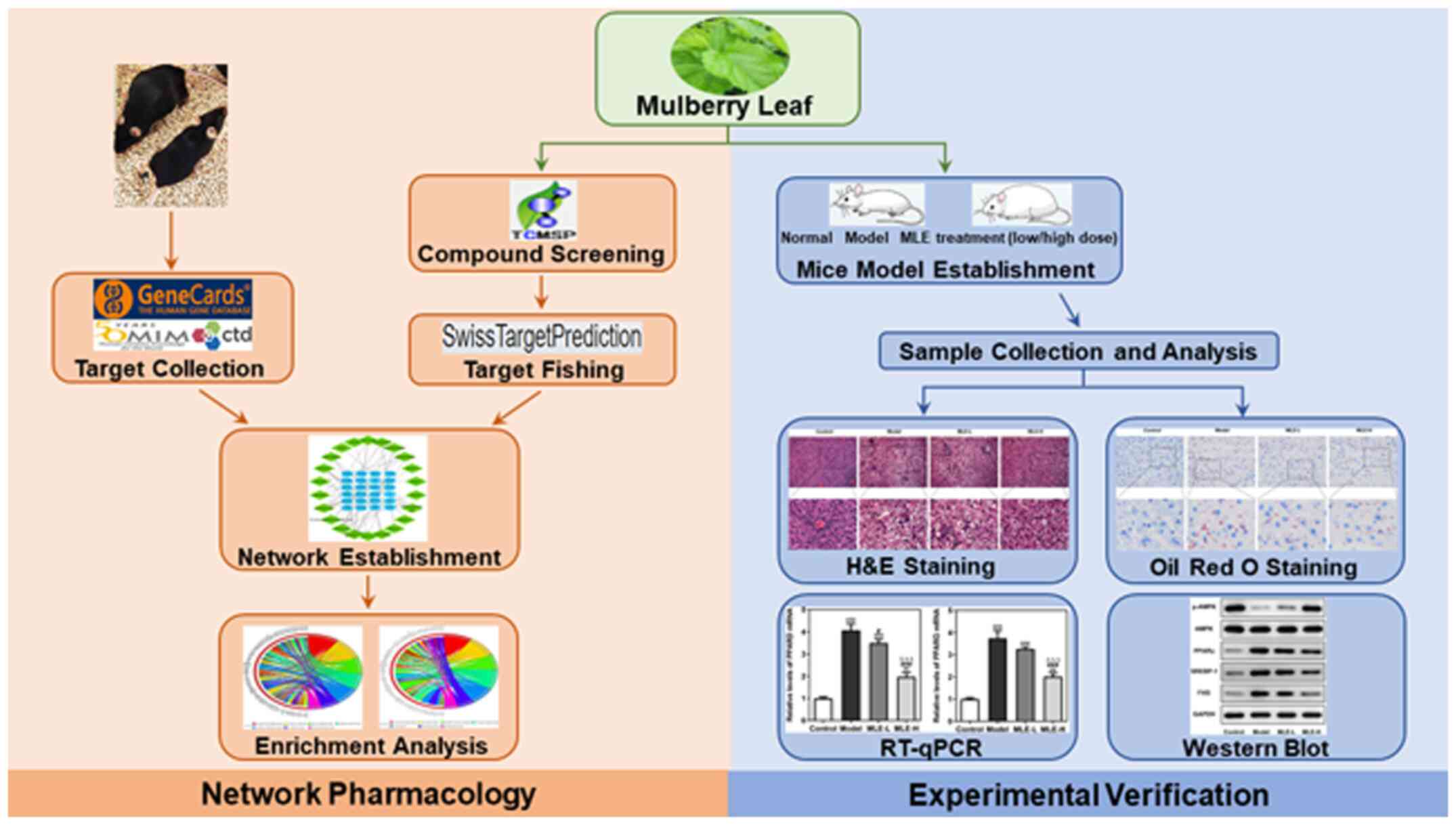
Introduction
Obesity is a chronic metabolic condition closely
associated with lifestyle (1).
According to previous epidemiological survey data, male and female
individuals with a BMI >28 kg/m2 in China account for
17.10 and 13.37% of the total population as of 2017, respectively,
where a clear upward trend can be observed (2). Obesity is a risk factor for type 2
diabetes (3), cardiovascular and
cerebrovascular diseases (4) and a
variety of cancers (5). To improve
human health and the quality of life, it is necessary to
fundamentally determine the pathogenesis of obesity to design
curative treatments.
Mulberry leaf extract (MLE) is a well-known herb
that is widely used as a medicine or for food consumption in China
(6). It has been previously
reported that MLE and its extracts can exert a variety of
therapeutic effects, including the pharmacological effects of
lowering blood glucose (7) and
blood lipid levels (8),
anti-diabetic (9), antioxidant
(10) and antitumor properties
(11). In a previous study, Zeni
and Dall'Molin (12) treated a
total of 33 hyperlipidemic Wistar rats with a water extract of MLE
following the oral administration of a cholesterol-rich (1 g/100 g
body weight) diet. After 14 days, the concentrations of
triglyceride (TG), total cholesterol (TC) and low-density
lipoprotein in the MLE treatment group were reduced compared with
those in the hyperlipidemic control group. In another study, Lee
et al (13) revealed that
the water extract of MLE could ameliorate atherosclerosis and
hypertension, hyperlipidemia and vascular dysfunction in rats with
atherosclerosis. In addition, MLE extracts have been reported to
alleviate non-alcoholic fatty liver disease in high-fat diet
(HFD)-fed mice by suppressing adipocytokines, inflammation and
oxidative stress (14). MLE also
markedly reduced obesity in mice induced by feeding on a high-fat
diet by increasing the activity of brown adipose tissues (15). Li et al (16) previously demonstrated that MLE
polyphenols and fibers confer synergistic effects on weight loss
and the regulation of the intestinal flora and metabolites.
However, to the best of our knowledge, the mechanism by which MLE
inhibits adipogenesis remains poorly understood.
In the present study, genetic or protein targets of
active components contained within MLE towards obesity were
identified using bioinformatics analysis. These target components
by MLE for the treatment of obesity were verified using network
pharmacology, including target prediction, protein-protein
interaction (PPI), ingredients-targets network, Gene Ontology (GO)
terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analyzes. Finally, in vivo experiments using a
mouse model of obesity induced by a high-fat diet were performed to
assess the effect of the key compounds derived from network
pharmacology on obesity.
Materials and methods
Experimental design
A detailed experimental design of the present study
is shown (Fig. 1).
Collection of chemical ingredients of
MLE
To determine the chemical ingredients in MLE, the
Traditional Chinese Medicine Systems Pharmacology (TCMSP; version
2.3; https://old.tcmsp-e.com/tcmsp.php) was performed by
searching with the key words ‘Sangye’ or ‘Mori Follum’. The
screening conditions were as follows: Oral bioavailability (OB)
≥30%; Drug-likeness (DL) ≥0.18 and Caco-2 permeability (Caco-2)
>0.
Targets of MLE and obesity
The herb targets were predicted by the
SwissTargetPrediction website (https://www.swisstargetprediction.ch/) through the
structures of active ingredients in MLE selected from TCMSP
(Fig. S1). The disease targets
were searched using ‘obesity’ as the key word through Online
Mendelian Inheritance in Man (OMIM; https://www.omim.org/), GeneCards (version 5.3;
https://www.genecards.org/) and
Comparative Toxicogenomics Database (CTD; https://ctdbase.org/).
Potential targets of MLE in the
treatment of obesity
The herb targets were intersected with the disease
targets to obtain the candidate targets of MLE in obesity by Venny
(version 2.1; https://bioinfogp.cnb.csic.es/tools/venny/index.html),
which were defined as MLE-obesity related targets. These targets
were uploaded into the Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database (version 11.0; https://string-db.org/) to establish a PPI network.
The ingredients-targets network was constructed by linking the
active compounds with their potential targets using Cytoscape
(version 3.7.2; https://cytoscape.org/).
GO gene enrichment analysis and KEGG
pathway annotation
To examine the biofunctions of the 234 genes, the
‘clusterProfiler’ package (17) in
R software (version 3.6.3) was used to perform functional
annotation, which included the three categories of GO (biological
processes, molecular functions and cellular components) and KEGG
enrichment analysis (https://www.r-project.org/). The parameters were set
at P=0.05 cutoff and q=0.05 cutoff. The results were visually
presented using the ‘GOplot’ package (18).
High-fat diet-induced obesity mouse
model
In total, 20 male C57BL/6 mice (4 weeks old;
20.01±0.24 g) were provided by Hunan SJA Laboratory Animal Co., Ltd
(Hunan, China). Mice were housed under a constant temperature
(22±2˚C), 50-70% relative humidity and 12-h light/dark cycle with
ad libitum access to food and clean water for 1 week. After
1 week of adaptation, 20 mice were randomly divided into the
following four groups (n=5): i) Normal control diet group
(Control); ii) obese high-fat diet group (Model); iii) obese mice
receiving a low dose (133 mg/kg) of MLE group; and iv) obese mice
receiving a high dose (666 mg/kg/day) of MLE group. The doses for
low-dose and high-dose of MLE was determined according to a
previous study (19).
MLE was freshly suspended in distilled water and
mice were administered via oral gavage five times a week not during
the weekend for 8 weeks. The Control and Model groups were given
the same volumes of distilled water. At the end of experiment at
week 8, mice were weighed and sacrificed using an intraperitoneal
injection of 150 mg/kg pentobarbital sodium, before their blood,
liver tissues and visceral white adipose tissues were collected
immediately and stored at -80˚C. The experimental protocols were
approved by the Institutional Animal Care and Use Committee of the
Hunan Future Health Technology Group Co., Ltd. (Changsha,
China).
Serum analysis
Blood was centrifuged at 500 x g for 15 min at 4˚C
to collect the serum. Commercial analysis kits (Abcam) were used to
detect the levels of serum TG (cat. no. ab65336) and TC (cat. no.
ab282928).
H&E staining
The liver tissues frozen in liquid nitrogen were
taken out, fixed in 10% neutral buffered formalin at room
temperature for 24 h before being embedded in paraffin wax. The
liver tissue sections (4 µm) were then cut and hydrated in a
decreasing ethanol gradient. All sections were deparaffinized with
xylene and stained with hematoxylin for 5 min at room temperature
and eosin for 3 min at room temperature. The sections were finally
observed under a light microscope (magnification, x200; OCT-HS100;
Canon, Inc.).
Oil red O staining
The liver tissue sections (4 µm) were stained with
Oil red O according to the manufacturer's protocol of the Oil Red O
Staining Kit (cat. no. C0157S; Beyotime Institute of
Biotechnology). Oil red O-stained sections were imaged using an
inverted light microscope (magnification, x400; Nikon
Corporation).
Western blot analysis
Total proteins were extracted from the visceral
white adipose tissues using RIPA reagent (Beyotime institute of
Biotechnology) on ice. The protein concentration was determined
using a BCA kit. Equal amounts (20 µg) of protein were loaded and
separated by 10% SDS-PAGE. After electrophoresis, proteins were
transferred onto PVDF membranes and the membranes were blocked with
5% non-fat milk for 2 h at 37˚C. Subsequently, the membranes were
immunoblotted with the following primary antibodies overnight at
4˚C: TNF-α (dilution, 1:1,000; cat. no. ab183218; Abcam), IL-1β
(dilution, 1:1,000; cat. no. ab234437; Abcam), NF-κB inhibitor α
(NFKBIA; dilution, 1:1,000; cat. no. ab32518; Abcam), inducible
nitric oxide synthase (iNOS; dilution, 1:1,000; cat. no. ab178945;
Abcam), AMP-activated protein kinase (AMPK; dilution, 1:2,000; cat.
no. 5832; Cell Signaling Technology, Inc.), phosphorylated (p-)
AMPK (dilution, 1:2,000; cat. no. 5759; Cell Signaling Technology,
Inc.), peroxisome proliferator activated receptor (PPAR)-γ (PPARG;
dilution, 1:500; cat. no. ab45036; Abcam), sterol regulatory
element-binding proteins (SREBP-1; dilution, 1:1,000; cat. no.
ab28481; Abcam), fatty acid synthase (FAS; dilution, 1:1,000; cat.
no. ab82419; Abcam) and GAPDH (dilution, 1:2,500; cat. no. ab9485;
Abcam). This was followed by incubation with the HRP-conjugated
secondary antibody (cat. no. 7074; dilution, 1:1,000; Cell
Signaling Technology, Inc.) at room temperature for 1 h. The
protein bands were visualized using enhanced chemiluminescence
reagent (Research-bio). Image Lab 3.0 software (Bio-rad
laboratories, inc.) was used to quantify the WB densitometry.
Reverse transcription-quantitative PCR
analysis
Total RNA from the visceral white adipose tissues
was isolated using the TRIzol® reagent (Thermo Fisher
Scientific, Inc.). Subsequently, total RNA was converted into cDNA
using PrimeScript 1st strand cDNA Synthesis Kit (cat. no. 6110A;
Takara Bio, Inc.). The reverse transcription condition was as
follows: 10 min at 30˚C and 40 min at 42˚C. Quantitative PCR was
performed using SYBR® Premix Ex Taq™ (Takara Bio, Inc.)
with an ABI Prism 7900 machine (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The internal control was GAPDH. Amplification
was performed as follows: 5 sec at 95˚C, followed by 40 cycles of
15 sec at 95˚C and 40 sec at 56˚C. The relative gene expression
levels of TNFα, PPARD, PPARG, fatty acid amide hydrolase (FAAH) and
hydroxysteroid 11-β dehydrogenase 1 (HSD11B1) were calculated using
the 2-ΔΔCq method (20).
Primers sequences were as follows: TNFα forward,
5'-AGCCCATGTTGTAGCAAACC-3' and reverse, 5'-GGAAGACCCCTCCCAGATAG-3';
PPARD forward, 5'-ACGCACCCTTTGTCATCC-3' and reverse,
5'-GAAGAGGCTGCTGAAGTTGG-3'; PPARG forward,
5'-GAGAAGGAGAAGCTGTTGGC-3' and reverse, 5'-ATGGCCACCTCTTTGCTCT-3';
FAAH forward, 5'-GCCTCAAGGAATGCTTCAGC-3' and reverse, 5'-
TGCCCTCATTCAGGCTCAAG-3'; HSD11B1 forward,
5'-AAGCAGACCAACGGGAGCATT-3' and reverse, 5'-
GGAGAAGAACCCATCCAGAGCA-3' and GAPDH forward,
5'-TCTTGCTCAGTGTCCTTGC-3' and reverse,
5'-CTTTGTCAAGCTCATTTCCTGG-3'.
Statistical analysis
Data are presented as the mean ± SD with five mice
per group. The significance of differences was determined by
one-way ANOVA followed by Tukey's post hoc test for multiple
comparisons using SPSS v22.0 (IBM Corp.). P<0.05 was considered
to indicate a statistically significant difference.
Results
Active components and potential target
proteins of MLE
TCMSP was used to retrieve the composition of MLE
and there were 26 active components when the filter conditions were
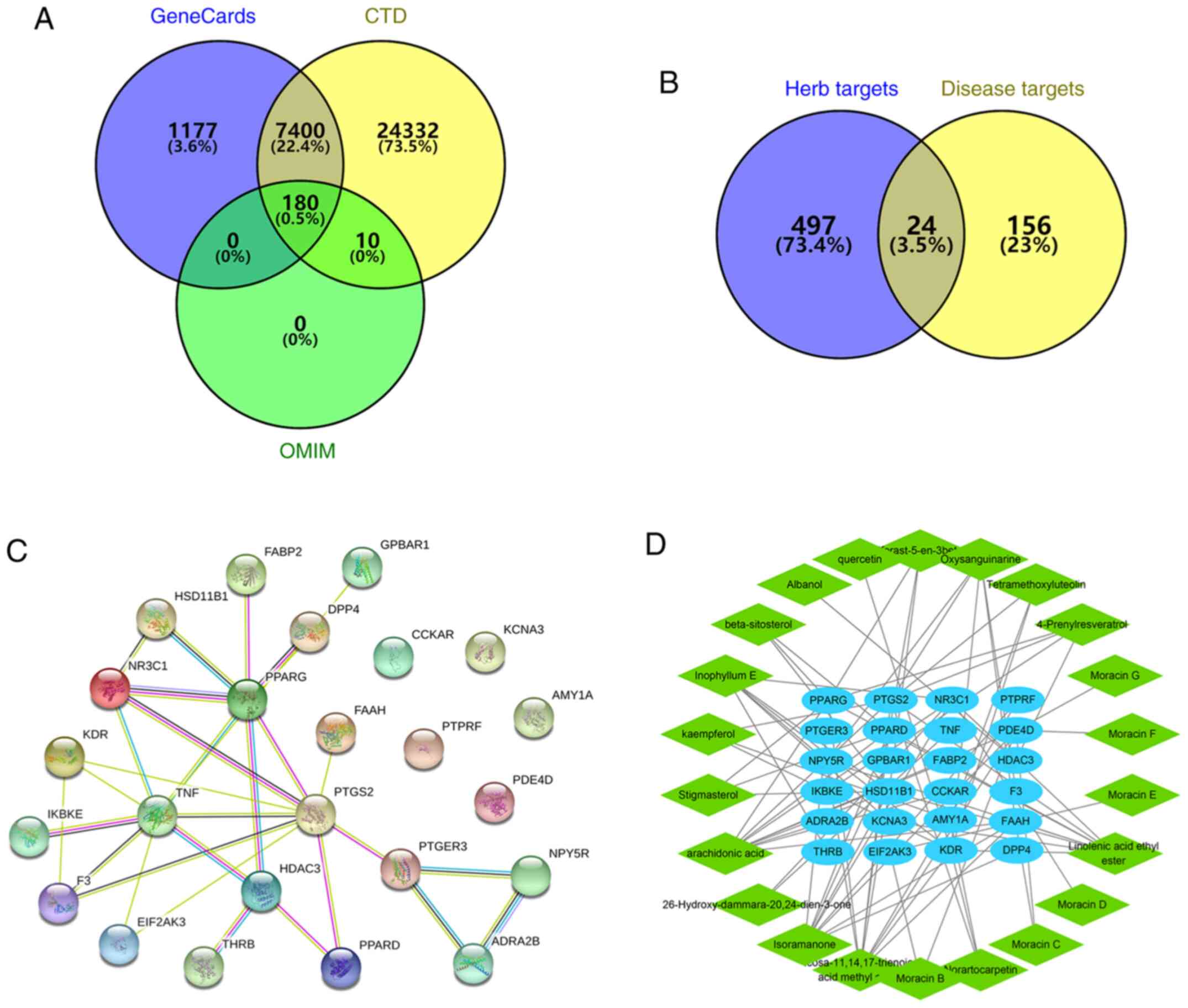
as follows: OB ≥30%, DL ≥0.18 and Caco2 >0 (Table I and Fig. S1). Using SwissTargetPrediction, 521
herb targets were predicted. From the analysis using OMIM,
GeneCards and CTD, 190 obesity-related genes were retrieved from
OMIM, 8,757 obesity-related genes were retrieved from GeneCards and
31,922 obesity-related genes were retrieved from CTD. The disease
targets were identified by the intersection of genes retrieved from
the three databases, where 180 disease targets were obtained for
subsequent analysis (Fig. 2A). The
herb targets were intersected with the disease targets to obtain 24
MLE-obesity-related targets by Venny (version 2.1; https://bioinfogp.cnb.csic.es/tools/venny/index.html;
Fig. 2B). As shown in Fig. 2C, a network of 24 interacting
proteins consisted of 34 edges. The average node degree was 2.83
and the PPI enrichment P-value was 5.49x10-12. TNF,
PPARG, prostaglandin-endoperoxide synthase 2 (PTGS2), nuclear
receptor subfamily 3 group C member 1 and histone deacetylase 3
(HDAC3) had high degree of the nodes. In addition, an
MLE-compounds-targets-obesity interaction network was constructed
to facilitate the understanding of the potential regulatory effects
of MLE holistically (Fig. 2D). In
total, four compounds had no corresponding targets found in the
database, therefore only the interaction network of 22 compounds
and their relevant targets was shown. In this network, arachidonic
acid had 10 targets, whereas isoramanone, icosa-11,14,17-trienoic
acid methyl ester and linolenic acid ethyl ester all had nine
targets each. These multi-target compounds may serve to be the key
active components of MLE. Fatty acid oxidation associated proteins
(HSD11B1), kinase insert domain receptor (KDR), phosphodiesterase
4D (PDE4D), PPARD, PPARG, PTGS2 and FAAH had high degree of the
nodes in the network, which may be the core candidate targets of
MLE.
 | Table IMol ID of the 26 active components
found in mulberry leaves. |
Table I
Mol ID of the 26 active components
found in mulberry leaves.
| Mol ID | Molecule name | Oral
bioavailability (%) | Drug likeness | Caco-2
permeability |
|---|
| MOL001771 |
Poriferast-5-en-3β-ol | 36.91 | 0.75 | 1.45 |
| MOL002773 | β-carotene | 37.18 | 0.58 | 2.25 |
| MOL003842 | Albanol | 83.16 | 0.24 | 0.41 |
| MOL003847 | Inophyllum E | 38.81 | 0.85 | 0.68 |
| MOL003850 |
26-Hydroxy-dammara-20,24-dien-3-one | 44.41 | 0.79 | 0.82 |
| MOL003851 | Isoramanone | 39.97 | 0.51 | 0.05 |
| MOL003856 | Moracin B | 55.85 | 0.23 | 0.83 |
| MOL003857 | Moracin C | 82.13 | 0.29 | 0.87 |
| MOL003858 | Moracin D | 60.93 | 0.38 | 1.03 |
| MOL003859 | Moracin E | 56.08 | 0.38 | 0.96 |
| MOL003860 | Moracin F | 53.81 | 0.23 | 0.81 |
| MOL003861 | Moracin G | 75.78 | 0.42 | 0.98 |
| MOL003862 | Moracin H | 74.35 | 0.51 | 0.87 |
| MOL003879 |
4-Prenylresveratrol | 40.54 | 0.21 | 0.9 |
| MOL000729 |
Oxysanguinarine | 46.97 | 0.87 | 1.08 |
| MOL000098 | Quercetin | 46.43 | 0.28 | 0.05 |
| MOL000358 | β-sitosterol | 36.91 | 0.75 | 1.32 |
| MOL000422 | Kaempferol | 41.88 | 0.24 | 0.26 |
| MOL000449 | Stigmasterol | 43.83 | 0.76 | 1.44 |
| MOL001439 | Arachidonic
acid | 45.57 | 0.2 | 1.2 |
| MOL001506 | Supraene | 33.55 | 0.42 | 2.08 |
| MOL003759 | Iristectorigenin
A | 63.36 | 0.34 | 0.54 |
| MOL003975 |
Icosa-11,14,17-trienoic acid methyl
ester | 44.81 | 0.23 | 1.52 |
| MOL006630 |
Norartocarpetin | 54.93 | 0.24 | 0.14 |
| MOL007179 | Linolenic acid
ethyl ester | 46.1 | 0.2 | 1.48 |
| MOL007879 |
Tetramethoxyluteolin | 43.68 | 0.37 | 0.96 |
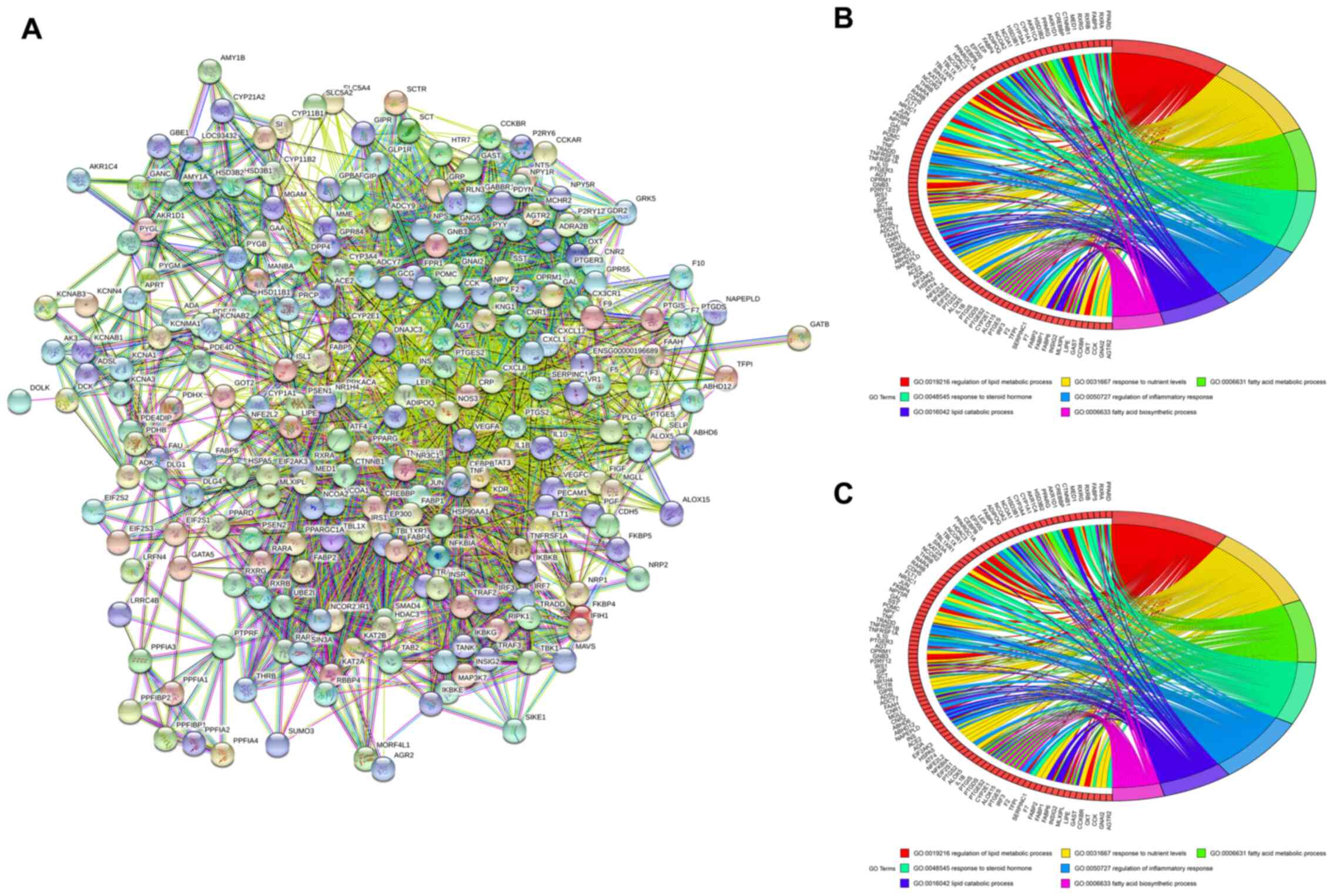
PPI, GO and KEGG analysis
There were 234 adjacent genes of 24 MLE-obesity
related targets identified by STRING. A PPI network analysis of the
234 adjacent genes of the 24 targets was conducted using STRING to
explore the potential interactions among them (Fig. 3A). GO functional enrichment analysis
was subsequently performed and the target enrichment was relatively
concentrated in the biological processes of ‘regulation of lipid
metabolic process’, ‘response to nutrient levels’, ‘fatty acid
metabolic process’, ‘response to steroid hormone’, ‘regulation of
inflammatory response’, ‘lipid catabolic process’ and ‘fatty acid
biosynthetic process’ (Fig. 3B).
Additionally, KEGG pathways of the 234 adjacent genes were analyzed
and shown in Fig. 3C. The pathways
included ‘adipocytokine signaling pathway’, ‘TNF signaling
pathway’, ‘Regulation of lipolysis in adipocytes’, ‘NF-κB signaling
pathway’, ‘insulin resistance’, ‘PPAR signaling pathway’ and ‘AMPK
signaling pathway’. In general, these biological processes and
signaling pathways were likely to be associated with the beneficial
effects of MLE against obesity.
MLE treatment of high-fat diet-induced
obese mice
The weight of mice was found to be significantly
increased in the Model group compared with that in the control
group but significantly reversed by both low- and high-dose MLE
treatment at 8 weeks (Fig. 4A). The
serum levels of TC and TG of the Model group were also
significantly upregulated, which were also significantly reversed
by both doses MLE treatment, although the effect of high-dose MLE
on the TC levels was more prominent (Fig. 4B). Hepatic histological examination
demonstrated that fat deposition (the white area represents fat)
was increased in the Model group compared with that in the Control
group. MLE treatment, especially at higher doses, was associated
with markedly reduced fat accumulation compared with that in the
Model group (Fig. 4C). According to
Oil red O staining, higher numbers of lipid droplets could be
observed in the liver of high-fat diet-induced obese mice compared
with those in the Control group, whilst addition of MLE markedly
decreased the hepatic lipid droplets at both doses (Fig. 4D).
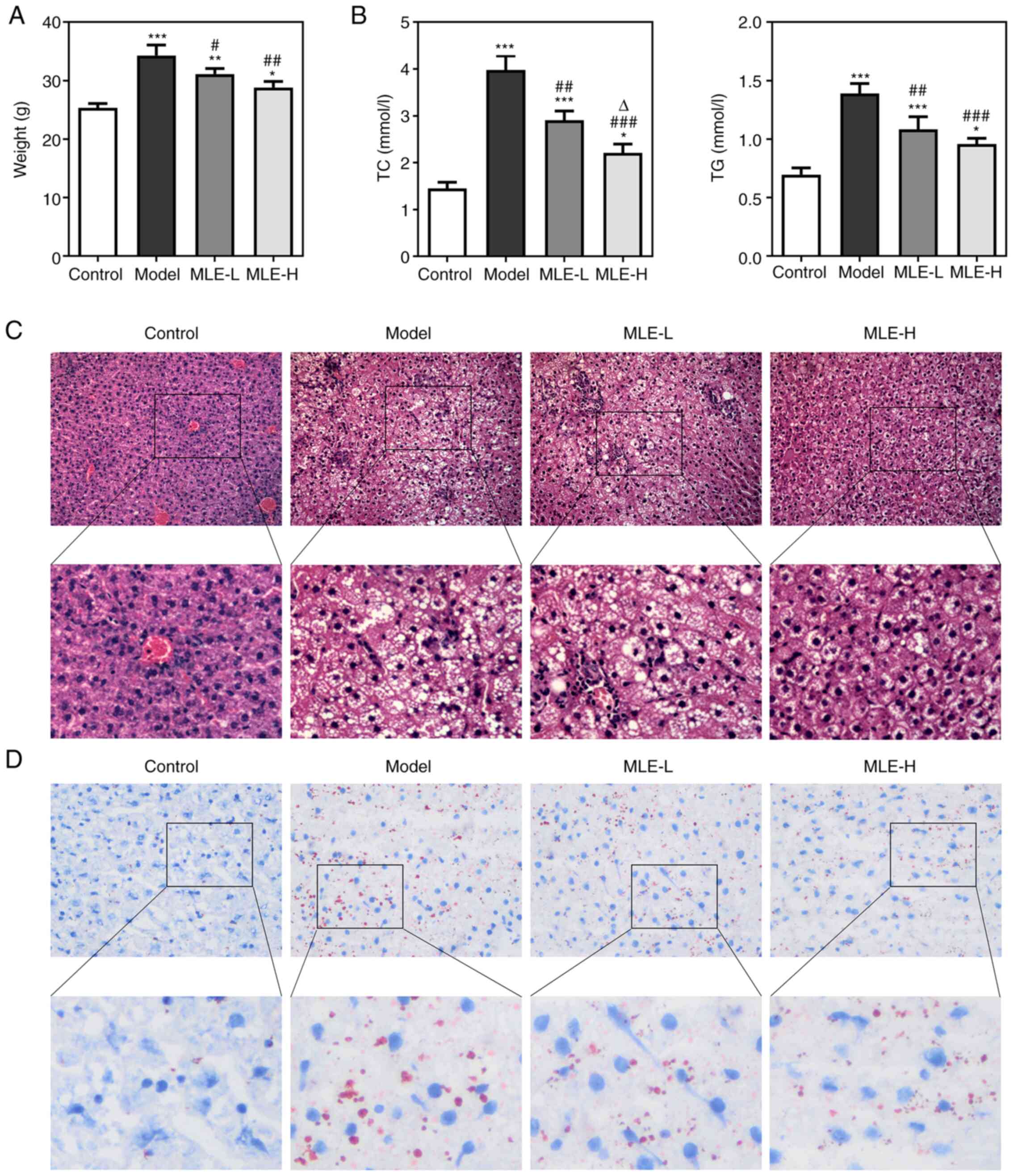 | Figure 4Effects of MLE on high-fat
diet-induced obese mice. (A) Mice body weight at 8 weeks. (B) The
levels of serum TC and TG were measured using TC and TG assay kits.
(C) Hepatic fat accumulation was detected by H&E staining.
Original (magnification, x200) and blow-up (magnification, x400).
(D) Liver lipid droplets were detected by Oil red O staining.
Original (magnification, x400) and blow-up (magnification, x800).
*P<0.05, **P<0.01 and
***P<0.001 vs. Control; #P<0.05,
##P<0.01 and ###P<0.001 vs. Model;
∆P<0.05 vs. MLE-L. TC, total cholesterol; TG,
triglyceride; MLE, Mulberry leaf extract; MLE-L, low dose MLE;
MLE-H, high dose MLE. |
Effects of MLE on inflammation,
lipogenesis, lipid catabolism, fatty acid oxidation and the
AMP-activated protein kinase (AMPK) signaling pathway
The inflammatory factors (TNF-α, IL-1β, NFKBIA and
iNOS), fat synthesis related proteins (PPARD and PPARG), lipid
catabolism related protein (FAAH) and fatty acid oxidation
associated proteins (HSD11B1) were chosen from KEGG analysis. The
protein expression levels of TNF-α, IL-1β, NFKBIA and iNOS in
visceral white adipose tissues were all significantly increased in
the Model group compared with those in the Control group, which
were all significantly reversed by treatment with of MLE in a
dose-dependent manner (Fig. 5A).
The mRNA expression levels of TNF-α, PPARD and PPARG were all
significantly increased, whereas those of FAAH and HSD11B1 were
significantly decreased in the visceral white adipose tissues of
mice in the Model group compared with those in the Control group
(Fig. 5B). However, these effects
were also markedly reversed by MLE, with significant reversals
observed on all mRNAs analyzed following high-dose MLE treatment
(Fig. 5B). The levels of
phosphorylated (p-)AMPK/AMPK were significantly reduced, whilst the
expression levels of PPARG, SREBP-1 and FAS were significantly
increased in the model group compared with those in the Control
group. Treatment with the high-dose MLE significantly reversed the
aforementioned changes in AMPK phosphorylation and PPARG, SREBP-1
and FAS expression (Fig. 5C).
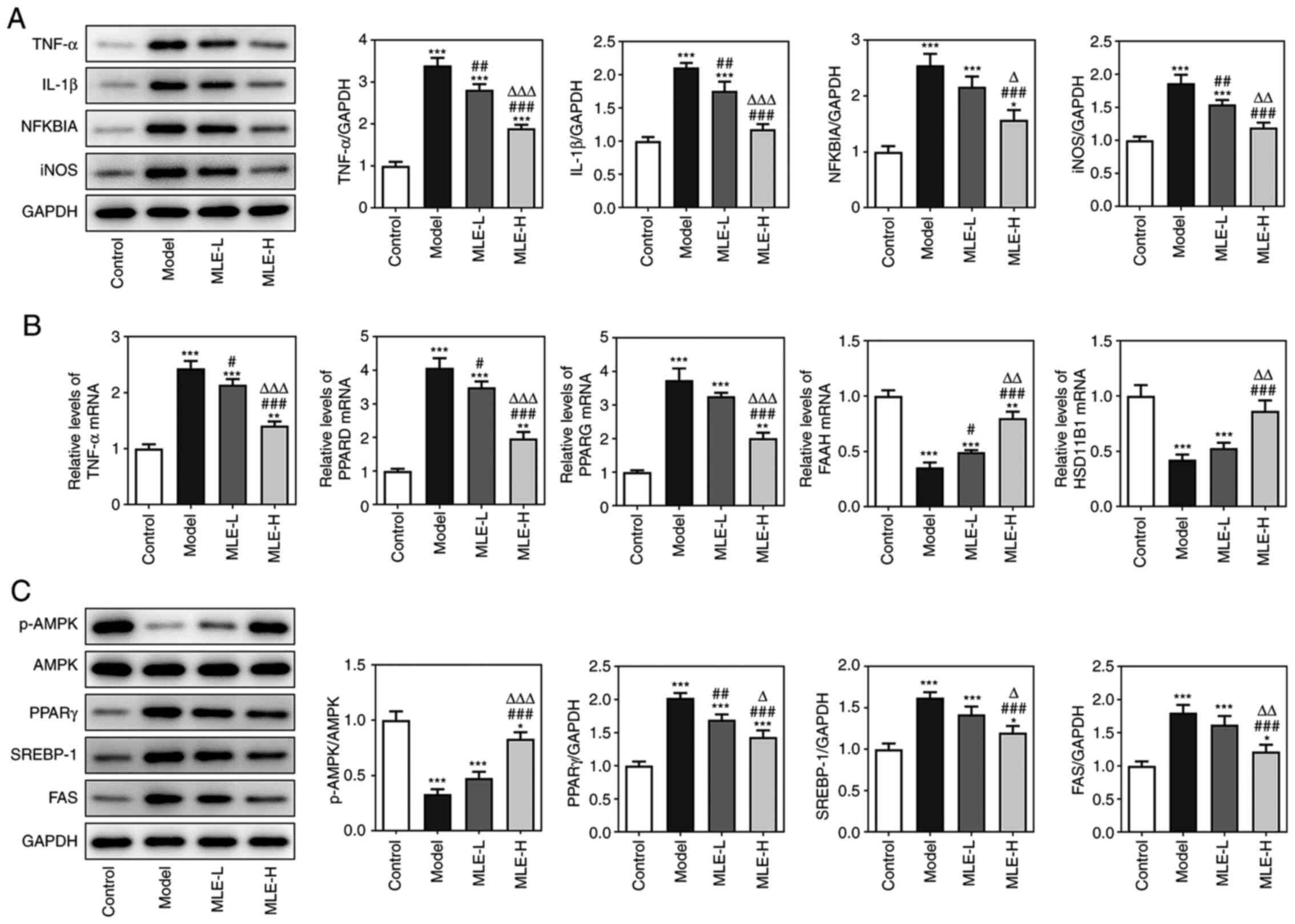 | Figure 5Effects of MLE on adipocyte
inflammation, lipogenesis, lipid catabolism, fatty acid oxidation
and AMPK signaling. (A) The protein expressions of inflammatory
factors TNF-α, IL-1β, NFKBIA and iNOS in adipose tissue were
detected by western blot analysis. (B) The mRNA expression of key
targets of MLE in the treatment of obesity was verified by reverse
transcription-quantitative PCR. (C) The activity of the AMPK
pathway in adipose tissue was detected by western blot analysis.
*P<0.05, **P<0.01 and
***P<0.001 vs. Control; #P<0.05,
##P<0.01 and ###P<0.001 vs. Model;
∆P<0.05, ∆∆P<0.01 and
∆∆∆P<0.001 vs. MLE-L. MLE, Mulberry leaf extract;
MLE-L, low dose MLE; MLE-H, high dose MLE; NFKBIA, NF-κB inhibitor
α; iNOS, inducible nitric oxide synthase; AMPK, 5'AMP-activated
protein kinase; PPAR, peroxisome proliferator activated receptor;
SREBP-1, Sterol regulatory element binding protein-1; FAS, fatty
acid synthase. |
Discussion
The effects of specific active components contained
within certain traditional Chinese medicines on mammalian cells and
gene expression remain unclear. Therefore, it is necessary to form
a knowledge base associated with these compounds (21). The emergence of big data in the
biomedical research field has promoted the establishment of network
pharmacology for the systematic analysis of drug targets.
Additionally, it changed the concept of drug discovery from ‘a
single target, a single drug’ to ‘multi-targets, multi-components’
(22). Network pharmacology is an
emerging discipline that is based on the statistical analysis of
data, virtual computing and the observation of
component-target-disease-pathway interactions retrieved from
network databases (23).
MLE have been previously demonstrated to alleviate
hyperglycemia, hyperlipidemia, obesity, oxidation and inflammation
(24-28).
Pharmacological analysis has indicated that the active components
in MLE include β-carotene, quercetin, β-sitosterol, kaempferol and
arachidonic acid, all of which can exert beneficial effects against
obesity (29-33).
Using network pharmacological analysis, we speculated that these
components can alleviate high-fat diet-induced obesity by targeting
proteins in various metabolic pathways. Furthermore, the majority
of the identified pathways are predominantly involved in glucolipid
metabolism, inflammation, lipogenesis and fatty acid oxidation,
which should be explored in future studies.
A previous study has revealed that obesity is
associated with chronic low-grade inflammation, such that insulin
signaling may be interfered by inflammatory factors, leading to
insulin resistance and metabolic disorders, including type 2
diabetes and metabolic syndrome, where impairments in insulin
signaling disrupt the entry of glucose into the adipocytes and
skeletal muscle cells (34).
Adipocytes in the adipose tissues of obese individuals are
frequently found to be enlarged, which is accompanied with
increased tissue vascularization, which affects the utilization of
oxygen and leads to oxygen deficiency in the adipose tissue
(35). In particular,
hypoxia-inducible factor-1α is associated with chronic inflammation
of the adipose tissue by promoting the recruitment of macrophages
and their transformation to the proinflammatory M1 type, which
releases a variety of proinflammatory cytokines, including TNF-α
and IL-6 in obesity (36). During
obesity, inflammation occurs not only in the adipose tissue, but
also in other tissues (37). It has
been previously reported that obese mice fed with a high-fat diet
exhibit intestinal inflammation, elevated levels of TNF-α and
activation of the NF-κB signaling pathway (38). In the present study, it was observed
that the protein expression levels of inflammatory factors TNF-α,
IL-1β, NFKBIA and iNOS were also increased in the adipose tissues
of obese mice. Additionally, ‘TNF signaling pathway’, ‘NF-κB
signaling pathway’ and ‘insulin resistance’ were identified by KEGG
pathway enrichment analysis, suggesting that MLE may exert
beneficial effects in the treatment of obesity through the
signaling pathways aforementioned.
The first indication for increased cytokine release
in obesity is increased TNF-α expression, the levels of which are
associated with the degree of adiposity and insulin resistance
(39). The rs2016520 polymorphism
of PPARD has been revealed to serve a leading role in the
development of abdominal obesity (40). In addition, obesity-susceptibility
gene transmembrane protein 18 can upregulate PPARG expression to
promote adipogenesis during obesity (41). FAAH is a primary catabolic regulator
of N-acylethanolamines, which activates G-protein-coupled receptors
within the endocannabinoid system (42). FAAH is associated with increased
BMI, increased triglyceride levels and reduced levels of
high-density lipoprotein cholesterol, which occurs during obesity
(42). Hydroxysteroid (11-β)
dehydrogenase type 1 (11-βHSD1) is a bidirectional enzyme encoded
by the HSD11B1 gene and is highly expressed in the liver and
adipose tissues (43). It can
convert inactive cortisone into its active form cortisol (43). Individuals with obesity typically
show decreased HSD11B1 gene expression in the intra-abdominal
visceral adipose tissue (VAT) (44). Therefore, these obesity-related
genes (TNF-α, PPARD, PPARG, FAAH and HSD11B1) were selected to
verify the effects of treatment with MLE on obesity. The results
indicated that the expression levels of TNF-α, PPARD and PPARG were
increased, whilst the expression levels of FAAH and HSD11B1 were
decreased by obesity, which were markedly reversed by MLE
treatment. The altered expression levels of TNF-α, PPARD, PPARG,
FAAH and HSD11B1 in obesity treated with MLE were consistent with
results from previous studies aforementioned.
AMPK is a cellular energy sensor that functions as a
serine/threonine specific protein kinase (45). It serves a key role in maintaining
the metabolic balance in the body (46,47).
Activation of AMPK can increase catabolism, reduce the expression
levels of regulators of lipid synthesis, including acetyl-CoA
carboxylase (ACC), FAS and SREBP, to reduce the rate of anabolism
whilst regulating lipid synthesis and utilization (48). PPARγ is a major transcription factor
that can modulate glucose and lipid metabolism, the expression of
which in adipose tissues has been previously associated with high
fat diet-induced adipocyte hypertrophy and insulin resistance
(49,50). SREBP-1c is a transcription factor
that regulates lipogenesis (51).
AMPK-knockout mice frequently develop metabolic complications, such
as dysregulated glucose homeostasis and insulin resistance, when
they are fed a high-fat diet (52).
However, activation of AMPK using A-769662 has been found to
alleviate metabolic dysfunction in mice fed a high-fat diet
(52). AMPK can reduce de
novo lipogenesis and promote fatty acid oxidation and
inhibition of ACC phosphorylation and SREBP-1c activation by AMPK
can inhibit fatty-acid synthesis (53). In the present study, it was observed
that p-AMPK/AMPK levels were decreased, whilst the expression
levels of PPARγ, SREBP-1 and FAS were increased by obesity, which
were markedly reversed by MLE treatment.
However, the present study has some limitations.
Although the present study suggested that MLE negatively regulated
inflammation, lipogenesis, lipid catabolism, fatty acid oxidation
and AMPK signaling, whether this effect was indeed mediated by
these signaling pathways should be improved using related
inhibitors, gene knockout or overexpression.
In conclusion, a network pharmacology approach was
used in the present study to identify active components of MLE and
their potential target proteins associated with obesity. The
results indicated that 24 targets associated with obesity were
screened to be targets for MLE, where seven related biological
processes and seven related signaling pathways were extracted by GO
and KEGG analyzes, respectively. Subsequently, an in vivo
experiment was used to verify the network pharmacology analysis and
revealed that the MLE-induced obesity improvement may be due to the
upregulation of FAAH and HSD11B1 and downregulation of TNF, PPARδ
and PPARγ.
Supplementary Material
Structures of the 26 active components
in Mulberry leaf extract downloaded from the Traditional Chinese
Medicine Systems Pharmacology database.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JD conceived and designed the study. GW performed
the experiments, analyzed the data and drafted the manuscript. JD
and GW confirm the authenticity of all raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocols of the present study were
approved by the Institutional Animal Care and Use Committee of the
Hunan Future Health Technology Group Co., Ltd. (Changsha,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Tsai AG and Bessesen DH: Obesity. Ann
Intern Med. 170:ITC33–ITC48. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jia A, Xu S, Ming J, et al: Epidemic
characteristics of obesity in China under various diagnostic
criteria. Chin J Diabetes Mellitus. 9:221–225. 2017.(In
Chinese).
|
|
3
|
Kim YJ, Lee HS, Kim YK, Park S, Kim JM,
Yun JH, Yu HY and Kim BJ: Association of metabolites with obesity
and Type 2 diabetes based on FTO genotype. PLoS One.
11(e0156612)2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lavie CJ, De Schutter A, Parto P, Jahangir
E, Kokkinos P, Ortega FB, Arena R and Milani RV: Obesity and
prevalence of cardiovascular diseases and Prognosis-The obesity
paradox updated. Prog Cardiovasc Dis. 58:537–547. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yang XD, Jiang S, Wang G, Zhang R, Zhang J
and Zhu JS: Link of obesity and gastrointestinal cancer: Crossroad
of inflammation and oxidative stress. J Biol Regul Homeost Agents.
29:755–760. 2015.PubMed/NCBI
|
|
6
|
Wang R, Li Y, Mu W, Li Z, Sun J, Wang B,
Zhong Z, Luo X, Xie C and Huang Y: Mulberry leaf extract reduces
the glycemic indexes of four common dietary carbohydrates. Medicine
(Baltimore. 97(e11996)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Park JM, Bong HY, Jeong HI, Kim YK, Kim JY
and Kwon O: Postprandial hypoglycemic effect of mulberry leaf in
Goto-Kakizaki rats and counterpart control Wistar rats. Nutr Res
Pract. 3:272–278. 2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yang X, Yang L and Zheng H: Hypolipidemic
and antioxidant effects of mulberry (Morus alba L.) fruit in
hyperlipidaemia rats. Food Chem Toxicol. 48:2374–2379.
2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Andallu B, Suryakantham V, Lakshmi
Srikanthi B and Reddy GK: Effect of mulberry (Morus indica
L.) therapy on plasma and erythrocyte membrane lipids in patients
with type 2 diabetes. Clin Chim Acta. 314:47–53. 2001.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Arabshahi-Delouee S and Urooj A:
Antioxidant properties of various solvent extracts of mulberry
(Morus indica L.) leaves. Food Chem. 102:1233–1240.
2007.
|
|
11
|
Ramadan EM, Abou-Taleb KA, Galal GF and
Abdel-Hamid SN: Antibacterial, antibiofilm and antitumor activities
of grape and mulberry leaves ethanolic extracts towards bacterial
clinical strains. Ann Agric Sci. 62:151–159. 2017.
|
|
12
|
Zeni AL and Dall'Molin M:
Hypotriglyceridemic effect of Morus alba L., Moraceae,
leaves in hyperlipidemic rats. Rev Bras Farmacogn. 20:130–133.
2010.
|
|
13
|
Lee YJ, Choi DH, Kim EJ, Kim HY, Kwon TO,
Kang DG and Lee HS: Hypotensive, hypolipidemic, and vascular
protective effects of Morus alba L. in rats fed an
atherogenic diet. Am J Chin Med. 39:39–52. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Peng CH, Lin HT, Chung DJ, Huang CN and
Wang CJ: Mulberry leaf extracts prevent obesity-induced NAFLD with
regulating adipocytokines, inflammation and oxidative stress. J
Food Drug Anal. 26:778–787. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sheng Y, Liu J, Zheng S, Liang F, Luo Y,
Huang K, Xu W and He X: Mulberry leaves ameliorate obesity through
enhancing brown adipose tissue activity and modulating gut
microbiota. Food Funct. 10:4771–4781. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li Q, Liu F, Liu J, Liao S and Zou Y:
Mulberry leaf polyphenols and fiber induce synergistic antiobesity
and display a modulation effect on gut microbiota and metabolites.
Nutrients. 11(1017)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An Rpackage for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ann JY, Eo H and Lim Y: Mulberry leaves
(Morus alba L.) ameliorate obesity-induced hepatic
lipogenesis, fibrosis, and oxidative stress in high-fat diet-fed
mice. Genes Nutr. 10(46)2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang RZ, Yu SJ, Bai H and Ning K:
TCM-Mesh: The database and analytical system for network
pharmacology analysis for TCM preparations. Sci Rep.
7(2821)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li S: Network target: A starting point for
traditional Chinese medicine network pharmacology. Zhongguo Zhong
Yao Za Zhi. 36:2017–2020. 2011.PubMed/NCBI(In Chinese).
|
|
23
|
Tang H, He S, Zhang X, Luo S, Zhang B,
Duan X, Zhang Z, Wang W, Wang Y and Sun Y: A network pharmacology
approach to uncover the pharmacological mechanism of XuanHuSuo
powder on osteoarthritis. Evid Based Complement Alternat Med.
2016(3246946)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kim GN, Kwon YI and Jang HD: Mulberry leaf
extract reduces postprandial hyperglycemia with few side effects by
inhibiting α-glucosidase in normal rats. J Med Food. 14:712–717.
2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lee HJ, Na YG, Han M, Pham TMA, Lee H, Lee
HK, Myung CS, Han JH, Kang JS, Kim KT and Cho CW: Statistical
design of sustained-release tablet garcinia cambogia extract and
bioconverted mulberry leaf extract for anti-obesity. Pharmaceutics.
12(932)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Aramwit P, Supasyndh O, Siritienthong T
and Bang N: Mulberry leaf reduces oxidation and C-reactive protein
level in patients with mild dyslipidemia. Biomed Res Int.
2013(787981)2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Huang J, Wang Y, Ying C, Liu L and Lou Z:
Effects of mulberry leaf on experimental hyperlipidemia rats
induced by high-fat diet. Exp Ther Med. 16:547–556. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tian S, Wang M, Liu C, Zhao H and Zhao B:
Mulberry leaf reduces inflammation and insulin resistance in type 2
diabetic mice by TLRs and insulin signalling pathway. BMC
Complement Altern Med. 19(326)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Marcelino G, Machate DJ, Freitas KC, Hiane
PA, Maldonade IR, Pott A, Asato MA, Candido CJ and Guimarães RCA:
β-Carotene: Preventive role for type 2 diabetes mellitus and
obesity: A review. Molecules. 25(5803)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jiang H, Horiuchi Y, Hironao KY, Kitakaze
T, Yamashita Y and Ashida H: Prevention effect of quercetin and its
glycosides on obesity and hyperglycemia through activating AMPKα in
high-fat diet-fed ICR mice. J Clin Biochem Nutr. 67:74–83.
2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kurano M, Hasegawa K, Kunimi M, Hara M,
Yatomi Y, Teramoto T and Tsukamoto K: Sitosterol prevents
obesity-related chronic inflammation. Biochim Biophys Acta Mol Cell
Biol Lipids. 1863:191–198. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang T, Wu Q and Zhao T: Preventive
effects of kaempferol on high-fat diet-induced obesity
complications in C57BL/6 mice. Biomed Res Int.
2020(4532482)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mak IL, Lavery P, Agellon S, Rauch F,
Murshed M and Weiler HA: Arachidonic acid exacerbates diet-induced
obesity and reduces bone mineral content without impacting bone
strength in growing male rats. J Nutr Biochem.
73(108226)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yaribeygi H, Farrokhi FR, Butler AE and
Sahebkar A: Insulin resistance: Review of the underlying molecular
mechanisms. J Cell Physiol. 234:8152–8161. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hammarstedt A, Gogg S, Hedjazifar S,
Nerstedt A and Smith U: Impaired adipogenesis and dysfunctional
adipose tissue in human hypertrophic obesity. Physiol Rev.
98:1911–1941. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Poblete JMS, Ballinger MN, Bao S,
Alghothani M, Nevado JB Jr, Eubank TD, Christman JW and Magalang
UJ: Macrophage HIF-1α mediates obesity-related adipose tissue
dysfunction via interleukin-1 receptor-associated kinase M. Am J
Physiol Endocrinol Metab. 318:E689–E700. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yao L, Bhatta A, Xu Z, Chen J, Toque HA,
Chen Y, Xu Y, Bagi Z, Lucas R, Huo Y, et al: Obesity-induced
vascular inflammation involves elevated arginase activity. Am J
Physiol Regul Integr Comp Physiol. 313:R560–R571. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ding S, Chi MM, Scull BP, Rigby R,
Schwerbrock NM, Magness S, Jobin C and Lund PK: High-fat diet:
Bacteria interactions promote intestinal inflammation which
precedes and correlates with obesity and insulin resistance in
mouse. PLoS One. 5(e12191)2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tzanavari T, Giannogonas P and Karalis KP:
TNF-alpha and obesity. Curr Dir Autoimmun. 11:145–156.
2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ding Y, Guo ZR, Wu M, Chen Q, Zhou ZY, Yu
H, Zhang LJ, Liu JC and Luo WS: Effects of PPARD-87T >C and
interactions with single nucleotide polymorphisms in PPARA and
PPARG on abdominal obesity. Zhonghua Yi Xue Za Zhi. 92:1517–1521.
2012.PubMed/NCBI(In Chinese).
|
|
41
|
Landgraf K, Klöting N, Gericke M, Maixner
N, Guiu-Jurado E, Scholz M, Witte AV, Beyer F, Schwartze JT, Lacher
M, et al: The obesity-susceptibility gene TMEM18 promotes
adipogenesis through activation of PPARG. Cell Rep.
33(108295)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang Y, Sonnenberg GE, Baye TM, Littrell
J, Gunnell J, DeLaForest A, MacKinney E, Hillard CJ, Kissebah AH,
Olivier M and Wilke RA: Obesity-related dyslipidemia associated
with FAAH, independent of insulin response, in multigenerational
families of Northern European descent. Pharmacogenomics.
10:1929–1939. 2009.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tomlinson JW, Moore JS, Clark PM, Holder
G, Shakespeare L and Stewart PM: Weight loss increases
11beta-hydroxysteroid dehydrogenase type 1 expression in human
adipose tissue. J Clin Endocrinol Metab. 89:2711–2716.
2004.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chedid MF, do Nascimento FV, de Oliveira
FS, de Souza BM, Kruel CR, Gurski RR, Canani LH, Crispim D and
Gerchman F: Interaction of HSD11B1 and H6PD polymorphisms in
subjects with type 2 diabetes are protective factors against
obesity: A cross-sectional study. Diabetol Metab Syndr.
11(78)2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhu XJ, Dai JQ, Tan X, Zhao Y and Yang WJ:
Activation of an AMP-activated protein kinase is involved in
post-diapause development of Artemia franciscana encysted embryos.
BMC Dev Biol. 9(21)2009.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Hardie DG: New roles for the LKB1->AMPK
pathway. Curr Opin Cell Biol. 17:167–173. 2005.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hardie DG: The AMP-activated protein
kinase pathway-new players upstream and downstream. J Cell Sci.
117:5479–5487. 2004.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Herzig S and Shaw RJ: AMPK: Guardian of
metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol.
19:121–135. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kubota N, Terauchi Y, Miki H, Tamemoto H,
Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et
al: PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy
and insulin resistance. Mol Cell. 4:597–609. 1999.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Jones JR, Barrick C, Kim KA, Lindner J,
Blondeau B, Fujimoto Y, Shiota M, Kesterson RA, Kahn BB and
Magnuson MA: Deletion of PPARgamma in adipose tissues of mice
protects against high fat diet-induced obesity and insulin
resistance. Proc Natl Acad Sci USA. 102:6207–6212. 2005.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Crewe C, Zhu Y, Paschoal VA, Joffin N,
Ghaben AL, Gordillo R, Oh DY, Liang G, Horton JD and Scherer PE:
SREBP-regulated adipocyte lipogenesis is dependent on substrate
availability and redox modulation of mTORC1. JCI insight.
5(e129397)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wu L, Zhang L, Li B, Jiang H, Duan Y, Xie
Z, Shuai L and Li J and Li J: AMP-Activated Protein Kinase (AMPK)
regulates energy metabolism through modulating thermogenesis in
adipose tissue. Front Physiol. 9(122)2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Jeon SM: Regulation and function of AMPK
in physiology and diseases. Exp Mol Med. 48(e245)2016.PubMed/NCBI View Article : Google Scholar
|