Introduction
Glioma is considered to be the most aggressive tumor
among the primary central nervous system tumors. It is classified
as astrocytoma, oligodendroglioma, ependymoma, medulloblastoma, and
glioblastoma (1,2). The tumor is clinically treated
primarily via surgical resection, chemoradiotherapy, gene therapy
and sometimes, also by using Chinese medicine (3-5),
while considering the patient's health condition, age, tumor type,
location, size, and clinical grade. Nonetheless, as glioma is
resistant to traditional chemotherapy, the mortality and relapse
rates remain high. The in-depth pathogenesis of glioma, including
aberrant activation of proto-oncogenes and inactivation of tumor
suppressors, remains to be elucidated (6-8).
Thus, probing the molecular mechanism and developing efficacious
methods for diagnosing and treating gliomas are important.
MicroRNAs (miRNAs/miRs), measuring 18 to 26
nucleotides, are a class of non-coding RNA molecules in eukaryotes
(9) that trigger the RNA-induced
silencing complex to degrade messenger RNAs (mRNAs) or curb
translation by pairing with their target genes (10). miRNAs have been highly conserved
during species evolution and those discovered in plants, animals
and fungi are expressed in specific tissues and at specific
developmental stages only. The tissue and timing-associated
specificities of miRNAs determine the distinctive functions of
tissues and cells, indicating the multiple roles that miRNAs play
in controlling developmental processes (11-14).
It has been verified by recent studies that miRNAs
have an indispensable impact on the pathogenesis of gliomas
(15); for example, intercellular
transfer of miRNAs via gap junctions influences glioma cell
proliferation (16). Mesenchymal
stem cell-derived exosomal miR-133b targets enhancer of zeste
homolog 2 via the Wnt/β-catenin signaling pathway (17) to curb glioma progression. In
addition, miR-3148 has been reported to hinder proliferation and
boost apoptosis in cervical cancer cells (18). Liu et al (19) pointed out that miR-3148 is
significantly downregulated in human glioma stem cells compared
with that in human neural stem cells. However, whether miR-3148
regulates glioma remains unknown.
In the current study, the downregulation of miR-3148
in human glioma tissues and cell lines was verified and the aim was
to investigate the role of miR-3148 in glioma.
Materials and methods
Ethical compliance
The present study was approved by the Ethics
Committee of the First People's Hospital of Jingmen (Hubei, China).
All population-based studies were carried out in accordance with
the World Medical Association's Declaration of Helsinki, and all
subjects provided written informed consent.
Specimen collection and
processing
Forty-eight surgically resected glioma specimens and
non-tumor tissues (2 cm away from the tumor) were collected from
patients (male=27, female=21; I/II 23, III/IV 25) at the First
People's Hospital of Jingmen between March 2012 and March 2014. The
median age of the patients was 46 years (range, 30 to 72 years).
These patients without tumors of other classes, autoimmune
diseases, viral hepatitis and those not undergoing preoperative
chemoradiotherapy before operation. Following surgical resection
under aseptic conditions, all specimens were immediately frozen in
liquid nitrogen and preserved at -80˚C until further use.
Taking the average expression level of miR-3148 as
the cutoff, 48 patients were split into high and low expression
groups for survival analysis. Data were analyzed by Fisher's exact
test.
Cell culture
U87 MG (glioblastoma of unknown origin, BNCC100646),
SHG-44, U251 and H4 glioma cell lines, and HEB normal cells (BeNa
Culture Collection) were cultured in an incubator containing
Dulbecco's Modified Eagle Medium (DMEM, Thermo Fisher Scientific,
Inc.) with 10% fetal bovine serum (FBS, Sigma-Aldrich; Merck KGaA),
at 37˚C and 5% CO2. The cell lines were authenticated
using STR profiling.
In total, 50 nM miR-3148 mimics (Sense,
5'-UGGAAAAAACUGGUGUGUGCUU-3'; Antisense,
5'-GCACACACCAGUUUUUUCCAUU-3') and the negative controls (Sense,
5'-UUCUCCGAACGUGUCACGUTT-3'; Antisense,
5'-ACGUGACACGUUCGGAGAATT-3') (Shanghai GenePharma Co., Ltd.) were
used to overexpress miR-3148 in U251 cells. 50 nM miR-3148
inhibitors (5'-AAGCACACACCAGUUUUUUCCA-3') and the equivalent NC
(5'-CAGUACUUUUGUGUAGUACAA-3') (Shanghai GenePharma Co., Ltd.) were
used to knock down miR-3148 in U87 MG cells. Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.) was
utilized to transfect these plasmids into U251 or U87 MG cells
(5x106 cells/well) following the manufacturer's
instructions. Following 48 h of transfection at 37˚C, cells were
collected (centrifugation at 4˚C at 12,000 x g for 15 min) and the
transfection efficiency was determined using RT-qPCR. Subsequent
experiments were performed 48 h after transfection.
Reverse transcription quantitative PCR
(RT-qPCR)
Sequestering of total RNA was achieved by means of
RNAzol reagent (Vigorous Biotechnology Beijing Co., Ltd.) as per
the manufacturer's instructions. Then, Moloney murine leukemia
virus reverse transcriptase (Promega Corporation) was used to
reverse transcribe total isometric RNA (2 µg), with the
transcription level normalized to the 18S rRNA level. Subsequently,
RT-qPCR was performed using an ABI 7300 RT-PCR system with
SYBR® Green RT-PCR Master mix (Toyobo Life Science). The
qPCR was conducted at 95˚C for 10 min followed by 40 cycles of 95˚C
for 30 sec and 60˚C for 1 min. The primers for CRNDE,
miR-3148 and DCUN1D1 were purchased from Guangzhou RiboBio
Co., Ltd. The primer sequences that were used are as follows:
miR-3148 forward (F), 5'-TGGAAAAAACTGGTGTGTGCTT-3'; miR-3148
reverse (R), 5'-GCTGTCAACGATACGCTACCTA-3'; DCUN1D1 F,
5'-AGGATCATTGGACAGGAAGAAGT-3'; DCUN1D1 R,
5'-TGCCAGGTCATCACAGAACTG-3'; GAPDH F, 5'-AGAAGGCTGGGGCTCATTTG-3';
and GAPDH R, 5'-AGGGGCCATCCACAGTCTTC-3' was used as an endogenous
control for mRNA. The expression of miRNA was normalized to U6 F,
5'-CTCGCTTCGGCAGCACA-3'; and U6 R, 5'-AACGCTTCACGAATTTGCGT-3'.
Finally, the relative content of specimens was examined using the
2-ΔΔCq method (20).
Each assay was averaged over three performances.
Cell proliferation
After culturing cells at 37˚C for 24 h in a 96-well
plate at a density of 2x103 cells/well, U87 MG and U251
cell lines were incubated at 37˚C for 2 h with 10 µl Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) solution. Then,
the optical density at 450 nm was measured and recorded.
The role of miR-3148 in glioma cell proliferation
was evaluated using Click-iT® EdU Imaging Kits
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. U87 MG and U251 cells were cultured in
96-well plates (8x103 cells/well) and incubated at room
temperature with 10 µl of EdU reagent for 3 h. At room temperature,
the cells were fixed with 4% formaldehyde for 20 min and washed
with formaldehyde in PBS. The cells were then incubated at room
temperature in 0.5% Triton X-100 (Sigma-Aldrich; Merck KGaA) for 5
min. The nuclei were then stained with a
4',6-diamidino-2-phenylindole solution (1 ml; Sigma-Aldrich; Merck
KGaA), which was added to each well and incubated for 25 min at
room temperature in the dark. The staining solution was then
removed by washing thrice with PBS. Finally, the stained cells were
imaged and counted under a fluorescence microscope (magnification,
x100) (CKX41-F32FL; Olympus, Beijing, China). Each assay was
performed in triplicates.
Cell migration
After being inoculated on a six-well plate, the
cells were transfected with miR-3148 mimics, inhibitors and NCs.
Approximately 100 µl serum-free medium containing cell suspension
was added to the upper chamber, and 600 µl medium containing 10%
FBS was added to the lower chamber. Following 36 h of transfection,
the cells were stained with 0.1% crystal violet staining solution
(Beyotime Institute of Biotechnology) at room temperature for 15
min and counted; 6 fields of view were selected in each well for
imaging under x40 magnification using a light microscope. Each
experiment was performed in triplicates.
Immunohistochemistry (IHC)
Tissues were sectioned at 5 µm and collected on
microscope slides (SuperFrost plus; Thermo Fisher Scientific,
Inc.). Sections were then re-hydrated and deparaffinized by
immersion in xylene (100% x2) followed by immersion in a graded
alcohol series (100% ethanol for 1 min twice; 95% ethanol for 1 min
and 70% ethanol for 1 min twice), ending with treatment distilled
water. Heat-induced antigen retrieval was performed in citrate
buffer, pH 6.0 (10 mM sodium citrate) containing 0.05% Tween-20
(Sigma-Aldrich; Merck KGaA) for 10 min at 90˚C, followed by
immersion in distilled water for 10 min and in PBS (thrice for 3
min). Sections were then incubated in PBS containing 0.3%
H2O2 at room temperature for 10 min, followed
by rinses in PBS (thrice for 3 min). Sections were incubated in PBS
containing 0.05% Triton X-100 (AppliChem GmbH) and 1% bovine serum
albumin (BSA; Sigma-Aldrich; Merck KGaA; PBS-TX-BSA) for 30 min at
room temperature. Incubation was performed in primary antibodies
made in rabbit against DCUN1D1 (PA5-83298; 1:50; Thermo
Fisher Scientific, Inc.) for 16 h at 4˚C. After rinsing in PBS
(thrice for 3 min), sections were incubated with horseradish
peroxidase (HRP)-conjugated secondary antibodies (goat anti-rabbit;
cat. no. A0208; 1:1,000; Beyotime Institute of Biotechnology) for
45 min at room temperature. Following rinses in PBS (thrice for 3
min), sections were incubated for 10 min in PBS containing
3,3'-diaminobenzidine (DAB, 25 mg/ml) and 0.05%
H2O2. Sections were then rinsed in PBS
(thrice for 3 min) and counterstained with hematoxylin (Histolab
Products AG).
In vivo xenograft tumor model
6 BALB/c athymic nude mice (female; 4-5 weeks old;
16-22 g) (National Laboratory Animal Center) were fed in sterile
housing cages at 25˚C, with a humidity of 45-55% and 12-h
light/dark cycle. The feed was sterilized by high temperature and
high pressure, and mice had free access to food and water. All the
nude mice were hypodermically injected with 1x106 U251
cells (miR-3148 mimics or miR-NC). Fifteen days later, mice were
euthanized via cervical dislocation, and the tumor tissues were
harvested and tumor weight was measured with an electronic scale.
Experiments in the present study were approved by the Ethics
Committee for Experimental Animals at First People's Hospital of
Jingmen and were conducted in the SPF Animal Laboratory at the
Medical College of Hubei University of Arts and Science.
H&E staining
Tumors were fixed in 4% paraformaldehyde overnight
at 4˚C. For staining, tissues were washed with water for several
hours, and then dehydrated in 70, 80 and 90% ethanol, xylene, and
other mixture for 15 min, and then in xylene I for 15 min, II for
15 min, until transparent. Samples were then placed in a mixture of
xylene and paraffin (1:1) for 15 min, and then paraffin was added
for 50-60 min. The tissues were then paraffin-embedded and sections
(4-µm thick) were mounted on slides, dewaxed and rehydrated. Then,
the sections were stained with hematoxylin for 4 min and eosin for
90 sec at room temperature and observed by an Olympus BX51 light
microscopy (Olympus Corporation).
Western blotting
Cells were lysed in RIPA buffer (Beyotime Institute
of Biotechnology) containing a protease inhibitor. Protein
concentration was measured using a Bicinchoninic Acid Protein Assay
kit (Thermo Fisher Scientific, Inc.). Next, the protein extracts
were separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (40 µg/lane) and transferred to polyvinylidene
fluoride) membranes (Sangon Biotech Co. Ltd.). The membranes were
blocked with 5% defatted milk, followed by overnight incubation at
4˚C with primary antibodies [anti-DCUN1D1 antibody (cat. no.
ab181233; 1:10,000; Abcam), anti-nuclear factor κ enhancer binding
protein (NF-κB) p50 antibody (cat. no. ab32360; 1:5,000; Abcam),
anti-NF-κB p65 antibody (cat. no. ab28856; 1:1,000; Abcam)] and
later with HRP-conjugated secondary antibodies (goat anti-rabbit,
A0208; 1:1,000; Beyotime Institute of Biotechnology) at room
temperature for 2 h. The protein levels were normalized to
glyceraldehyde 3-phosphate dehydrogenase (GAPDH; cat. no. ab8245;
1:1,000; Abcam).
Dual-luciferase reporter gene
assay
The binding sites between miR-3148 and DCUN1D1 were
predicted using miRDB (http://mirdb.org/custom.html). The 3'-UTR sequence of
DCUN1D1 interplayed with miR-3148 and pGL3 promoter vectors
(Promega Corporation) were injected with full-length DCUN1D1
or a mutant sequence with the predicted target sites. After seeding
on a 24-well plate, the cells were co-transfected with 5 ng
pRL-SV40 (Promega Corporation), a Renilla luciferase vector,
using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37˚C for 48 h. 48 h after transfection,
Dual-Luciferase® Reporter Assay System (Promega
Corporation) was used for activity measurement.
Statistical analysis
SPSS 20.0 (IBM Corp.) and GraphPad Prism 5 (GraphPad
Software, Inc.) statistical software were used for data assessment.
For normally distributed data with equal variance, the difference
was evaluated by two-tailed Student's t-test (two group
comparisons) or one-way ANOVA followed by the Bonferroni post hoc
test (multigroup comparisons) as appropriate. The paired t-test was
only performed to detect the differential expression of miR-3148
and DCUN1D1 in cancer tissues compared with adjacent non-malignant
tissues. The unpaired Student's t test were performed for assessing
the significance of other between-group differences. For
non-normally distributed data or data with unequal variances, the
difference was evaluated by a nonparametric Mann-Whitney U test
(two group comparisons) or the Kruskal-Wallis test followed by the
Bonferroni post hoc test (multigroup comparisons). The average
expression level was used as the cutoff and log-rank analysis was
used for survival analysis. Data are presented as the mean ± SD.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characteristics and expression of
miR-3148 in glioma
Liu et al (19) have reported that miR-3148 is
evidently decreased in glioma. However, no studies have reported
whether miR-3148 plays a role in glioma. Here, 48 clinical
specimens of human glioma were collected to examine miR-3148
expression levels in cancerous and non-cancerous tissues. miR-3148
was expressed in glioma tissues at a lower level compared with that
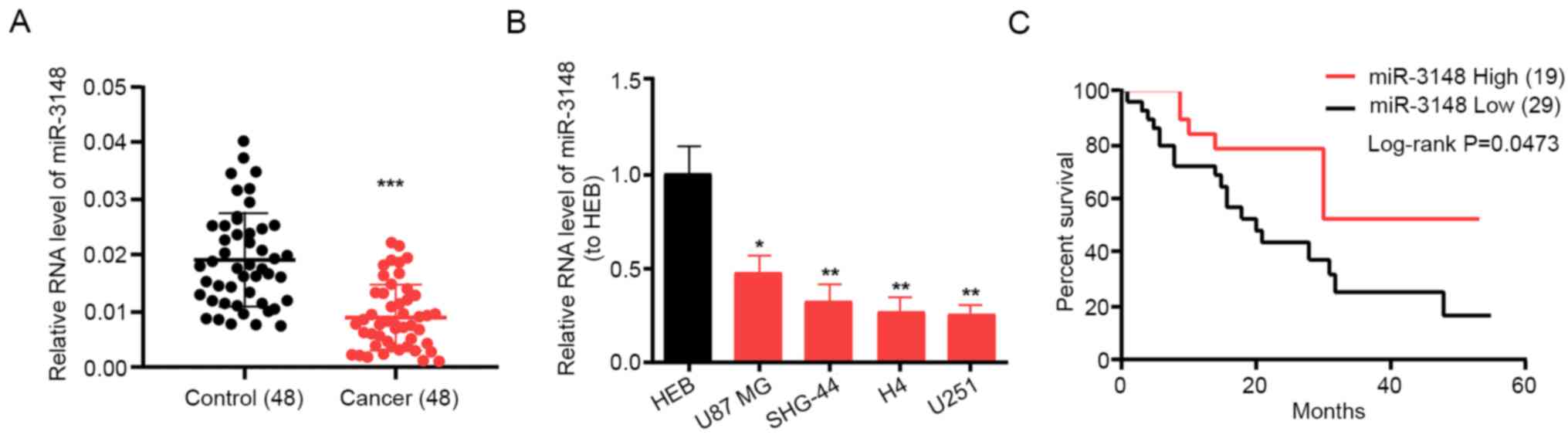
in non-tumor tissues (Fig. 1A).
Additionally, miR-3148 was expressed in glioma cells at a notably
lower level compared with that in the HEB cell line. miR-3148
expression reached the maximum in the U87 MG cell line and was the
minimum in the U251 cell line (Fig.
1B); therefore, these two cell lines were selected as research
models for the subsequent assays. Furthermore, patients with high
miR-3148 expression had a higher survival rate compared with those
with low miR-3148 expression (Fig.
1C). The average expression level was used as the cutoff. These
results suggest that miR-3148 is downregulated in gliomas and is
associated with survival and prognosis.
Biological changes post treatment with
miR-3148 mimics or inhibitors
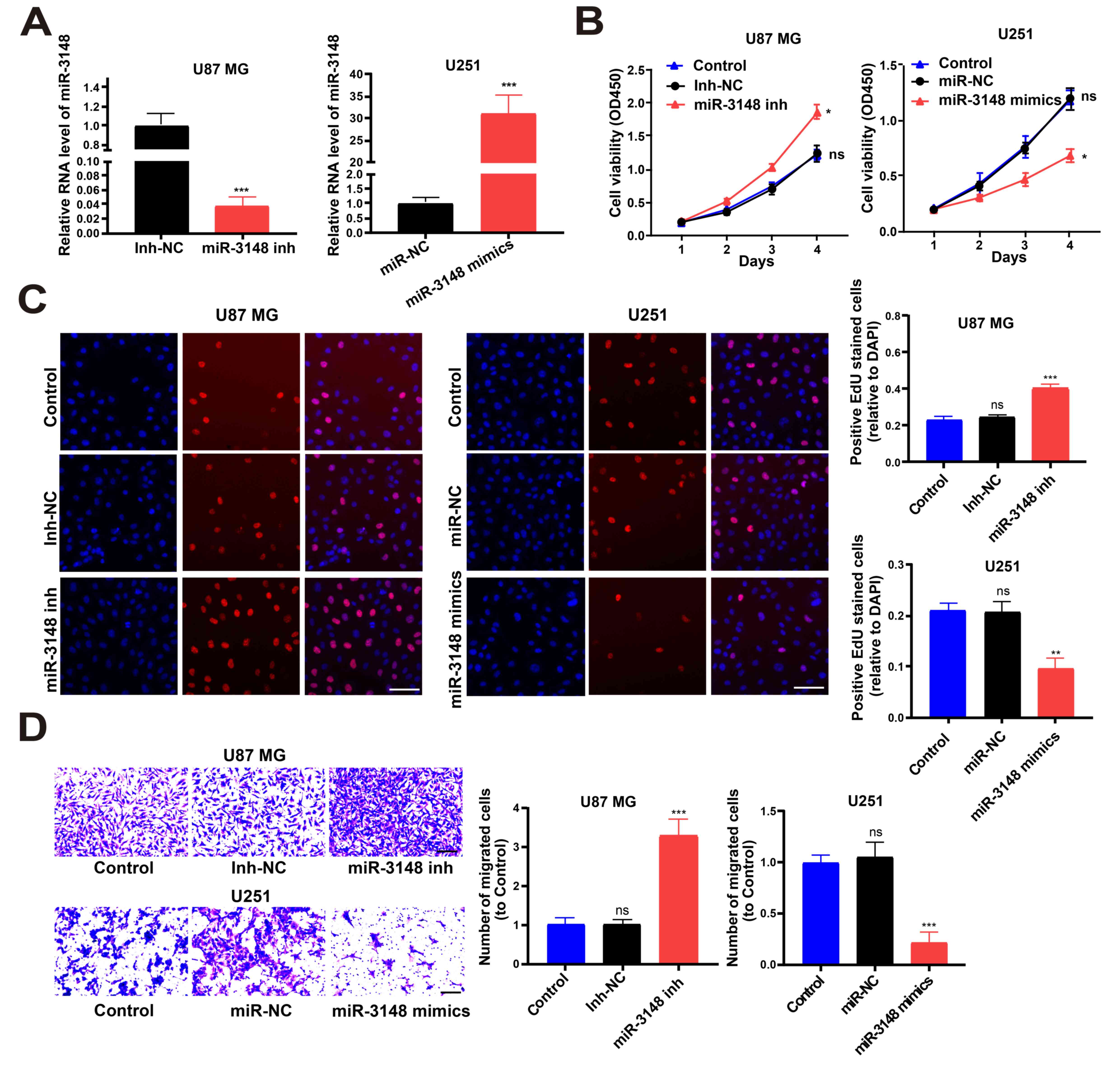
miRNA mimics were utilized to increase miR-3148
expression in U251 cell lines and miRNA inhibitors were utilized to
decrease miR-3148 expression in U87 MG cell lines (Fig. 2A). CCK-8 and EdU assays showed that
overexpression of miR-3148 inhibited glioma cell proliferation,
while downregulated miR-3148 expression level boosted it (Fig. 2B and C). In addition, the Transwell assay
revealed that cell migration was boosted by lowering miR-3148
expression level and was impeded by overexpression of miR-3148
(Fig. 2D). Hence, miR-3148
suppressed the proliferation and migration of glioma cells.
miR-3148 inhibits glioma growth in
vivo
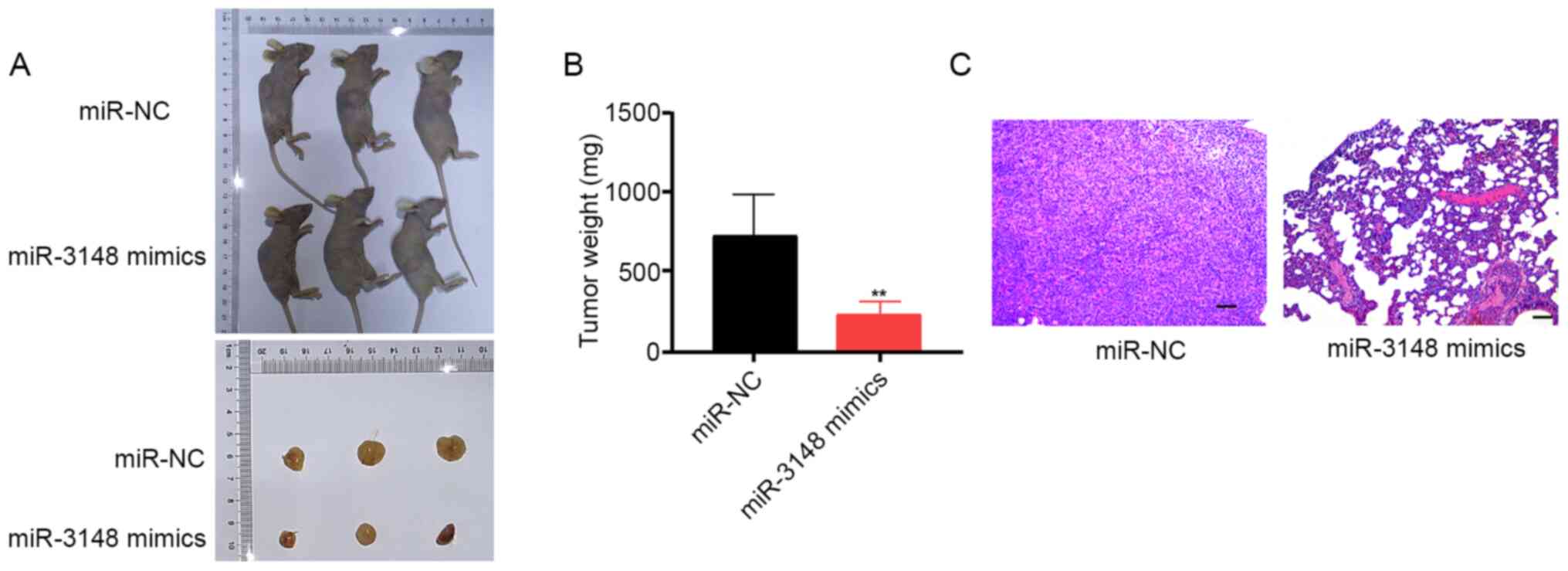
An in vivo xenograft tumor model was
constructed to verify the functions of miR-3148 in glioma
development. Post miR-3148 mimic transfection, U251 cells were
subcutaneously inoculated, and miR-3148 overexpression was observed
to impede tumor growth in vivo. The tumor volume and weight
in the miR-3148 mimic group were significantly lower compared with
those in the miR-NC group (Fig. 3A
and B). H&E staining of tumor
cells in the miR-NC and miR-3148 mimic groups verified that
miR-3148 lowered the rate of occurrence of lung metastasis in nude
mice (Fig. 3C). These data revealed
that miR-3148 inhibits tumor growth in vivo.
miR-3148 targets DCUN1D1 and regulates
its expression
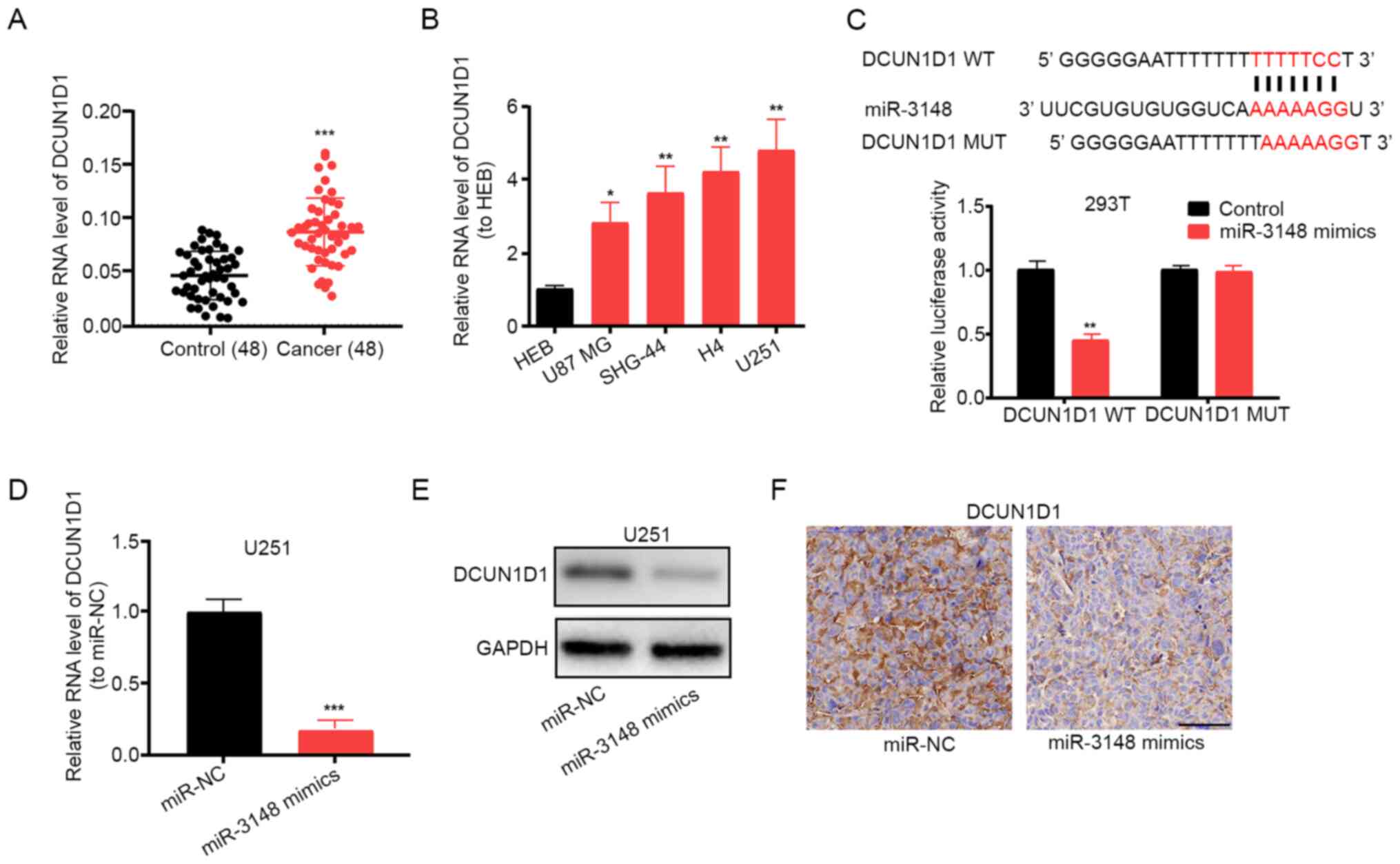
Using miRDB, DCUN1D1 was identified as the
target gene of miR-3148. Importantly, DCUN1D1 has been
reported to promote glioma formation and malignant progression in
mice (21). Using RT-qPCR assay,
increased expression levels of DCUN1D1 were detected in
glioma tissues and cell lines (Fig.
4A and B). The luciferase
reporter gene assay showed that miR-3148 did bind to DCUN1D1
(Fig. 4C). Additionally, the mRNA
and protein expression levels of DCUN1D1 decreased following
miR-3148 overexpression in U251 cells (Fig. 4D and E). DCUN1D1 expression was also
found to be inhibited by miR-3148 overexpression by
immunohistochemical staining of DCUN1D1 in xenograft tumor
tissues (Fig. 4F). The
aforementioned results indicate that miR-3148 targets and regulates
DCUN1D1 expression.
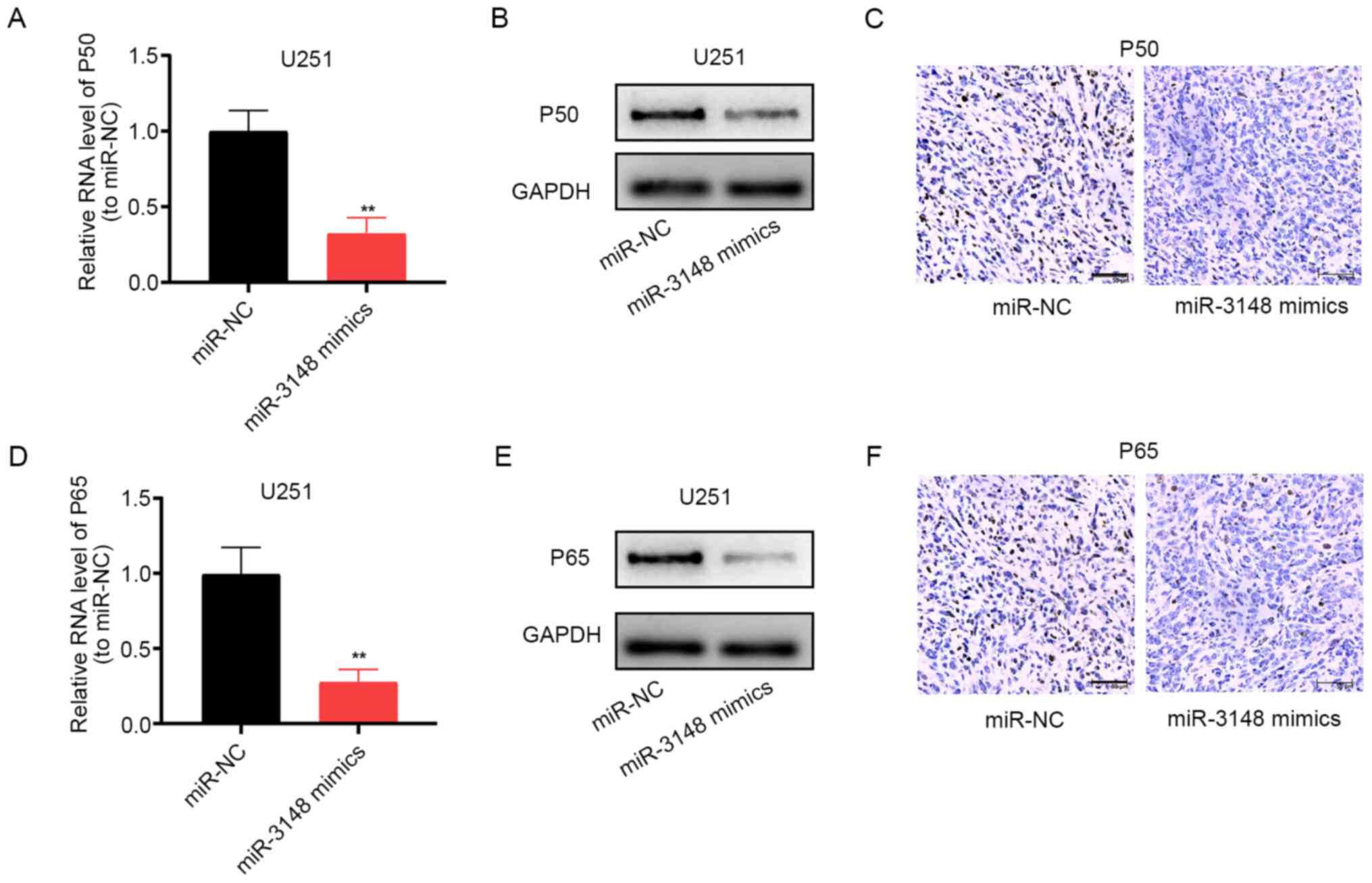
miR-3148 blocks the NF-κB pathway
The aberrantly activated NF-κB pathway has been
implicated in glioma development (22-24).
Following upregulation of miR-3148 expression, the mRNA and protein
expression levels of p50 was markedly decreased (Fig. 5A-C), and those of p65 showed the
same trend (Fig. 5D-F). Thus,
miR-3148 can block the NF-κB pathway, impeding the development of
glioma. Furthermore, the upregulation of the expression of
DCUN1D1 stimulates the NF-κB pathway (Fig. S1).
Discussion
In the process of cancer development, miRNAs are
dysregulated and function as oncogenes or tumor suppressors by
promoting or impeding tumor cell proliferation and invasion,
respectively (25-27).
miRNA deregulation is also known to be a key mechanism in glioma
pathogenesis (28-30).
The present study aimed to determine the function of
miR-3148 in the onset of glioma using tissue specimens. Decreased
miR-3148 expression was observed in glioma tissues compared with
non-tumor tissues. The lower the expression level of miR-3148, the
lower the overall survival rate of patients with glioma.
Thereafter, glioma cells were transfected with miR-3148 mimics and
inhibitors, and functional bioassays were used to examine the role
of miR-3148. The present study showed that miR-3148 inhibits the
proliferation and migration of tumor cells.
The mechanisms of action of miRNAs can be divided
into two categories. Firstly, miRNAs bind closely to the open
reading frame of mRNA to form a double-stranded structure, which
leads to the degradation of mRNA. Secondly, miRNAs bind to the
3'-UTR of mRNA, which decreases the stability of mature mRNA and
inhibits the post transcriptional translation of mRNA (31).
Bioinformatics analysis and the dual-luciferase
reporter assay were performed to confirm that DCUN1D1 was a
target gene of miR-3148, which was highly expressed in tumor cells
and tissues. DCUN1D1 expression was decreased when miR-3148
was upregulated. A previous study demonstrated that DCUN1D1
is an important component of the neddylation E3 complex (32) and can promote the nuclear
translocation and assembly of this complex (33). Several studies have also suggested
that DCUN1D1 has oncogenic activity in human cancer. For
instance, DCUN1D1 is highly conserved and is activated by
its amplification in squamous cell carcinomas (34). DCUN1D1 can also induce
extracellular matrix invasion by activating matrix
metalloproteinase 2, and thus, it is important for cancer
metastasis (35). Recently,
Broderick et al (21) showed
that DCUN1D1 promotes glioma formation and malignant
progression in mice. These findings indicate that DCUN1D1 is
targeted and downregulated by miR-3148, and this function of
miR-3148 could lead to the possible inhibition of glioma
progress.
The NF-κB/Rel class of proteins include NF-κB2
p52/p100, NF-κB1 p50/p105, c-rel, RelA/p65 and RelB proteins. The
NF-κB/Rel protein can act as polymerization transcription factor 2,
control gene expression and affect biological processes, including
B cell and lymphoid organ formation, inflammation, innate and
adaptive immunity, and stress reactions (36). The activation of the NF-κB pathway
has recently been determined to be closely associated with the
onset of glioma (37-39).
The present study proved that miR-3148 was involved in the
mechanism of glioma by modulating the NF-κB pathway.
The present study had several limitations, for which
the following measures are suggested for future work. Firstly, a
stereotactic method in situ is required to validate the
conclusion. Secondly, more target genes should be used that
interact with miR-3148. Thirdly, further studies are required to
determine whether other factors affect glioma cell proliferation
and migration.
In conclusion, miR-3148 is expressed in glioma
tissues at a low level and is associated with overall survival.
Furthermore, high miR-3148 expression potentially curbs the onset
of glioma by modulating DCUN1D1 and suppresses the NF-κB
pathway.
Supplementary Material
Overexpression of DCUN1D1
stimulates the NF-κB pathway. (A) Transfection efficiency. (B) P50
mRNA levels were assessed by RT-qPCR. (C) P50 protein levels after
transfection were examined via western blotting, with GAPDH as an
internal control. (D) P65 mRNA levels were assessed by RT-qPCR. (E)
P65 protein levels were analyzed following transfection via western
blotting, with GAPDH as an internal control. Data are presented as
the mean ± SD. **P<0.01. RT-qPCR, reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QX and XC performed the experiments and generated
data. BC and QX made substantial contributions to the conception
and design of the present study. QX and XC conducted data analysis
and interpretation of data. All authors contributed to the drafting
and revision of the manuscript. All authors read, revised and
approved the manuscript and agreed to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved. QX and BC confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First People's Hospital of Jingmen (Hubei, China;
approval no. 2012018). All population-based studies were carried
out in accordance with the World Medical Association's Declaration
of Helsinki, and all subjects provided written informed
consent.
Animal experiments in the present study were
approved by the Ethics Committee for Experimental Animals at First
People's Hospital of Jingmen and were conducted in the SPF Animal
Laboratory at the Medical College of Hubei University of Arts and
Science.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rynkeviciene R, Simiene J, Strainiene E,
Stankevicius V, Usinskiene J, Kaubriene EM, Meskinyte I, Cicenas J
and Suziedelis K: Non-coding RNAs in glioma. Cancers (Basel).
11(17)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen Y, Bao C, Zhang X, Lin X, Huang H and
Wang Z: Long non-coding RNA HCG11 modulates glioma progression
through cooperating with miR-496/CPEB3 axis. Cell Prolif.
52(e12615)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zheng Y, Lu S, Xu Y and Zheng J: Long
non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells
by sponging miR-15a/b-5p to upregulate the expression of HDGF and
activating wnt/beta-catenin signaling pathway. Int J Biol Macromol.
128:521–530. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhou Z, Huang R, Chai R, Zhou X, Hu Z,
Wang W, Chen B, Deng L, Liu Y and Wu F: Identification of an energy
metabolism-related signature associated with clinical prognosis in
diffuse glioma. Aging (Albany NY). 10:3185–3209. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Cheng M, Zhang ZW, Ji XH, Xu Y, Bian E and
Zhao B: Super-enhancers: A new frontier for glioma treatment.
Biochim Biophys Acta Rev Cancer. 1873(188353)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kundu M, Das S, Dhara D and Mandal M:
Prospect of natural products in glioma: A novel avenue in glioma
management. Phytother Res. 33:2571–2584. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Yan C, Wang J, Yang Y, Ma W and Chen X:
Molecular biomarker-guided anti-angiogenic targeted therapy for
malignant glioma. J Cell Mol Med. 23:4876–4882. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cao H, Li X, Wang F, Zhang Y, Xiong Y and
Yang Q: Phytochemical-mediated glioma targeted treatment: Drug
resistance and novel delivery systems. Curr Med Chem. 27:599–629.
2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Orellana EA, Li C, Lisevick A and Kasinski
AL: Identification and validation of microRNAs that synergize with
miR-34a-a basis for combinatorial microRNA therapeutics. Cell
Cycle. 18:1798–1811. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pereira TD, Brito JAR, Guimarães ALS,
Gomes CC, de Lacerda JC, de Castro WH, Coimbra RS, Diniz MG and
Gomez RS: MicroRNA profiling reveals dysregulated microRNAs and
their target gene regulatory networks in cemento-ossifying fibroma.
J Oral Pathol Med. 47:78–85. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Witwer KW and Halushka MK: Toward the
promise of microRNAs-enhancing reproducibility and rigor in
microRNA research. RNA Biol. 13:1103–1116. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Seo HA, Moeng S, Sim S, Kuh HJ, Choi SY
and Park JK: MicroRNA-based combinatorial cancer therapy: Effects
of microRNAs on the efficacy of anti-cancer therapies. Cells.
9(29)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Koufaris C: Human and primate-specific
microRNAs in cancer: Evolution, and significance in comparison with
more distantly-related research models: The great potential of
evolutionary young microRNA in cancer research. Bioessays.
38:286–294. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sakaue S, Hirata J, Maeda Y, Kawakami E,
Nii T, Kishikawa T, Ishigaki K, Terao C, Suzuki K, Akiyama M, et
al: Integration of genetics and miRNA-target gene network
identified disease biology implicated in tissue specificity.
Nucleic Acids Res. 46:11898–11909. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yan D, Hao C, Xiao-Feng L, Yu-Chen L,
Yu-Bin F and Lei Z: Molecular mechanism of notch signaling with
special emphasis on microRNAs: Implications for glioma. J Cell
Physiol. 234:158–170. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Peng Y, Wang X, Guo Y, Peng F, Zheng N, He
B, Ge H, Tao L and Wang Q: Pattern of cell-to-cell transfer of
microRNA by gap junction and its effect on the proliferation of
glioma cells. Cancer Sci. 110:1947–1958. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xu H, Zhao G, Zhang Y, Jiang H, Wang W,
Zhao D, Hong J, Yu H and Qi L: Mesenchymal stem cell-derived
exosomal microRNA-133b suppresses glioma progression via
wnt/β-catenin signaling pathway by targeting EZH2. Stem Cell Res
Ther. 10(381)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang H and Xie Y: BRD7-mediated miR-3148
inhibits progression of cervical cancer by targeting
wnt3a/β-catenin pathway. Reprod Sci. 27:877–887. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu S, Yin F, Zhang J, Wicha MS, Chang AE,
Fan W, Chen L, Fan M and Li Q: Regulatory roles of miRNA in the
human neural stem cell transformation to glioma stem cells. J Cell
Biochem. 115:1368–1380. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Broderick SR, Golas BJ, Pham D, Towe CW,
Talbot SG, Kaufman A, Bains S, Huryn LA, Yonekawa Y, Carlson D, et
al: SCCRO promotes glioma formation and malignant progression in
mice. Neoplasia. 12:476–484. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tan C, Liu L, Liu X, Qi L, Wang W, Zhao G,
Wang L and Dai Y: Activation of PTGS2/NF-κB signaling pathway
enhances radiation resistance of glioma. Cancer Med. 8:1175–1185.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Geeviman K, Babu D and Babu PP:
Pantoprazole induces mitochondrial apoptosis and attenuates NF-κB
signaling in glioma cells. Cell Mol Neurobiol. 38:1491–1504.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ius T, Ciani Y, Ruaro ME, Isola M,
Sorrentino M, Bulfoni M, Candotti V, Correcig C, Bourkoula E,
Manini I, et al: An NF-κB signature predicts low-grade glioma
prognosis: A precision medicine approach based on patient-derived
stem cells. Neuro Oncol. 20:776–787. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hou G, Xu W, Jin Y, Wu J, Pan Y and Zhou
F: MiRNA-217 accelerates the proliferation and migration of bladder
cancer via inhibiting KMT2D. Biochem Biophys Res Commun.
519:747–753. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang R, Sun Y, Yu W, Qiao M, Jiang R, Guan
W and Wang L: Downregulation of miRNA-214 in cancer-associated
fibroblasts contributes to migration and invasion of gastric cancer
cells through targeting FGF9 and inducing EMT. J Exp Clin Cancer
Res. 38(20)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hetta HF, Zahran AM, Shafik EA, El-Mahdy
RI, Mohamed NA, Nabil EE, Esmaeel HM, Alkady OA, Elkady A, Mohareb
DA, et al: Circulating miRNA-21 and miRNA-23a expression signature
as potential biomarkers for early detection of non-small-cell lung
cancer. Microrna. 8:206–215. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhou Q, Liu J, Quan J, Liu W, Tan H and Li
W: MicroRNAs as potential biomarkers for the diagnosis of glioma: A
systematic review and meta-analysis. Cancer Sci. 109:2651–2659.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cheng W, Ren X, Zhang C, Han S and Wu A:
Expression and prognostic value of microRNAs in lower-grade glioma
depends on IDH1/2 status. J Neurooncol. 132:207–218.
2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xiong W, Ran J, Jiang R, Guo P, Shi X, Li
H, Lv X, Li J and Chen D: miRNA-320a inhibits glioma cell invasion
and migration by directly targeting aquaporin 4. Oncol Rep.
39:1939–1947. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
Kim AY, Bommeljé CC, Lee BE, Yonekawa Y,
Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, et al: SCCRO
(DCUN1D1) is an essential component of the E3 complex for
neddylation. J Biol Chem. 283:33211–33220. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Huang G, Kaufman AJ, Ramanathan Y and
Singh B: SCCRO (DCUN1D1) promotes nuclear translocation and
assembly of the neddylation E3 complex. J Biol Chem.
286:10297–10304. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sarkaria I, O-charoenrat P, Talbot SG,
Reddy PG, Ngai I, Maghami E, Patel KN, Lee B, Yonekawa Y, Dudas M,
et al: Squamous cell carcinoma related oncogene/DCUN1D1 is highly
conserved and activated by amplification in squamous cell
carcinomas. Cancer Res. 66:9437–9444. 2006.PubMed/NCBI View Article : Google Scholar
|
|
35
|
O-charoenrat P, Sarkaria I, Talbot SG,
Reddy P, Dao S, Ngai I, Shaha A, Kraus D, Shah J, Rusch V, et al:
SCCRO (DCUN1D1) induces extracellular matrix invasion by activating
matrix metalloproteinase 2. Clin Cancer Res. 14:6780–6789.
2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Williams LM and Gilmore TD: Looking down
on NF-κB. Mol Cell Biol. 40:e00104–e00120. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhou L, Deng ZZ, Li HY, Jiang N, Wei ZS,
Hong MF, Chen XD, Wang JH, Zhang MX, Sh YH, et al: TRIM31 promotes
glioma proliferation and invasion through activating NF-κB pathway.
Onco Targets Ther. 12:2289–2297. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhou Y, Tan Z, Chen K, Wu W, Zhu J, Wu G,
Cao L, Zhang X, Zeng X, Li J and Zhang W: Overexpression of SHCBP1
promotes migration and invasion in gliomas by activating the NF-κB
signaling pathway. Mol Carcinog. 57:1181–1190. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hai L, Liu P, Yu S, Yi L, Tao Z, Zhang C,
Abeysekera IR, Li T, Tong L, Ma H, et al: Jagged1 is clinically
prognostic and promotes invasion of glioma-initiating cells by
activating NF-κB(p65) signaling. Cell Physiol Biochem.
51:2925–2937. 2018.PubMed/NCBI View Article : Google Scholar
|