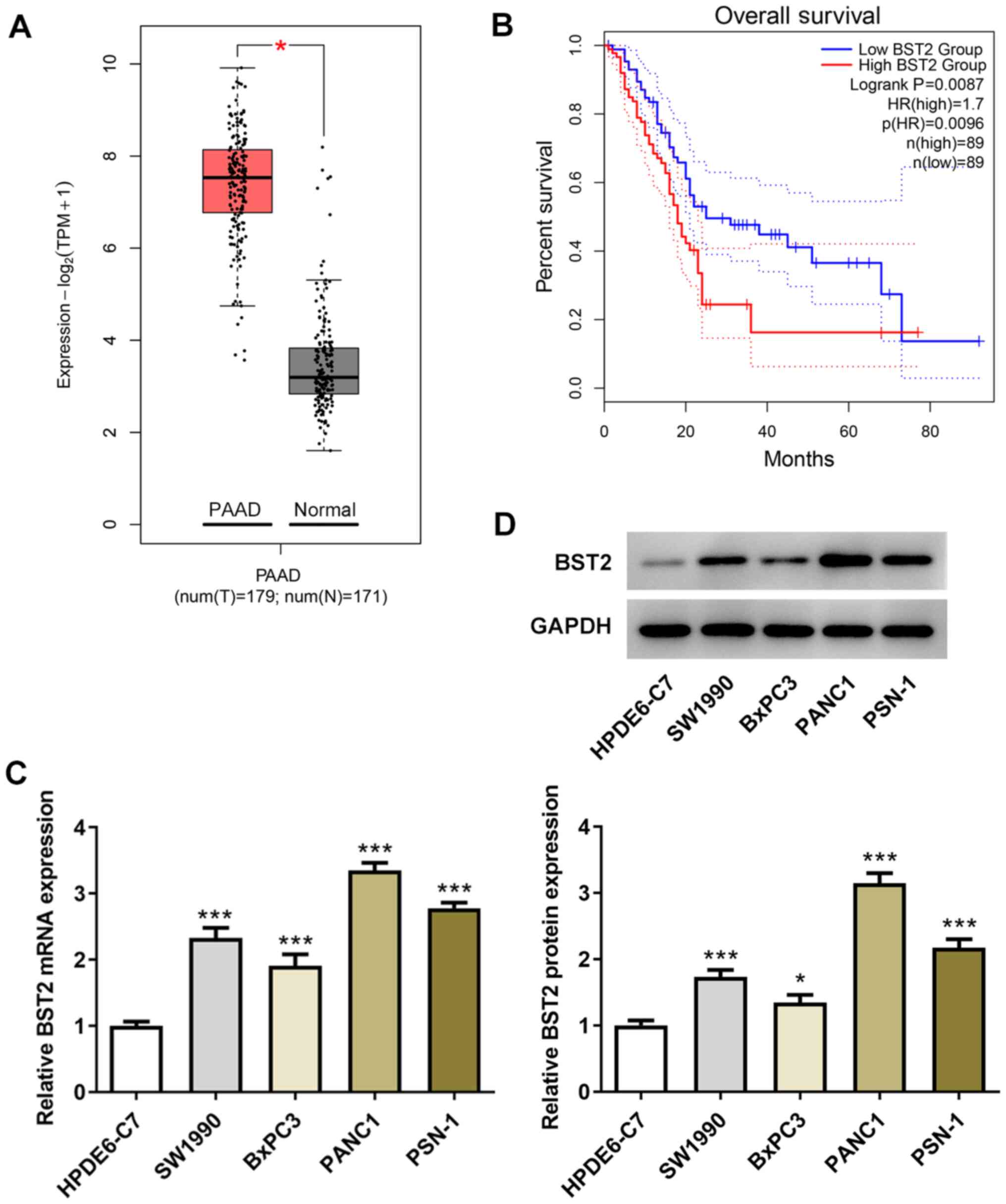
BST2 is highly expressed in pancreatic
cancer cell lines
The GEPIA database was analyzed for 179 pancreatic
adenocarcinoma samples and 171 normal pancreatic tissue samples.
Based on the results of the bioinformatics analysis showing that
BST2 was upregulated in pancreatic cancer (Fig. 1A) and the negative correlation
between BST2 expression and overall survival (Fig. 1B), BST2 expression levels in
non-cancerous pancreatic duct epithelial cells and pancreatic
cancer cell lines were examined for comparison. It was observed
that both the mRNA and protein expression levels of BST2 were
elevated to varying degrees in SW1990, BxPC3, PANC1 and PSN-1 cells
in comparison with HPDE6-C7 cells (Fig. 1C and D). Among the four pancreatic cancer cell
lines, PANC1 cells exhibited the highest level of BST2 expression
and were therefore selected for the subsequent experiments.
Inhibition of pancreatic cancer cell
proliferation and migration by BST2 knockdown
BST2 interference was conducted by siRNA plasmid
transfection in PANC1 cells, and siBST2-1 plasmid exerted a more
significant knockdown effect on BST2 expression compared with
siBST2-2 (Fig. 2A); hence,
siBST2-1 was selected for the following assays. In PANC1 cells
transfected with siBST2-1, the number of colonies was notably lower
compared with that in the siNC and control groups (Fig. 2B). Moreover, CCK-8 assay detected
reduced absorbance in cells transfected with siBST2-1, indicating
that knockdown of BST2 reduced cell proliferation (Fig. 2C). The greater scratch width at 24
h in cells transfected with siBST2-1 in the wound healing assay
demonstrated that BST2 knockdown could prevent pancreatic cancer
cell migration to a significant extent (Fig. 2D). Furthermore, Transwell assay
demonstrated that BST2 knockdown could markedly prevent pancreatic
cancer cell invasion (Fig. 2D).
Additionally, the markers of proliferation (Ki67 and PCNA) and
migration (MMP2 and MMP9) in PANC1 cells were found to be expressed
at a markedly lower level following BST2 interference (Fig. 2E). These results collectively
suggested that BST2 depletion may lead to reduced pancreatic cancer
cell proliferation and migration.
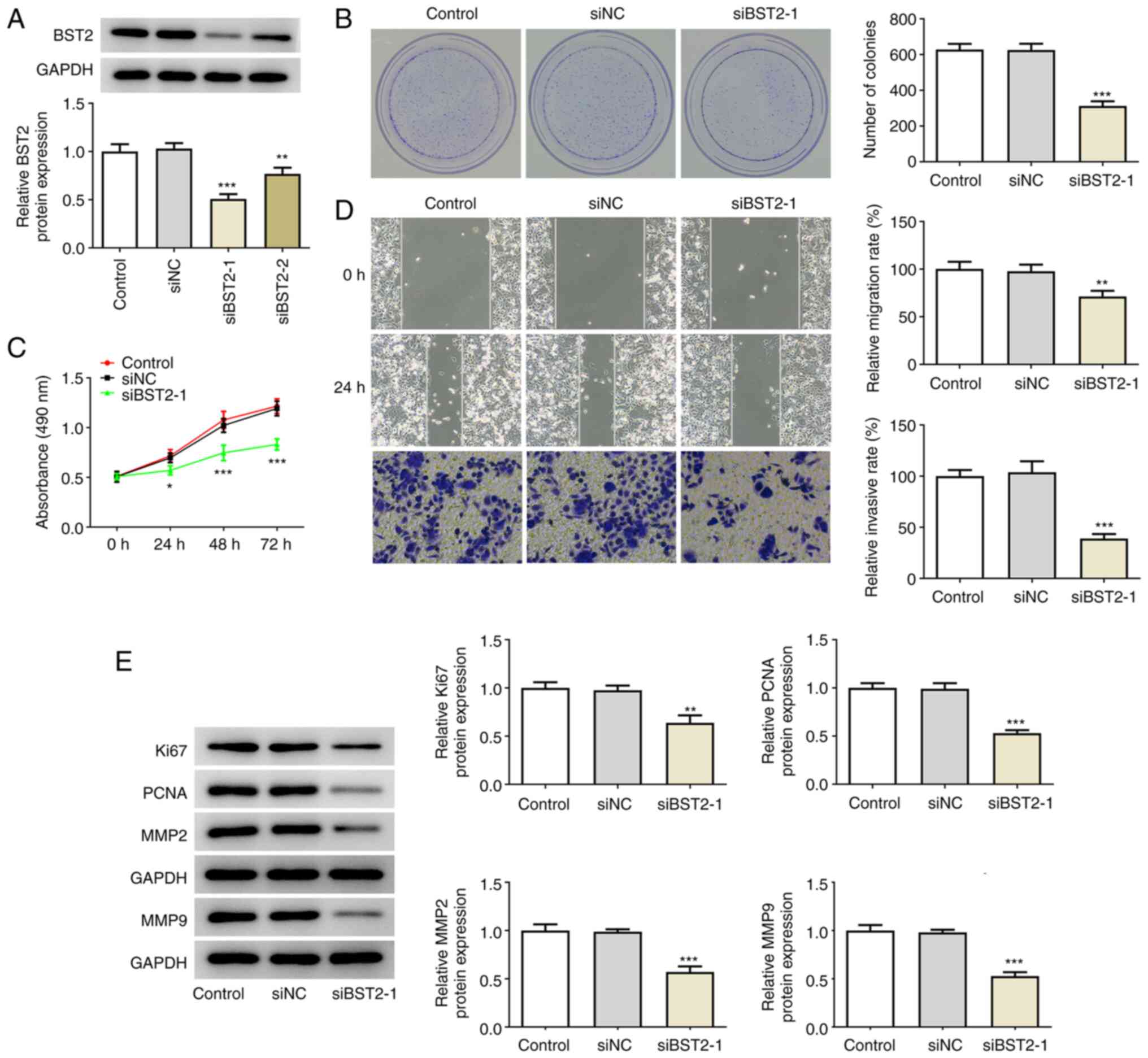 | Figure 2BST2 knockdown inhibits pancreatic
cancer cell proliferation and migration. (A) Plasmid interference
efficacy of siBST2-1 and siBST2-2, detected by reverse
transcription-quantitative PCR analysis. (B) PANC1 colony-forming
capacity before and after interference with BST2, detected by
colony formation assay (magnification, x10). (C) PANC1
proliferation rate before and after interference with BST2,
detected by Cell Counting Kit-8 assay. (D) Analysis of PANC1 cell
migration and invasion by wound healing (top and middle rows) and
Transwell (bottom row) assays, respectively (magnification, x100).
(E) Proliferation and migration markers assayed by western
blotting. *P<0.05, **P<0.01,
***P<0.001 vs. siNC. BST2, bone marrow stromal cell
antigen 2; si, small interfering RNA; NC, negative control. |
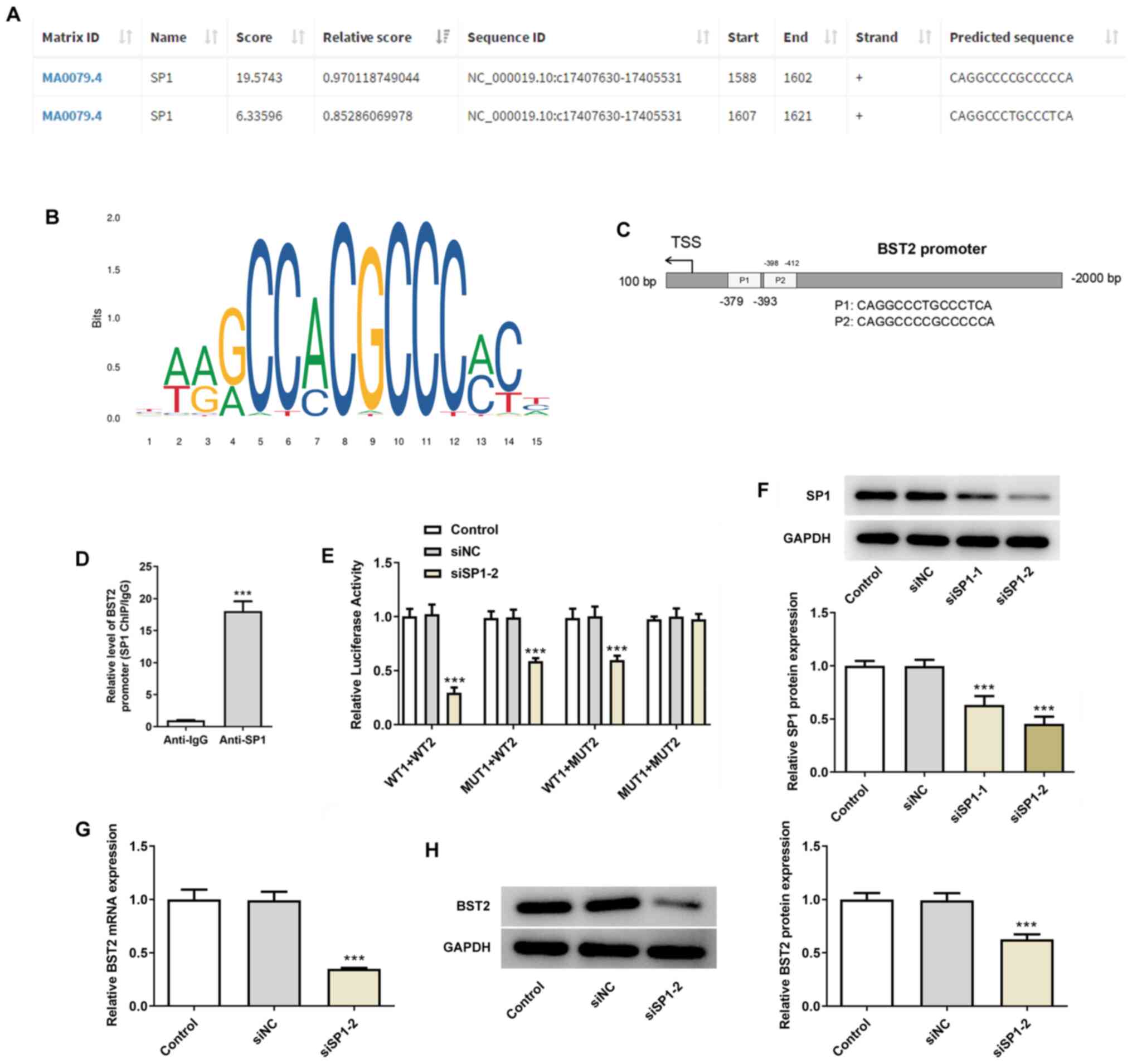
Binding interaction between SP1 and
BST2 promoter
The predicted sequence of SP1 (score >5) and the
transcription factor DNA motif logo are shown in Fig. 3A and B. In view of the high probability of SP1
binding to the BST2 promoter (Fig.
3C), ChIP (Fig. 3D) and
dual-luciferase reporter (Fig. 3E)
assays were performed for verification purposes. The results
revealed a binding interaction between the SP1 and BST2 promoter.
Western blotting confirmed superior interferential efficacy of
siSP1-2 compared with siSP1-1 (Fig.
3F); hence, siSP1-2 was used in the dual-luciferase reporter
assay. Furthermore, BST2 mRNA and protein expression levels were
found to be significantly lower in the siSP1-2 group compared with
those in the siNC group (Fig. 3G
and H). Thus, SP1 may regulate
BST2 expression in pancreatic cancer cells through their binding
interaction.
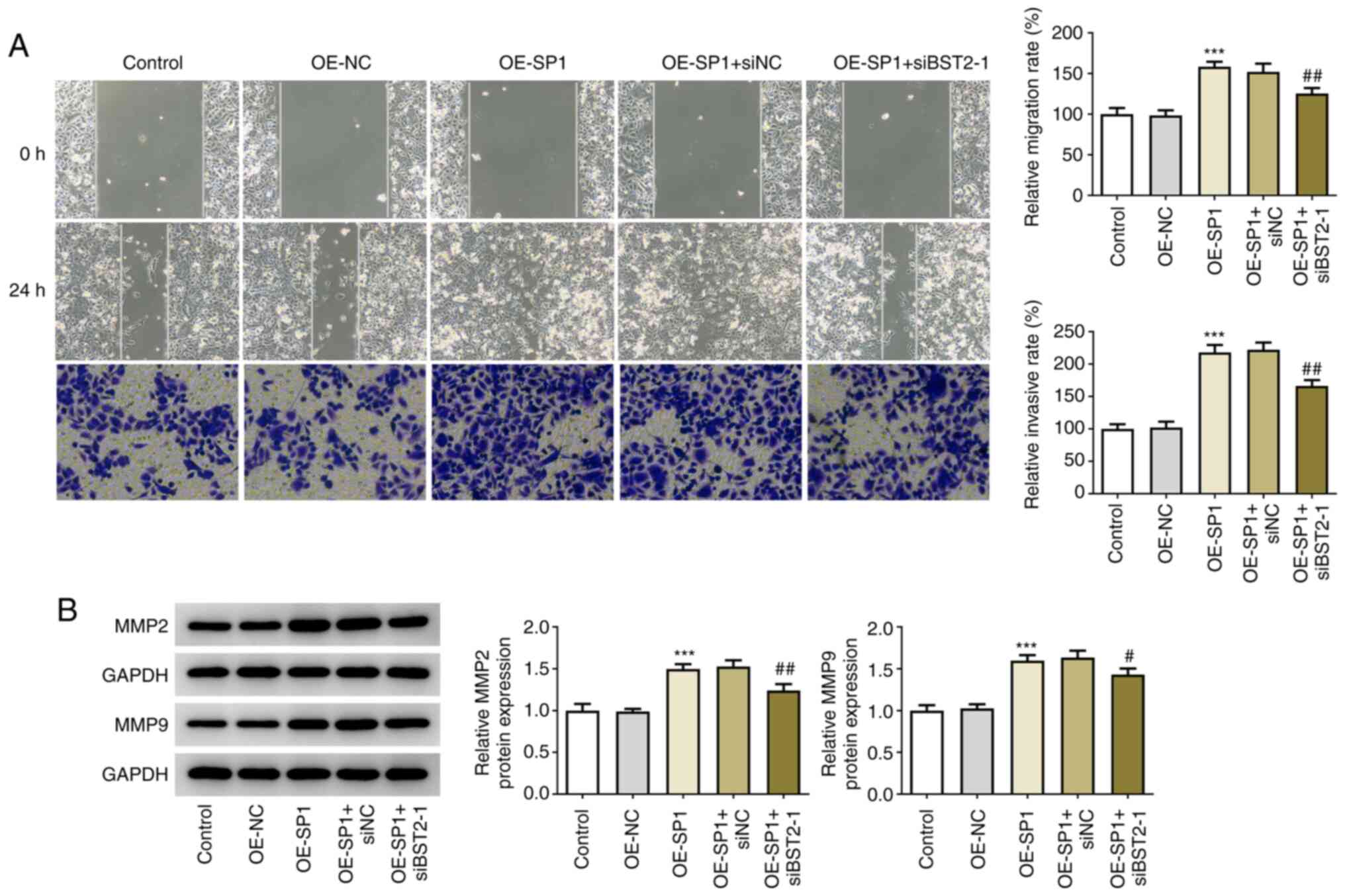
SP1 overexpression-induced cell
proliferation and migration are rescued by BST2 knockdown
These next assays were performed to detect the
activation of BST2 by SP1 at the oncogenic transcriptional
activation level in pancreatic cancer. Increased SP1 expression was
verified by western blotting in PANC1 cells transfected with OE-SP1
(Fig. 4A). In subsequent colony
formation assays, it was observed that the number of colonies
increased after overexpression of SP1 compared with OE-NC, and that
co-transfection of OE-SP1 and siBST2-1 decreased the colony number
compared with the OE-SP1+siNC group (Fig. 4B). Additionally, the absorbance
level (450 nm) was the highest in the OE-SP1 and OE-SP1+siNC
groups, the lowest in the OE-NC and control groups, and
intermediate in the OE-SP1+siBST2-1 group (Fig. 4C), suggesting that the promoting
effect of SP1 overexpression on PANC1 proliferation was diminished
by BST2 knockdown. Furthermore, the high expression level of Ki67
and PCNA in PANC1 cells transfected with OE-SP1 declined after the
knockdown of BST2 (Fig. 4D).
Therefore, BST2 may be activated by SP1 in pancreatic cancer to
promote cell proliferation.
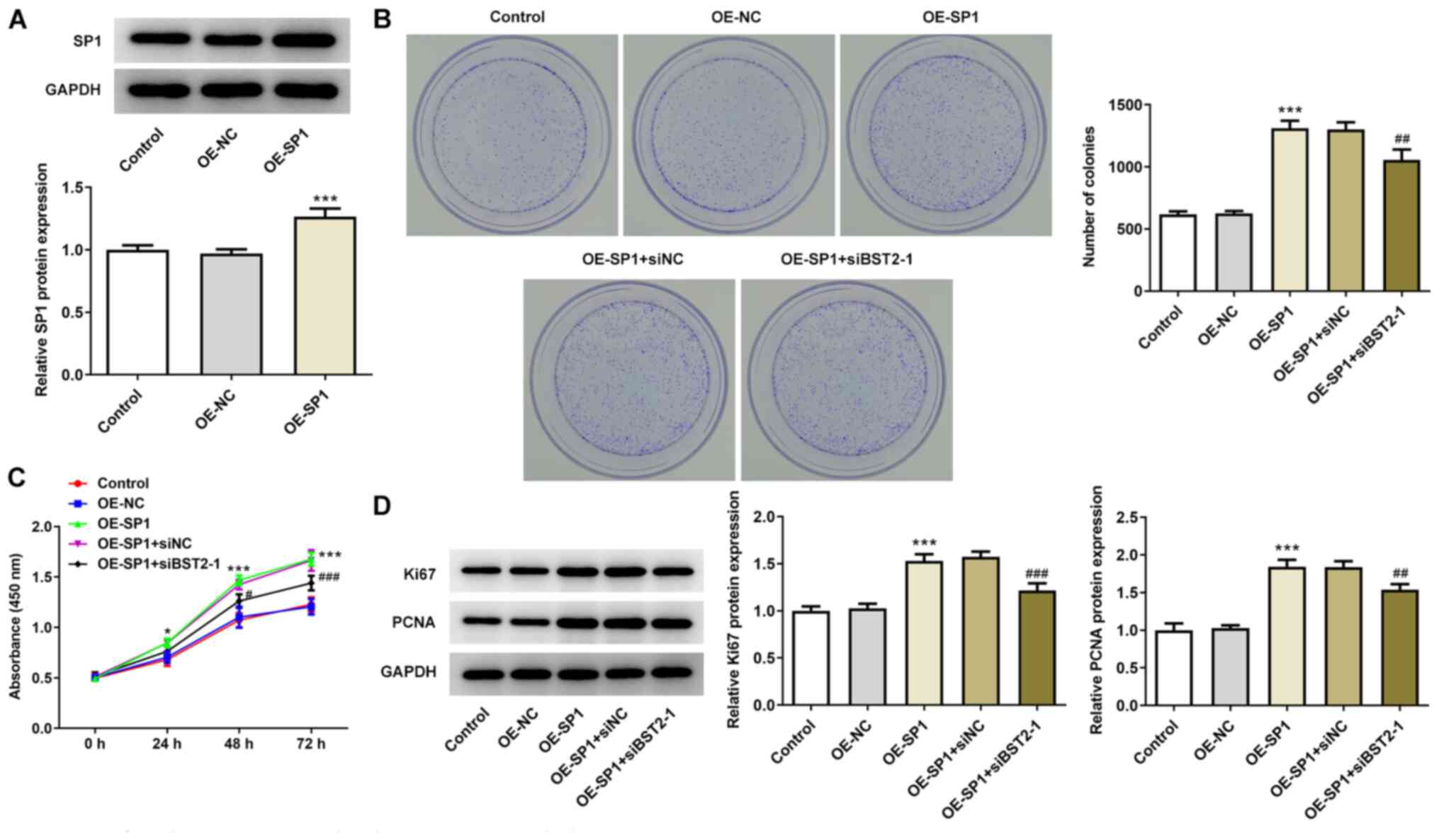 | Figure 4BST2 knockdown reverses SP1
overexpression-induced cell proliferation. (A) SP1 expression in
cells transfected with OE-NC or OS-SP1, detected by western
blotting. (B) PANC1 cell colony-forming capacity after transfection
with OE-NC/OE-SP1 or co-transfection with OE-SP1 and siNC/siBST2-1,
detected by colony formation assay (magnification, x10). (C) PANC1
cell proliferation rate after transfection with OE-NC/OE-SP1 or
co-transfection with OE-SP1 and siNC/siBST2-1, detected by Cell
Counting Kit-8 assay. (D) Expression of proliferation markers
assayed by western blotting. *P<0.05,
***P<0.001 vs. OE-NC; #P<0.05,
##P<0.01, ###P<0.001 vs. OE-SP1 + siNC.
SP1, specificity protein 1; BST2, bone marrow stromal cell antigen
2; OE, overexpression; NC, negative control; si, small interfering
RNA; PCNA, proliferating cell nuclear antigen |
Following transfection of OE-SP1, a notably higher
migration rate was observed in PANC1 cells, which then decreased
following interference with BST2, as shown by the results of the
wound healing assay (Fig. 5A). The
expression levels of migration markers, MMP2 and MMP9, which were
elevated by SP1 overexpression, were also found to be downregulated
by co-transfection with OE-SP1 and siBST2-1 (Fig. 5B). These results revealed a
potential promoting effect of SP1-activated BST2 on pancreatic
cancer cell migration.
Discussion
Pancreatic cancer is a malignant tumor originating
from the pancreatic duct epithelium and it has the highest degree
of malignancy and the highest rate of mortality among
gastrointestinal diseases (19).
Pancreatic cancer can develop local invasion and distant metastasis
in its early stages; therefore, ~80% of patients are at an advanced
stage at the time of diagnosis (1,20).
Surgery is currently the only possible curative option, but radical
resection is suitable for <10% of patients (21,22).
It has been confirmed that the main causes of death in patients
with pancreatic cancer are tumor metastasis and early recurrence
(23). Therefore, the focus of
pancreatic cancer research is to identify tumor markers and
therapeutic molecular targets associated with pancreatic cancer
metastasis and to further elucidate the biological mechanisms
implicated in tumor development.
BST2 is expressed in not only normal human tissues,
but also in a variety of tumor cells, and is involved in the
regulation of cancer cell proliferation, migration and invasion
(24). A microarray analytic
data-driven study reported that tetraspanin-8 and BST2 were
expressed at abnormally high levels in CD166-positive pancreatic
cancer cell lines and in a mouse model with significantly
accelerated tumor growth (25). In
the present study, the results analyzed in the GEPIA database also
showed a direct association between BST2 expression and pancreatic
cancer. Upregulated BST2 expression was observed in all four
pancreatic cancer cell lines compared with the non-cancerous
pancreatic duct epithelial cells. Moreover, multiple pathways and
interactive activities have been explored in previous studies to
determine the mechanisms of action of BST2 in cancer progression.
Xu et al (26) investigated
the role of BST2 in hepatocellular carcinoma (HCC) and found that
high BST2 expression in HCC tissues was positively associated with
tumor growth, possibly via activation of the NF-κB pathway. Liu
et al (27) demonstrated
that BST2 downregulation by microRNA (miR)-760 was favorable for
the repression of gastric cancer cell survival and migration. An
earlier study by Liu et al (28) also elucidated the association
between BST2-induced NF-κB activation and increased proliferation
and migration of gastric cancer cells. The present study
demonstrated that knockdown of BST2 in the PANC1 pancreatic cancer
cell line effectively mitigated the proliferative and migratory
cell behaviors.
The JASPAR database of transcription factor binding
prediction was consulted to describe the action of BST2 on PANC1
cell proliferation and migration from a mechanistic perspective.
The transcription factor SP1, a proven oncogene in various types of
tumors, was predicted to have a matching sequence on the BST2
promoter. The results of the ChIP and dual-luciferase reporter
assays in the present study verified the binding relationship
between SP1 and the BST2 promoter. Zhang et al (29) reported that the oncogenic effects
of SP1 in HCC in vitro were mediated through transcriptional
upregulation of RAS guanyl-releasing protein 1. Targeted inhibition
of SP1 by miR-502-5p in gastric cancer cells decreased the levels
of cell proliferation, invasion and migration (30). Moreover, in a previous study, SP1
was found to be targeted by miR-529, thereby suppressing the
malignant behaviors and epithelial-to-mesenchymal transition of
pancreatic cancer cells (31).
lncRNA-LINC00514 upregulation mediated via SP1 acts as an oncogene
in metastatic osteosarcoma by regulating miR-708 expression
(32). As a transcription factor,
SP1 activates lncRNA SNGH7 to promote ovarian tumorigenesis
(33). The present study
demonstrated SP1 overexpression-induced PANC1 cell proliferation
and migration, while BST2 knockdown weakened this effect of SP1
overexpression.
The results of the present study supplemented the
current understanding of the role of BST2 in cancer progression
with corroborating evidence of increased proliferation and
migration of pancreatic cancer cells via SP1-activated BST2. The
findings of the present study may provide a useful guideline for
the identification of the prognostic and therapeutic value of SP1
and BST2 in future relevant research and clinical trials. However,
a limitation of this study was the insufficient discussion on the
downstream regulatory mechanisms of BST2. Therefore, further
research is required in the future.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the National
Nature Science Foundation of China (grant no. 81071985).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CL, YH and JC conceived and designed the study, and
acquired and interpreted the data. CL was a major contributor in
writing the manuscript. All authors confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620.
2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ilic M and Ilic I: Epidemiology of
pancreatic cancer. World J Gastroenterol. 22:9694–9705.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mizrahi JD, Surana R, Valle JW and Shroff
RT: Pancreatic cancer. Lancet. 395:2008–2020. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Qian L, Yu S, Chen Z, Meng Z, Huang S and
Wang P: Functions and clinical implications of exosomes in
pancreatic cancer. Biochim Biophys Acta Rev Cancer. 1871:75–84.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang L, Sanagapalli S and Stoita A:
Challenges in diagnosis of pancreatic cancer. World J
Gastroenterol. 24:2047–2060. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yang J, Ren B, Yang G, Wang H, Chen G, You
L, Zhang T and Zhao Y: The enhancement of glycolysis regulates
pancreatic cancer metastasis. Cell Mol Life Sci. 77:305–321.
2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Rouanet M, Lebrin M, Gross F, Bournet B,
Cordelier P and Buscail L: Gene Therapy for Pancreatic Cancer:
Specificity, Issues and Hopes. Int J Mol Sci.
18(1231)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kurtanich T, Roos N, Wang G, Yang J, Wang
A and Chung EJ: Pancreatic Cancer Gene Therapy Delivered by
Nanoparticles. SLAS Technol. 24:151–160. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wu J, Li Z, Zeng K, Wu K, Xu D, Zhou J and
Xu L: Key genes associated with pancreatic cancer and their
association with outcomes: A bioinformatics analysis. Mol Med Rep.
20:1343–1352. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Arnaud F, Black SG, Murphy L, Griffiths
DJ, Neil SJ, Spencer TE and Palmarini M: Interplay between ovine
bone marrow stromal cell antigen 2/tetherin and endogenous
retroviruses. J Virol. 84:4415–4425. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gong S, Osei ES, Kaplan D, Chen YH and
Meyerson H: CD317 is over-expressed in B-cell chronic lymphocytic
leukemia, but not B-cell acute lymphoblastic leukemia. Int J Clin
Exp Pathol. 8:1613–1621. 2015.PubMed/NCBI
|
|
14
|
Wang W, Nishioka Y, Ozaki S, Jalili A, Abe
S, Kakiuchi S, Kishuku M, Minakuchi K, Matsumoto T and Sone S:
HM1.24 (CD317) is a novel target against lung cancer for
immunotherapy using anti-HM1.24 antibody. Cancer Immunol
Immunother. 58:967–976. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Malsy M, Graf B and Almstedt K: The active
role of the transcription factor Sp1 in NFATc2-mediated gene
regulation in pancreatic cancer. BMC Biochem. 20(2)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Safe S, Nair V and Karki K:
Metformin-induced anticancer activities: Recent insights. Biol
Chem. 399:321–335. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Nair V, Pathi S, Jutooru I, Sreevalsan S,
Basha R, Abdelrahim M, Samudio I and Safe S: Metformin inhibits
pancreatic cancer cell and tumor growth and downregulates Sp
transcription factors. Carcinogenesis. 34:2870–2879.
2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hogendorf P, Durczyński A and Strzelczyk
J: Metastatic Pancreatic Cancer. J Invest Surg. 31:151–152.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ferrone CR, Brennan MF, Gonen M, Coit DG,
Fong Y, Chung S, Tang L, Klimstra D and Allen PJ: Pancreatic
adenocarcinoma: The actual 5-year survivors. J Gastrointest Surg.
12:701–706. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hackert T, Klaiber U, Pausch T, Mihaljevic
AL and Büchler MW: Fifty Years of Surgery for Pancreatic Cancer.
Pancreas. 49:1005–1013. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Das S and Batra SK: Pancreatic cancer
metastasis: Are we being pre-EMTed? Curr Pharm Des. 21:1249–1255.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ishikawa J, Kaisho T, Tomizawa H, Lee BO,
Kobune Y, Inazawa J, Oritani K, Itoh M, Ochi T, Ishihara K, et al:
Molecular cloning and chromosomal mapping of a bone marrow stromal
cell surface gene, BST2, that may be involved in pre-B-cell growth.
Genomics. 26:527–534. 1995.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fujiwara K, Ohuchida K, Sada M, Horioka K,
Ulrich CD III, Shindo K, Ohtsuka T, Takahata S, Mizumoto K, Oda Y,
et al: CD166/ALCAM expression is characteristic of tumorigenicity
and invasive and migratory activities of pancreatic cancer cells.
PLoS One. 9(e107247)2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu X, Wang Y, Xue F, Guan E, Tian F, Xu J
and Zhang H: BST2 Promotes Tumor Growth via Multiple Pathways in
Hepatocellular Carcinoma. Cancer Invest. 38:329–337.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu W, Li Y, Feng S, Guan Y and Cao Y:
MicroRNA-760 inhibits cell viability and migration through
down-regulating BST2 in gastric cancer. J Biochem. 168:159–170.
2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu W, Cao Y, Guan Y and Zheng C: BST2
promotes cell proliferation, migration and induces NF-κB activation
in gastric cancer. Biotechnol Lett. 40:1015–1027. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang X, Zhuang H, Han F, Shao X, Liu Y,
Ma X, Wang Z, Qiang Z and Li Y: Sp1-regulated transcription of
RasGRP1 promotes hepatocellular carcinoma (HCC) proliferation.
Liver Int. 38:2006–2017. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Peng X, Wu M, Liu W, Guo C, Zhan L and
Zhan X: miR-502-5p inhibits the proliferation, migration and
invasion of gastric cancer cells by targeting SP1. Oncol Lett.
20:2757–2762. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Xue L, Shen Y, Zhai Z and Zheng S: miR 539
suppresses the proliferation, migration, invasion and epithelial
mesenchymal transition of pancreatic cancer cells through targeting
SP1. Int J Mol Med. 45:1771–1782. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mi LD, Sun CX, He SW and Du GY:
SP1-Induced Upregulation of lncRNA LINC00514 Promotes Tumor
Proliferation and Metastasis in Osteosarcoma by Regulating miR-708.
Cancer Manag Res. 12:3311–3322. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Bai Z, Wu Y, Bai S, Yan Y, Kang H, Ma W,
Zhang J, Gao Y, Hui B, Ma H, et al: Long non-coding RNA SNGH7 Is
activated by SP1 and exerts oncogenic properties by interacting
with EZH2 in ovarian cancer. J Cell Mol Med. 24:7479–7489.
2020.PubMed/NCBI View Article : Google Scholar
|