Introduction
Hepatocellular carcinoma (HCC) is one of the most
common cancer types and a primary cause of cancer-associated
mortality worldwide. HCC accounted for 9% of all cancer-related
deaths worldwide in 2019 as per the World Health Organization
report (1). Although considerable
progress has been made in the diagnosis and treatment of liver
cancers, the lack of good staging methods and efficient diagnostic
markers results in poor prognosis (2-4).
Therefore, there is an urgent requirement to identify novel
molecular markers and related pathways that underlie the
tumorigenesis and progression of HCC.
Recently, circular RNAs (circRNAs) have received
increasing attention due to their potential function in gene
regulation (5-7).
circRNAs are non-coding RNAs with conserved, stable, abundant and
tissue/developmental stage-specific expression (8,9).
circRNAs are able to mediate rolling circle translation, regulate
the transcription of host genes, participate in the formation of
alternatively spliced mRNAs and serve as microRNA (miRNA/miR)
sponges (8,10-12).
circRNAs have key roles in regulating the biological development,
pathogenesis and progression of numerous cancer types. Accumulating
evidence has indicated that circRNAs are involved in normal
physiological processes and in the development of various diseases,
including liver cancer (13,14).
In addition, aberrant expression of certain circRNAs has been
demonstrated to be related to numerous diseases, including cancer,
cartilage degradation, heart disease, neurodegenerative diseases
and stroke (15-17).
However, only a small amount of studies examined the circRNA
expression pattern in peripheral blood mononuclear cells (PBMCs)
and this has not been thoroughly studied in liver cancers (18).
Numerous studies have reported on the expression of
circRNAs in cancer patients (19-21),
but the findings are frequently inconsistent and no reliable
biomarkers have been identified. These challenges may be attributed
to several points and the key factors include individual
differences in sampling and detection methods. The sample type
constitutes another essential factor, as the commonly used
specimens are tumor tissues. It is well known that tumor tissues
are complex and contain multiple types of cells, such as immune
cells, cancer cells, endothelial cells, fibroblasts and
inflammatory cells (22,23). This may be a major reason for the
inconsistencies in previous results. Heterogeneity may be
eliminated in most studies by analyzing large lumps of tumor
tissues (24,25). In the present study, PBMC samples
were collected and examined using RNA-sequencing (RNA-seq), and
differentially expressed circRNAs (DE-circRNAs) were identified.
PBMCs have relatively simple components that reduce tumor
heterogeneity considerably (26).
In the present study, alterations in circRNA expression patterns in
HCC were detected using RNA-seq. A Differential expression profile
of circRNAs was identified in PBMCs from 19 clinical patients with
HCC and 19 healthy controls. A total of 40 circRNAs with
significant differential expression were identified. Considering
that circRNA function is related to the function of the parent
gene, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analyses were performed to predict the
potential functions of the DE-circRNAs in HCC. Based on the results
of the sequencing, a Bioinformatics analysis and reverse
transcription-quantitative (RT-q)PCR, certain functions of these
DE-circRNAs in HCC were revealed. Furthermore, the potential of the
selected circRNAs as biomarkers was assessed.
Materials and methods
Patients and sample collection
A total of 22 healthy individuals and 28 patients
with HCC who visited Shenzhen Traditional Chinese Medicine Hospital
(Shenzhen, China) from July 2018 to May 2019 were enrolled in the
study and fresh blood samples were collected in EDTA tubes after
obtaining written informed consent. All patients with HCC were
diagnosed according to Guidelines for Diagnosis and Treatment of
Primary Liver Cancer in China (2017 Edition) (27) and most of the patients received
surgery or interventional therapy before blood sampling. Patients
were excluded if they were pregnant or lactating, had a previous
history of or currently undergoing for other malignant tumors, had
cardiac disease or serious and active infections. However, patients
with hepatitis B virus (HBV) and hepatitis C virus (HCV) infections
were not excluded. Individuals in the control group were enrolled
from The Health Examination Center of Shenzhen Traditional Chinese
Medicine Hospital at random during the same period, who were
selected from the Health Examination Center database with recorded
medical history after completing the informed consent form. All
available information and clinical characteristics of the patients
with HCC are listed in Table SI.
The samples of 19 controls and 19 patients with HCC were measured
using RNA-seq and assayed by reverse transcription-quantitative
PCR. The patients with HCC were aged 14-82 years, with an average
age of 55±18 years, whereas the healthy controls were aged 21-75
years, with an average age of 40±15 years. Among the 28 patients
with HCC enrolled in this study, 20 were male and 8 were female. Of
the 22 controls, 17 were male and 5 were female. The study was
approved by the Ethics Committee of the Shenzhen Traditional
Chinese Medicine Hospital (Shenzhen, China). All experiments were
performed in accordance with the relevant guidelines and
regulations established by the Ethics Committee.
Processing of blood and RNA
extraction
Peripheral blood (2 ml, with EDTA anticoagulant) was
collected from patients and matched healthy controls. Ficoll-Paque
PREMIUM (Cytiva) was used for the isolation of PBMCs according to
the manufacturer's instructions. Total RNA was isolated using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
reagent following standard procedures as previously described
(28). The quality of RNA was
determined by measuring the absorbance at 260 nm (A260)
and the A280 using a NanoDrop ND-1000 (Thermo Fisher
Scientific, Inc.), and RNA integrity was determined based on the
RNA integrity number derived from the peak area of 28s RNA, 18s RNA
and 5s RNA (RIN ≥7; Agilent 2100 RIN Beta Version Software; Agilent
Technologies, Inc.).
RNA-seq
A total of 2 µg of RNA sample from each subject was
prepared as the input for RNA-seq. In the first step, ribosomal RNA
(rRNA) was removed using Epicenter Ribozero™ rRNA Removal kit
(Epicenter; Illumina, Inc.) and rRNA-free components were cleaned
by ethanol precipitation. Next, the RNAs were submitted for library
preparation according to the method described in a previous study
(29). It was prepared using the
VAHTS Total RNA-seq (H/M/R) Library Prep kit for Illumina (cat. no.
NR603-01; Vazyme Biotech Co., Ltd.) according to the manufacturer's
protocols. In brief, the following steps, including fragmentation,
reverse transcription, adaptor ligation and preamplification, were
performed sequentially. Finally, the libraries were purified,
quality-tested and quantified using the Agilent Bioanalyzer 2100
system (Agilent Technologies, Inc.). The libraries were sequenced
on Illumina HiSeq X Ten System by using HiSeq X Ten Reagent Kit
(cat. no. FC-501-2501) with 2x150 bp paired-end technology.
Bioinformatics analysis
The quality of raw sequencing data in fastq format
was assessed using FastQC software (v 0.11.5; https://www.bioinformatics.babraham.ac.uk/projects/download.html#fastqc).
SOAPnuke software (v2.0) was used to filter low-quality data
(30). BWA software (v0.6;
http://bio-bwa.sourceforge.net/bwa.shtml) was used to
align the clean reads with high quality to the human reference
genome (GRCh38/hg38) (31,32) with default parameters. The unaligned
sequencing data were then subjected to circRNA analysis by
recognition of the reverse splicing event using the CIRI2 program
(33). The CIRI algorithm
identified circRNA candidates using two-step filtering. First, the
paired chiastic clipping signals were collected and then the
false-positive junctions were removed.
Reads per million mapped reads was used for
quantification of circRNA expression (34). P-values for the differences in gene
expression were adjusted by false discovery rate using the
Benjamini-Hochberg procedure. P<0.05 and |log2
(fold-change)| >1 were used as the threshold for filtering the
DE-circRNAs. The R package DESeq (1.10.1) was used to analyze the
differential circRNA expression. Functional annotation and pathway
enrichment of the parent genes of the DE-circRNAs were performed
using KEGG Orthology-based annotation system (KOBAS 3.0) (35). GO and KEGG annotation and enrichment
were also performed for the parent genes and corresponding target
genes of circRNAs using the KOBAS software (corrected P-value
<0.05) (36-38).
To investigate the correlation between circRNAs and
mRNAs in patients with HCC, interactions among the DE-circRNAs were
studied and the location of the related parent genes on exons and
their corresponding target miRNAs were identified. TargetScan
(version no 7.1; http://www.targetscan.org/vert_71/) and miRanda
(v3.3a; http://www.microrna.org/microrna/home.do) were used to
predict the target miRNAs of these circRNAs. Cytoscape (v3.8.0;
https://cytoscape.org/) was used to illustrate
the co-expression network (39,40).
RT-qPCR
Total RNA extracted from PBMCs and cell lines was
subjected to RT-qPCR validation. Specific divergent primers
spanning the back-splice junction sites of the circRNAs were
designed. To detect the expression levels of the selected circRNAs,
cDNA was synthesized using the PrimeScript™ RT reagent kit (Takara
Biotechnology Co., Ltd.) by incubating at 37˚C for 15 min and then
85˚C for 5 sec to terminate the reaction, with a final volume of 20
µl. TB Green Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) was
used for qPCR analyses on ABI 7500 thermal cycler according to the
manual instruction (Thermo Fisher Scientific, Inc.). After
predenaturing at 95˚C for 2 min, the reaction system were submitted
to 94˚C for 15 sec and 60˚C, 45 sec, for 40 cycles. The internal
control gene GAPDH was measured at the same time. The expression of
different circRNAs was compared using the 2-ΔΔCq method
(41). The primers (synthesized by
Sangon Biotech) used in the present study are listed in Table I.
 | Table IPrimers for qPCR of the selected
circRNAs. |
Table I
Primers for qPCR of the selected
circRNAs.
| Name | Direction | Sequence
(5'-3') |
|---|
|
hsa_circ_0017586 | F |
GGATTTGCAAGGCTAAGTGC |
| | R |
CCGTAGTAAGGGTTTCGATCC |
|
hsa_circ_0008304 | F |
TTTGACTTCCACAAATGCAGA |
| | R |
AAAAACCTTCGTCTTCAATAATTC |
|
hsa_circ_0001181 | F |
AACTGCCATTCAATGCACAA |
| | R |
GTCGGGAAGTTCAGTGGAAA |
|
hsa_circ_0073239 | F |
TGACTTCCGTTGGACTGATG |
| | R |
AGATGGGGTGGTTGTCACAT |
|
hsa_circ_0070190 | F |
TTATGGAGGGAAACCCATCA |
| | R |
TACGCACGAGGAAAACTGTG |
|
hsa_circ_0000369 | F |
CAACACGGAAGTGCTGTTGT |
| | R |
GTTGACCCTCACTGGCTGAT |
| GAPDH | F |
ACAACTTTGGTATCGTGGAAGG |
| | R |
GCCATCACGCCACAGTTTC |
Cell culture and transfection
To further explore the potential diagnostic roles of
the DE-circRNAs in liver cancer, the human liver cancer cell line
HepG2 was obtained from the Cell Bank of the Chinese Academy of
Sciences. The cell line was authenticated via short tandem repeat
testing by the Chinese Academy of Sciences.
Cells were cultured in DMEM (Corning, Inc.)
supplemented with 10% (v/v) fetal bovine serum (Biological
Industries) at 37˚C in a humidified atmosphere with 5%
CO2.
Following confirmation by RT-qPCR, Homo
sapiens (hsa)_circ_0001181 was selected for further study. To
knockdown the expression of endogenous hsa_circ_0001181 in HepG2
cells, the hsa_circ_0001181 siRNA (corresponding double stranded
oligo sequences are as follows, sense,
5'-CACCGAAGCTGGTTGATGATAATTCGAAAATTATCATCAACCAGC-3'; antisense,
5'-AAAAGCTGGTTGATGATAATTTTCGAATTATCATCAACCAGCTTC-3') was packaged
with a pGLV2-U6 lentivector packaging plasmid mix (Sangon Biotech
Co., Ltd.) in a 293T packaged cell line (cat. no. CRL-11268; ATCC).
The routine culture conditions are as follows: DMEM + 10% FBS at
37˚C and 5% CO2. When packaging, one tube contained 20
µg lentiviral plasmid, 15 µg packaging vector (pCMV-dR8.9; Addgene,
Inc.) and 10 µg envelope (pCMV-VSV-G; Addgene, Inc.) were prepared,
then 500 µl Trans-EZ solutions (Sangon Biotech Co., Ltd.) was
diluted with 4.5 ml Opti-MEM medium (cat. no. 31985; Gibco; Thermo
Fisher Scientific, Inc.). The Trans-EZ solution was then added into
the plasmid mix to obtain the transfection complex, it was
incubated 20 min at room temperature; the mixture was added to 293T
cell line and cultured for about 48 h at 37˚C and 5%
CO2. The lentiviral particles were isolated by
ultracentrifugation (4˚C, 82700 x g and 2 h). An empty vector was
transfected as the negative controls. HepG2 cells were then
transfected with the lentiviral vector particles (MOI=8). After
incubation at 37˚C for 24-36 h, RT-qPCR was also performed to
confirm the knockdown efficiency.
Proliferation assay
Proliferation of the transfected cells was measured
using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies,
Inc.) according to the manufacturer's protocol. In total, ~5,000
cells were seeded into each well and after incubation at 37˚C for
24, 48 or 72 h in a 96-well microplate (Corning, Inc.), CCK-8 stain
(5 µl) and 100 µl of fresh medium were added to each well, followed
by incubation at 37˚C for 2 h. The absorbance was measured at 450
nm using a Multiskan FC microplate reader (Thermo Fisher
Scientific, Inc.). HepG2 cells without any transfection served as
the blank control and cells transfected with the empty vector were
used as the negative control (NC) (42).
Statistical analysis
Data were analyzed using SPSS software (version
18.0; SPSS, Inc.) and graphs were generated using GraphPad Prism
8.0 (GraphPad Software, Inc.). Comparisons between the HCC and
control groups were performed using Student's t-test. The results
of the cell proliferation assay were compared between different
groups by one-way ANOVA using Tukey's test. RT-qPCR data are
presented as the mean ± standard deviation of at least three
independent tests. Statistical significance was set at P<0.05.
Receiver operating characteristic (ROC) curve analysis was
performed to evaluate the diagnostic accuracy of the different
genes analyzed.
Results
CircRNA profiling of PBMCs from
patients with HCC and normal controls
A total of 1,807.83 M raw reads with a mean length
of 300 bp was obtained. Quality control analysis indicated that
93.64-95.19% of the sequencing data had a Q score of >30. The
mean GC content of the clean reads for PBMCs from HCCs and controls
ranged from 45.02 to 47.99%. Due to the high quality of the
sequencing data obtained, the present results are reliable.
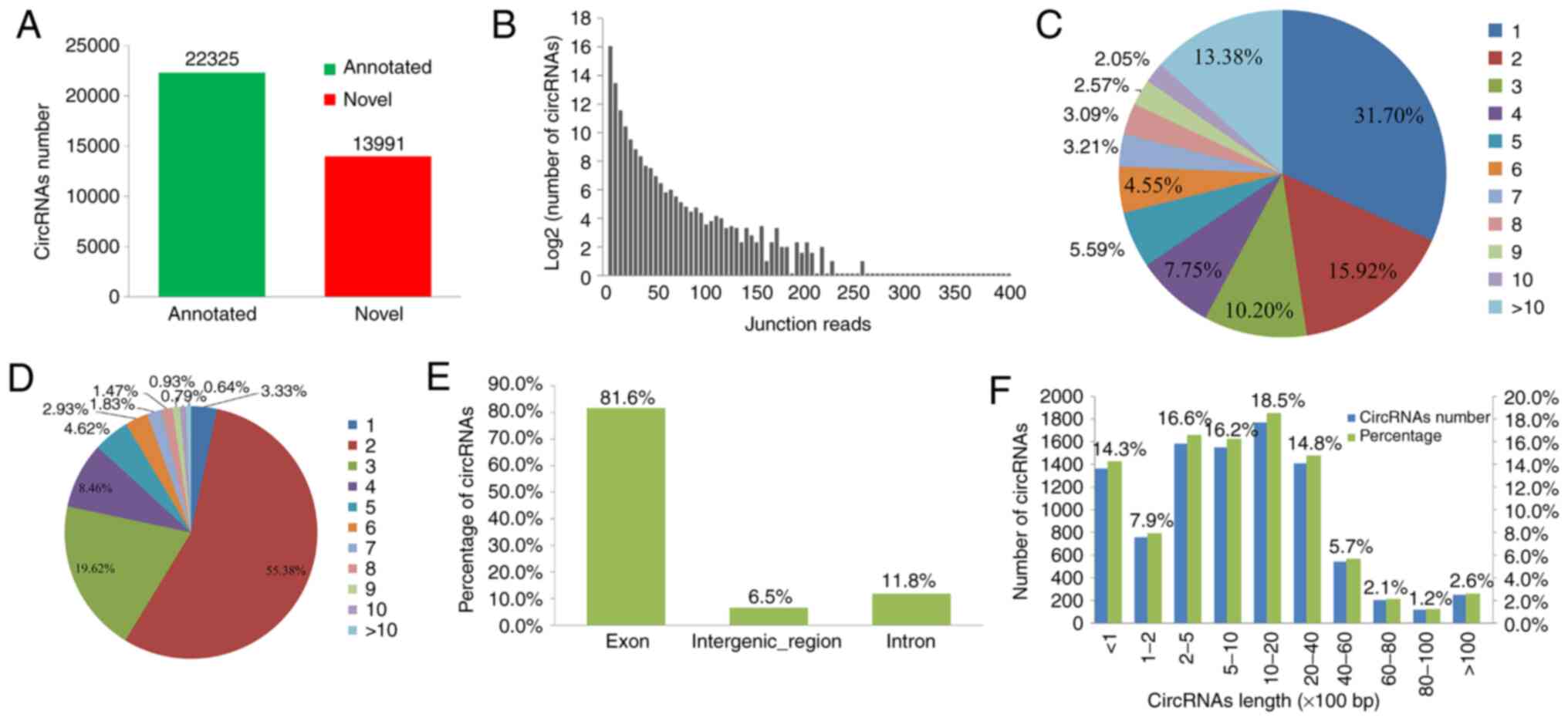
A total of 36,316 unique circRNAs were sequenced
from the PBMC samples by counting the junction reads (count number
>2) and a total of 2,239-9,897 circRNAs were identified from
these 38 PBMC samples. Of these, 22,325 circRNAs were already
listed in the circBase that were identified in previous studies and
13,991 novel circRNAs were identified in the present study. The
host genes of the identified circRNAs were distributed across all
chromosomes. Most of them contained two or more unique junction
reads (Fig. 1A and B).
Multiple circRNAs may be generated from one gene as
a result of the alternative back-splicing mechanism (43). In the present study, it was analyzed
whether alternative back-splicing is related to circRNA diversity
in PBMCs from patients with HCC. As presented in Fig. 1C, two or more circRNAs were produced
from 68.3% of the host genes. Of note, >10 circRNAs were
generated from certain genes. Most of the circRNAs were identified
from the exonic regions and consisted of 2-5 exons. Among them, the
circRNAs consisted of 2, 3, 4, 5 and 6 exons accounted for 55.38,
19.62, 8.46, 4.62 and 2.93% respectively (Fig. 1D and E). In addition, the lengths of most 71.8%
circRNAs ranged between 200 and 6,000 nucleotides (Fig. 1F).
Detection of dysregulated circRNAs in
PBMCs from patients with HCC
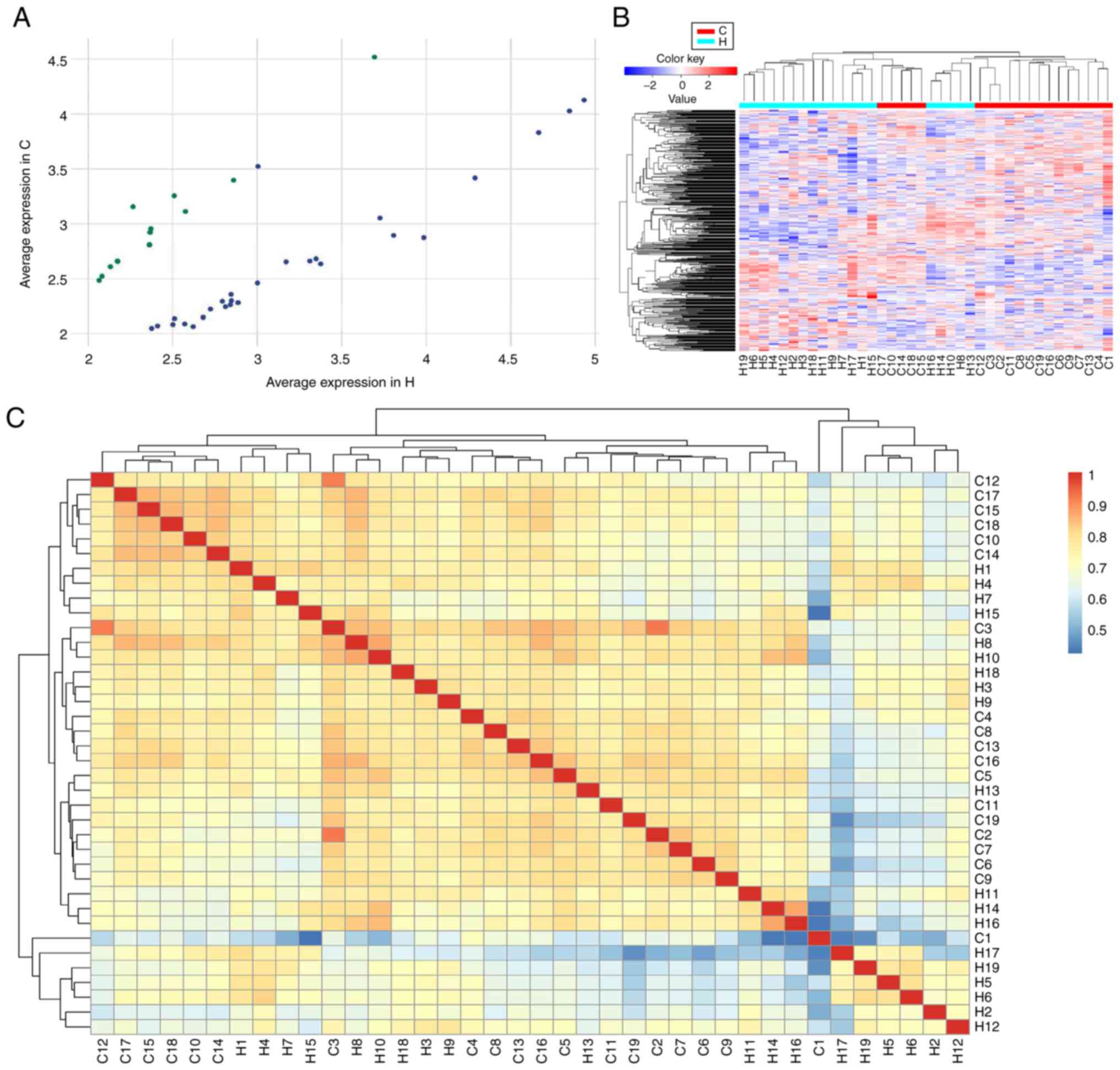
Among the circRNAs identified, only 608 and 474
circRNAs were co-expressed in all control and HCC samples,
respectively. Of the co-expressed circRNAs, 27 upregulated and 13
downregulated circRNAs were determined to be differentially
expressed between the HCC and healthy controls with a fold-change
of ≥2.0 and P<0.05 by scatter plot filtering (Fig. 2A and Table II). Hierarchical clustering
analysis of the abnormally expressed circRNAs suggested that the
circRNA expression pattern was significantly different between
patients with HCC and healthy controls (Fig. 2B).
 | Table IIDifferentially expressed of circRNAs
in peripheral blood mononuclear cells from patients with
hepatocellular carcinoma. |
Table II
Differentially expressed of circRNAs
in peripheral blood mononuclear cells from patients with
hepatocellular carcinoma.
| CircRNA | Circbase ID | Host gene
symbol | Direction of
regulation | Adj P-value | Location |
|---|
|
CHR6:31238850|31323369 | Novel | HLA-C | Up |
3.91617x10-12 | n/a |
|
CHR13:50571210|50618904 |
hsa_circ_0100694 | n/a | Up |
8.68089x10-5 | Intergenic
region |
|
CHR7:128218007|128278951 |
hsa_circ_0133256 | n/a | Up |
8.68089x10-5 | Intergenic
region |
|
CHR10:5815805|5842668 |
hsa_circ_0017586 | GDI2 | Up | 0.000268902 | Exon |
|
CHR3:141811903|141820683 |
hsa_circ_0008304 | TFDP2 | Down | 0.000327428 | Exon |
|
CHR21:30693542|30702014 |
hsa_circ_0001181 | BACH1 | Up | 0.000359788 | Exon |
|
CHR5:82832826|82850857 |
hsa_circ_0073239 | VCAN | Up | 0.000409673 | Exon |
|
CHR4:79747191|79772210 |
hsa_circ_0070190 | BMP2K | Up | 0.000745549 | Exon |
|
CHR11:128628010|128642880 |
hsa_circ_0000369 | FLI1 | Up | 0.000950703 | Exon |
|
CHR10:28408580|28420620 |
hsa_circ_0093467 | MPP7 | Up | 0.000987788 | Exon |
|
CHR8:22332467|22333137 |
hsa_circ_0083619 | PPP3CC | Down | 0.00124762 | Exon |
|
CHR9:35546427|35548532 |
hsa_circ_0002702 | RUSC2 | Up | 0.002681018 | Exon |
|
CHR16:19627436|19659204 |
hsa_circ_0005699 | VPS35L | Up | 0.002681018 | Exon |
|
CHR2:228356263|228389631 |
hsa_circ_0058514 | AGFG1 | Up | 0.002889102 | Exon |
|
CHR12:10530731|10531304 | Novel | KLRK1 | Down | 0.002895133 | Exon |
|
CHR5:82815168|82818128 |
hsa_circ_0073239 | VCAN | Up | 0.003273089 | Exon |
|
CHR4:17963526|17974508 |
hsa_circ_0069285 | LCORL | Down | 0.003518927 | Exon |
|
CHR13:21732061|21746820 |
hsa_circ_0007547 | SKA3 | Up | 0.004447874 | Exon |
|
CHR12:66597491|66611015 |
hsa_circ_0005505 | IRAK3 | Up | 0.005338426 | Exon |
|
CHR5:82832826|82838087 |
hsa_circ_0073239 | VCAN | Up | 0.005492673 | Exon |
|
CHR9:99271955|99327765 |
hsa_circ_0087636 | CDC14B | Up | 0.005492673 | Exon |
|
CHR5:150838349|150859050 |
hsa_circ_0074623 | SLC36A1 | Up | 0.005492673 | Exon |
|
CHR1:211526581|211527809 |
hsa_circ_0007234 | TRAF5 | Down | 0.005492673 | Exon |
|
CHR4:146767108|146770713 |
hsa_circ_0003187 | ZNF827 | Down | 0.005492673 | Exon |
|
CHR6:145992375|146007432 |
hsa_circ_0130925 | EPM2A | Down | 0.005492673 | Exon |
|
CHR5:130590788|130591885 | Novel | CDC42SE2 | Down | 0.005492673 | Intron |
|
CHR13:95686859|95715113 |
hsa_circ_0008463 | ABCC4 | Up | 0.005715187 | Exon |
|
CHR11:108200941|108202764 |
hsa_circ_0024235 | ATM | Down | 0.005715187 | Exon |
|
CHR18:2890559|2892484 |
hsa_circ_0004658 | EMILIN2 | Up | 0.006202165 | Exon |
|
CHR2:174819601|174820960 |
hsa_circ_0002642 | SP3 | Up | 0.006497297 | Exon |
|
CHR1:46032241|46032692 |
hsa_circ_0000067 | AKR1A1 | Down | 0.006946213 | Exon |
|
CHR9:117116916|117117565 | Novel | AKNA | Down | 0.006977218 | Intron |
|
CHR11:129979324|129992408 |
hsa_circ_0000372 | APLP2 | Up | 0.007668903 | Exon |
|
CHR20:57574398|57574864 | Novel | CTSZ | Up | 0.007894677 | Intron |
|
CHR7:27668990|27672064 |
hsa_circ_0003958 | HIBADH | Down | 0.007894677 | Exon |
|
CHR8:67484717|67485741 |
hsa_circ_0136924 | MYBL1 | Down | 0.007894677 | Exon |
|
CHR10:75830428|75834661 |
hsa_circ_0018881 | VCL | Up | 0.008421014 | Exon |
|
CHR14:91947920|91957146 |
hsa_circ_0003045 | PPP4R3A | Up | 0.00909621 | Exon |
|
CHR3:150834125|150845771 |
hsa_circ_0067735 | MED12L | Up | 0.009420242 | Exon |
|
CHR15:50923614|50941082 |
hsa_circ_0035249 | TRPM7 | Up | 0.009420242 | Exon |
The significantly dysregulated mRNAs are presented
in the heatmap of the inter-sample correlation (Pearson's
correlation coefficient). As indicated in Fig. 2C, the circRNA expression levels were
significantly different between the HCC and control groups, whereas
a slight variation was observed within each group.
Construction of the circRNA-miRNA-mRNA
interaction network
circRNAs inhibit miRNA-mediated gene regulation by
serving as miRNA sponges (44). The
potential target miRNAs of HCC-specific circRNAs were identified
using miRanda. Cytoscape was used to generate a circRNA-miRNA
interaction network through the predicted miRNAs and circRNAs
(39,40). All abnormally expressed circRNAs
were predicted based on complementary sequences of miRNAs. A total
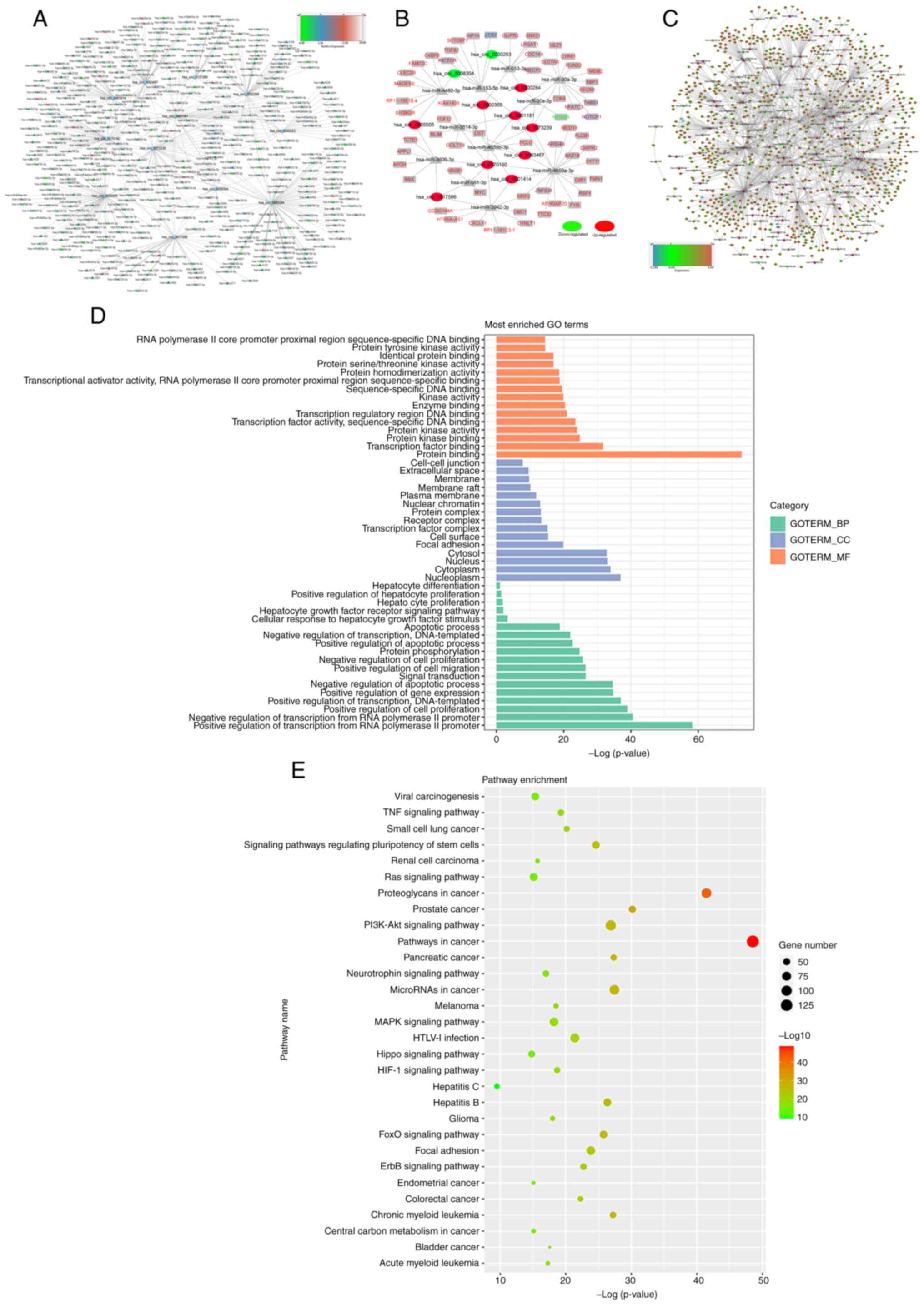
of 514 miRNAs that are able to bind to circRNAs were identified.
The complete network of 11 DE-circRNAs (circular nodes) and target
miRNA (triangular nodes) interactions are delineated in Fig. 3A, where different colors of the
miRNAs presented in the figure indicate differential expression in
HCC compared with normal controls by miRNA sequencing. Increasing
expression levels of the miRNAs is represented as changes in color
increased from green to blue to red, as indicated in the legend bar
on the top right corner of the figure.
To study the potential functions of the circRNAs, a
regulatory network of circRNA-miRNA-mRNA was generated based on
these circRNAs. Fig. 3B indicates
the top 150 circRNA-miRNA-mRNA interactions with significant
P-values (P<0.05). The results suggested that a single circRNA
is able to interact with several miRNAs by acting as an antagonist.
For instance, >50 miRNAs were predicted to bind to
hsa_circ_0070190 and hsa_circ_0001181. Furthermore, both these
circRNAs contained binding sites for hsa-miR-4659b-3p,
hsa-miR-3614-3p and hsa-miR-4659a-3p. Different circRNAs with the
same miRNA binding site may interact with one miRNA and regulate
the expression level of a target gene in HCC. As presented in
Fig. 3C, miRNA/mRNA interactions
were predicted using the miRNA database (Fig. 3C).
As shown in Fig. 3D,
functional annotation indicated that these abnormally expressed
circRNAs have important roles in ‘nucleoplasm’, ‘cytoplasm’ and
‘nucleus’ of cellular components; ‘positive regulation of
transcription from RNA polymerase II promoter’, ‘negative
regulation of transcription from RNA polymerase II promoter’ and
‘positive regulation of cell proliferation’ of biological processes
and molecular functions, including ‘protein binding’,
‘transcription factor binding’ and ‘protein kinase binding’. It was
indicated that the target mRNAs were involved in cancer-related
pathways, including the TNF, MAPK and the PI3K-AKT signaling
pathways, which were implicated in carcinogenesis and the cell
cycle (Fig. 3E). In addition,
several pathways associated with HCC were found, including pathways
in cancer, TNF signaling pathway, hepatitis B, hepatitis C and
hepatocyte differentiation (Fig.
3E).
Validation of the DE-circRNAs
To verify the sequencing and prediction results, the
expression levels of the top six circRNAs annotated in circBase
were analyzed, including five upregulated and one downregulated
circRNA from the 28 clinical cases with HCC and 22 healthy controls
by RT-qPCR. GAPDH was used as an internal control. The results
indicated that the relative expression levels of hsa_circ_0017586,
hsa_circ_0001181, hsa_circ_0073239, hsa_circ_0070190 and
hsa_circ_0000369 in PBMCs from patients with HCC were significantly
higher compared with those in healthy controls, whereas
hsa_circ_0008304 was downregulated in patients with HCC compared
with that in the controls (Fig.
4A). These results are consistent with the results of the
RNA-seq analysis (Fig. 4B).
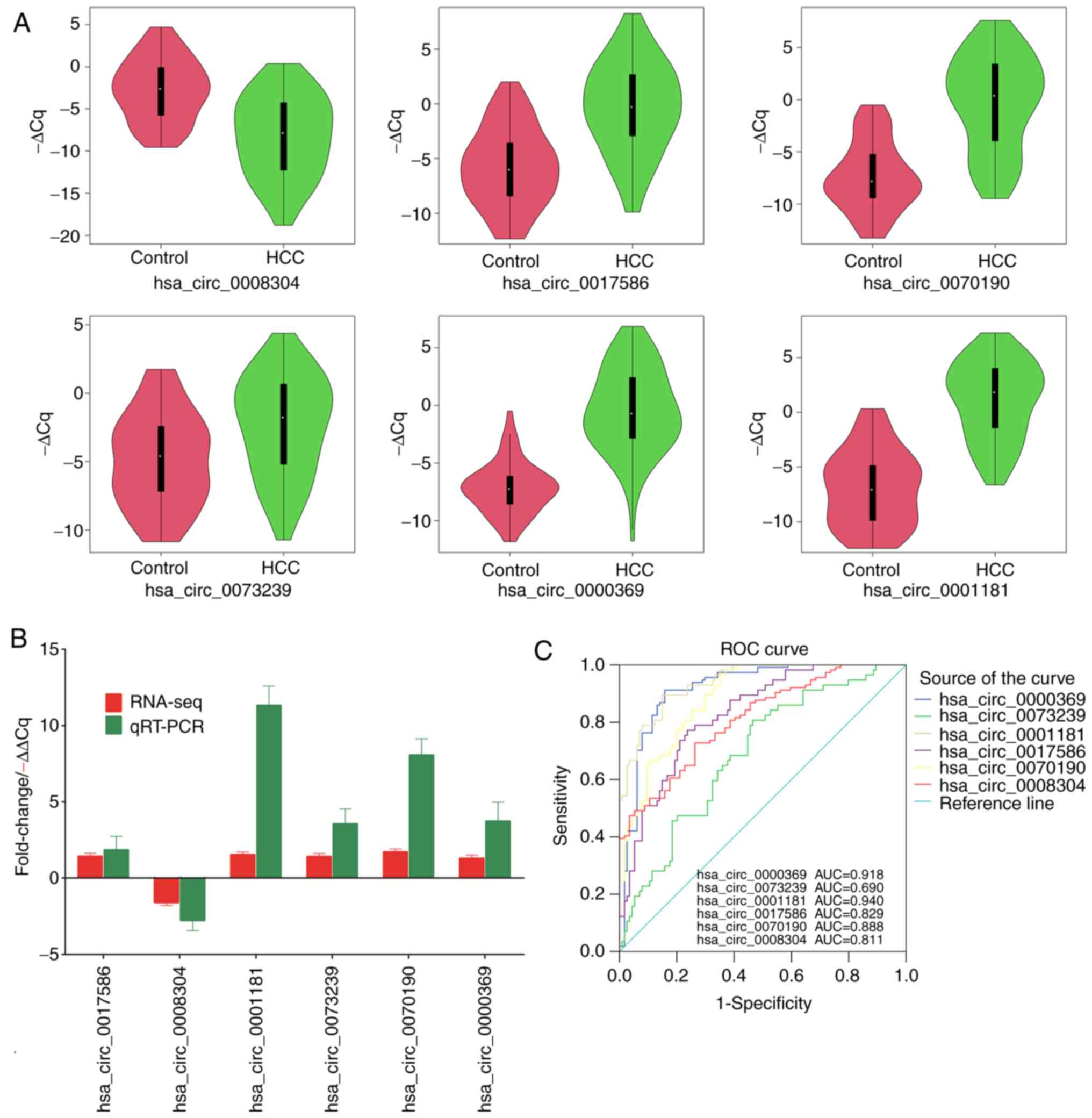 | Figure 4Validation of the expression of
significant transcripts by RT-qPCR and ROC analysis of the
circRNAs. (A) Violin plots displaying the expression levels of the
six circRNAs measured by RT-qPCR. Significant differences were
present between HCC and healthy controls (P<0.001). (B)
Comparison of RNA-seq results and RT-qPCR results in the six
circRNAs. (C) ROC curve analysis of hsa_circ_0017586,
hsa_circ_0008304, hsa_circ_0001181, hsa_circ_0073239,
hsa_circ_0070190 and hsa_circ_0000369. AUC values are provided on
the graphs. ROC, receiver operating characteristic; AUC, area under
the ROC curve; RT-qPCR, reverse transcription-quantitative PCR;
circRNA, circular RNA; hsa, Homo sapiens; HCC,
hepatocellular carcinoma; Cq, quantification threshold;
Fold-change, the ratio of normalized sequencing counts between HCC
group and control group; RT-qPCR, reverse
transcription-quantitative PCR. |
ROC curve analysis was performed to evaluate the
potential of the circRNAs as diagnostic biomarkers in all 50
samples (28 HCC and 22 controls) collected for the present study.
As presented in Fig. 4C, the area
under the ROC curve to differentiate patients with HCC from healthy
controls was 0.690 (95% CI: 0.622-0.758) for hsa_circ_0073239,
0.888 (95% CI: 0.847-0.928) for hsa_circ_0070190, 0.918 (95% CI:
0.881-0.956) for hsa_circ_0000369, 0.811 (95% CI: 0.757-0.865) for
hsa_circ_0008304, 0.829 (95% CI: 0.777-0.881) for hsa_circ_0017586
and 0.940 (95% CI: 0.913-0.967) for hsa_circ_0001181, indicating
the potential of these circRNAs as diagnostic markers for HCC.
Among them, hsa_circ_0001181 and hsa_circ_0000369 had the highest
potential as diagnostic markers and may be used for diagnosing
HCC.
circRNAs may be involved in the
regulation of HCC cell growth
The results of the CCK-8 proliferation assay
(Fig. 5A) were analyzed by ANOVA.
It was indicated that significant differences were present between
different groups and different times (P<0.001), suggesting that
the knockdown of hsa_circ_0001181 expression in HepG2 cells
significantly reduced cell growth compared to the blank (no
transfection) and NC (transfected with empty pGLV2-U6 lentivector)
groups, particularly at 48 and 72 h. There was no significant
difference between the blank and NC groups (P=0.056), suggesting
that cell proliferation was specifically affected by
hsa_circ_0001181 and not by the vector. RT-qPCR was performed to
confirm the knockdown efficiency and the results indicated that the
expression level of hsa_circ_0001181 was significantly
downregulated following hsa_circ_0001181 silencing (Fig. 5B).
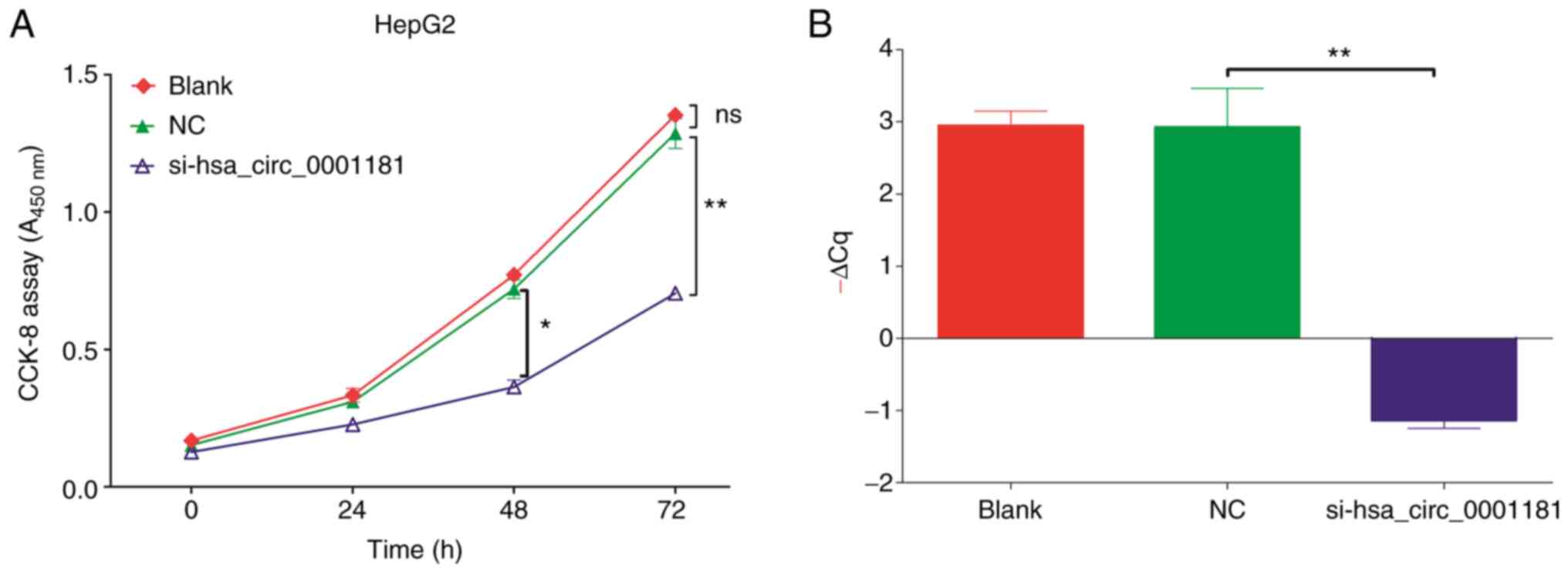 | Figure 5(A) The proliferation of HepG2 cells
was evaluated using a CCK-8 assay with transfection of
si-hsa_circ_0001181 after 0, 24, 48 and 72 h. The A450
nm was measured to reflect the amount of viable cells. The
cell proliferation level of the si-hsa_circ_0001181 group was
significantly lower than that of the blank or NC group after 48 h
and highly significantly lower after 72 h. However, the blank group
and NC group exhibited no significant difference. (B) Reverse
transcription-quantitative PCR detection of the circRNA knockdown
efficiency, -ΔCq was used to denote the expression level.
*P<0.05, **P<0.01. ns, no significance;
A450 nm, absorbance at 450 nm; NC, negative control;
si-hsa_circ_0001181, small inhibitory RNA targeting Homo
sapiens circRNA 0001181; circRNA, circular RNA; -ΔCq,
quantification threshold. |
Discussion
CircRNAs were once thought of as by-products of
aberrant RNA splicing, but with the development of RNA-seq and
bioinformatics technologies in the past few decades, a number of
circRNAs have been functionally characterized, revealing their
ability to adsorb to miRNA (45)
and regulate the expression of the target gene(s) (46). Studies have also indicated that
circRNAs may be translated into functional proteins (47). In addition, studies have revealed
that circRNAs are highly abundant, stable and resistant to
exonuclease (8,48). Furthermore, differences in circRNAs
exist among different tissues and diseases (49,50).
Thus, circRNAs have demonstrated great potential for application in
early diagnosis and as therapeutic targets, and it is worthwhile
investigating their mechanistic involvement in disease occurrence
and development.
Several studies on cancers have focused on PBMCs
(51-53).
Certain studies have also reported an association with
immunological diseases. PBMCs consist of monocytes and lymphocytes,
which originate from the innate and adaptive immune systems.
Changes in gene expression in PBMCs are generally associated with
the status of tumors and tumor-related immune diseases (54), suggesting that the DE-circRNAs in
PBMCs may be important potential biomarkers for the diagnosis and
prognosis of liver diseases.
In the present study, circRNAs were identified in
PBMCs from 19 patients with HCC and 19 controls using RNA-seq.
Profiling of the DE-circRNAs led to the identification of 40
DE-circRNAs between HCC and healthy controls. Among them, 27 were
upregulated and 13 were downregulated and of these, certain
selected DE-circRNAs were confirmed by RT-qPCR. For the ROC
analysis, 50 PBMCs samples were involved, which includes 38 samples
were used for sequencing and another 12 PBMCs samples collected. It
is a limitation of the present study that there was no separate
cohort for RT-qPCR validation and further patients with HCC will be
collected for future study. Among the DE-circRNAs, hsa_circ_0001181
and hsa_circ_000369 demonstrated a high diagnostic potential in
patients with HCC and represent potential diagnostic biomarkers for
HCC prediction. A CCK-8 assay was performed and the results
indicated that silencing of hsa_circ_0001181 reduced the
proliferation of HepG2 cells. This suggests that circRNA 0001181
may be involved in the genesis and development of HCC and may thus
serve as a potential therapeutic target for HCC.
To investigate the potential function of these
circRNAs profiled, a circRNA-miRNA-mRNA interaction network was
constructed. Most of the circRNAs were newly identified to be
related to HCC. As one of the types of competing endogenous RNAs,
circRNAs have important roles in regulating gene expression through
sequestering miRNAs. Numerous miRNAs in the network were studied
previously, such as hsa-miR-30a-3p, which had been reported to be
related to numerous cancer types, including lung (55), breast (56), prostatic (57) and pancreatic cancer (58). It has also been reported to be
related to hepatic astrocyte fibrosis and autophagy (59). It is the target of hsa-circ-0001181,
hsa-circ-0073239 and hsa-circ-0000284, which were determined to be
significantly dysregulated in HCC in the present study. It is
inferred that these circRNAs may have important roles in the
genesis and development of tumors in numerous cancer types.
In the present study, >100,000 circRNAs were
profiled from PBMCs of patients with HCC and only those with counts
of >2 were considered to be reliable circRNAs for the follow-up
experiments. Differential expression analysis was performed only
for these co-expressed circRNAs and this may be the main reason why
only 40 DE-circRNAs were identified in the present study.
Although only a limited number of DE-circRNAs were
identified in the present study, most of them were detected for the
first time in patients with HCC and they were confirmed by RT-qPCR.
Furthermore, GO functional annotation results indicated that these
DE-circRNAs were mainly enriched in protein binding, regulation of
cell proliferation, signal transduction, protein phosphorylation,
hepatocyte differentiation, among others, and were mainly involved
in cancer-associated pathways related to HCC. These included
pathways in cancer, the TNF signaling pathway, ErbB signaling
pathway, MAPK signaling pathway, hepatitis B, hepatitis C and
hepatocyte differentiation, which indicates that the host genes of
these circRNAs may participate in the regulation of HCC development
through related molecular functions or signaling pathways. Ni et
al (60) reported that lncRNA
inhibits HCC progression through CUL4A-mediated LATS1
ubiquitination and increased YAPS127 phosphorylation. Wang et
al (61) indicated that SIRT4
inhibits glutamine metabolism and increases the level of adenosine
diphosphate/adenosine monophosphate (AMP) and blocks the mTOR
signaling pathway through phosphorylation of AMP kinase α that has
an antitumor effect in HCC.
In the present study, circRNA expression patterns in
PBMCs from HCC were determined by RNA-seq, indicating that patients
with HCC had distinct circRNA expression signatures when compared
to healthy controls. ROC analysis indicated that hsa_circ_0073239,
hsa_circ_007090, hsa_circ_0008304, hsa_circ_0017586,
hsa_circ_0001181 and hsa_circ_0000369 may potentially be used as
biomarkers for HCC diagnosis. In addition, silencing of
hsa_circ_0001181 reduced the growth of HepG2 cells, suggestive of
its potential therapeutic target of HCC.
Supplementary Material
Clinicopathological data of patients
with HCC and healthy controls.
Acknowledgements
Not applicable.
Funding
This research was funded by the Shenzhen Science and Technology
Project and Sanming Project of Medicine in Shenzhen (grant nos.
JCYJ20170817094901026, JCYJ20180302173542393 and
SZSM201612074).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI project repository
(accession no. PRJNA754685; https://dataview.ncbi.nlm.nih.gov/object/PRJNA754685?reviewer=ofg2bodnrahhf5de9utin5g22k).
Authors' contributions
XZ and ZH contributed to the conception, design,
performing of experiments and analysis of the data. XS, RH and WM
contributed to sample collection and cell culture. WZ, SX, BZ, LZ
and QL contributed to sample collection and CCK-8 assay. QG, LZ and
ZH contributed to the bioinformatics analysis. WF and RH
contributed to the acquisition of data and cell culture. ZH and WF
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol was established, according
to the ethical guidelines of the Helsinki Declaration and was
approved by the Human Ethics Committee of Shenzhen Traditional
Chinese Medicine Hospital (Shenzhen, China). Written informed
consent was obtained from individual or guardian participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Balogh J, Victor D III, Asham EH,
Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM and Monsour
HP Jr: Hepatocellular carcinoma: A review. J Hepatocell Carcinoma.
3:41–53. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia
X, Zhao W, Huai W, Qiu Y, Sun L and Han L: MiR-424-5p reversed
epithelial-mesenchymal transition of anchorage-independent HCC
cells by directly targeting ICAT and suppressed HCC progression.
Sci Rep. 4(6248)2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chan KM, Kuo CF, Hsu JT, Chiou MJ, Wang
YC, Wu TH, Lee CF, Wu TJ, Chou HS and Lee WC: Metformin confers
risk reduction for developing hepatocellular carcinoma recurrence
after liver resection. Liver Int. 37:434–441. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wu TJ, Chang SS, Li CW, Hsu YH, Chen TC,
Lee WC, Yeh CT and Hung MC: Severe hepatitis promotes
hepatocellular carcinoma recurrence via NF-kB pathway-mediated
epithelial-mesenchymal transition after resection. Clin Cancer Res.
22:1800–1812. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15(409)2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ren S, Xin Z, Xu Y, Xu J and Wang G:
Construction and analysis of circular RNA molecular regulatory
networks in liver cancer. Cell Cycle. 16:2204–2211. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang M, Gu B, Yao G, Li P and Wang K:
Circular RNA expression profiles and the pro-tumorigenic function
of circRNA_10156 in hepatitis B virus-related liver cancer. Int J
Med Sci. 17:1351–1365. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhang P, Ming Y, Ye Q and Niu Y:
Comprehensive circRNA expression profile during ischemic
postconditioning attenuating hepatic ischemia/reperfusion injury.
Sci Rep. 9(264)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jiang L, Wang X, Zhan X, Kang S, Liu H,
Luo Y and Lin L: Advance in circular RNA modulation effects of
heart failure. Gene X. 5(100036)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ren S, Lin P, Wang J, Yu H, Lv T, Sun L
and Du G: Circular RNAs: Promising molecular biomarkers of human
aging-related diseases via functioning as an miRNA sponge. Mol Ther
Methods Clin Dev. 18:215–229. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Joshi SS and Coffin CS: Hepatitis B virus
lymphotropism: Emerging details and challenges. Biotechnol Genet
Eng Rev. 34:139–151. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Szilágyi M, Pös O, Márton É, Buglyó G,
Soltész B, Keserű J, Penyige A, Szemes T and Nagy B: Circulating
cell-free nucleic acids: Main characteristics and clinical
application. Int J Mol Sci. 21(6827)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ho-Xuan H, Glažar P, Latini C, Heizler K,
Haase J, Hett R, Anders M, Weichmann F, Bruckmann A, Van den Berg
D, et al: Comprehensive analysis of translation from overexpressed
circular RNAs reveals pervasive translation from linear
transcripts. Nucleic Acids Res. 48:10368–10382. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Que F, Wang H, Luo Y, Cui L, Wei L, Xi Z,
Lin Q, Ge Y and Wang W: Comprehensive analysis of differentially
expressed circRNAs reveals a colorectal cancer-related ceRNA
network. Comput Math Methods Med. 2020(7159340)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Albini A, Magnani E and Noonan DM: The
tumor microenvironment: Biology of a complex cellular and tissue
society. Q J Nucl Med Mol Imaging. 54:244–248. 2010.PubMed/NCBI
|
|
23
|
Grade M, Ghadimi BM, Varma S, Simon R,
Wangsa D, Barenboim-Stapleton L, Liersch T, Becker H, Ried T and
Difilippantonio MJ: Aneuploidy-dependent massive deregulation of
the cellular transcriptome and apparent divergence of the
Wnt/beta-catenin signaling pathway in human rectal carcinomas.
Cancer Res. 66:267–282. 2006.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Navin NE: The first five years of
single-cell cancer genomics and beyond. Genome Res. 25:1499–1507.
2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Rugbjerg P and Sommer MOA: Overcoming
genetic heterogeneity in industrial fermentations. Nat Biotechnol.
37:869–876. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hashimoto K, Kouno T, Ikawa T, Hayatsu N,
Miyajima Y, Yabukami H, Terooatea T, Sasaki T, Suzuki T, Valentine
M, et al: Single-cell transcriptomics reveals expansion of
cytotoxic CD4 T cells in supercentenarians. Proc Natl Acad Sci USA.
116:24242–24251. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bureau of Medical Administration National
Health, Family Planning Comission of the People's Republic of
China. Diagnosis, management, and treatment of hepatocellular
carcinoma (V2017). Zhonghua Gan Zang Bing Za Zhi. 25:886–895.
2017.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
28
|
Marioni JC, Mason CE, Mane SM, Stephens M
and Gilad Y: RNA-seq: An assessment of technical reproducibility
and comparison with gene expression arrays. Genome Res.
18:1509–1517. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jia E, Zhou Y, Liu Z, Wang L, Ouyang T,
Pan M, Bai Y and Ge Q: Transcriptomic profiling of circular RNA in
different brain regions of Parkinson's disease in a mouse model.
Int J Mol Sci. 21(3006)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y,
Li S, Li Y, Ye J, Yu C, Li Z, et al: SOAPnuke: A MapReduce
acceleration-supported software for integrated quality control and
preprocessing of high-throughput sequencing data. Gigascience.
7:1–6. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang L, Liu C and Dong S: PipeMEM: A
framework to speed Up BWA-MEM in spark with low overhead. Genes
(Basel). 10(886)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Giannoulatou E, Park SH, Humphreys DT and
Ho JW: Verification and validation of bioinformatics software
without a gold standard: A case study of BWA and Bowtie. BMC
Bioinformatics. 15 (Suppl 16)(S15)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16(4)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11(R106)2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li S, Chen L, Xu C, Qu X, Qin Z, Gao J, Li
J and Liu J: Expression profile and bioinformatics analysis of
circular RNAs in acute ischemic stroke in a South Chinese Han
population. Sci Rep. 10(10138)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11(R14)2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36:D480–D484. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5(R1)2003.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang Z, Fan Y, Xie F, Zhou H, Jin K, Shao
L, Shi W, Fang P, Yang B, van Dam H, et al: Breast cancer
metastasis suppressor OTUD1 deubiquitinates SMAD7. Nat Commun.
8(2116)2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Tang X, Ren H, Guo M, Qian J, Yang Y and
Gu C: Review on circular RNAs and new insights into their roles in
cancer. Comput Struct Biotechnol J. 19:910–928. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xu T, Wu J, Han P, Zhao Z and Song X:
Circular RNA expression profiles and features in human tissues: A
study using RNA-seq data. BMC Genomics. 18 (Suppl
6)(680)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16(94)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Shen Y, Bu L, Li R, Chen Z, Tian F, Lu N,
Ge Q, Bai Y and Lu Z: Screening effective differential expression
genes for hepatic carcinoma with metastasis in the peripheral blood
mononuclear cells by RNA-seq. Oncotarget. 8:27976–27989.
2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Li Z, Yang X, Zhang Y, Yang X, Cui X,
Zhang Y, Gong W, Bai H, Liu N, Tang Z, et al: A human peripheral
blood mononuclear cell (PBMC) engrafted humanized xenograft model
for translational immuno-oncology (I-O) research. J Vis Exp
2019.
|
|
53
|
Barbon CM, Janec KJ, Kelner RH, Norton JE
and Guinan EC: Alloanergization method for inducing allospecific
hyporesponsiveness in PBMC exposed to allostimulation in vitro in
the context of costimulatory molecule blockade. Methods Mol Biol.
1899:103–118. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cheng WC, Saleh F, Abuaisha Karim B,
Hughes FJ and Taams LS: Comparative analysis of immune cell subsets
in peripheral blood from patients with periodontal disease and
healthy controls. Clin Exp Immunol. 194:380–390. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yuan Y, Zheng S, Li Q, Xiang X, Gao T, Ran
P, Sun L, Huang Q, Xie F, Du J and Xiao C: Overexpression of
miR-30a in lung adenocarcinoma A549 cell line inhibits migration
and invasion via targeting EYA2. Acta Biochim Biophys Sin
(Shanghai). 48:220–228. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J,
Cheng L, Fan Z, Yuan B, Tian P, et al: miR-30a suppresses breast
cancer cell proliferation and migration by targeting Eya2. Biochem
Biophys Res Commun. 445:314–319. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wang T, Chen G, Ma X, Yang Y, Chen Y, Peng
Y, Bai Z, Zhang Z, Pei H and Guo W: MiR-30a regulates cancer cell
response to chemotherapy through SNAI1/IRS1/AKT pathway. Cell Death
Dis. 10(153)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Yang C, Zhang JJ, Peng YP, Zhu Y, Yin LD,
Wei JS, Gao WT, Jiang KR and Miao Y: A Yin-Yang 1/miR-30a
regulatory circuit modulates autophagy in pancreatic cancer cells.
J Transl Med. 15(211)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Chen J, Yu Y, Li S, Liu Y, Zhou S, Cao S,
Yin J and Li G: MicroRNA-30a ameliorates hepatic fibrosis by
inhibiting Beclin1-mediated autophagy. J Cell Mol Med.
21:3679–3692. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Ni W, Zhang Y, Zhan Z, Ye F, Liang Y,
Huang J, Chen K, Chen L and Ding Y: A novel lncRNA uc.134 represses
hepatocellular carcinoma progression by inhibiting CUL4A-mediated
ubiquitination of LATS1. J Hematol Oncol. 10(91)2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Wang YS, Du L, Liang X, Meng P, Bi L, Wang
YL, Wang C and Tang B: Sirtuin 4 depletion promotes hepatocellular
carcinoma tumorigenesis through regulating
adenosine-monophosphate-activated protein kinase alpha/mammalian
target of rapamycin axis in mice. Hepatology. 69:1614–1631.
2019.PubMed/NCBI View Article : Google Scholar
|