Introduction
Abdominal aortic aneurysm (AAA) is a degenerative
vascular disease caused by the progressive weakening and dilation
of abdominal aorta with a diameter >3-cm (1). As a complex and serious vascular
surgery disease, AAA has an extremely high mortality rate,
especially in populations aged >65 years (2). Immune inflammatory response,
extracellular matrix degradation and vascular smooth muscle cell
(VSMC) dysfunction are suggested to be important factors in AAA
formation (3). However, the exact
regulatory mechanisms underlying AAA remain unclear. In addition,
more studies and experiments are still needed to explore how to
prevent AAA formation and progression earlier.
Non-coding RNA (ncRNA) is composed of diverse RNA
transcripts including microRNA (miRNA), long non-coding RNA
(lncRNA) and circular RNA (circRNA), a number of which are known to
act as transcriptional regulators (4). miRNAs that are 21-23 nucleotides in
length can specifically bind with messenger RNAs (mRNAs) and
reversely regulate the expression of genes (5). By contrast, lncRNAs and circRNAs can
function as competing endogenous RNAs (ceRNAs) and sponge miRNAs,
thus offsetting the negative regulation of miRNAs on target mRNAs
(6). The role of the ceRNA network
is extensive and has been observed in the regulation of disease
progression, including cardiovascular problems (7). This builds a notable basis for
exploring pathogenetic mechanisms of AAA through constructing a
ceRNA network mediated via lncRNAs or circRNAs.
In fact, to the best of our knowledge, the majority
of experimental studies mainly focus on the effects of
lncRNA-miRNA-mRNA axes on AAA risk (8-12).
The evolution of novel computational tools is a rapidly growing
field of interest, which provides putative predictions of ceRNA
interactions (7). Based on
bioinformatics analysis, Tian et al (13) constructed lncRNA-miRNA-mRNA
networks for AAA to reveal six novel lncRNA-miRNA-mRNA axes (five
upregulated and one downregulated) closely associated with the
pathogenesis of AAA. Current evidence has confirmed that circRNAs
may have effects on AAA development through modulating endothelial
cells, macrophages and VSMCs (14). Zhao et al (15) reported that circRNA cerebellar
degeneration-related protein 1 antisense RNA is downregulated in
AAA and is associated with the upregulation of miR-7, resulting in
the low-degree expression of cytoskeleton-associated protein 4 and
dysregulation of VSMCs. Yue et al (16) identified that circRNA core-binding
factor subunit β is a sponge of miR-28-5p that is highly expressed
in AAA and that induces the downregulation of glutamate ionotropic
receptor AMPA type subunit 4 and LY6/PLAUR domain-containing 3,
both of which are linked with VSMC apoptosis. However, data on
circRNAs in human AAA are limited, and there is still a lack of
systematic analysis on circRNA-miRNA-mRNA interaction networks in
AAA based on bioinformatics analysis.
The current bioinformatics study aimed to establish
AAA-associated circRNA-miRNA-mRNA networks and to identify the
center ceRNA axes. Subsequently, the expression levels of mRNAs
involved in center axes were verified by reverse
transcription-quantitative PCR (RT-qPCR). In addition, the
potential biological functions of circRNA-miRNA-mRNA axes that are
closely linked with AAA were explored and the clinical significance
of key ceRNA-associated mRNAs was evaluated. The results of the
present study may aid the improved understanding of AAA
pathogenesis, especially in terms of ceRNA implication; it may also
provide some potential molecular targets and novel ideas for
further experiments to explore novel therapeutic approaches for
AAA.
Materials and methods
Microarray datasets
First, appropriate public datasets of human AAA were
collected by using Gene Expression Omnibus (GEO) database in
National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/). Overall, three
microarray datasets (GSE7084, GSE57691 and GSE144431) were included
in the present study (Table SI)
(17-19).
In GSE7084, seven AAA samples and eight healthy samples were
involved for mRNA expression profiles by GPL2507 platform. In
GSE57691, 49 AAA samples and 10 healthy samples were included for
mRNA expression profiles using GPL10558 platform. In GSE144431,
four AAA samples and four healthy samples were involved for circRNA
expression profiles by GPL21825 platform. Principal component
analysis (PCA) was utilized to reduce the dimension and assess the
data availability of aforementioned datasets (20). The PCA score plots of dimension
(Dim) 1 and 2 were visualized using R studio (version 4.0.3)
(21). Superior group
differentiation is demonstrated by higher Dim scores, indicating
the availability of datasets.
Differential expression analysis
Steps of differential expression analysis of each
dataset are presented below. The differentially expressed genes
(DEGs) and differentially expressed circRNAs (DECs) were detected
by applying the limma R package (version 3.46.0; https://bioconductor.org/packages/limma/) with the
statistical threshold of P<0.05 and |log2fold change
(FC)| >1.5 in R studio (version 4.0.3) (21). The DEGs and DECs were annotated
according to the corresponding platform information, respectively.
Pheatmap R package (version 1.0.12; https://cran.r-project.org/web/packages/pheatmap/index.html)
was used to generate heatmaps in R studio (version 4.0.3) (21). DEGs in GSE7084 and GSE57691 or DECs
in GSE144431 have been annotated as official symbols already.
However, due to the different sequencing chips used, GSE7084 and
GSE57691 may have different official symbols for the same DEGs.
Therefore, the public database for Annotation, Visualization and
Integrated Discovery Online tool (DAVID; version 6.8; https://david.ncifcrf.gov/) was used to convert the
DEGs of GSE7084 and GSE57691 into uniform Entrez_Gene_ID (22). In this way, the Entrez_Gene_ID of
DEGs could be overlapped between GSE7084 and GSE57691 to obtain the
common up and downregulated DEGs. However, this process was not
necessary for GSE144431 since GSE144431 only contained information
for DECs, which cannot be overlapped with DEG datasets.
Construction of circRNA-miRNA-mRNA
networks and center axis exploration
Subsequently, online tools were used to predict the
potential target miRNAs of DEGs and DECs, respectively. The
miRNA-circRNA pairs were predicted through the CircInteractome
database (version 1.0) (https://circinteractome.nia.nih.gov/index.html)
(23). The miRWalk database
(version 2.0) (http://mirwalk.umm.uni-heidelberg.de/) (24) and TargetScan database (version 7.2)
(http://www.targetscan.org) were used to
predict miRNA-mRNA pairs (25).
Furthermore, miRNA-circRNA and miRNA-mRNA pairs were overlapped to
select common target miRNAs of DEGs and DECs. Finally, upregulated
circRNAs/mRNAs and downregulated circRNAs/mRNAs were selected to
construct two different ceRNA networks, respectively. Cytoscape
software (version 3.6.1) (26) was
used for the visualization of ceRNA networks. The molecular complex
detection (MCODE) method (a plugin in Cytoscape; version 2.0.0) was
performed to identify highly interconnected clusters in ceRNA
networks (27), and the
circRNA-miRNA-mRNA axes involved in the clusters were considered as
center axes. The criterion for MCODE analysis was set up as default
parameters.
Patient sample collection and RT-qPCR
validation
RT-qPCR validation was conducted on DEGs in the
center ceRNA axes. A total of four patients with AAA had large and
non-ruptured cases and full-thickness aneurysmal abdominal aortic
tissues were collected from them during the open surgical repair,
whereas four control samples were derived from non-aneurysmal
abdominal aortas of organ donors during kidney transplantation. The
study was approved by the Ethics Committee of the First Hospital of
China Medical University (approval no. 2020-146-2; Shenyang, China)
and was carried out in accordance with the Declaration of Helsinki
(28). Written informed consent
was obtained from each patient. Characteristics of AAA and control
subjects are presented in Table
SII. All patients with AAA were males with ages of 68.0±7.8
years, whereas control individuals had the age distribution of
58.3±3.9 years, with and one female and three males. Sample
collection took place in the First Hospital of China Medical
University between July 2020 and April 2021. Patients with AAA were
diagnosed using CT angiography and underwent open surgery.
Individuals with congenital disorders, malignant tumors,
hematological diseases, autoimmune diseases, severe organ failure
or previous aortic surgery were excluded.
Whole RNA in samples was extracted from tissues
using RNAiso Plus (Takara Bio, Inc.). The gDNA Eraser with strong
DNA decomposition activity can remove genomic DNA at 42˚C for 2 min
and the samples treated by gDNA Eraser can be directly synthesized
into cDNA by reverse transcription reaction at 37˚C for 15 min
using PrimeScript™ RT Reagent kit (Takara Bio, Inc.). RT-qPCR was
performed with the SYBR® Premix Ex Taq™ II (Takara Bio,
Inc.) according to the manufacturer's protocol. RT-qPCR
thermocycling conditions included: Initial denaturation step at
95˚C for 2 min, followed by 40 cycles of 10 sec at 95˚C, 20 sec at
58˚C and 30 sec at 72˚C. β-actin was used as the reference gene to
calculate relative fold difference using 2-ΔΔCq method
(29). Primer sequences for CNN1,
CD8a molecule (CD8A) and β-actin are presented in Table SIII. Gene-specific primers were
designed with Primer Express Software 3.0 (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and purchased from Beijing Genomics
Institute (https://www.genomics.cn/).
Statistical analysis
Quantitative data are expressed as means ± standard
deviation and were analyzed using SPSS 17.0 statistical software
(SPSS, Inc.). The unpaired t-test was applied for comparisons
between AAA and control groups. P<0.05 was considered to
indicate a statistically significant difference.
Functional and pathway enrichment for
DEGs
The results of Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways (https://www.genome.jp/kegg/) and Gene Ontology (GO)
terms (https://geneontology.org/) including
biological processes (BP), cellular component (CC) and molecular
function (MF) enrichment analyses for common DEGs were obtained
using the DAVID online tool (version 6.8; https://david.ncifcrf.gov/) (22). The threshold was P<0.05. GOplot
R package (version 1.0.2) (30)
was used for the visualization of the enrichment analysis results
and z-score was introduced to evaluate the results.
Construction of
circRNA-miRNA-mRNA-biological function networks
Based on the previous GO analysis, DEG-GO terms
pairs and circRNA-miRNA-mRNA-biological function networks were
constructed using Cytoscape software (version 3.6.1) (26). Similarly, upregulated circRNA/mRNA
and downregulated circRNAs/mRNAs were selected to construct two
separate networks.
Receiver operating characteristic
(ROC) analysis
To further explore the clinical diagnostic value of
circRNA-miRNA-mRNA-biological process network-associated DEGs, ROC
analysis was conducted through ROC R package (version 1.68.1;
https://bioconductor.org/packages/release/bioc/html/ROC.html)
in R studio (version 4.0.3) (21).
The data were derived from gene mRNA expression levels in each
sample of GSE7084 and GSE57691 with a further normalization.
Results
Identification of DEGs and DECs
The workflow of the present study is presented in
Fig. 1. PCA confirmed a clear
separation between AAA and healthy groups of three datasets
(Fig. S1A). Fig. S1B indicates the 643 DEGs, 454 DEGs
and 142 DECs in GSE7084, GSE57691 and GSE144431, respectively.
Matrix metallopeptidase 9 (MMP9) is the most upregulated DEG and
protein phosphatase 1 regulatory subunit 3C (PPP1R3C) is the most
downregulated DEG in the heatmap of GSE7084 (Fig. S1C). For the heatmap of GSE57691,
Ig kappa chain V-III region HAH-like (LOC642113) is the most
upregulated DEG and integrin subunit alpha 8 (ITGA8) is the most
downregulated DEG (Fig. S1D). In
the heatmap of GSE144431, hsa_circ_0001588 represents the most
upregulated DEC and hsa_circ_0092291 represents the most
downregulated DEC (Fig. S1E).
Tables SIV and SV present a total of 643 DEGs and 454
DEGs in GSE7084 and GSE57691, respectively. Finally, a total of 135
common DEGs (62 upregulated and 73 downregulated) between GSE7084
and GSE57691 (Fig. S1F and
Table SVI) and 142 DECs (64
upregulated and 78 downregulated) in GSE144431 (Table SVII) were identified for the
construction of ceRNA networks.
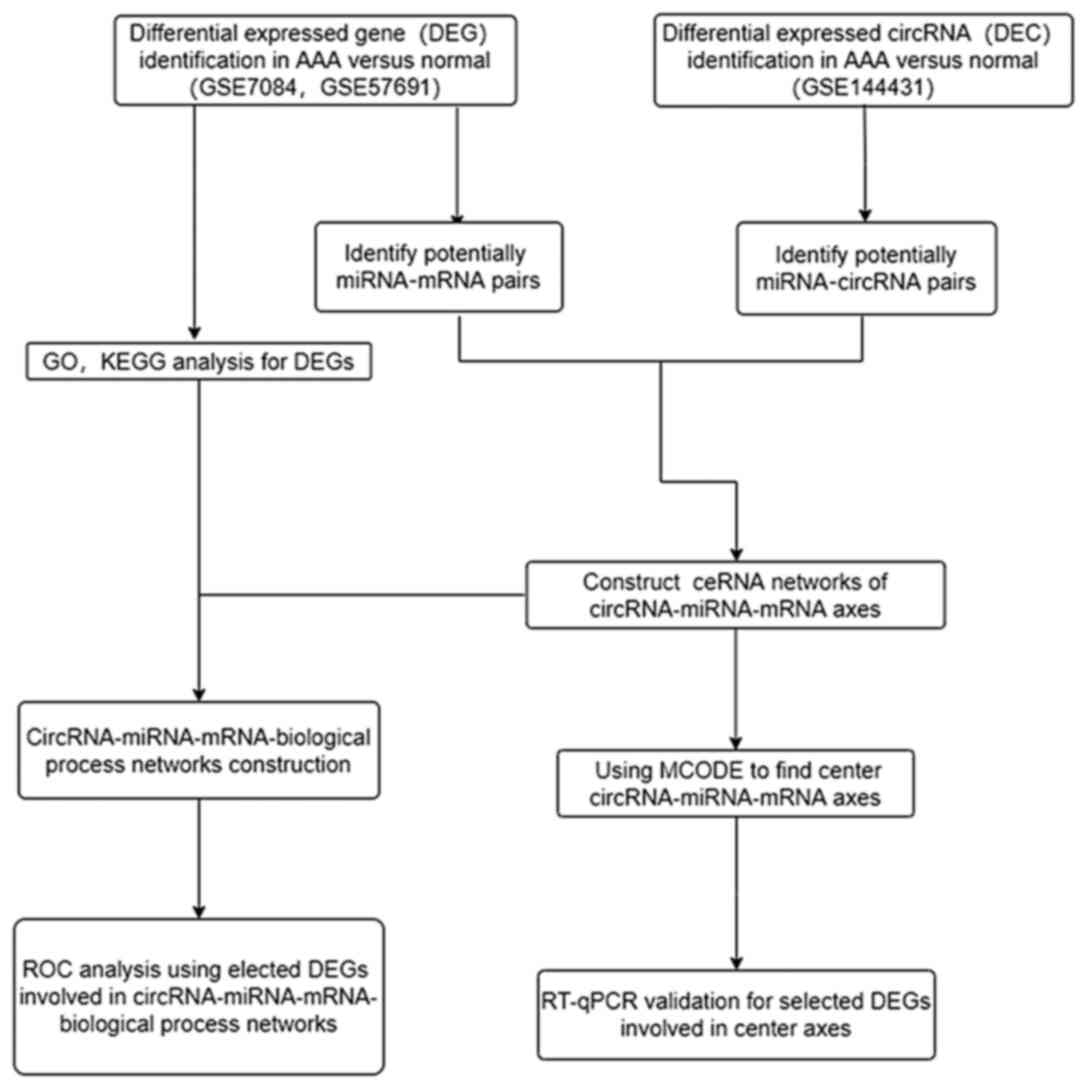 | Figure 1Workflow for the present study. DEGs,
differentially expressed genes; AAA, abdominal aortic aneurysm;
circRNA, circular RNA; miRNA, microRNA; mRNA, messenger RNA; GO,
Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes;
ceRNA, competing endogenous RNA; RT-qPCR, reverse
transcription-quantitative; ROC, receiver operating
characteristic. |
Construction of ceRNA regulatory
networks in AAA
To explore the effect of DECs on DEGs mediated by
binding with miRNAs, upregulated and downregulated ceRNA networks
were constructed. As presented in Fig. S2, the upregulated ceRNA networks
contained four circRNA-miRNA-mRNA axes [CD8A, IL-10 receptor
subunit α (IL10RA), semaphorin 4A (SEMA4A) and cell migration
inducing hyaluronan-binding protein (CEMIP)] and downregulated
ceRNA networks contained eight circRNA-miRNA-mRNA axes [atonal bHLH
transcription factor 8 (ATOH8), prune homolog 2 with BCH domain
(PRUNE2), calponin 1 (CNN1), zinc finger and BTB domain-containing
16 (ZBTB16), sclerostin (SOST), transmembrane protein 47 (TMEM47),
F-box protein 32 (FBXO32) and nuclear factor IA (NFIA)]. In
addition, Fig. S3 displays a
total of 12 circRNA-miRNA-mRNA networks constructed based on every
single DEG.
Identification of center ceRNA axes
and RT-qPCR validation
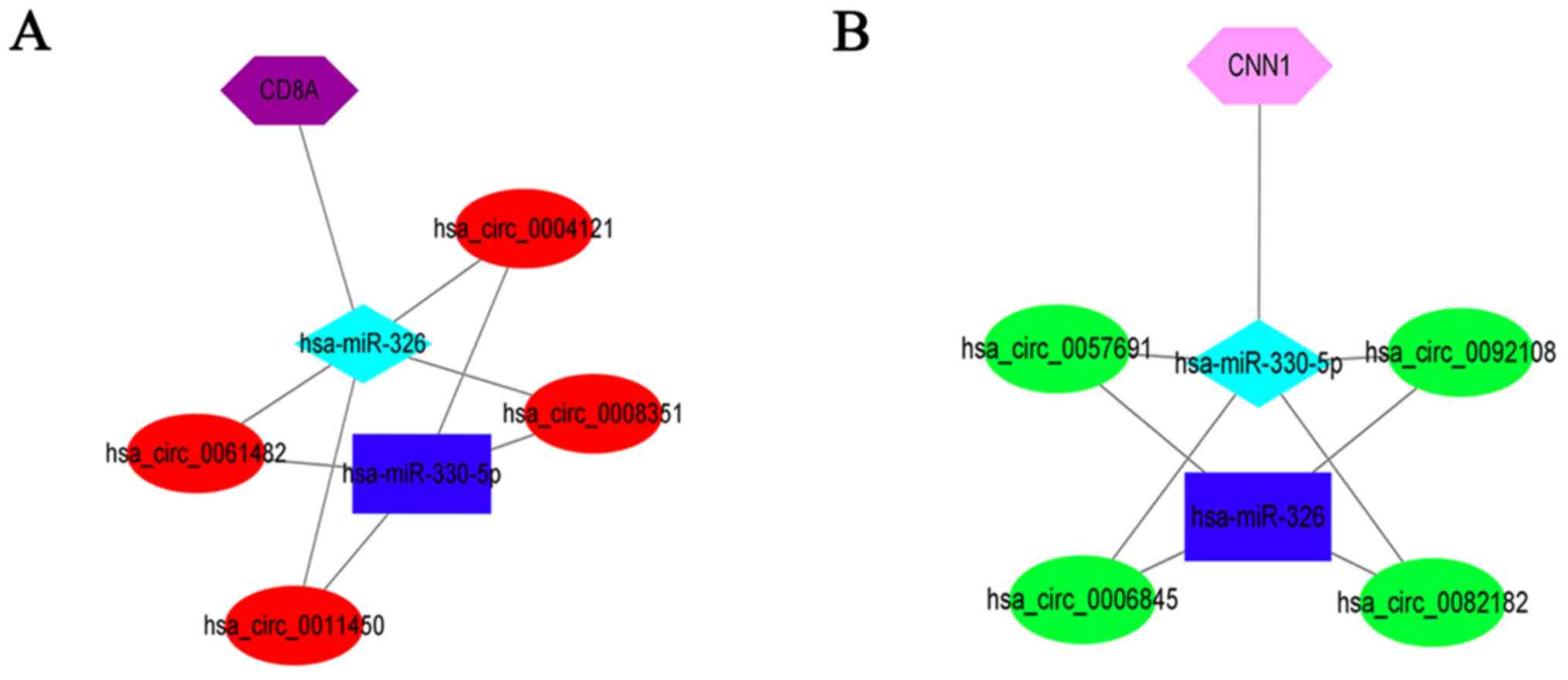
Based on MCODE algorithm,
(hsa_circ_0057691/0092108/0006845/0082182)-miR-330-5p-CNN1 and
(hsa_circ_0061482/0011450/0008351/0004121)-miR-326-CD8A were
regarded as two center axes (Fig.
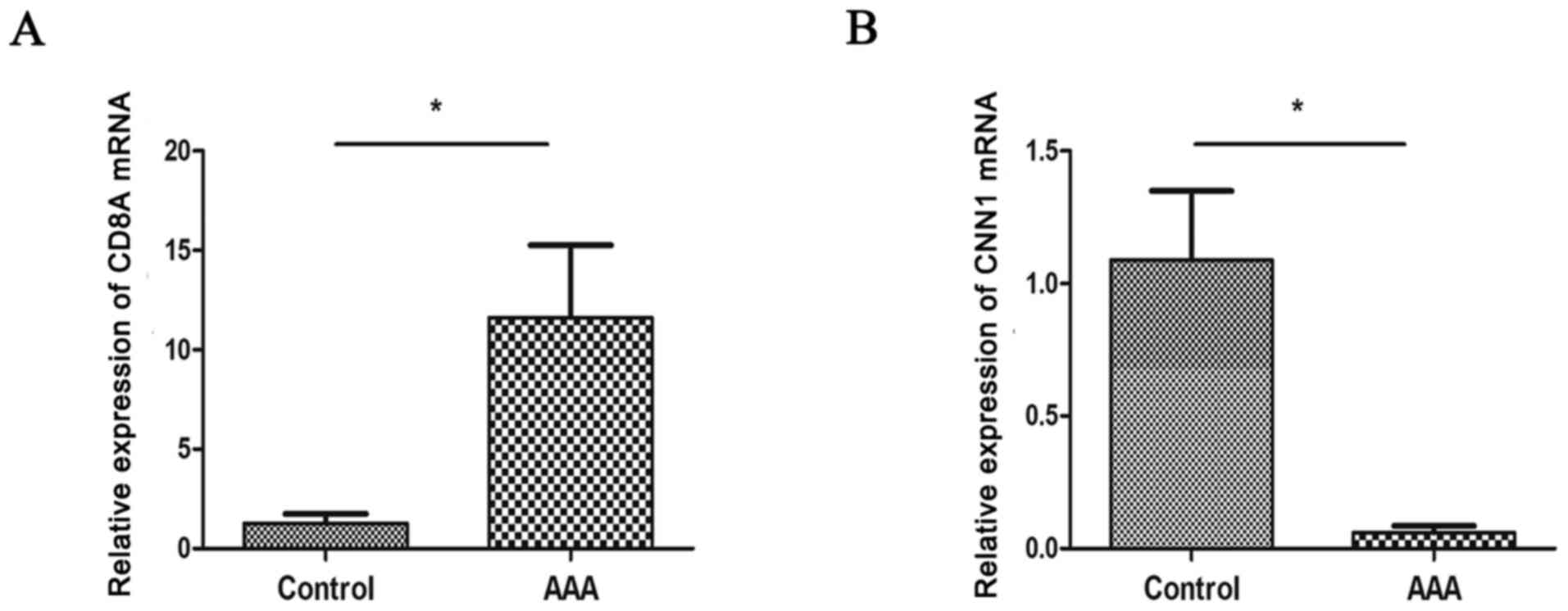
2). RT-qPCR analysis further displayed that patients with AAA
had significantly lower expression of CNN1 and higher expression of
CD8A compared with controls (P<0.05; Fig. 3), which was consistent with the
results of microarray analysis.
Functional prediction of DEGs in
AAA
To explore the potential biological function
characteristics of detected common DEGs, GO terms and KEGG pathway
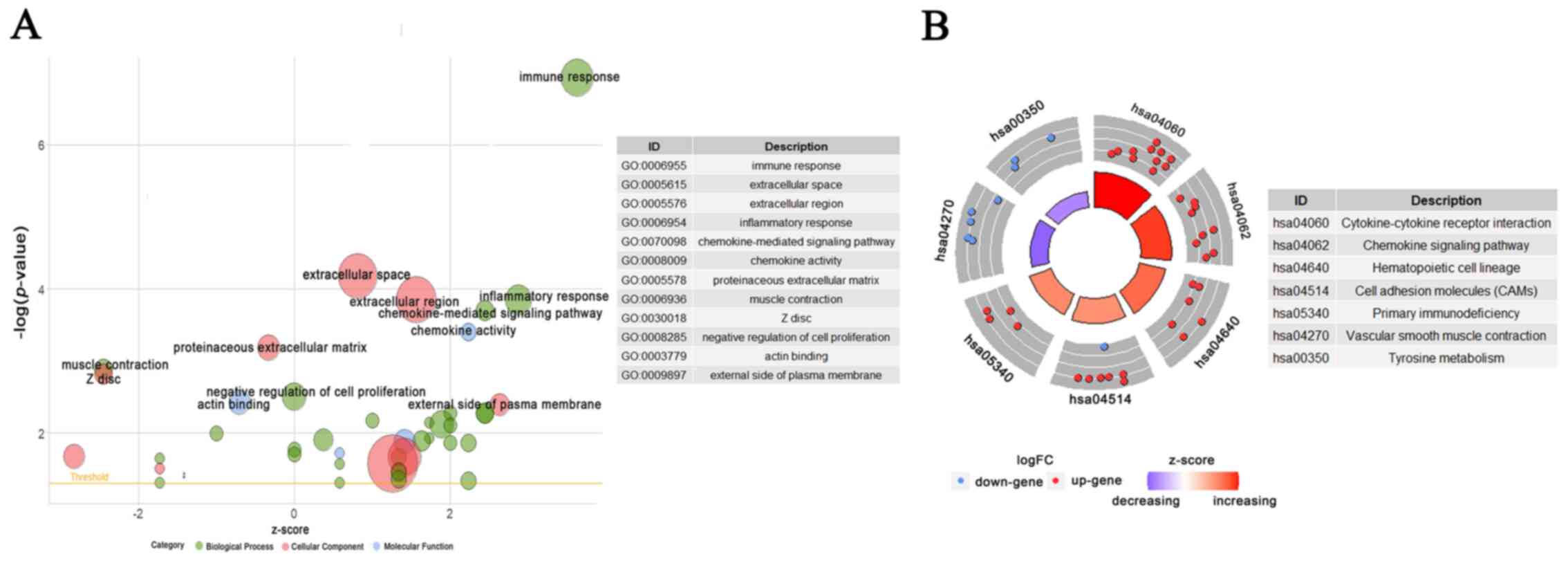
analyses were conducted. A total of 45 GO terms and 11 KEGG
pathways were obtained. GO analysis suggested that DEGs were mainly
involved in ‘immune response’ and ‘inflammatory response’ of BP,
‘extracellular region’ and ‘Z disc’ of CC and ‘chemokine activity’
and ‘actin binding’ of MF. KEGG pathways indicated that DEGs were
associated with ‘cytokine-cytokine receptor interaction’,
‘chemokine signaling pathway’ and ‘vascular smooth muscle
contraction’. A total of 12 important GO terms and seven crucial
KEGG pathways were visualized into Bubble plot and Circular plot,
respectively (Fig. 4). All GO
terms and KEGG pathways can be obtained in Tables SVIII and SIX. Based on the z-score, GO terms such
as ‘immune response’, ‘inflammatory response’ and ‘chemokine
activity’ were upregulated and ‘muscle contraction’, ‘Z disc’ and
‘actin binding’ were downregulated. And KEGG pathways such as
‘cytokine-cytokine receptor interaction’ and ‘chemokine signaling
pathway’ were upregulated, while ‘vascular smooth muscle
contraction’ and ‘tyrosine metabolism’ were downregulated.
The potential biological functions of
ceRNA axes in AAA
By combining GO terms and ceRNA networks, we could
identify the potential biological functions of 12
circRNA-miRNA-mRNA axes. Finally, nine axes (four upregulated and
five downregulated) were linked with GO terms (Fig. 5). Overall, four upregulated axes
(SEMA4A, CEMIP, IL10RA and CD8A) interacted with 13 GO terms,
including the ‘immune response’, ‘regulation of immune response’,
‘T cell activation’, ‘positive regulation of cell migration’,
‘extracellular region’, ‘external side of plasma membrane’,
‘integral component of plasma membrane’, ‘coreceptor activity’,
‘negative chemotaxis’, ‘regulation of cell shape’, ‘extracellular
space’, ‘plasma membrane’, ‘response to lipopolysaccharide’
(Fig. 5B). Furthermore, five
downregulated axes (CNN1, ZBTB16, SOST, TMEM47 and FBXO32)
interacted with 11 GO terms, including ‘focal adhesion’, ‘Z disc’,
‘actin binding’, ‘proteinaceous extracellular matrix’, ‘negative
regulation of cell proliferation’, ‘extracellular region’,
‘response to mechanical stimulus’, ‘extracellular space’, ‘positive
regulation of fat cell differentiation’, ‘plasma membrane’,
‘cell-cell junction’ (Fig.
5A).
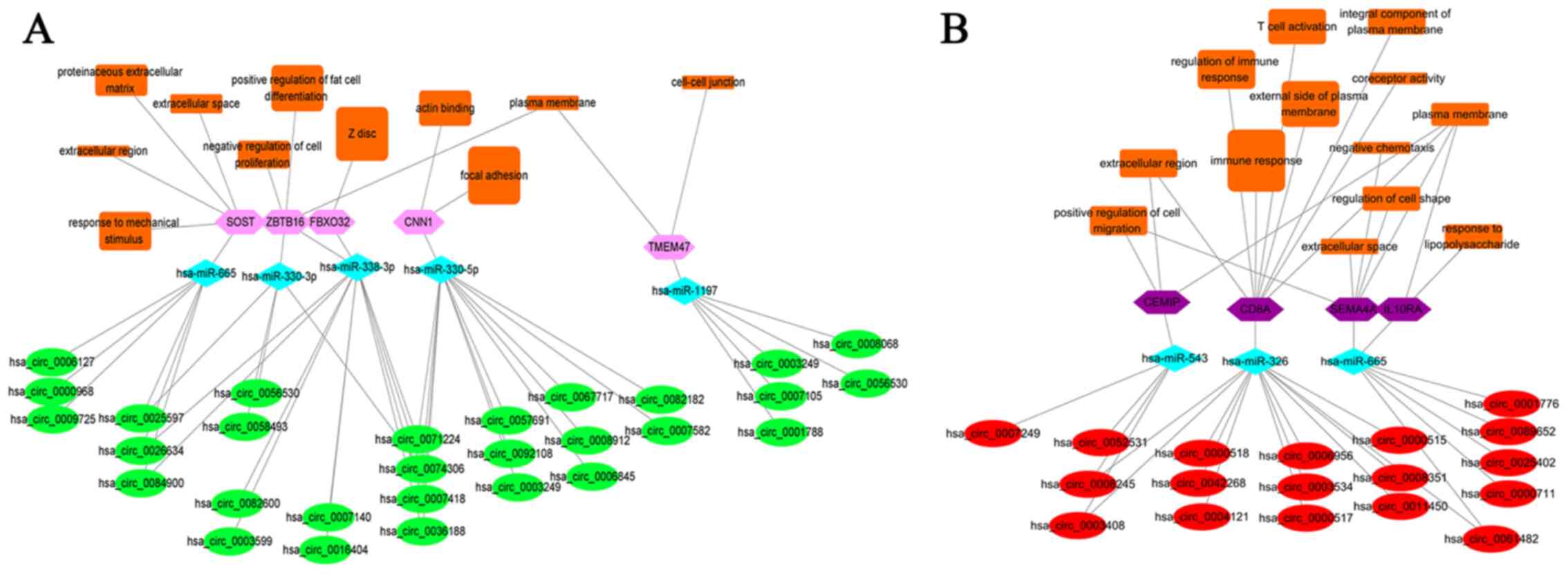 | Figure 5Construction of circRNA-miRNA-mRNA
biological function networks. (A) Networks for five downregulated
axes. (B) Networks for four upregulated axes. Green ellipse,
downregulated circRNAs; Pink hexagon, downregulated mRNAs; Red
ellipse, upregulated circRNAs; Purple hexagon, upregulated mRNAs;
light blue diamond, common target miRNAs of circRNAs and mRNAs;
Orange rectangle, GO terms. The larger the rectangle, the more
positive or negative z-score for GO terms in upregulated or
downregulated axes. circRNA, circular RNA; miRNA/miR, microRNA;
mRNA, messenger RNA; CNN1, calponin 1; SOST, sclerostin; TMEM47,
transmembrane protein 47; ZBTB16, zinc finger and BTB
domain-containing 16; FBXO32, F-box protein 32; CEMIP, cell
migration inducing hyaluronan-binding protein; IL10RA, interleukin
10 receptor subunit α; SEMA4A, semaphorin 4A; CD8A, CD8a
molecule. |
ROC analysis of ceRNA related
DEGs
A total of nine DEGs involved in
circRNA-miRNA-mRNA-biological function networks were selected for
ROC analysis to explore their clinical diagnostic value. Table I presents the basic information for
these nine DEGs. The area under the curve (AUC) and 95% confidence
interval for every DEG are listed in Table II, and ROC curves are displayed in
Fig. 6. As a result, all selected
DEGs had AUCs >0.80 with a high sensitivity and specificity.
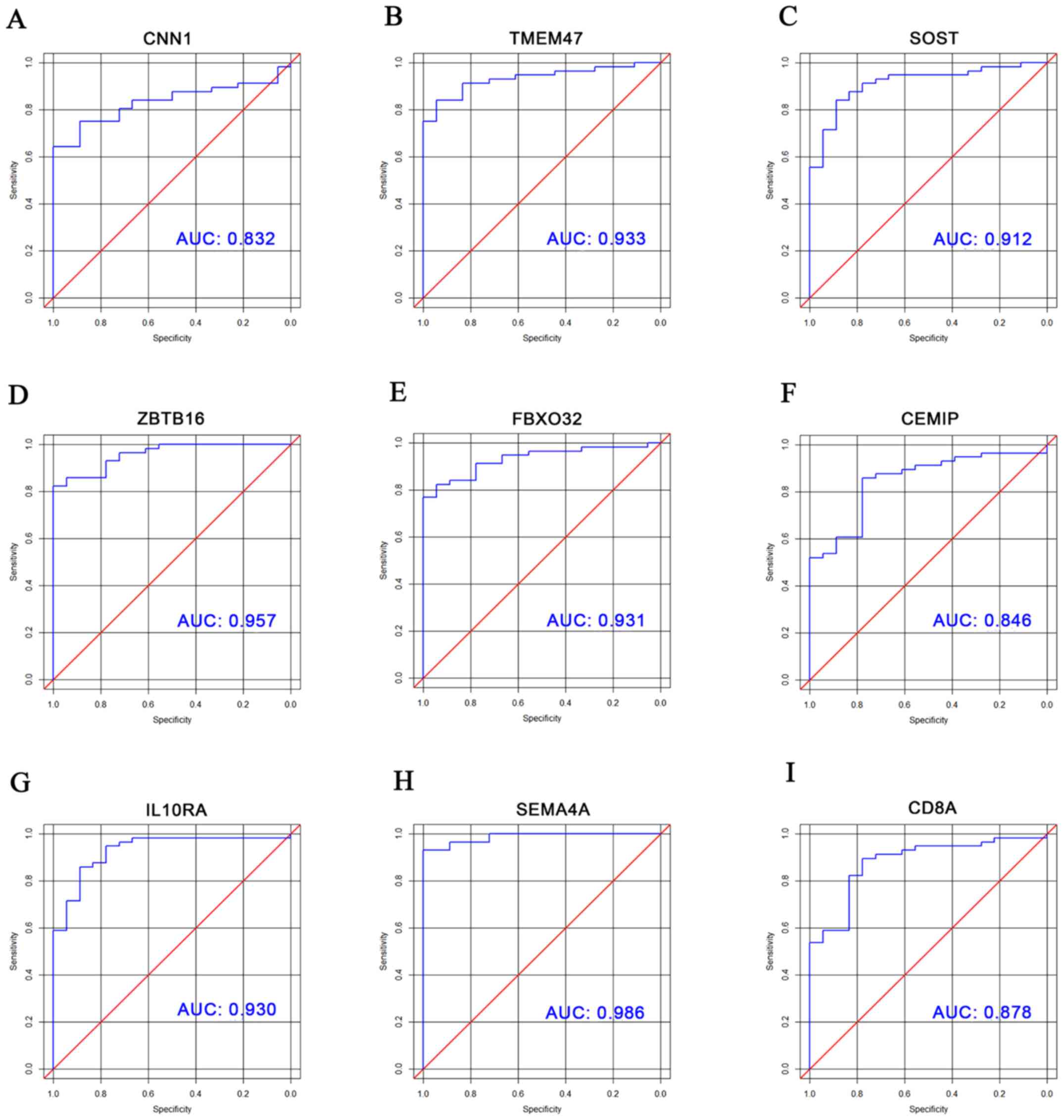 | Figure 6ROC curves for DEGs involved in
circRNA-miRNA-mRNA-biological process axes. ROC curves for (A)
CNN1, (B) TMEM47, (C) SOST, (D) ZBTB16, (E) FBXO32, (F) CEMIP, (G)
IL10RA, (H) SEMA4A and (I) CD8A, respectively. circRNA, circular
RNA; miRNA/miR, microRNA; mRNA, messenger RNA; CNN1, calponin 1;
SOST, sclerostin; TMEM47, transmembrane protein 47; ZBTB16, zinc
finger and BTB domain-containing 16; FBXO32, F-box protein 32;
CEMIP, cell migration inducing hyaluronan-binding protein; IL10RA,
interleukin 10 receptor subunit α; SEMA4A, semaphorin 4A; CD8A,
CD8a molecule; ROC, receiver operating characteristic; DEGs,
differentially expressed genes. |
 | Table IDifferentially expressed genes in
circular RNA-micro RNA-messenger RNA-biological process
networks. |
Table I
Differentially expressed genes in
circular RNA-micro RNA-messenger RNA-biological process
networks.
| Gene ID | Gene symbol | Gene full name | Group |
|---|
| 1264 | CNN1 | Calponin 1 | Down |
| 50964 | SOST | Sclerostin | Down |
| 83604 | TMEM47 | Transmembrane
protein 47 | Down |
| 7704 | ZBTB16 | Zinc finger and BTB
domain-containing 16 | Down |
| 114907 | FBXO32 | F-box protein
32 | Down |
| 57214 | CEMIP | Cell migration
inducing hyaluronan-binding protein | Up |
| 3587 | IL10RA | Interleukin 10
receptor subunit alpha | Up |
| 64218 | SEMA4A | Semaphorin 4A | Up |
| 925 | CD8A | CD8a molecule | Up |
 | Table IIAUC and 95% CI for differentially
expressed genes in circular RNA-micro RNA-messenger RNA-biological
process networks. |
Table II
AUC and 95% CI for differentially
expressed genes in circular RNA-micro RNA-messenger RNA-biological
process networks.
| Gene symbol | AUC | 95% CI |
|---|
| CNN1 | 0.83 | 0.742-0.923 |
| TMEM47 | 0.93 | 0.877-0.988 |
| SOST | 0.91 | 0.842-0.982 |
| ZBTB16 | 0.96 | 0.917-0.998 |
| FBXO32 | 0.93 | 0.875-0.986 |
| CEMIP | 0.85 | 0.751-0.942 |
| IL10RA | 0.93 | 0.865-0.994 |
| SEMA4A | 0.99 | 0.968-1.000 |
| CD8A | 0.88 | 0.792-0.964 |
Discussion
To the best of our knowledge, few in-depth
bioinformatics studies on the key circRNAs of ceRNA networks
involved in AAA pathogenesis have been conducted. Although some GEO
datasets in the present study have been used to perform other
analyses (2,13,31,32),
there are no studies exploring circRNA-miRNA-mRNA networks for AAA
with these datasets. For the first time, the present study screened
crucial circRNA-miRNA-mRNA axes through a series of bioinformatics
tools and validated some key molecular targets. The results may
deepen the understanding of molecular mechanisms in AAA formation
and offer potential candidate biomarkers for AAA.
First, 135 DEGs (62 upregulated and 73
downregulated) and 142 DECs (64 upregulated and 78 downregulated)
from GEO database were identified. Then, miRNAs of DEGs and DECs
were predicted using online tools and the intersections were
revealed. Finally, 12 circRNA-miRNA-mRNA axes were discovered to
construct ceRNA networks. Moreover, CNN1 and CD8A associated axes
were revealed to be in the top significant modules based on MCODE
algorithm. RT-qPCR analysis further verified that human AAA tissues
had lower expression of CNN1 and higher expression of CD8A compared
with controls. KEGG enrichment analysis revealed that DEGs were
mainly enriched in ‘cytokine-cytokine receptor interaction’,
‘chemokine signaling pathway’ and ‘vascular smooth muscle
contraction’. By linking circRNA-miRNA-mRNA axes to the
corresponding GO terms, a total of nine axes (CNN1, ZBTB16, SOST,
TMEM47, FBXO32, SEMA4A, CEMIP, IL10RA and CD8A) were identified
with potential biological functions. Further ROC analyses were
conducted on these DEGs, which indicated that they might be
potential predictors of AAA with significant diagnostic value.
Notably, CNN1 and CD8A related axes were also matched to GO terms,
in which CNN1 was associated with ‘focal adhesion’ and ‘actin
binding’, while CD8A was associated with ‘immune response’ and ‘T
cell activation’, further suggesting the important role of these
two axes in AAA disease.
Calponin is considered as the marker of VSMC
differentiation and contractility (33,34).
As a notable homologous gene of calponin, CNN1 has been
demonstrated to be downregulated in intracranial aneurysm and
thoracic aortic aneurysm (35-37).
A previous study showed that CNN1 deficiency can lead to decreased
contractile force in VSMCs, indicating the loss of elasticity in
blood vessel (38). Recently,
Lesiak et al (39) found
that the number of CNN1-positive cells is reduced in all layers of
human AAA. However, there are limited data concerning the role of
CNN1 associated ceRNAs in AAA.
Therefore, to the best of our knowledge, the present
study was the first to reveal that CNN1 was downregulated in AAA
specimens and targeted by miR-330-5p and four circRNAs
(hsa_circ_0057691/0092108/0006845/0082182). The present study
provided evidence that CNN1 could be a key molecule in AAA
formation from the perspective of ceRNA theory. As for CD8A, its
expression can be mainly detected in CD8+ cytotoxic T
cells (40). Previous research
revealed that CD8+ T cells can induce the death of VSMCs
by secreting various cytotoxic mediators, thus promoting AAA
formation (41), whereas
CD8+ T cell depletion reduces the development of AAA
(42). A bioinformatics study
indicated that CD8A expression is inhibited by the overexpression
of miR-370-3p, leading to the loss of CD8+ T cell immune
function and sepsis occurrence (43). Another previous bioinformatics
analysis suggested that CD8A expression is upregulated in large AAA
(18). The present research
further observed novel circRNA-miRNA pairs
(hsa_circ_0061482/0011450/0008351/0004121-miR-326) targeting CD8A
which was highly expressed in AAA tissues.
Except for the CNN1 and CD8A associated axes, there
were three upregulated axes (CEMIP, SEMA4A and IL10RA) and four
downregulated axes (SOST, ZBTB16, FBXO32 and TMEM47) associated
with GO terms, which were significantly enriched in positive
regulation of cell migration, extracellular region, plasma
membrane, extracellular space, regulation of cell shape, negative
chemotaxis/response to lipopolysaccharide, response to mechanical
stimulus, extracellular region, proteinaceous extracellular matrix,
extracellular space, negative regulation of cell proliferation,
positive regulation of fat cell differentiation, Z disc, plasma
membrane and cell-cell junction. Notably, these biological
processes are also important processes involved in the
pathophysiological mechanisms of AAA. These results indicated that
these ceRNA networks may be potential molecular factors involved in
AAA pathogenesis. It is of great importance to intervene these
pathways during the initial stages to effectively reduce the
development of AAA.
For upregulated axes-associated DEGs, CEMIP can
regulate the proliferation and migration of VSMCs in
atherosclerosis and is associated with the carcinogenesis and
progression of cancer (44,45).
In intervertebral disc degeneration, circ_001653-silencing can
downregulate CEMIP by upregulating miR-486-3p to alleviate nucleus
pulposus apoptosis and metabolic imbalance of extracellular matrix
(46). SEMA4A is generally
regarded as a key gene in positive regulation of immune-cell
activation, differentiation and migration (47). As a part of IL10 receptors, IL10RA
is associated with anti-inflammatory response and can be inhibited
by miR-142-5p to induce inflammatory bowel disease (48). For downregulated axes-associated
DEGs, SOST has been discovered to be downregulated in human aortic
aneurysm and its overexpression can prevent aortic aneurysm
development by inhibiting the Wnt signaling pathway (49). In murine models of ankylosing
spondylitis, SOST activation can be suppressed by miR-96 to improve
osteoblast differentiation and bone formation (50). ZBTB16 has the potential to enhance
mitochondrial respiratory function and promote brown adipocyte
changes (51). FBXO32 has been
reported to be targeted by various miRNAs (miR-1-3p, miR-29a/b-3p
and miR-133a/b-3p) to induce muscle wasting in diabetic murine
models (52). TMEM47 can organize
the epithelial cell junction maturation and morphogenesis by
influencing localization of a subset of tight junction proteins and
associated actomyosin structures (53). However, the role of these DEGs and
the relevant regulatory networks in AAA occurrence are largely
unknown. The present study provided evidence for future studies
that concentrate on how circRNA-miRNA-mRNA networks take part in
AAA.
The present study had some limitations. First,
public datasets for analyzing AAA associated circRNA and miRNA
expression profiles were limited. Second, there was a lack of
AAA-associated clinical information in the public database. Third,
the sample size of the collected AAA and control tissues were small
because of the prevalence of endovascular aneurysm repair over open
surgical repair, in addition to the difficulty in obtaining healthy
aortic samples. This also restricted the further validation of CNN1
and CD8A expression on protein level. In addition, further in
vitro and in vivo experiments should be performed to
verify the present results, especially the causal regulatory
relationships among circRNA, miRNA and mRNA.
In summary, the present study revealed novel
circRNA-miRNA-mRNA axes and their potential biological functions
implicated in AAA pathogenesis. CNN1 and CD8A associated ceRNA axes
were revealed to be center axes, and the expression levels of CNN1
and CD8A were also validated using RT-qPCR. To the best of our
knowledge, the present study is the first in-depth bioinformatics
study to identify potential novel biomarkers for AAA in light of
comprehensive analysis of circRNA-miRNA-mRNA networks; therefore,
these findings may provide clues to demonstrate the molecular
mechanisms and therapeutic targets for AAA in terms of ceRNA
theory.
Supplementary Material
Identification of DEGs and DECs. (A)
PCA of three datasets. (B) Blue bar represents the number of
upregulated DEGs or DECs, and orange bar represents the number of
downregulated DEGs or DECs. Hierarchical clustering and heatmap
analysis of DEGs and DECs are shown in (C) GSE7084, (D) GSE57691
and (E) GSE144431. (F) Venn diagrams present the overlap of up and
downregulated DEGs between GSE7084 and GSE57691. DEGs,
differentially expressed genes; DECs, differentially expressed
circRNAs; PCA, principal component analysis; AAA, abdominal aortic
aneurysm.
Construction of ceRNA networks. (A)
Upregulated circRNAs/mRNAs. (B) The downregulated circRNAs/mRNAs.
Red ellipse, upregulated circRNAs; Purple hexagon, upregulated
mRNAs; Green ellipse, downregulated circRNAs; Pink hexagon,
downregulated mRNAs; light blue diamond, common target miRNAs of
circRNAs and mRNAs; Dark blue rectangle, predicted miRNAs of
circRNAs or mRNAs; miRs, microRNA; IL10RA, interleukin 10 receptor
subunit α; SEMA4A, semaphorin 4A; CEMIP, cell migration inducing
hyaluronan-binding protein; ATOH8, atonal bHLH transcription factor
8; PRUNE2, prune homolog 2 with BCH domain; CNN1, calponin 1;
ZBTB16, zinc finger and BTB domain-containing 16; SOST, sclerostin;
TMEM47, transmembrane protein 47; FBXO32, F-box protein 32; NFIA,
nuclear factor IA.
A total of 12 ceRNA networks based on
every single DEG. For downregulated DEGs: (A) ATOH8, (B) ZBTB16,
(C) TMEM47, (D) SOST, (E) PRUNE2, (F) NFIA, (G) FBXO32 and (H)
CNN1. For upregulated DEGs: (I) SEMA4A, (J) CEMIP, (K) IL10RA and
(L) CD8A. Green ellipse, downregulated circRNAs; Pink hexagon,
downregulated mRNAs; Red ellipses, upregulated circRNAs; Purple
hexagon, upregulated mRNAs; light blue diamond, common target
miRNAs of circRNAs and mRNAs; Dark blue rectangle, predicted miRNA
of cirRNAs or mRNAs. FBXO32, F-box protein 32; TMEM47,
transmembrane protein 47; CEMIP, cell migration inducing
hyaluronan-binding protein; IL10RA, interleukin 10 receptor subunit
α; SEMA4A, semaphorin 4A; ATOH8, atonal bHLH transcription factor
8; PRUNE2, prune homolog 2 with BCH domain; CNN1, calponin 1;
ZBTB16, zinc finger and BTB domain-containing 16; SOST, sclerostin;
NFIA, nuclear factor IA.
Information of the three
datasets.
Characteristics of AAA and control
subjects used for RT-qPCR.
Primer sequences of CNN1 and
CD8A.
All differentially expressed geness of
GSE7084.
All differentially expressed geness of
GSE57691.
All common differentially expressed
genes between GSE7084 and GSE57691.
All differentially expressed genes of
GSE144431.
GO term enrichment analysis for
differentially expressed genes (P<0.05).
KEGG pathway enrichment analysis for
differentially expressed genes (P<0.05).
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National Natural
Science Foundation of China (grant nos. 82001828 and 81871373) and
Natural Science Foundation of Liaoning Province (grant no.
2020-BS-102).
Availability of data and materials
The sequencing datasets generated and/or analyzed
during the current study are available in the GEO database
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7084;
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57691;
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144431).
The non-sequencing datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TL designed the research, performed the experiments
and wrote the manuscript. TW contributed to drafting the article
and performing the bioinformatics analysis. LY participated in the
data analysis and interpretation. CM was involved in study
conception and revising the manuscript critically for important
intellectual content. TL and CM confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Hospital of China Medical University (approval no.
2020-146-2) and was carried out in accordance with the Declaration
of Helsinki. Written informed consent was obtained from each
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sakalihasan N, Michel JB, Katsargyris A,
Kuivaniemi H, Defraigne JO, Nchimi A, Powell JT, Yoshimura K and
Hultgren R: Abdominal aortic aneurysms. Nat Rev Dis Primers.
4(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen S, Yang D, Lei C, Li Y, Sun X, Chen
M, Wu X and Zheng Y: Identification of crucial genes in abdominal
aortic aneurysm by WGCNA. PeerJ. 7(e7873)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wu QY, Cheng Z, Zhou YZ, Zhao Y, Li JM,
Zhou XM, Peng HL, Zhang GS, Liao XB and Fu XM: A novel STAT3
inhibitor attenuates angiotensin II-induced abdominal aortic
aneurysm progression in mice through modulating vascular
inflammation and autophagy. Cell Death Dis. 11(131)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Li W, Xu C, Guo J, Liu K, Hu Y, Wu D, Fang
H, Zou Y, Wei Z, Wang Z, et al: Cis- and Trans-Acting
Expression Quantitative Trait Loci of Long Non-Coding RNA in 2,549
Cancers With Potential Clinical and Therapeutic Implications. Front
Oncol. 10(602104)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Vishnoi A and Rani S: miRNA Biogenesis and
Regulation of Diseases: An Overview. Methods Mol Biol. 1509:1–10.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang S, Shen S, Yang Z, Kong X, Liu F and
Zhen Z: Coding and Non-coding RNAs: Molecular Basis of
Forest-Insect Outbreaks. Front Cell Dev Biol. 8(369)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ala U: Competing Endogenous RNAs,
Non-Coding RNAs and Diseases: An Intertwined Story. Cells.
9(9)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sun Y, Zhong L, He X, Wang S, Lai Y, Wu W,
Song H, Chen Y, Yang Y, Liao W, et al: lncRNA H19 promotes vascular
inflammation and abdominal aortic aneurysm formation by functioning
as a competing endogenous RNA. J Mol Cell Cardiol. 131:66–81.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lin H, You B, Lin X, Wang X, Zhou D, Chen
Z, Chen Y and Wang R: Silencing of long non-coding RNA Sox2ot
inhibits oxidative stress and inflammation of vascular smooth
muscle cells in abdominal aortic aneurysm via microRNA-145-mediated
Egr1 inhibition. Aging (Albany NY). 12:12684–12702. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tian Z, Sun Y, Sun X, Wang J and Jiang T:
LINC00473 inhibits vascular smooth muscle cell viability to promote
aneurysm formation via miR-212-5p/BASP1 axis. Eur J Pharmacol.
873(172935)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xia Q, Zhang L, Yan H, Yu L, Shan W and
Jiang H: LUCAT1 contributes to MYRF-dependent smooth muscle cell
apoptosis and may facilitate aneurysm formation via the
sequestration of miR-199a-5p. Cell Biol Int. 44:755–763.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang B, Wang X, Ying C, Peng F, Xu M, Chen
F and Cai B: Long Noncoding RNA SNHG16 Facilitates Abdominal Aortic
Aneurysm Progression through the miR-106b-5p/STAT3 Feedback Loop. J
Atheroscler Thromb. 28:66–78. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tian L, Hu X, He Y, Wu Z, Li D and Zhang
H: Construction of lncRNA-miRNA-mRNA networks reveals functional
lncRNAs in abdominal aortic aneurysm. Exp Ther Med. 16:3978–3986.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Han Y, Zhang H, Bian C, Chen C, Tu S, Guo
J, Wu Y, Böckler D and Zhang J: Circular RNA Expression: Its
Potential Regulation and Function in Abdominal Aortic Aneurysms.
Oxid Med Cell Longev. 2021(9934951)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhao F, Chen T and Jiang N:
CDR1as/miR-7/CKAP4 axis contributes to the pathogenesis of
abdominal aortic aneurysm by regulating the proliferation and
apoptosis of primary vascular smooth muscle cells. Exp Ther Med.
19:3760–3766. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yue J, Zhu T, Yang J, Si Y, Xu X, Fang Y
and Fu W: circCBFB-mediated miR-28-5p facilitates abdominal aortic
aneurysm via LYPD3 and GRIA4. Life Sci. 253(117533)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lenk GM, Tromp G, Weinsheimer S, Gatalica
Z, Berguer R and Kuivaniemi H: Whole genome expression profiling
reveals a significant role for immune function in human abdominal
aortic aneurysms. BMC Genomics. 8(237)2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Biros E, Gäbel G, Moran CS, Schreurs C,
Lindeman JH, Walker PJ, Nataatmadja M, West M, Holdt LM,
Hinterseher I, et al: Differential gene expression in human
abdominal aortic aneurysm and aortic occlusive disease. Oncotarget.
6:12984–12996. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou M, Shi Z, Cai L, Li X, Ding Y, Xie T
and Fu W: Circular RNA expression profile and its potential
regulative role in human abdominal aortic aneurysm. BMC Cardiovasc
Disord. 20(70)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jolliffe IT and Cadima J: Principal
component analysis: A review and recent developments. Philos Trans
A Math Phys Eng Sci. 374(20150202)2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
RStudio Team: RStudio: Integrated
Development for R. RStudio, Inc. Boston, MA, 2015. http://www.rstudio.com/. Accessed June 1, 2021.
|
|
22
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sticht C, De La Torre C, Parveen A and
Gretz N: miRWalk: An online resource for prediction of microRNA
binding sites. PLoS One. 13(e0206239)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4(4)2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4(2)2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
World Medical Association Declaration of
Helsinki. Ethical principles for medical research involving human
subjects. JAMA. 310:2191–2194. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Δ Δ C(T)) Method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu Y, Wang X, Wang H and Hu T:
Identification of key genes and pathways in abdominal aortic
aneurysm by integrated bioinformatics analysis. J Int Med Res.
48(300060519894437)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Oh CK, Ko Y, Park JJ, Heo HJ, Kang J, Kwon
EJ, Kang JW, Lee Y, Myung K, Kang JM, et al: FRZB as a key molecule
in abdominal aortic aneurysm progression affecting vascular
integrity. Biosci Rep. 41(41)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Luo Y and Huang C: CircSFMBT2 facilitates
vascular smooth muscle cell proliferation by targeting
miR-331-3p/HDAC5. Life Sci. 264(118691)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Guo X, Li D, Chen M, Chen L, Zhang B, Wu T
and Guo R: miRNA-145 inhibits VSMC proliferation by targeting CD40.
Sci Rep. 6(35302)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Liu R and Jin JP: Calponin isoforms CNN1,
CNN2 and CNN3: Regulators for actin cytoskeleton functions in
smooth muscle and non-muscle cells. Gene. 585:143–153.
2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Gong J, Zhou D, Jiang L, Qiu P, Milewicz
DM, Chen YE and Yang B: In Vitro Lineage-Specific Differentiation
of Vascular Smooth Muscle Cells in Response to SMAD3 Deficiency:
Implications for SMAD3-Related Thoracic Aortic Aneurysm.
Arterioscler Thromb Vasc Biol. 40:1651–1663. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cheng Q, Li Z, Wang R, Zhang H, Cao H,
Chen F, Li H, Xia Z, Feng S, Zhang H, et al: Genetic Profiles
Related to Pathogenesis in Sporadic Intracranial Aneurysm Patients.
World Neurosurg. 131:e23–e31. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Feng HZ, Wang H, Takahashi K and Jin JP:
Double deletion of calponin 1 and calponin 2 in mice decreases
systemic blood pressure with blunted length-tension response of
aortic smooth muscle. J Mol Cell Cardiol. 129:49–57.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lesiak M, Augusciak-Duma A, Stepien KL,
Fus-Kujawa A, Botor M and Sieron AL: Searching for new molecular
markers for cells obtained from abdominal aortic aneurysm. J Appl
Genet. 62:487–497. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ma K, Qiao Y, Wang H and Wang S:
Comparative expression analysis of PD-1, PD-L1, and CD8A in lung
adenocarcinoma. Ann Transl Med. 8(1478)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sagan A, Mikolajczyk TP, Mrowiecki W,
MacRitchie N, Daly K, Meldrum A, Migliarino S, Delles C, Urbanski
K, Filip G, et al: T Cells Are Dominant Population in Human
Abdominal Aortic Aneurysms and Their Infiltration in the
Perivascular Tissue Correlates With Disease Severity. Front
Immunol. 10(1979)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li G, Zhou H, He Y, Sun S, Wu X and Yuan
H: Ulinastatin Inhibits the Formation and Progression of
Experimental Abdominal Aortic Aneurysms. J Vasc Res. 57:58–64.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chen J, Lin M and Zhang S: Identification
of key miRNA mRNA pairs in septic mice by bioinformatics analysis.
Mol Med Rep. 20:3858–3866. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xue Q, Wang X, Deng X, Huang Y and Tian W:
CEMIP regulates the proliferation and migration of vascular smooth
muscle cells in atherosclerosis through the WNT-beta-catenin
signaling pathway. Biochem Cell Biol. 98:249–257. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen Y, Li L and Zhang J: Cell migration
inducing hyaluronidase 1 (CEMIP) activates STAT3 pathway to
facilitate cell proliferation and migration in breast cancer. J
Recept Signal Transduct Res. 41:145–152. 2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Cui S and Zhang L: circ_001653 Silencing
Promotes the Proliferation and ECM Synthesis of NPCs in IDD by
Downregulating miR-486-3p-Mediated CEMIP. Mol Ther Nucleic Acids.
20:385–399. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ito D and Kumanogoh A: The role of Sema4A
in angiogenesis, immune responses, carcinogenesis, and retinal
systems. Cell Adhes Migr. 10:692–699. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Duijvis NW, Moerland PD, Kunne C, Slaman
MMW, van Dooren FH, Vogels EW, de Jonge WJ, Meijer SL, Fluiter K
and Te Velde AA: Inhibition of miR-142-5P ameliorates disease in
mouse models of experimental colitis. PLoS One.
12(e0185097)2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Krishna SM, Seto SW, Jose RJ, Li J, Morton
SK, Biros E, Wang Y, Nsengiyumva V, Lindeman JH, Loots GG, et al:
Wnt Signaling Pathway Inhibitor Sclerostin Inhibits Angiotensin
II-Induced Aortic Aneurysm and Atherosclerosis. Arterioscler Thromb
Vasc Biol. 37:553–566. 2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ma S, Wang DD, Ma CY and Zhang YD:
MicroRNA-96 promotes osteoblast differentiation and bone formation
in ankylosing spondylitis mice through activating the Wnt signaling
pathway by binding to SOST. J Cell Biochem. 120:15429–15442.
2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wei S, Zhang M, Zheng Y and Yan P: ZBTB16
Overexpression Enhances White Adipogenesis and Induces Brown-Like
Adipocyte Formation of Bovine White Intramuscular Preadipocytes.
Cell Physiol Biochem. 48:2528–2538. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Gerlinger-Romero F, Yonamine CY, Junior
DC, Esteves JV and Machado UF: Dysregulation between TRIM63/FBXO32
expression and soleus muscle wasting in diabetic rats: Potential
role of miR-1-3p, -29a/b-3p, and -133a/b-3p. Mol Cell Biochem.
427:187–199. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Dong Y and Simske JS: Vertebrate
Claudin/PMP22/EMP22/MP20 family protein TMEM47 regulates epithelial
cell junction maturation and morphogenesis. Dev Dyn. 245:653–666.
2016.PubMed/NCBI View Article : Google Scholar
|