Introduction
Fluorescence in situ hybridization (FISH) is
a well-established technique in pathology laboratories for the
identification of recurrent tumor-specific chromosomal
translocations and copy number variations, assisting in clinical
diagnosis and the selection of treatment strategies (1). FISH offers distinct advantages over
other molecular diagnostic methods. Unlike reverse
transcription-polymerase chain reaction (RT-PCR) or next-generation
sequencing (NGS), FISH utilizes fluorophore-coupled probes that
specifically bind to complementary sequences and provides detailed
intracellular localization of target genes, which is particularly
important when evaluating tissues with small tumor volumes or
tumors consisting of multiple mixed cells (2,3). For
gene rearrangement, a FISH break-apart probe is capable of
detecting all possible rearrangements involving a common breakpoint
region despite corresponding fusion partners or variants. By
contrast, the RT-PCR technique requires multiple pairs of primers
to identify certain known rearrangements, which is time-consuming
and relatively inefficient (4).
There are also limitations with FISH, which may
result in experimental failure or diagnostic errors. The lack of
standard interpretation guidelines for multiple FISH probes leads
to difficulties in explaining atypical, abnormal signal patterns,
such as isolated or unbalanced signals (5). Furthermore, it is necessary for
clinical laboratories to establish an analytical normal cutoff
value for individual probes, which may vary among different
institutions. Another challenge for FISH detection is the sample
quality, which is predominantly determined by the tissue processing
procedure and tissue component. Fixation time, storage,
decalcifying agents, and collagen and extracellular matrix
abundance may influence the intensity of FISH signals and therefore
hinder pathological diagnosis (4).
In clinical practice, one obstacle for FISH is that not all
pathology archives are desirable. Unsatisfactory paraffin blocks or
FFPE sections always lead to interpretation failure and
difficulties in clinical diagnosis. To solve this problem,
conventional FISH procedures urgently require to be improved.
Heat-induced antigen retrieval (HIAR) is an
effective method widely applied in immunohistochemistry to unmask
antigens in FFPE sections (6).
Prompted by this simple technique, the present study provided a
modified FISH protocol. In the pretreatment step, HIAR with either
citrate or Tris-EDTA buffer was applied to poor-quality FFPE
sections that failed in the conventional pathology workflow, and
the detection validity and signal intensity markedly improved. In
addition, evaluation data acquired from two HIAR-assisted FISH
methods were compared and the same interpretation results were
obtained. Overall, this protocol is sufficient for enhancing FISH
signals and produces reliable results.
Materials and methods
Tissue samples
Among 328 archived tumor tissues sent to the
National Cancer Center/National Clinical Research Center for
Cancer/Cancer Hospital and Shenzhen Hospital (Shenzhen, China) for
FISH detection from January 2020 to May 2021, seven FFPE sections
failed in conventional FISH experiments and HIAR-FISH was performed
for these samples to assess the efficacy of the modified protocol.
Furthermore, four adequately handled specimens were randomly
selected to evaluate whether HIAR would affect the original signal
pattern. The median age of the patients (9 male and 2 female) was
45 years. Our study cohort consisted of nine cases of soft tissue
sarcomas and two cases of microphthalmia family translocation renal
cell carcinoma. All samples were excision tissues and sectioned at
4-µm thickness for detection. Patient clinical and pathological
characteristics are summarized in Table I.
 | Table IClinicopathological information of the
cases. |
Table I
Clinicopathological information of the
cases.
| Case no. | Sex | Age, years | Site | Specimen type | Diagnosis |
|---|
| 1 | M | 30 | Right shoulder | Excision | Clear cell
sarcoma |
| 2 | F | 34 | Left kidney | Excision | MiT family
translocation renal cell carcinoma |
| 3 | M | 56 | Right kidney | Excision | MiT family
translocation renal cell carcinoma |
| 4 | M | 27 | Left foot | Excision | Synovial sarcoma |
| 5 | M | 33 | Right thigh | Excision | Myxoid
liposarcoma |
| 6 | M | 45 | Right thigh | Excision | Myxoid
liposarcoma |
| 7 | M | 55 | Right thigh | Excision | Pleomorphic
liposarcoma |
| 8 | M | 44 | Chest wall | Excision | High-grade
fibrosarcoma |
| 9 | M | 54 | Retroperitoneal | Excision | Well-differentiated
liposarcoma |
| 10 | M | 48 | Left thigh | Excision | Low-grade fibromyxoid
sarcoma |
| 11 | F | 53 | Abdominal wall | Excision | Undifferentiated
small round cell sarcoma |
FISH protocol
FISH experiments were performed on the interphase
nuclei of 4-µm FFPE sections using commercial probe kits, including
synovial sarcoma translocation chromosome 18 (SS18), Ewing sarcoma
breakpoint region 1 gene (EWSR1), DNA damage inducible transcript 3
(DDIT3), transcription factor binding to IGHM enhancer 3 (TFE3) and
murine double minute-2 (MDM2). The detailed probe information is
listed in Table II, and all
probes were diluted 1:10 with LSI/WCP hybridization buffer (Abbott
Molecular, Inc.).
 | Table IIInformation on the probes. |
Table II
Information on the probes.
| Probe | Type | Chromosome
location | Manufacturer | Lot no. |
|---|
| SS18 | Break-apart | 18q11.2 | Abbott Molecular,
Inc. | 30-608250/R3 |
| EWSR1 | Break-apart | 22q12 | Abbott Molecular,
Inc. | 30-608248/R4 |
| DDIT3 | Break-apart | 12q13 | Abbott Molecular,
Inc. | 30-608246/R3 |
| TFE3 | Break-apart | Xp11.23 | ZytoVision | Z-2109 |
| MDM2/CEP12 | Amplification | 12q15 | Abbott Molecular,
Inc. | 30-608268/R2 |
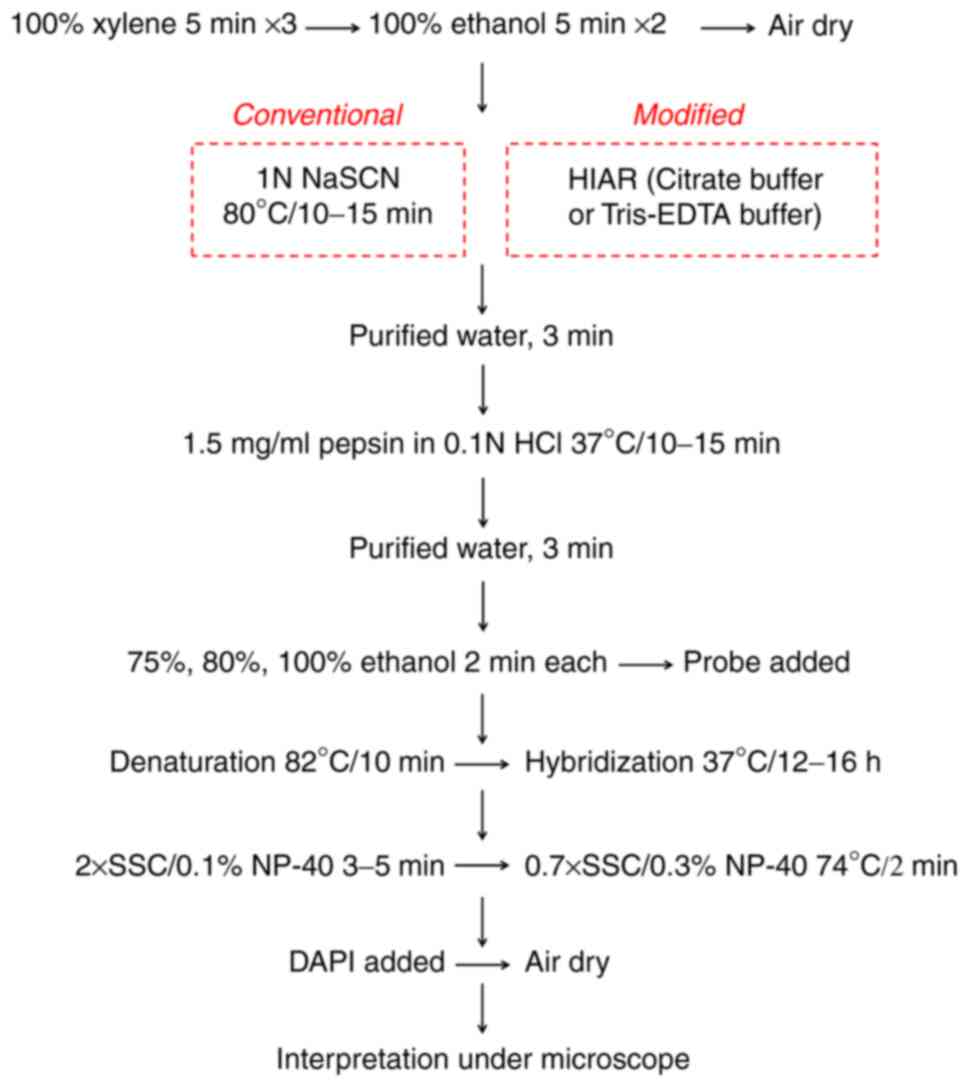
Conventional FISH was performed using a Vysis
paraffin pretreatment IV & post hybridization wash buffer kit
(Abbott Molecular, Inc.) following the manufacturer's protocol, as
illustrated in Fig. 1. In brief,
the FFPE slides were dewaxed in 100% xylene, dehydrated in 100%
ethanol and dried in air. For tissue pretreatment, the slides were
immersed in pretreatment solution containing 1 N sodium thiocyanate
(NaSCN) at 80˚C for 10-15 min and then soaked in purified water for
3 min. Subsequently, the excess water along the edges of the slides
was removed with a paper towel, followed by enzymatic digestion
using pepsin buffer (1.5 mg/ml) for 10-15 min at 37˚C. The slides
were then immersed in purified water for 3 min at room temperature.
After ethanol gradient dehydration, diluted probes were applied to
the target tissue areas and the slides were processed with the
ThermoBrite Denaturation/Hybridization System (Abbott Molecular,
Inc.) using the following program: 10 min at 82˚C and 12-16 h at
37˚C. The next day, the slides were rapidly washed with 2X saline
sodium citrate buffer (SSC)/0.1% nonidet P (NP)-40 at ambient
temperature for 3-5 min and 0.7X SSC/0.3% NP-40 at 74˚C for 2 min.
After air-drying in the dark, the tissues were counterstained using
DAPI to visualize nuclei. FISH signals were viewed with suitable
filter sets under a fluorescence microscope (magnification, x600;
Leica Microsystems GmbH) and the images were captured using a
Bioview Allegro Plus System (Abbott Molecular, Inc.) with the same
parameter settings.
HIAR in FISH
HIAR was applied for slides exhibiting
unsatisfactory signals in conventional FISH (Fig. 1). In the present study, two AR
solutions of different pH values were used: Citrate buffer (pH=6.0)
and Tris-EDTA buffer (pH=9.0). In detail, AR solution was added
into a pressure cooker on a hotplate. The lid of the cooker was
simply rested on top at this point. After boiling, slides were
rapidly immersed in AR solution and the cooker lid was secured.
Once the cooker reached the full pressure, conditions were
maintained for 2 min. Subsequently, the pressure cooker was placed
in an empty sink and the pressure valve was carefully released.
Once depressurized, the lid was removed and slides were cooled at
room temperature for 60 min.
FISH signal enumeration
For each case, the slide adequacy was evaluated
according to three criteria: i) The background should be dark and
relatively free of fluorescence particles; ii) the fluorescence
signals under both channels should appear unequivocal, bright and
easily identified; and iii) the nuclei morphology should be intact
and distinguishable. Only slides with these features were
interpreted.
A minimum of 50 non-overlapping nuclei were counted
for each case independently by two pathologists (16 and 2 years of
experience, respectively) in a blinded manner. For break-apart
probes targeting EWSR1, DDIT3, SS18 and TFE3, the typical
rearrangement pattern was 1F/1G/1O (1 fusion, 1 green, 1 orange)
and the cell was considered positive when at least one set of green
and orange signals were separated at ≥2 signal diameters apart.
Cells without rearrangement exhibited fused or adjacent (distance
<2 signal diameters) green and orange signals. The cutoff value
for each break-apart probe was 15% in our laboratory, which was
established as recommended by the American College of Medical
Genetics guidelines (1,7). For the MDM2/CEP12 probe,
amplification was considered to have occurred in cells with an MDM2
(orange)/CEP12 (green) ratio ≥2.0.
Statistical analysis
To quantify the hybridization efficiency of
different FISH protocols, three captured views of individual
samples were randomly selected and quantification of i) the total
cell number, ii) number of cells with clear fluorescence signals
(either green or orange), and iii) number of cells conforming to
interpretation criteria above was performed. The percentage of
cells is presented as the mean ± standard error of the mean. All
analyses were performed using GraphPad Prism software 5.0 (GraphPad
Software, Inc.).
Results
HIAR markedly promotes FISH efficiency
in detecting fluorescence signals
In conventional FISH experiments using commercial
pretreatment kits, all seven pathology samples failed to meet the
interpretation standard because the density ratio of signal to
background was low in either the orange or green channel, making it
impossible to identify signal patterns. To obtain satisfactory
results, HIAR was introduced to replace the pretreatment step. Of
the seven samples, five were processed with both citrate buffer and
Tris-EDTA buffer, while the remaining two samples were only treated
with Tris-EDTA buffer due to the lack of FFPE slides provided by
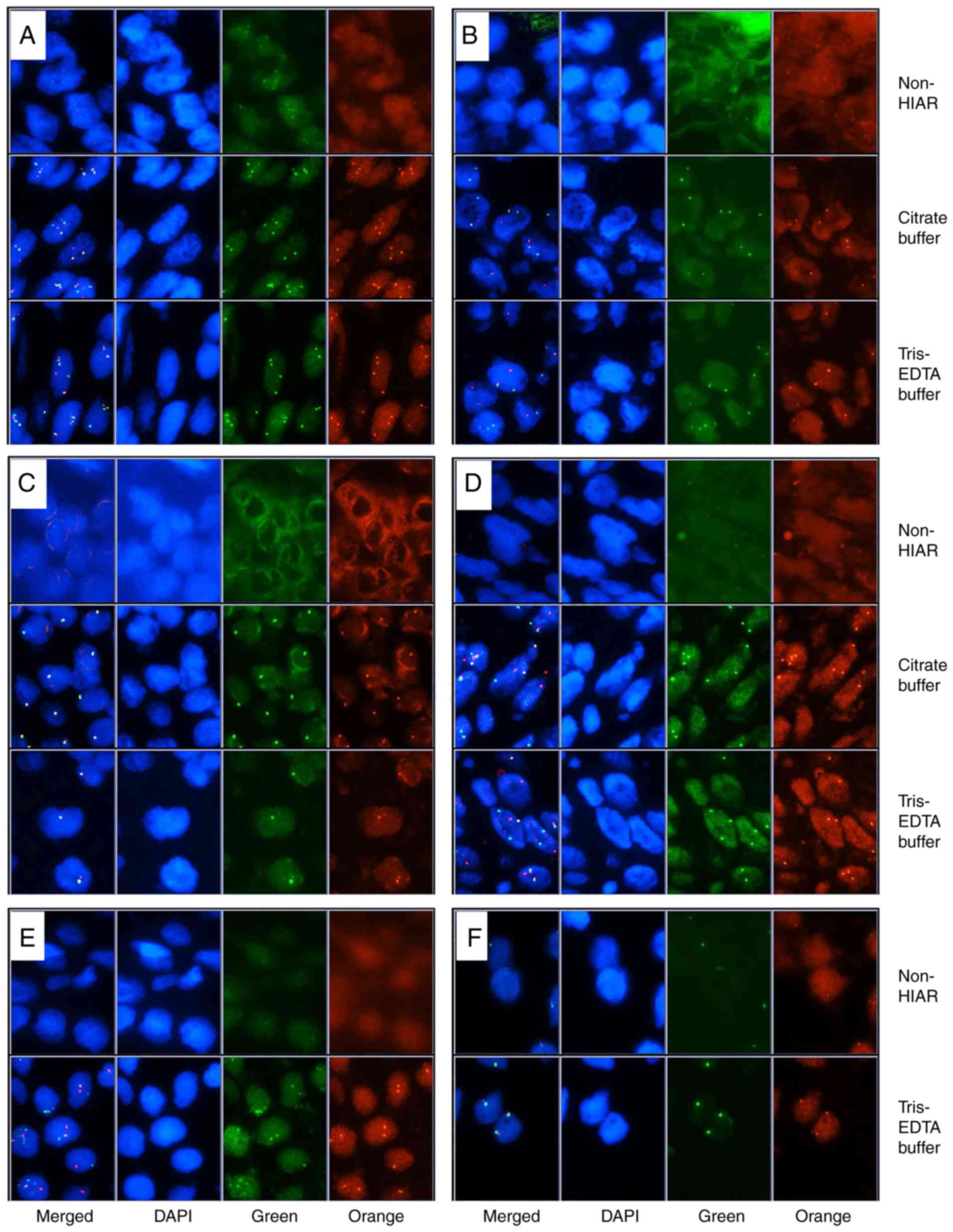
external medical centers. As illustrated in Fig. 2A-F, FISH analysis was successfully
performed on six of the slides, which yielded clear and bright
fluorescence signals in both the orange and green channels with
intact cell nucleus morphology. The other sample hybridized with
DDIT3 only exhibited a marginal improvement after HIAR using either
HIAR buffer (data not shown). To directly illustrate the enhanced
hybridization efficiency after antigen retrieval, the ratio of
cells satisfying the aforementioned interpretation criteria were
quantified as indicated, as well as cells exhibiting fluorescence
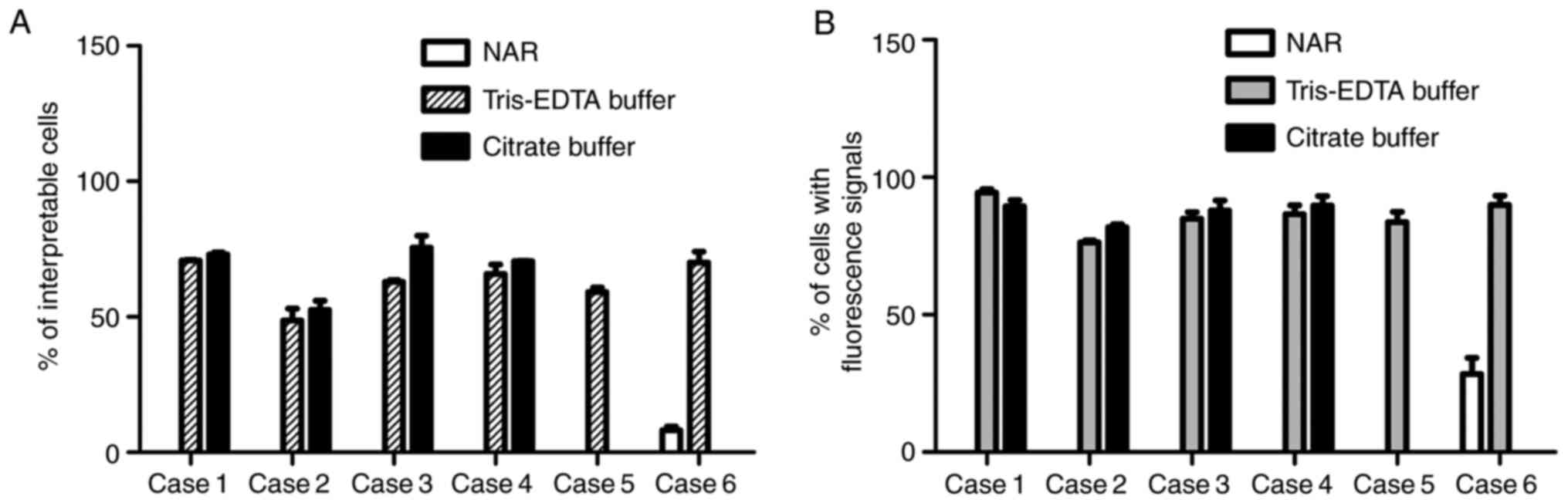
signals, either orange or green. As depicted in Fig. 3A, only case 6 contained several
interpretable cells (<10%) after the conventional FISH
experiment, but the retrieval treatment with either Tris-EDTA or
citrate buffer significantly increased the ratio of non-overlapped
cells clearly exhibiting both green and orange signals
(P<0.001). Furthermore, the signal retrieval effect of the two
buffers was compared and no significant difference was observed in
cases 1-4. Similar results were obtained after calculating the
percentage of cells exhibiting either a green or an orange signal
(Fig. 3B). These data indicated an
encouraging effect of HIAR in facilitating probe binding and
fluorescence signal restoration for poor-quality FFPE slides.
Consistent interpretation results
obtained with two HIAR-FISH methods
After assessing the signal quality of HIAR-FISH, two
experienced pathologists interpreted the FISH slides separately to
compare the results from the two assays. The detailed counting
results and final diagnosis are listed in Table III. For slides 1-4, stable
results were obtained from two HIAR-FISH experiments, suggesting
that these two AR solutions significantly amplified fluorescence
intensity without influencing signal patterns. In addition, the
interpretation results for all six samples provided by the two
pathologists had high consistency.
 | Table IIIFluorescence in situ
hybridization interpretation results of poor-quality FFPE
sections. |
Table III
Fluorescence in situ
hybridization interpretation results of poor-quality FFPE
sections.
| Case/probe | HIER buffer | Pathologist 1 | Pathologist 2 |
|---|
| 1/EWSR1 | Tris-EDTA buffer | Positive | Positive |
| | | (1G/1O/1-3F, 30%;
3-4F, 52%; other, 18%) | (1G/1O/1-3F, 22%;
3-4F, 52%; other, 26%) |
| | Citrate buffer | Positive | Positive |
| | | (1G/1O/1-3F, 28%;
3-4F, 44%; other, 28%) | (1G/1O/1-3F, 30%;
3-4F, 40%; other, 30%) |
| 2/TFE3 | Tris-EDTA buffer | Positive | Positive |
| | | (1G/1O/1F, 72%; 2F,
12%; other, 16%) | (1G/1O/1F, 64%; 1-2F,
8%; other, 28%) |
| | Citrate buffer | Positive | Positive |
| | | (1G/1O/1F, 62%,
1G/1O, 20%; 1-2F, 8%; other, 10%) | (1G/1O/1F, 52%,
1G/1O, 1-2F, 12%; other, 24%) |
| 3/TFE3 | Tris-EDTA
buffer | Negative | Negative |
| | | (1F, 82%; 2F,
18%) | (1F, 86%; 2F,
14%) |
| | Citrate buffer | Negative | Negative |
| | | (1F, 78%; 2F,
22%) | (1F, 82%; 2F,
18%) |
| 4/SS18 | Tris-EDTA
buffer | Positive | Positive |
| | | (1G/1O/1F, 24%;
atypical break-apart patterns, 60%; 1-2F, 16%) | (1G/1O/1F, 20%;
atypical break-apart patterns, 60%; 1-2F, 20%) |
| | Citrate buffer | Positive | Positive |
| | | (1G/1O/1F, 20%;
atypical break-apart patterns, 58% 1-2F, 22%) | (1G/1O/1F, 20%;
atypical break-apart patterns, 58%; 1-2F, 22%) |
| 5/DDIT3 | Tris-EDTA
buffer | Positive | Positive |
| | | (1G/1O/1F, 48%; 2F,
30%; other, 22%) | (1G/1O/1F, 44%;
1-2F, 36%; other, 20%) |
| | Citrate buffer | N/A | N/A |
| 6/MDM2 | Tris-EDTA
buffer | Negative | Negative |
| | | (MDM2/cell,
1.6a; CEP12/cell,
1.7b; MDM2/CEP12,
0.94c) | (MDM2/cell, 1.66;
MDM2/CEP12, 1.0) |
| | Citrate buffer | N/A | N/A |
HIAR does not affect the signal of
adequately handled samples
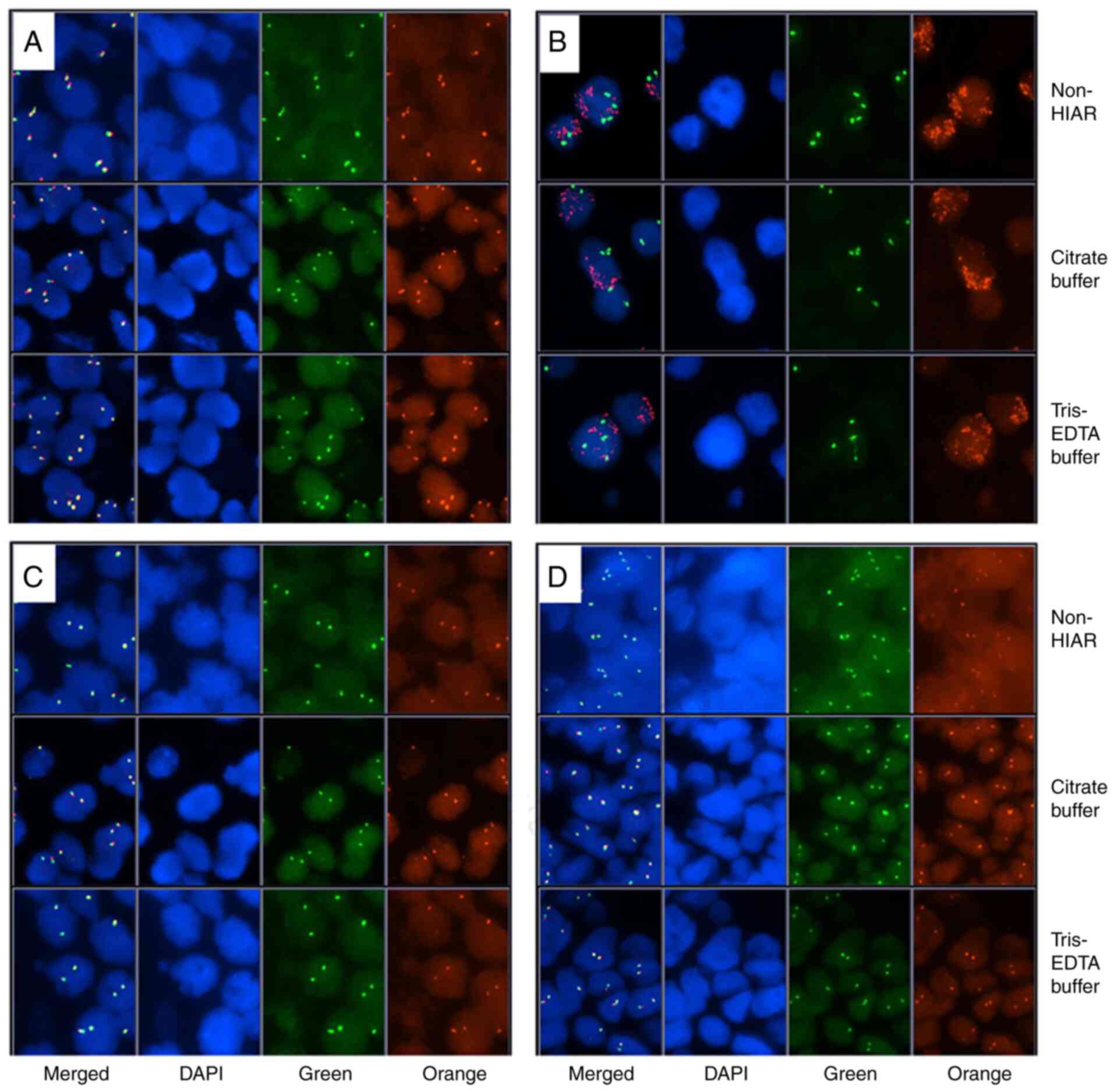
To evaluate the effect of HIAR on well-handled
samples, 4 specimens exhibiting adequate signals after conventional
FISH were randomly selected and HIAR was applied to them. First,
the signal intensity of these slides was compared and no
significant difference was observed between conventional and
HIAR-FISH using either citrate or Tris-EDTA buffer (Fig. 4A-D). The two pathologists then
interpreted the FISH slides and similar results were obtained after
introducing HIAR (Table IV).
These data indicated that HIAR did not influence the results of
FISH in adequately handled samples.
 | Table IVFluorescence in situ
hybridization interpretation results of adequately handled
samples. |
Table IV
Fluorescence in situ
hybridization interpretation results of adequately handled
samples.
| Case/probe | HIER buffer | Pathologist 1 | Pathologist 2 |
|---|
| 8/SS18 | Tris-EDTA
buffer | Negative | Negative |
| | | (2F, 78%; 3F, 8%;
other, 14%) | (2F, 80%; 3F, 10%;
other, 10%) |
| | Citrate buffer | Negative | Negative |
| | | (2F, 80%; 3F, 8%;
other, 12%) | (2F, 76%; 3F, 12%;
other, 12%) |
| 9/MDM2 | Tris-EDTA
buffer | Positive | Positive |
| | | (MDM2/cell,
21.8a; CEP12/cell,
2.2b; MDM2/CEP12,
9.91c) | (MDM2/cell, 23.3;
CEP12/cell, 2.4; MDM2/CEP12, 9.71) |
| | Citrate buffer | Positive | Positive |
| | | (MDM2/cell, 22.6;
CEP12/cell, 2.5; MDM2/CEP12, 9.04) | (MDM2/cell, 22.9;
CEP12/cell, 1.9; MDM2/CEP12, 12.05) |
| 10/DDIT3 | Tris-EDTA
buffer | Negative | Negative |
| | | (2F, 64%; 3F,
10%;1F, 10%; other, 16%) | (2F, 68%; 3F, 10%;
1F, 12%; other, 10%) |
| | Citrate buffer | Negative | Negative |
| | | (2F, 55%; 3F, 16%;
1F, 12%; other, 17%) | (2F, 63%; 3F, 12%;
1F, 8%; other, 17%) |
| 11/EWSR1 | Tris-EDTA
buffer | Negative | Negative |
| | | (2F, 80%; 1F, 12%;
1G/1O/1F, 2%; other, 6%) | (2F, 75%; 1F, 14%;
1G/1O/1F, 3%; other, 8%) |
| | Citrate buffer | Negative | Negative |
| | | (2F, 79%; 1F, 8%;
1G/1O/1F, 3%; other, 10%) | (2F, 78%; 1F, 15%;
1G/1O/1F, 2%; other, 5%) |
Discussion
Over the decades, precise genome testing has
achieved rapid progress, demonstrating great value in definitive
and differential diagnoses as well as guidance for therapy for
multiple tumor types. FISH is a useful pathological tool for
detecting chromosome rearrangement, amplification, gain or deletion
and polysomy (8). In the present
study, five specific DNA probes extensively used in pathological
diagnosis were applied, including EWSR1, TFE3, SS18, DDIT3 and
MDM2. Chromosomal rearrangement involving the EWSR1 (22q12)
prevalently occurs in Ewing sarcoma, and EWSR1 mostly fuses with
FLI1 (11q24) and ERG (21q22) (9).
Chromosomal translocation of TFE3 (Xp11.2) is an important
characteristic of alveolar soft part sarcoma and microphthalmia
transcription factor family translocation renal cell carcinoma
(4,10). Recurrent DDIT3 (12q13)
rearrangement is commonly regarded as a diagnostic marker for
myxoid liposarcoma with high specificity and sensitivity (11). SS18 (18q11) rearrangement occurs in
90% of synovial sarcomas but has not yet been detected in other
sarcomas (12). MDM2 gene
amplification is an important index for classifying atypical
lipomatous tumor/well-differentiated liposarcoma and the
amplification level may influence tumor dedifferentiation and
progression (13,14).
In laboratory practice, successful FISH detection
first requires appropriate sample handling, including fixation,
embedding and preservation. Poorly processed FFPE sections
frequently fail with regard to signal observation and clinical
diagnosis. During tissue fixation, formaldehyde produces inter- and
intra-molecular crosslinks in proteins and nucleic acids. Methylene
bridges are assumed to be the major structure of those crosslinks
and are stable between molecules (15). Inappropriate fixation procedures,
such as over-fixation, lead to excessive formation of methylene
bridges and a large proportion of nucleic acids may be trapped in
protein-protein crosslinks, hampering probe binding to targeted DNA
sequences (16). Several
troubleshooting tips have been proposed to solve this problem. One
recommended method is adjusting the pretreatment and/or digestion
time. Conventionally, NaSCN and pepsin are used as pretreatment and
digestion reagents, respectively, to cleave crosslinks and increase
tissue permeability. In the present study, slides were routinely
immersed in NaSCN at 80˚C for 10-15 min and subsequently, tissues
were digested with pepsin at 37˚C for 10-15 min. To obtain clear
fluorescence signals in the present study, the pretreatment and
digestion time was first prolonged in increments of 3-5 min.
However, extending the pretreatment time resulted in only minor
signal intensity enhancement and long-term (20-25 min) soaking in
NaSCN solution always resulted in swollen nuclei, which were
difficult to distinguish due to nucleus overlap, particularly in
dense connective tissues. Similarly, limited improvement was
observed after increasing the duration of digestion. By contrast,
excessive digestion led to morphological degradation of nuclei and
other strategies were required to be sought to resolve this
issue.
Several studies have used a microwave oven during
the DNA-DNA hybridization process. In a previous study,
intermittent microwave irradiation at 42˚C with a 3-sec
irradiation/2-sec stop cycle for 1 h, followed by overnight
incubation at 42˚C was employed (17). According to their report, among
paraffin blocks yielding no signal in regular FISH, >95%
produced acceptable signals following this modified protocol and
scanning electron microscopy revealed a looser nuclear matrix in
samples exposed to microwave radiation, suggesting a possible
mechanism underlying the enhanced probe binding activity (17). In addition, Weise et al
(18) compared several different
microwave settings during probe hybridization and recommended a
treatment of 4-5 times of microwave exposure within 30 min at 600
W. Soriani et al (19)
applied the rapid FISH approach by Weise et al (18) to detect chromosome
t(15;17)(q24;q21) in acute promyelocytic leukemia and obtained
appreciable results.
Prompted by the marked effect of microwave-produced
heat in signal retrieval during DNA hybridization, HIAR may be
proposed for increasing cell permeability and therefore enhancing
specific probe binding ability. It is now generally accepted that
the removal of intra-molecular crosslinks depends on the total
amount of heat energy applied during the retrieval process instead
of the heating device utilized (20). Heating undermines the gel-like
structure formed by crosslinks in proteins and nucleic acids,
facilitating probe penetration into nuclei and subsequent DNA
binding (21). Unlike intermittent
microwave irradiation, requiring multiple manual operations and
careful temperature monitoring, the present method is relatively
easy to follow since HIAR is already widely utilized in
immunohistochemistry.
The present study has potential limitations. Up to
now, only seven samples for which conventional FISH failed were
collected and modified FISH was performed on them. Furthermore, due
to the limited number of FFPE slides that were provided for each
patient, only one necessary probe was applied to each specimen to
guarantee the pathological diagnosis and comparison of signal
retrieval efficiency between HIAR- and microwave-assisted FISH was
not possible. For cases exhibiting atypical signal patterns, gene
rearrangement cannot be validated using other methods, such as
RT-PCR or NGS, as no extra sections were available for further
assessment. These limitations may be addressed in future research
after establishing a larger cohort. However, the current data are
encouraging for the use of HIAR in FISH to enhance signal
intensity, particularly for challenging specimens in clinical
practice.
In summary, HIAR, which has been applied routinely
in our laboratory for tumor tissues yielding weak or no signal
after a standard FISH workflow, is a reliable tool in clinical FISH
detection with high performance in augmenting fluorescence
signals.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science
Foundation of China (grant no. 82002829) and the Sanming Project of
Medicine in Shenzhen (grant no. SZSM201812076).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QY, CZ, WH and LL conceived the study. QY and CZ
conducted the experiments and interpreted the results. QY performed
the literature search, analyzed the data and drafted the
manuscript. WH and LL revised the work for important intellectual
content. WH and LL confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of the National Cancer Center/National Clinical Research
Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese
Academy of Medical Sciences and Peking Union Medical College
(Shenzhen, China; approval no. 2019-39-2). Written informed consent
was obtained from each patient included in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gu J, Smith JL and Dowling PK:
Fluorescence in situ hybridization probe validation for clinical
use. Methods Mol Biol. 1541:101–118. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tanas MR, Rubin BP, Tubbs RR, Billings SD,
Downs-Kelly E and Goldblum JR: Utilization of fluorescence in situ
hybridization in the diagnosis of 230 mesenchymal neoplasms: An
institutional experience. Arch Pathol Lab Med. 134:1797–1803.
2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sirvent N, Coindre JM, Maire G, Hostein I,
Keslair F, Guillou L, Ranchere-Vince D, Terrier P and Pedeutour F:
Detection of MDM2-CDK4 amplification by fluorescence in situ
hybridization in 200 paraffin-embedded tumor samples: Utility in
diagnosing adipocytic lesions and comparison with
immunohistochemistry and real-time PCR. Am J Surg Pathol.
31:1476–1489. 2007.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vargas AC, Selinger C, Satgunaseelan L,
Cooper WA, Gupta R, Stalley P, Brown W, Soper J, Schatz J, Boyle R,
et al: FISH analysis of selected soft tissue tumors: Diagnostic
experience in a tertiary center. Asia Pac J Clin Oncol. 15:38–47.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Papp G, Mihály D and Sápi Z: Unusual
signal patterns of break-apart FISH probes used in the diagnosis of
soft tissue sarcomas. Pathol Oncol Res. 23:863–871. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Magaki S, Hojat SA, Wei B, So A and Yong
WH: An introduction to the performance of immunohistochemistry.
Methods Mol Biol. 1897:289–298. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mascarello JT, Hirsch B, Kearney HM,
Ketterling RP, Olson SB, Quigley DI, Rao KW, Tepperberg JH,
Tsuchiya KD and Wiktor AE: Working Group of the American College of
Medical Genetics Laboratory Quality Assurance Committee. Section E9
of the American college of medical genetics technical standards and
guidelines: Fluorescence in situ hybridization. Genet Med.
13:667–675. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bertram S and Schildhaus HU: Fluorescence
in situ hybridization for the diagnosis of soft-tissue and bone
tumors. Pathologe. 41:589–605. 2020.PubMed/NCBI View Article : Google Scholar : (In German).
|
|
9
|
Grünewald TGP, Cidre-Aranaz F, Surdez D,
Tomazou EM, de Álava E, Kovsar H, Sorensen PH, Delattre O and
Dirksen U: Ewing sarcoma. Nat Rev Dis Primers. 4(5)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wyvekens N, Rechsteiner M, Fritz C, Wagner
U, Tchinda J, Wenzel C, Kuithan F, Horn LC and Moch H: Histological
and molecular characterization of TFEB-rearranged renal cell
carcinomas. Virchows Arch. 474:625–631. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cheng L, Zhang S, Wang L, MacLennan GT and
Davidson DD: Fluorescence in situ hybridization in surgical
pathology: principles and applications. J Pathol Clin Res. 3:73–99.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Stacchiotti S and Van Tine BA: Synovial
sarcoma: Current concepts and future perspectives. J Clin Oncol.
36:180–187. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ware PL, Snow AN, Gvalani M, Pettenati MJ
and Qasem SA: MDM2 copy numbers in well-differentiated and
dedifferentiated liposarcoma: Characterizing progression to
high-grade tumors. Am J Clin Pathol. 141:334–341. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Miura Y, Keira Y, Ogino J, Nakanishi K,
Noguchi H, Inoue T and Hasegawa T: Detection of specific genetic
abnormalities by fluorescence in situ hybridization in soft tissue
tumors. Pathol Int. 62:16–27. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shi SR, Shi Y, Taylor CR and Gu J: New
dimensions of antigen retrieval technique: 28 Years of development,
practice, and expansion. Appl Immunohistochem Mol Morphol.
27:715–721. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yamashita S and Katsumata O: Heat-induced
antigen retrieval in immunohistochemistry: Mechanisms and
applications. Methods Mol Biol. 1560:147–161. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sugimura H: Detection of chromosome
changes in pathology archives: An application of microwave-assisted
fluorescence in situ hybridization to human carcinogenesis studies.
Carcinogenesis. 29:681–687. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Weise A, Liehr T, Claussen U and Halbhuber
KJ: Increased efficiency of fluorescence in situ hybridization
(FISH) using the microwave. J Histochem Cytochem. 53:1301–1303.
2005.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Soriani S, Mura C, Panico AR, Scarpa AM,
Recchimuzzo P, Dadati R, Farioli R, De Canal G, Mura MA and Cesana
C: Rapid detection of t(15;17)(q24;q21) in acute promyelocytic
leukaemia by microwave-assisted fluorescence in situ hybridization.
Hematol Oncol. 35:94–100. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Yamashita S: Heat-induced antigen
retrieval: Mechanisms and application to histochemistry. Prog
Histochem Cytochem. 41:141–200. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Stumptner C, Pabst D, Loibner M, Viertler
C and Zatloukal K: The impact of crosslinking and non-crosslinking
fixatives on antigen retrieval and immunohistochemistry. N
Biotechnol. 52:69–83. 2019.PubMed/NCBI View Article : Google Scholar
|