Introduction
Ischemic stroke (IS) is a neurological disease
caused by stenosis or obstruction of the cerebral artery. The main
clinical manifestations of this condition are ischemia, anoxia or
necrosis in the cerebral tissue of the perfused area (1). The acute phase of IS is characterized
by a high degree of immune-inflammatory activation that is
accompanied by increased plasma levels of cytokines, adhesion
molecules and selectins (2). The
immune response to acute IS is a major pathophysiological factor
that is locally initiated in occluded and hypoperfused vessels and
proceeds in the ischemic brain parenchyma (3). Moreover, a higher frequency of
proinflammatory genes in subjects with IS may explain the
immune-inflammatory activation of the acute phase of stroke
(4). 5-lipoxygenase activating
protein/ 5-lipoxygenase (FLAP/5-LO) is a key inflammatory mediator
of the metabolic pathway, playing an important role in the
biogenesis of leukotriene (LT) (5-7).
FLAP, encoded by the arachidonate 5-lipoxygenase-activating protein
(ALOX5AP) gene, can adjust the biogenesis of LT, activating
neutrophils and monocytes to increase the adhesion and permeability
of the internal vascular wall, contributing to atherosclerosis
(8).
Ström et al (9) reported that knockout of the
ALOX5AP gene in mice was associated with decreased LT
production and amelioration of stroke damage. Xu et al
(10) identified the correlation
between ALOX5AP overexpression and the development of
hypertensive stroke in rats. Domingues-Montanari et al
(11) previously reported that the
mRNA expression levels of ALOX5AP in IS cases were
significantly higher compared with in control subjects. However,
the specific regulation that led to these higher expression levels
of ALOX5AP in IS cases was not clearly detailed.
DNA methylation and microRNA (miR/miRNA) regulation
are two important forms of epigenetic regulation (12,13).
DNA methylation mainly takes place at the cytosine base of a CpG
dinucleotide in differentiated mammalian cells. Higher DNA
methylation levels of the gene promoter region typically result in
transcriptional silencing (14). In
addition, miRNAs can either prevent the translation or promote the
degradation of mRNA by binding to its 3'-untranslated region
(3'-UTR). Accumulating evidence has revealed that miRNAs may act as
essential mediators of posttranscriptional gene silencing during
the development of IS (15).
However, it is not clear whether some specific miRNAs affect the
expression of the ALOX5AP gene in the peripheral blood of
patients with IS. Consequently, miRanda (http://www.microrna.org/microrna/home.do) was used to
predict miRNAs targeting the ALOX5AP gene in the current
study, and two potential miRNAs, miR-335 and mir-495 were
identified, which may mediate the development of IS by the
involvement of inflammatory responses in the arterial wall.
Therefore, in the present study, the effects of the
epigenetic mechanisms such as DNA methylation and miRNA regulation,
on the expression levels of the ALOX5AP gene in peripheral
blood samples of patients with IS were investigated. Initially,
higher expression levels of the ALOX5AP gene in patients
with IS were identified by reverse transcription-quantitative PCR
(RT-qPCR). Subsequently, the effects of DNA methylation on the
expression of the ALOX5AP gene were investigated in
peripheral blood samples of patients with IS. Finally, in
vitro luciferase assays were performed to estimate the effects
of miR-335 and miR-495 on ALOX5AP gene expression.
Materials and methods
Study populations
The study protocols were approved by the Ethics
Committee on Human Research of Zhengzhou University (affiliated to
both hospitals in the present study) and written informed consent
was obtained from each participant. A total of 150 IS patients were
recruited at the Department of Neurology in the First Affiliated
Hospital of Henan University of Chinese Medicine and at the First
People Hospital of Zhengzhou (Zhengzhou, China) from March 2017 to
March 2018 (95 males and 55 females; mean age, 60.76±12.83 years).
These subjects were included in the initial study. A total of 50
patients with IS (30 males and 20 females; mean age, 62.42±13.04
years) were recruited from April 2017 to July 2018 in the second
cohort. IS was diagnosed by the diagnostic criteria of IS revised
in the 4th session of the National Conference on Cerebrovascular
Disease (16). Brain imaging was
performed by computed tomography and/or magnetic resonance imaging.
Ancillary diagnostic investigations and standardized blood tests
were also performed. Patients with cerebral hemorrhage, atrial
fibrillation, hyperthyroidism, cardio embolic stroke, venous
thrombosis, peripheral vascular diseases, liver disorders or kidney
diseases were excluded from the study. All patients were initial
stroke cases. Fasting blood samples from the patients were
collected within 24 h following stroke symptom onset.
The control groups consisted of 150 (population 1:
80 males and 70 females; mean age, 60.83±13.44 years) and 50
(population 2: 28 males and 22 females; mean age, 58.00±13.41
years) subjects selected from the same demographic area. These
subjects were well matched with the patients according to age and
sex. Individuals who had cancer, cerebrovascular, cardiovascular,
hepatic and renal diseases were excluded. All participants were
from the Henan Han population.
Detection of ALOX5AP expression
levels
Total RNA was extracted from peripheral blood using
the RNA Prep Pure Blood kit (Tiangen Biotech Co., Ltd.) according
to the manufacturer's recommendations and quantified using
NanoDrop™ 2000 (Thermo Fisher Scientific, Inc.). Total RNA (300 ng
for each participant) was reverse transcribed into cDNA using the
Fast King RT kit (Tiangen Biotech Co., Ltd.) according to the
manufacturer's recommendations. RT-qPCR amplification was performed
with the SYBR Green PCR Master Mix kit (Takara Bio, Inc.). The
following primer pairs were used for the qPCR: ALOX5AP
forward, 5'-CCTGAAGCAAACATGGATCA-3', and reverse,
5'-GCTCCACTTTATGGGCAAAG-3' and β-actin forward,
5'-TGGCACCCAGCACAATGAA-3' and reverse,
5'-CTAAGTCATAGTCCGCCTAGAAGCA-3'. The reaction was performed on the
7500 Fast Real-time PCR system (Thermo Fisher Scientific, Inc.)
with the following program: Initial denaturation at 95˚C for 30
sec; 40 cycles of 95˚C for 3 sec, 60˚C for 30 sec and 95˚C for 15
sec; and dissolution curve at 60˚C for 30 sec and at 95˚C for 15
sec. β-actin was used as the internal reference gene. Relative
expression levels were calculated using the 2-∆∆Cq
method (17).
Detection of miR-335 and miR-495
expression levels
miRNAs in peripheral blood plasma were extracted
using the miRcute Serum/Plasma miRNA Isolation kit (Tiangen Biotech
Co., Ltd.) according to the manufacturer's instructions. A total of
10 fmol external control (Tiangen Biotech Co., Ltd.) was added into
200 µl plasma prior to extraction of plasma miRNAs. Subsequently,
miRNAs were reverse transcribed into cDNA using the miRcute Plus
miRNA First-Strand cDNA Synthesis kit (Tiangen Biotech Co., Ltd.)
according to the manufacturer's instructions. miRanda (http://www.microrna.org/microrna/home.do) was used to
predict miRNAs targeting the ALOX5AP gene. The primers of
the miRNAs were designed and synthesized by Tiangen Biotech Co.,
Ltd. And the sequences were as follows: miR-335 (Forward:
5'-ACACTCCAGCTGGGTCAAGAGCAATAACGAAA-3', Reverse:
5'-CTCAACTGGTGTCGTGGA-3'); miR-495 (Forward:
5'-GCGAAACAAACATGGTGC-3', Reverse: 5'-GCAGGGTCCGAGGTATTC-3'); the
synthetic C. Elegans oligonucleotide, cel-miR-39 was used as
external control (5'-UCACCGGGUGUAAAUCAGCUUG-3').
RT-qPCR amplification was performed using the
miRcute Plus miRNA qPCR Detection kit (Tiangen Biotech Co., Ltd.).
The PCR reaction was performed using the 7500 Fast Real-time PCR
System (Thermo Fisher Scientific, Inc.) with the following program:
Initial denaturation at 95˚C for 15 min, followed by five cycles of
94˚C for 20 sec, 65˚C for 30 sec and 72˚C for 34 sec without
collecting fluorescent signals and 40 cycles of 94˚C for 20 sec and
60˚C for 34 sec during which fluorescent signals were collected.
The dissolution curve was drawn at 60˚C for 30 sec and at 95˚C for
15 sec. The relative expression levels were calculated using the
2-∆∆Cq method.
Prediction of miRNA that targets
ALOX5AP
The miRanda (http://www.microrna.org/microrna/home.do) database was
used to predict the miRNA that targets ALOX5AP. To evaluate binding
stability and conservation, the potential miRNAs that may bind to
ALOX5AP were selected based on the mirSVR and PhastCons scores
assigned by the computational target prediction algorithm using
MiRanda software. mirSVR represented the thermodynamic score
(≤-0.1). The lower the score, the stronger the miRNA-mRNA binding
stability. Phastcons represented the conservative score. The higher
the score, the greater the miRNA-mRNA binding possibility. Based on
these two scores, the miRNAs most likely to target ALOX5AP
were selected.
DNA methylation
DNA methylation analyses were performed on 17 CpG
sites located in the promoter of the ALOX5AP gene. Genomic
DNA from peripheral blood samples was extracted using the TIANamp
Blood DNA kit (Tiangen Biotech Co., Ltd.) according to the
manufacturer's instructions and quantified with NanoDrop 2000
(Thermo Fisher Scientific, Inc.). Bisulfite conversion of DNA was
manipulated with the EZ DNA Methylation-Gold kit (Zymo Research
Corp.) according to the manufacturer's recommendations.
Subsequently, RT-qPCR was performed in a 20-µl amplification
reaction system including 2 µl DNA sample, 1X reaction buffer
(Takara Bio, Inc.), 2 mM Mg2+, 0.2 mM dNTP, 0.1 µM of
each primer and 1 unit of HotStarTaq polymerase (Takara Bio, Inc.).
The cycling program was as follows: 95˚C for 2 min; 11 cycles of
94˚C for 20 sec, 63-0.5˚C per cycle for 40 sec, 72˚C for 1 min and
24 cycles of 94˚C for 20 sec, 65˚C for 30 sec, 72˚C for 1 min; 72˚C
for 2 min. Subsequently, the samples were mixed with a specific tag
sequence. A total of 20 µl mixture was prepared for each reaction
including 1X reaction buffer (Q5; New England BioLabs, Inc.), 0.3
mM dNTP, 0.25 µM forward primer, 0.25 µM index primer, 1 unit of
Q5TM DNA polymerase (New England BioLabs, Inc.) and 1 µl diluted
template. The following temperature conditions were used: 98˚C for
30 sec; 11 cycles of 98˚C for 10 sec, 65˚C for 30 sec, 72˚C for 30
sec and 72˚C for 5 min. Subsequently, the samples were loaded onto
the Illumina NextSeq 500 (Illumina, Inc.) in order to analyze the
results. Sequencing was run using the 2x300 bp paired-end mode,
with an average target area sequencing depth of >200X in all
samples. The quality-control analysis of sequence reads was
performed using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Illumina sequence files were converted into FASTQ format. All reads
and the reference sequences of the target region were compared by
NCBI Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Reads that
covered 90% of the target sequence or 90% bases that could
completely cover the target region were classified as effective
reads.
Methylation and Haplotype
analysis
CpG islands were detected using CpGplot software
(http://emboss.bioinformatics.nl/cgi-bin/emboss/cpgplot).
The CpG island with the most CpG sites was selected from the
ALOX5AP gene promoter region for subsequent DNA methylation
sequencing. The CpG8 site in the promoter region of the
ALOX5AP gene, also known as the rs4073259 site, is
considered to be a CpG-SNP. This was genotyped, and the methylation
levels of individuals with different genotypes at this site were
compared at different CpG sites.
Dual luciferase assay
The wild-type (WT) or mutant 3'-UTRs of
ALOX5AP containing the predicted miR-335-5p or miR-495-3p
binding sites were synthesized and cloned into the pmirGLO vectors
according to the manufacturer's instructions (Genepharm, Inc.).
miR-335 mimics (5'-UCAAGAGCAAUAACGAAAAAUGUAUUUUU
CGUUAUUGCUCUUGAUU-3'; 20 µM), miR-335 inhibitor
(5'-ACAUUUUUCGUUAUUGCUCUUGA-3'; 20 µM), miR-495 mimics
(5'-AAACAAACAUG GUGCACUUCUUGAAGUGCACCAUGUU UGUUUUU-3'; 20 µM),
miR-495 inhibitor (5'-AAGAAGUGCACCAUGUUUGUUU-3'; 20 µM), negative
control (NC) mimics (forward, 5'-UUCUCCGAACGUGUCACGUTT-3' and
reverse, 5'-ACGUGACACGUUCGGAGAATT-3'; 20 µM) and inhibitor control
(5'-CAGUACUUUUGUGUAGUACAA-3'; 20 µM) were also synthesized
(Genepharm, Inc.). Luciferase reporter assays were performed using
293T cells purchased from ATCC, which were cultured in DMEM
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Hyclone; Cytiva) at 37˚C in a humidified incubator containing
5% CO2. 293T cells were plated at a density of
2x105 cells/well in a 24-well plate and co-transfected
with plasmid (4 µl, 1 µg/µl) and miRNA mimics (3 µl, 20 nM), mimic
negative control (3 µl, 20 nM), inhibitor (3 µl, 20 nM) and
inhibitor control (3 µl, 20 nM). The transfection was performed
using 2 µl simplefect transfection reagent (Polyplus;
PolyPlus-transfection, Inc according to the manufacturer's
recommendations and the cells were cultured for 48 h. To confirm
whether ALOX5AP was the direct target of miR-335 and
miR-495, the 3'-UTR of ALOX5AP was synthesized and cloned
into the luciferase reporter plasmid to construct dual luciferase
reporter gene plasmids including the miR-ALOX5AP 3'-UTR-wildtype
and miR-ALOX5AP 3'-UTR mutant. The dual luciferase reporter gene
plasmids were co-transfected into 293T cells with miR-335 or
miR-335 NC or miR-495 or miR-495 NC, with a blank group being
transfected with the dual luciferase reporter gene plasmids alone.
Luciferase activity was assayed using the Dual-Luciferase Reporter
Assay system (Promega Corporation). Firefly luciferase expression
levels were adjusted with reference to Renilla luciferase
activity. Three independent experiments were performed for each
reporter.
Cell transfection
miRNA mimics, mimic NC, inhibitor and inhibitor
control were obtained from GenePharma, Inc. For the different
transfection groups, 293T cells were seeded into a 6-well plate at
a density of 5x105 cells/well. After culture for 24 h,
cells were first transfected with 5 µl miRNA mimics, inhibitor or
corresponding mimics NC or inhibitor NC at room temperature. The
transfection was performed using the RiboFECT CP transfection
reagent (Guangzhou RiboBio Co., Ltd.) according to the
manufacturer's protocol. At 48 h post-transfection, the cells were
harvested for subsequent experiments. The overexpression/knockdown
efficiency was determined by RT-qPCR.
Statistical analysis
All statistical analyses were performed using SPSS
21.0 (IBM Corp.). Quantitative variables were expressed as the mean
± SD and analyzed using Student's t-test. Pearson's χ2
test was used to assess the differences between qualitative
variables. Statistical analyses of the methylation levels and mRNA
expression levels among different genotypes were performed using
one-way ANOVA with Bonferroni's correction. The correlation between
ALOX5AP mRNA expression level and methylation level was calculated
using Pearson's correlation analysis. Unpaired Student's T-test and
the Mann-Whitney U test were used for haplotype analysis of CpG
loci of the ALOX5AP gene promoter region between ischemic
stroke cases and controls. P<0.05 was considered to indicate a
statistically significant difference. Study power of sample sizes
was evaluated using PASS 15.0 software (NCSS, LLC), with the
results showing a power >70%.
Results
Clinical characteristics of
subjects
The clinical characteristics of the study
populations are shown in Table I.
The levels of total cholesterol and homocysteine in IS cases were
significantly higher compared with in controls (P<0.05). IS
cases exhibited higher percentage of diabetes and hypertension in
the two populations (P<0.05).
 | Table ICharacteristics of two study
populations. |
Table I
Characteristics of two study
populations.
| | Population 1 | Population 2 |
|---|
|
Characteristics | IS cases
(n=150) | Controls
(n=150) | P-value | IS cases
(n=50) | Controls
(n=50) | P-value |
|---|
| Sex
(male/female) | 95/55 | 80/70 | 0.079 | 30/20 | 28/22 | 0.685 |
| Age (mean ± SD,
years) | 60.76±12.83 | 60.83±13.44 | 0.961 | 62.42±13.04 | 58.80±13.66 | 0.178 |
| Total cholesterol
(mmol/l) | 4.81±0.78 | 4.52±1.10 | 0.007a | 4.78±0.84 | 4.36±1.04 | 0.030a |
| Total triglyceride
(mmol/l) | 1.66±0.77 | 1.49±0.77 | 0.057 | 1.60±0.83 | 1.58±1.01 | 0.902 |
| Homocysteine
(µmol/l) | 17.95±8.80 | 14.71±4.02 |
<0.001a | 19.05±7.88 | 15.59±7.34 | 0.025a |
| Diabetes (n,
%) | 30 (20.0%) | 13 (8.6%) | 0.005a | 27 (54.0%) | 11 (22.0%) | 0.001a |
| Hypertension (n,
%) | 79 (52.7%) | 21 (14.0%) |
<0.001a | 39 (78.0%) | 26 (52.0%) | 0.006a |
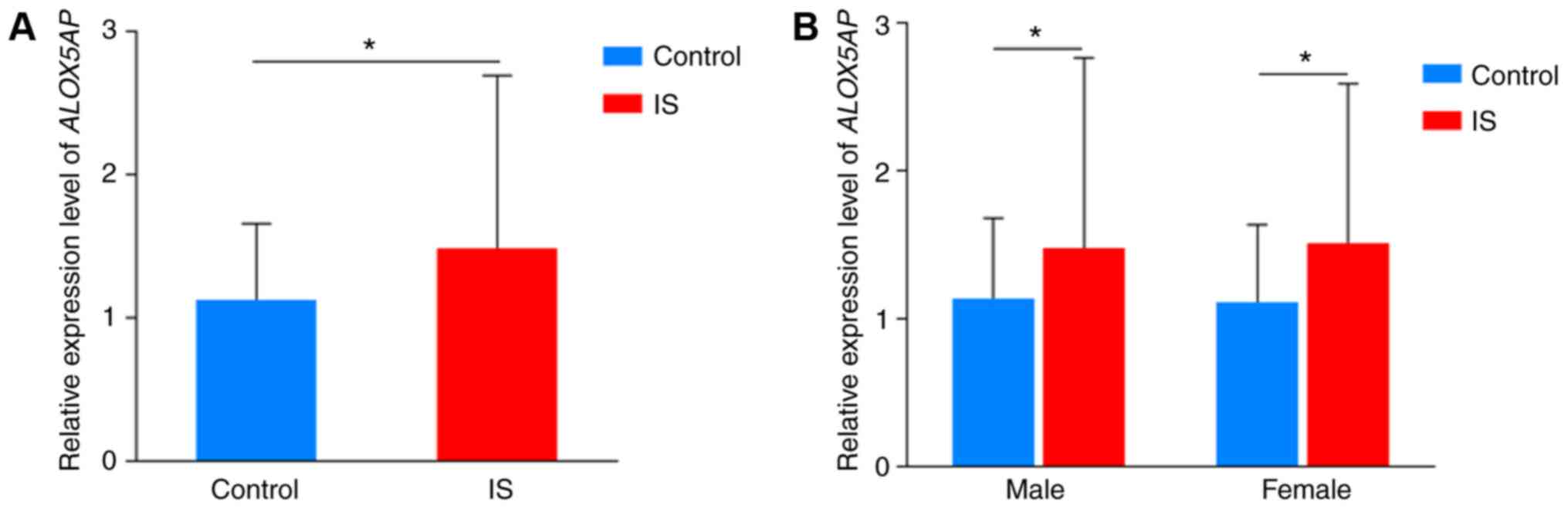
mRNA expression levels of ALOX5AP in
IS cases and controls
The results of mRNA expression levels of
ALOX5AP are shown in Fig. 1.
The mRNA expression levels of ALOX5AP in the IS cases
(1.49±1.21) were significantly higher compared with in controls
(1.13±0.53; (P<0.05). The mRNA expression levels of IS cases
(males, 1.48±1.29; females, 1.51±1.08) were significantly higher
compared with in the control subjects (males, 1.14±0.55; females,
1.11±0.52; P<0.05).
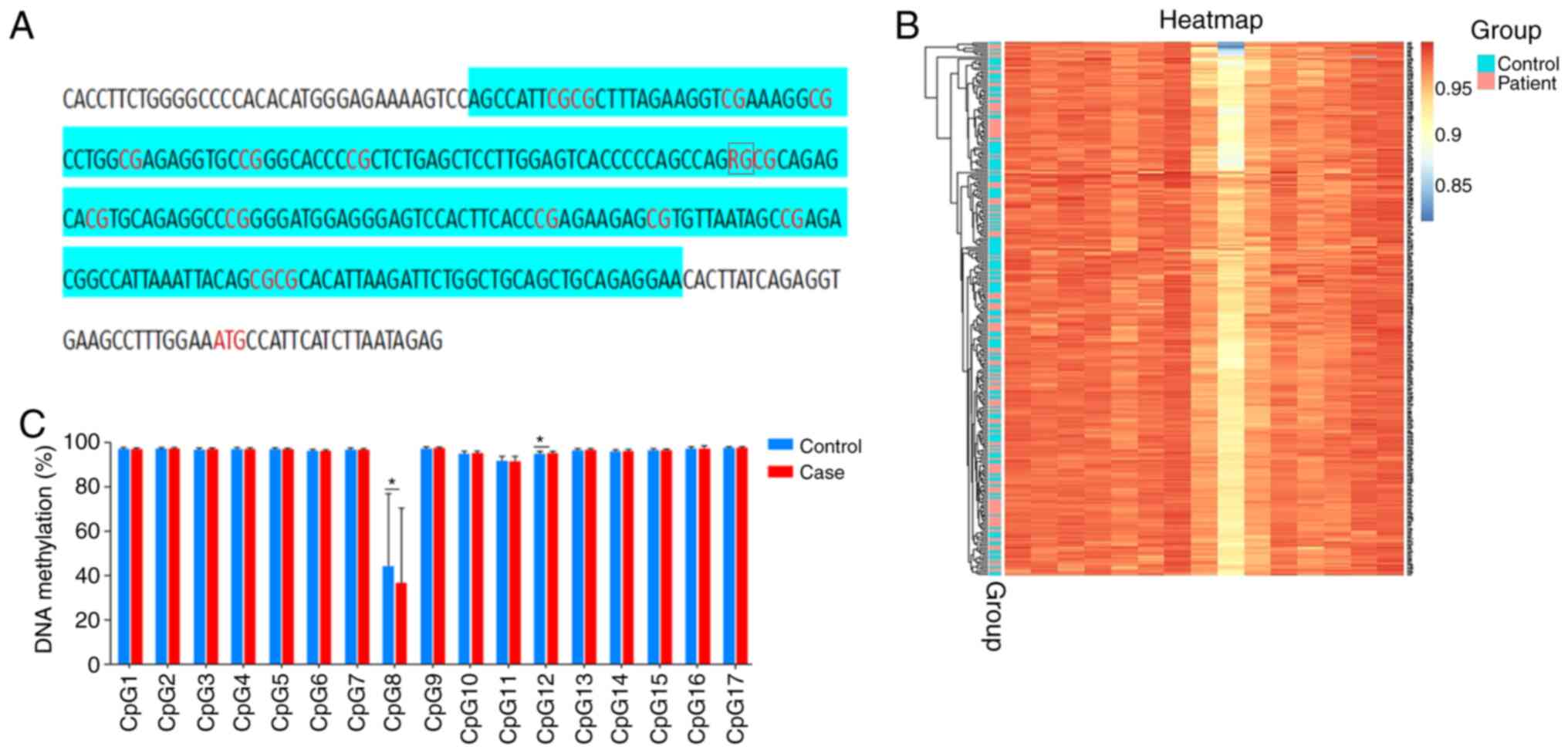
Bioinformatics screening of CpG island
region of ALOX5AP
The methylation status of the 17 CpG sites located
in the promoter region of ALOX5AP was assessed. The sequence
of the CpG Island is shown in Fig.
2A, with the CpG8 site being a CpG-single nucleotide
polymorphism (SNP) site.
Comparison of methylation levels
between IS cases and control subjects
The levels of DNA methylation of the ALOX5AP
gene are shown in Fig. 2B. The
results indicated that hypermethylation levels were noted in both
IS cases and control subjects. No significant differences were
noted in IS cases and control subjects with regard to the mean DNA
methylation levels in the ALOX5AP promoter region (controls,
93.62±1.94; IS, 93.19±1.97). The mean methylation level between
males and females demonstrated no significant difference (Fig. S1). A Pearson correlation analysis
of the mRNA expression of ALOX5AP and the methylation level
of 150 patients with IS was performed. The results revealed that
the methylation levels of the ALOX5AP promoter were
inversely correlated with ALOX5AP expression in IS patients
(r=-0.175; P=0.032; Fig. S1).
Comparison of methylation levels at 17
CpG loci between IS cases and control subjects
The DNA methylation levels of 17 CpG loci of the
ALOX5AP gene were measured by MethyTarget sequencing
(Fig. 2C). A total of 16 CpG loci
were hypermethylated compared to those of the CpG8 site.
Differential DNA methylation was noted on the CpG8 (controls,
42.70±32.56; IS, 37.00±33.67) and CpG12 (controls, 95.37±0.93; IS,
95.62±0.71) sites between patients with IS and control subjects
(P<0.05).
Methylation haplotype analysis of the
CpG island region
Haplotype analysis was applied to determine whether
the haplotypes of the CpG island were significantly different
between IS cases and control subjects. As shown in Table II, seven haplotypes were noted,
which exhibited significantly higher frequency in the IS case group
compared with the control subject group (P<0.05).
 | Table IIHaplotype analysis of CpG loci of the
ALOX5AP gene promoter region between ischemic stroke cases
and controls. |
Table II
Haplotype analysis of CpG loci of the
ALOX5AP gene promoter region between ischemic stroke cases
and controls.
| No. | Target | Haplotype | P-value
(t-test) | P-value (U
test) |
|---|
| 1 | ALOX5AP |
ccccctctccccccccc |
0.041458a |
0.01790895a |
| 2 | ALOX5AP |
ccccccccccccccccc | 0.05102 |
0.03765977a |
| 3 | ALOX5AP |
ccccccctcctcccccc |
0.0160988a |
0.03038263a |
| 4 | ALOX5AP |
ccccccctcccccctcc | 0.062201 |
0.02451332a |
| 5 | ALOX5AP |
ccccccccccctccccc |
0.0182789a | 0.05970357 |
| 6 | ALOX5AP |
ctccccctccccccccc |
0.04632a |
0.01672627a |
| 7 | ALOX5AP |
tcccccctccccccccc |
0.0035237a |
0.0083393a |
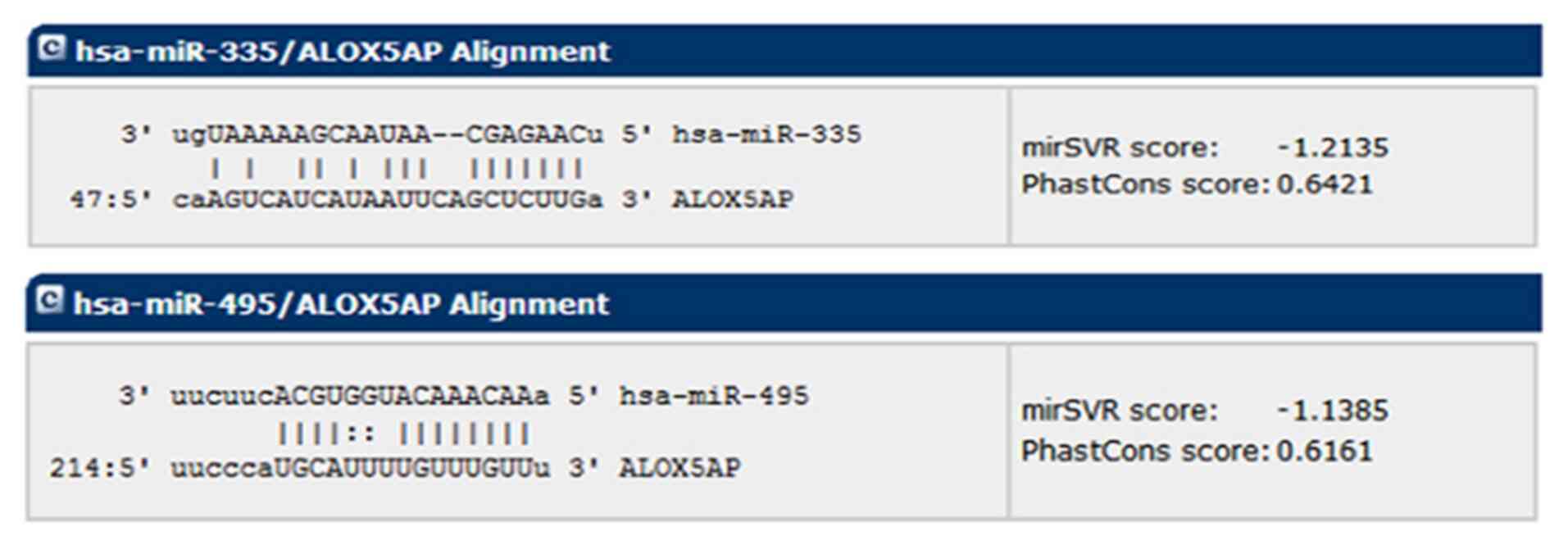
Post-transcriptional regulation of the
ALOX5AP gene by miRNAs as predicted by bioinformatic analysis
miRanda (http://www.microrna.org/microrna/home.do), mirSVR
scores and PhastCons scores were used to screen miR-335 and miR-495
as two miRNAs that could potentially target ALOX5AP
(Fig. 3).
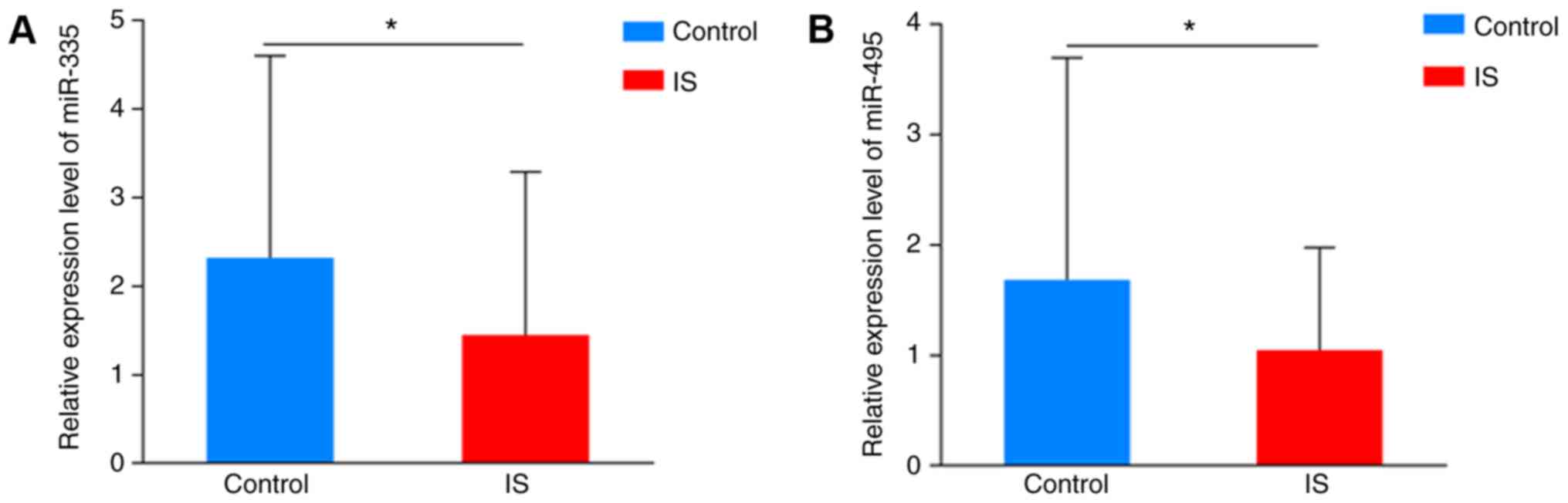
Comparison of the expression levels of
miR-335 and miR-495 in peripheral blood plasma
The expression levels of miR-335 and miR-495 are
shown in Fig. 4. The IS cases
exhibited significantly lower expression levels of miR-335
(1.49±1.84 vs. 2.32±2.28; P=0.047) and miR-495 (1.05±0.93 vs.
1.69±2.01; P=0.043) compared with control subjects .
miR-335 and miR-495 binds to the
3'-UTR region of the ALOX5AP gene
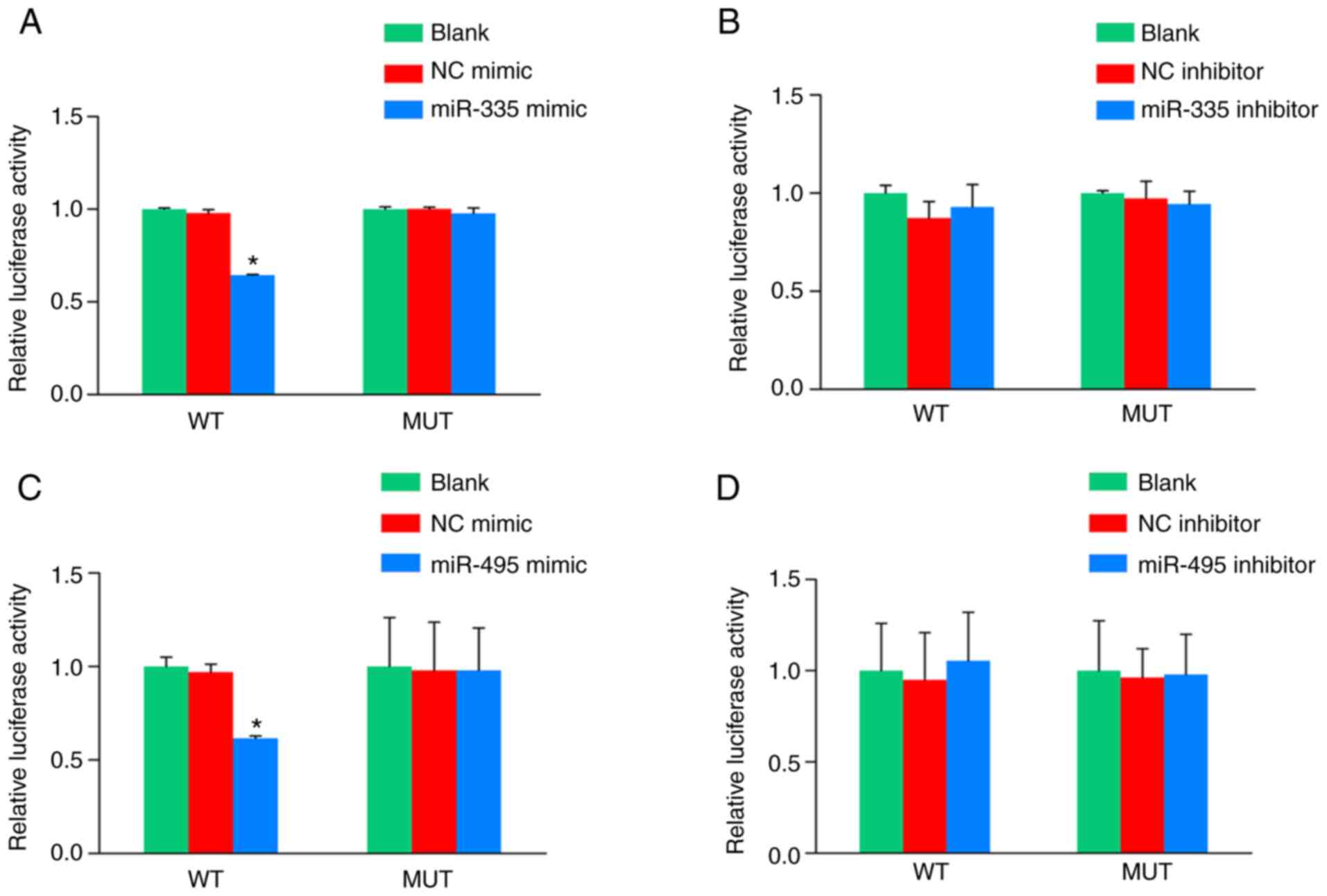
The results indicated that the relative luciferase
activity of 293T cells transfected with miR-335 mimics and
miR-335-ALOX5AP-WT-pmirGLO was decreased in comparison with cells
transfected with NC mimics and miR-335-ALOX5AP-WT-pmirGLO (Fig. 5A), while there were no differences
between miR-335 mimics and NC mimics transfected cells in the
mutation group (Fig. 5B).
Similarly, the relative luciferase activity of 293T cells
transfected with miR-495 mimics and miR-495-ALOX5AP-WT-pmirGLO was
decreased compared with the cells transfected with NC mimics and
miR-495-ALOX5AP-WT-pmirGLO (Fig.
5C), while there were no differences between miR-495 mimics and
NC mimics transfected cells in the mutation group (Fig. 5D).
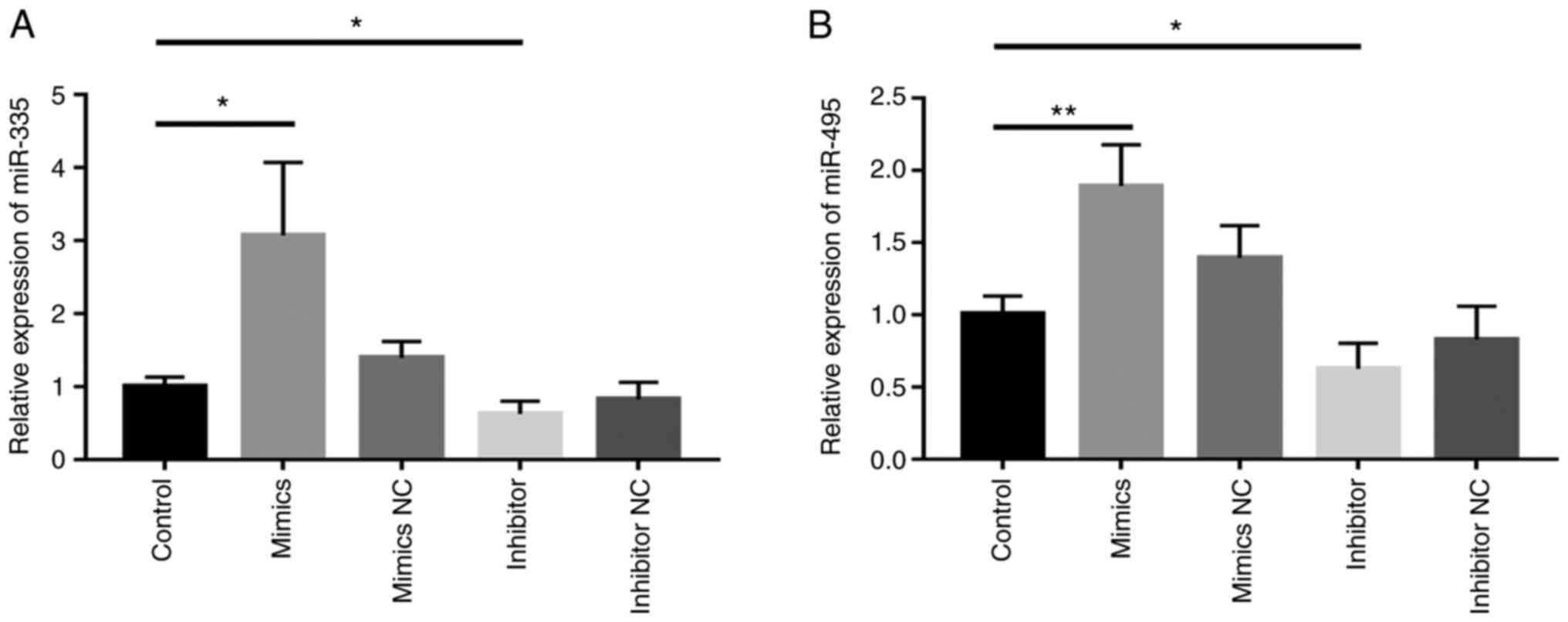
Effects of miR-335 and miR-495 on
ALOX5AP expression in 293T cells
293T cells were first transfected with miR-335
mimics, miR-495 mimics, miR-335 inhibitor, miR-495 inhibitor,
corresponding mimics NC or inhibitor NC. The
overexpression/knockdown efficiency was determined by RT-qPCR. The
results demonstrated that transfection of miR-335 mimics or miR-495
mimics led to a significant increase in the expression levels of
miR-335 (2.423±0.466 vs. 1.005±0.072; P=0.039) or miR-495
(1.893±0.164 vs. 1.005±0.072; P=0.005), respectively, compared with
controls. Meanwhile, transfection of miR-335 inhibitor (0.626±0.102
vs. 1.005±0.072; P=0.038) or miR-495 inhibitor (0.715±0.048 vs.
1.006±0.080; P=0.036) resulted in a significant decrease in miR-335
or miR-495 expression, respectively, compared with corresponding
negative controls (Fig. 6).
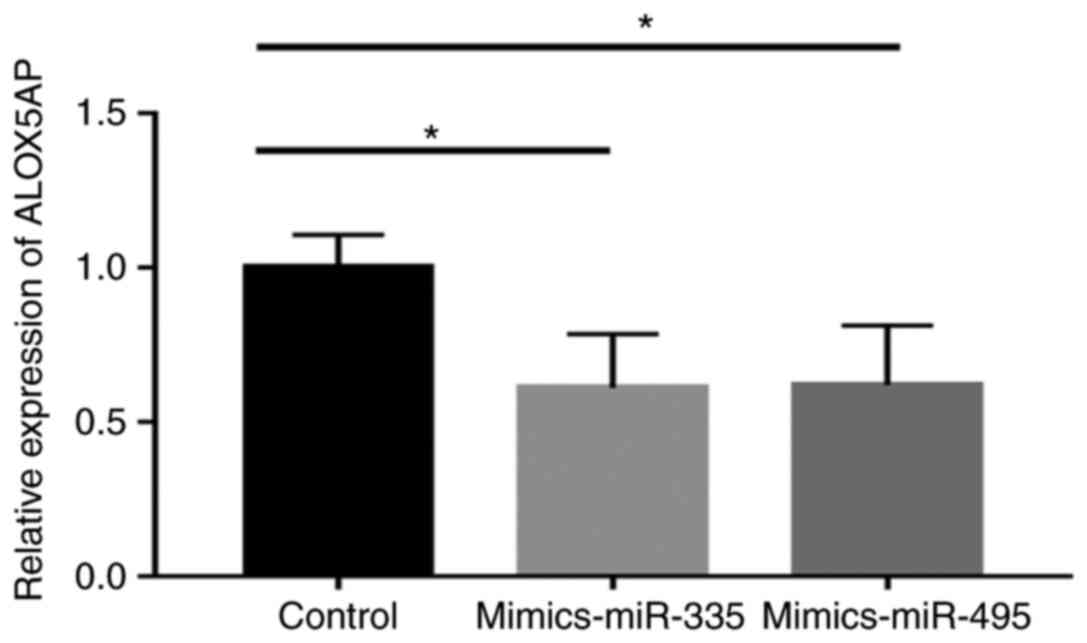
Compared with the control group, ALOX5AP expression in 293T
cells was significantly reduced (miR-335, 0.612±0.100 vs.
1.004±0.059, P=0.028; miR495, 0.6208±0.111 vs. 1.004±0.059,
P=0.038; Fig. 7).
Discussion
In the present study, a case-control study was
designed to investigate the epigenetic regulation mechanisms,
including DNA methylation and miRNA regulation, on the expression
levels of the ALOX5AP gene in patients with IS. In 2005,
Moore et al (18) detected
the gene expression levels of monocytes in peripheral blood samples
of 20 patients with IS and 20 control subjects using
oligonucleotide microarray technologies. The results indicated
significant changes in the expression levels of several genes in
monocytes and the differences in gene expression levels were
consistent with the central nervous system (CNS) response of IS
subjects. In 2007, Sharp et al (19) confirmed that the expression levels
of genes in white blood cells (WBCs) and monocytes in peripheral
blood samples of patients with IS were altered. Baird (20) discovered that WBCs could migrate and
infiltrate to the infarction and subsequently induce inflammatory
response and specific changes in gene expression levels of patients
with stroke. This study utilized peripheral blood samples of
patients with stroke and provided information on the CNS
microenvironment of the examined subjects (21). In the present study, RT-qPCR was
used to detect the mRNA expression levels of the ALOX5AP
gene in the peripheral blood of 150 patients with IS and 150
control subjects. The results indicated that the expression levels
of the ALOX5AP gene in the IS group were significantly
higher compared with in the control subjects. The mRNA expression
levels of IS cases in different sexes were higher compared with the
control subjects. In 2010, Domingues-Montanari et al
(11) demonstrated a difference in
the expression levels of the ALOX5AP gene between IS and the
control groups (P=0.003), which was consistent with the present
findings. The ALOX5AP gene product, FLAP, has been
implicated in the regulation of LTs and is recognized as an
important signaling molecule in a variety of inflammatory
responses. It is also implicated in the progression of
atherosclerosis. Increased FLAP activity leads to the accumulation
of LTs on fatty deposits of the arterial wall (22). The subsequent breakdown of these
deposits by the immune system may subsequently lead to the
development of atherosclerosis and may confer an increased risk of
stroke. Hence, the present study inferred that the increasing
expression of the ALOX5AP gene increased FLAP production in
patients with IS; FLAP combined with 5-LO upregulated the
biosynthesis of LT to promote the occurrence of inflammatory
reactions, participate in atherosclerosis and eventually lead to IS
(23). However, the specific
mechanism of the regulation of the ALOX5AP gene expression
of IS patients remains unclear.
In order to investigate the differences in the
expression levels of ALOX5AP in IS, MethyTarget sequencing
was used to detect the methylation levels of the promoter region of
150 patients with IS and of 150 healthy control subjects. The
results indicated that the hypermethylation levels were present in
both IS cases and control subjects. The overall methylation levels
between the two groups did not reveal significant differences. The
DNA methylation levels of 17 CpG loci of the ALOX5AP gene
were compared between the IS and the control groups and the data
demonstrated that the remaining 16 CpG loci were hypermethylated,
with the exception of the CpG8 site. Differential DNA methylation
was noted on the CpG8 and CpG12 sites between patients with IS and
control subjects. A total of seven methylation haplotypes were
noted with high frequency in the IS group. Significant differences
were noted between the IS and the control groups. A Pearson
correlation analysis of the mRNA expression of ALOX5AP and
methylation level of 150 patients with IS was performed. The
results indicated that the methylation levels of the ALOX5AP
promoter correlated inversely with ALOX5AP expression in IS
subjects (r=-0.175, P=0.032; Fig.
S1). However, the hypermethylation of the promoter region of
the ALOX5AP gene could not inhibit the expression levels of
the gene. The introduction or removal of CpG dinucleotides, which
are possible sites of DNA methylation, has been suggested as a
potential epigenetic mechanism by which SNPs can affect gene
function (24). Of note, the
methylation levels of CpG8 were significantly different from other
sites due to the generation of a CpG site. In a previous study
conducted by our group, IS-associated CpG-SNP rs4073259 correlated
with differential DNA methylation, presenting the phenomenon of
allele-specific methylation which may elucidate the phenotypic
effects of certain genetic variants (25).
In 2013, Cao et al (26) demonstrated that the hypermethylation
of the promoter region of the phospholipid-transporting ATPase
ABCA1 gene was more frequent in males than in female subjects. In
2015, Lin et al (27)
discovered that the methylation levels of the estrogen receptor-α
gene were significantly lower in female patients with IS compared
with control subjects, probably due to a potential protective
mechanism against neurological damage in females. Therefore, the
average DNA methylation levels of the ALOX5AP gene were
analyzed according to sex and no significant differences were noted
(Fig. S2). The results of the
present study were different from those reported in the study by
Lin et al (27), possibly
due to population differences, phenotypic heterogeneity and a small
sample size.
Bioinformatics analysis predicted that two miRNAs,
miR-335 and miR-495, are most likely to target the ALOX5AP
gene and downregulate its expression. Initially, the expression
levels of miR-335 and miR-495 were analyzed in the plasma of 50
patients with IS and in the 50 corresponding control subjects. The
results indicated that the expression levels of miR-335 and miR-495
in the IS group were significantly lower compared with the control
subjects. Therefore, it was hypothesized that binding of miR-335
and miR-495 to the 3'-UTR region of the ALOX5AP gene may
inhibit the expression levels of this gene. To confirm this
hypothesis, a WT and mutant reporter gene plasmid was constructed
containing miR-335 and miR-495 seed regions. The luciferase
activity of the different groups was assessed and the potential of
miR-335 and miR-495 to target the ALOX5AP gene was examined
by co-transfection of miRNA mimics, mimic negative control,
inhibitor, inhibitor negative control and recombinant plasmids. The
results indicated that the luciferase activity of the experimental
group with co-transfected miRNA mimic and WT reporter gene plasmid
was significantly lower compared with the other experimental
groups, suggesting that miR-335 and miR-495 could specifically bind
to the 3'-UTR of the ALOX5AP gene, thereby downregulating
its expression.
In summary, the present study demonstrated that the
mRNA expression levels of ALOX5AP were significantly higher
in the IS cases compared with in control subjects. However, the
methylation levels of the ALOX5AP promoter correlated
inversely with ALOX5AP expression in IS cases. miR-335 and
miR-495 are lowly expressed in the peripheral blood plasma of IS
patients, act on the 3'-UTR region of ALOX5AP gene by
negatively regulating its post-transcriptional expression, leading
to the increased expression of ALOX5AP gene in the IS group.
The differential regulation mechanism of the gene expression is an
extremely complex process involving various factors. The specific
differentiation mechanism of the ALOX5AP gene expression in
IS requires further studies.
The present study includes several limitations.
Firstly, the expression levels of the FLAP protein were not
explored in patients with IS and control subjects. Secondly, no
significant differences were noted in the overall methylation
levels of the promoter region of the ALOX5AP gene between IS
cases and control subjects. Thirdly, a potential selection bias may
have been present since the cases and controls were recruited from
the same hospitals.
Supplementary Material
The methylation levels of the
ALOX5AP promoter correlated inversely with ALOX5AP
expression in IS subjects. The abscissa represents the mean
methylation levels of the ALOX5AP gene and the abscissa
represents the mRNA level of ALOX5AP. ALOX5AP;
arachidonate 5-lipoxygenase-activating protein.
Mean DNA methylation of the
ALOX5AP promoter region in the peripheral blood of IS group
and control group between different sexes. ALOX5AP;
arachidonate 5-lipoxygenase-activating protein.
Acknowledgements
Not applicable.
Funding
The research reported in this publication was supported by a
grant from the National Science Foundation of China-Henan United
fund (grant no.U2004114).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XB, LX, XW and ZZ performed the experiments. HLZ, DY
and BZ conducted the statistical analysis. XZ, YW, SY and YL were
involved in study implementation and participant recruitment. YH,
ZZ and XW wrote the manuscript. YH, HZ, YX and HD conceived the
study and participated in its design and coordination. XB and LX
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study protocols were approved by the Ethics
Committee on Human Research of Zhengzhou University and informed
written consent was obtained from each participant. All experiments
were performed in accordance with relevant guidelines and
regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hankey GJ: Stroke. Lancet. 389:641–654.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tuttolomondo A, Pecoraro R, Casuccio A, Di
Raimondo D, Buttà C, Clemente G, Corte VD, Guggino G, Arnao V,
Maida C, et al: Peripheral frequency of CD4+
CD28- cells in acute ischemic stroke: Relationship with
stroke subtype and severity markers. Medicine (Baltimore).
94(e813)2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tuttolomondo A, Pinto A, Corrao S, Di
Raimondo D, Fernandez P, Di Sciacca R, Arnao V and Licata G:
Immuno-inflammatory and thrombotic/fibrinolytic variables
associated with acute ischemic stroke diagnosis. Atherosclerosis.
203:503–508. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tuttolomondo A, Di Raimondo D, Pecoraro R,
Casuccio A, Di Bona D, Aiello A, Accardi G, Arnao V, Clemente G,
Corte VD, et al: KIRIIND (KIR Infectious and Inflammatory Diseases)
Collaborative Group. HLA and killer cell immunoglobulin-like
receptor (KIRs) genotyping in patients with acute ischemic stroke.
J Neuroinflammation. 16(88)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rådmark O, Werz O, Steinhilber D and
Samuelsson B: 5-Lipoxygenase, a key enzyme for leukotriene
biosynthesis in health and disease. Biochim Biophys Acta.
1851:331–339. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jala VR and Haribabu B: Leukotrienes and
atherosclerosis: New roles for old mediators. Trends Immunol.
25:315–322. 2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Colazzo F, Gelosa P, Tremoli E, Sironi L
and Castiglioni L: Role of the cysteinyl leukotrienes in the
pathogenesis and progression of cardiovascular diseases. Mediators
Inflamm. 2017(2432958)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Fan Y, Chen H, Li A, Shi Y, Zhang Y, Feng
Q, Sun Y, Zheng H and He Y: A promoter polymorphism (rs17222919,
-1316T/G) of ALOX5AP gene is associated with decreased risk
of ischemic stroke in two independent Chinese populations. PLoS
One. 10(e0122393)2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ström JO, Strid T and Hammarström S:
Disruption of the alox5ap gene ameliorates focal ischemic stroke:
Possible consequence of impaired leukotriene biosynthesis. BMC
Neurosci. 13(146)2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xu X, Sheng W and Liu L: Association
between mRNA level of Pde4d and Alox5ap and hypertensive stroke as
well as hypertension in rats. Zhonghua Yi Xue Yi Chuan Xue Za Zhi.
24:491–494. 2007.PubMed/NCBI(In Chinese).
|
|
11
|
Domingues-Montanari S, Fernández-Cadenas
I, del Rio-Espinola A, Corbeto N, Krug T, Manso H, Gouveia L,
Sobral J, Mendioroz M, Fernández-Morales J, et al: Association of a
genetic variant in the ALOX5AP with higher risk of ischemic
stroke: A case-control, meta-analysis and functional study.
Cerebrovasc Dis. 29:528–537. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Matouk CC and Marsden PA: Epigenetic
regulation of vascular endothelial gene expression. Circ Res.
102:873–887. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chauhan G and Debette S: Genetic Risk
Factors for Ischemic and Hemorrhagic Stroke. Curr Cardiol Rep.
18(124)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yong D, Chao X, Hui WJ, Yu SY, Ye ZJ and
Gang C: Recent progress in epigenetics. Sci Sin. 1:3–15. 2017.
|
|
15
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lindsay P, Furie KL, Davis SM, Donnan GA,
Norrving B and et al: World Stroke Organization global stroke
services guidelines and action plan. Int J Stroke. Suppl A100:4–13.
2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Moore DF, Li H, Jeffries N, Wright V,
Cooper RA Jr, Elkahloun A, Gelderman MP, Zudaire E, Blevins G, Yu
H, et al: Using peripheral blood mononuclear cells to determine a
gene expression profile of acute ischemic stroke: A pilot
investigation. Circulation. 111:212–221. 2005.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sharp FR, Xu H, Lit L, Walker W, Pinter J,
Apperson M and Verro P: Genomic profiles of stroke in blood.
Stroke. 38 (Suppl):S691–S693. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Baird AE: Blood genomics in human stroke.
Stroke. 38 (Suppl):S694–S698. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Asano S, Chantler PD and Barr TL: Gene
expression profiling in stroke: Relevance of blood-brain
interaction. Curr Opin Pharmacol. 26:80–86. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li Y, Li Z, Zhang X, Yan C, Kang J, Liang
Z, Liu S, Feng X and Han Y: Association of ALOX5AP
haplotypes with susceptibility to coronary artery disease in a
Chinese Han population. Eur J Intern Med. 23:e119–e123.
2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bruno F, Spaziano G, Liparulo A, Roviezzo
F, Nabavi SM, Sureda A, Filosa R and D'Agostino B: Recent advances
in the search for novel 5-lipoxygenase inhibitors for the treatment
of asthma. Eur J Med Chem. 153:65–72. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen Z, Zheng J, Liu W, Yang K, Li K,
Huang B, Zhu R, Lu X and Li L: The SG13S114 polymorphism of the
ALOX5AP gene is associated with ischemic stroke in
Europeans: A meta-analysis of 8062 subjects. Neurol Sci.
38:579–587. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shi Y, Xu L, Feng Q, Li A, Jia J, Xu Y,
Yang D, Zhang Y, Zhang X, Zhao H, et al: Allele-specific
methylation contributed by CpG-SNP is associated with regulation of
ALOX5AP gene expression in ischemic stroke. Neurol Sci.
39:1717–1724. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cao XL, Yin RX, Huang F, Wu JZ and Chen
WX: Chromosome 9p21 and ABCA1 genetic variants and their
interactions on coronary heart disease and ischemic stroke in a
Chinese Han population. Int J Mol Sci. 17(586)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lin HF, Hsi E, Liao YC, Chhor B, Hung J,
Juo SH and Lin RT: Demethylation of circulating estrogen receptor
alpha gene in cerebral ischemic stroke. PLoS One.
10(e0139608)2015.PubMed/NCBI View Article : Google Scholar
|