Introduction
Talaromycosis is a severe deep mycosis caused by
Talaromyces marneffei, which was first isolated from a
bamboo rat in Vietnam in 1956 by Capponi et al (1). T. marneffei is a thermally
dimorphic endemic fungus. When cultured on SDA medium at 25˚C, the
colony appears velvety gray-green, and diffuses a red pigment into
the culture medium; however, at 37˚C, the spores convert to a
pathogenic yeast phase, and no diffusing pigment is produced
(2). T. marneffei
infections are usually initiated by the inhalation of dormant
spores, which are produced outside of the host during the
differentiation of the hyphal growth form. In the lungs, host
innate immune cells recognize these propagules (3,4).
Notably, T. marneffei causes disseminated infection in
immunocompromised patients, particularly in individuals of
Southeast Asian descent and southern China (5). The fungus is one of the leading
causes of death among immunocompromised patients. For infected
patients, early clinical diagnosis is difficult, and the
effectiveness of antifungal therapy is often limited, resulting in
high rates of mortality and morbidity (6,7). To
date, the underlying immunological mechanisms involved in the
recognition and control of T. marneffei are unclear.
Innate immunity represents the first line of defense
for hosts against microbes. Upon invasion, pathogens are suppressed
by the host innate immune system through the recognition of
pathogen-associated molecular patterns (PAMPs) by pattern
recognition receptors (PRRs). It is well established that NOD-like
receptors (NLRs), Toll-like receptors (TLRs), C-type lectin
receptors (CLRs) and retinoic acid-inducible gene I-like receptors
are the best characterized PRRs for sensing different types of
PAMPs (8,9). Among these, CLRs are one of the most
important PRRs that detect fungi in the innate immune system
(10). Specifically, the CLRs
consists of dendritic cell (DC)-associated C-type lectin-1
(Dectin-1, CLEC7A), Dectin-2 (CLEC4N), mannose receptor (CD206),
macrophage-inducible C-type lectin (CLEC4E), macrophage C-type
lectin (CLEC4D), melanin-sensing C-type lectin (CLEC1A) and
DC-specific intercellular adhesion molecule-3-grabbing nonintegrin
(CD209) (11). As previously
reported, CLRs are primarily expressed on myeloid cells, including
macrophages, DCs and neutrophils (12). Of interest is Dectin-1, which is a
key protein involved in the CLR-mediated antifungal signaling
pathway, recruiting additional proteins to form a multiprotein
complex capable of activating the NF-κB inflammatory pathway.
Dectin-1 is a type II transmembrane protein, which recognizes
β-1,3-glucans in the cell wall of various pathogenic fungi
(13). It may stimulate several
cellular responses via the spleen tyrosine kinase (Syk)/CARD9
signaling pathway, such as phagocytosis, the production of
cytokines and the respiratory burst (14). Furthermore, it is a major
recognition receptor for various types of fungi, including species
of Aspergillus, Candida, Histoplasma and
Cryptococcus, among others (15).
Despite Dectin-1 playing an important role in
regulating host immunity and fungal infection, the activation of
Dectin-1 induced by T. marneffei infection remains to be
elucidated. The aim of the current study was to explore role and
mechanism of Dectin-1-mediated signaling pathway in T.
marneffei infection using THP-1 macrophages.
Materials and methods
Reagents and antibodies
RPMI-1640 medium, FBS, penicillin-streptomycin
solution and β-mercaptoethanol were purchased from Gibco (Thermo
Fisher Scientific, Inc.). TRIzol® reagent was obtained
from Invitrogen (Thermo Fisher Scientific, Inc.). PMA and puromycin
were purchased from Sigma-Aldrich (Merck KGaA). One Step TB Green™
Prime Script™ RT-PCR kit II was purchased from Takara Bio, Inc.
RIPA lysis buffer was purchased from Beijing Solarbio Science &
Technology Co., Ltd. BSA, BCA assay and blocking buffer were
purchased from Beyotime Institute of Biotechnology. ECL was
purchased from Bio-Rad Laboratories, Inc. Dectin-1 short hairpin
(sh)RNA lentiviral particles and scramble shRNA lentiviral
particles encoding a GFP sequence were constructed by Shanghai
GeneChem Co., Ltd. Antibodies against Dectin-1 (cat. no. 60128),
NF-κB p65 (cat. no. 8242), phosphorylated (p)-NF-κB p65 (cat. no.
3033), IκBα (cat. no. 4814S), p-IκBα (cat. no. 9246), Syk (cat. no.
2712), p-Syk (cat. no. 2710) and anti-rabbit IgG (H + L), Alexa
Fluor® 555-conjugated anti-rabbit IgG (cat. no. 4413)
were purchased from Cell Signaling Technology, Inc.; horseradish
peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody
(cat. no. L3012 was purchased from Signalway Antibody LLC;
HRP-conjugated goat anti-mouse IgG secondary antibody (cat. no.
ab6789) was purchased from Abcam; and anti-β-actin (cat. no.
66009-1-Ig) was purchased from ProteinTech Group, Inc.
Cell culture and maintenance
The human monocyte cell line THP-1 (The Cell Bank of
Type Culture Collection of The Chinese Academy of Sciences) was
cultured in RPMI-1640 medium supplemented with 10% inactivated FBS,
0.05 mM β-mercaptoethanol and 100 U/ml penicillin-streptomycin
solution in a humidified atmosphere containing 5% CO2 at
37˚C. THP-1 cells could be differentiated into macrophages by
treatment with 100 ng/ml PMA at 37˚C for 48 h. The cells were then
cultured in medium without PMA.
Fungal culture
T. marneffei [CMCC(F)B33r; The Chinese
Academy of Medical Sciences and Peking Union Medical College]
strains were cultivated on potato dextrose agar slants and
incubated for 3-7 days at 25 or 37˚C. Fungal spores or hyphae were
harvested by washing the plates with sterile PBS with 0.01%
Tween-80 (PBST) solution. The hyphae were ground to obtain 20-40 µm
fragments. The suspension was then gently filtered through a 40-µm
nylon pore mesh cell strainer. Subsequently, the spores or hyphae
were thoroughly washed, centrifuged for 5 min at 2,000 x g at room
temperature, resuspended in sterile PBST, and adjusted to a
concentration of 1x108 CFU/ml with RPMI-1640 medium. As
required, spores and hyphae were killed by heating at 65˚C in water
for 2 h (16). Finally, the
suspension was stored at 4˚C for use within 48 h. THP-1 macrophages
were incubated with heat-killed T. marneffei spores or
hyphae (25:1, fungi to cell) cultured at 37˚C in a 5%
CO2 atmosphere for 0, 1, 2, 4, 8 and 12 h. THP-1
macrophages with Dectin-1 expression silencing using a
shRNA-Dectin-1 interference and transfected with an ineffective
interfering sequence were incubated with heat-killed T.
marneffei spores (25:1, fungi to cell) and cultured at 37˚C in
a 5% CO2 atmosphere for the indicated time periods.
PCR array
This study used a customized PCR array from CT
Bioscience to analyze the expression of key genes that participated
in T. marneffei-induced immune response. In brief, THP-1
macrophages were harvested after treatment with or without T.
marneffei spores or hyphae for 8 h. A total of 88 potential
genes that may be involved in T. marneffei-induced cellular
antifungal immune responses were selected as the target mRNAs.
After RNA isolation, 1 µg total RNA was used for reverse
transcription using an RT kit (cat. no. CTB101; CT Bioscience) in a
20 µl volume, according to the manufacturer's protocols. The PCR
array employed SYBR Green I-based quantitative PCR (qPCR) (cat. no.
CTB103; CT Bioscience) to quantify gene expression level. qPCR was
performed in a Roche LightCycler s480-II instrument (Roche
Diagnostics) under the following thermocycling conditions: 10 min
at 93˚C, followed by 45 cycles of 10 sec at 93˚C and 30 sec at
60˚C.Gene specific primers were pre-deposited into wells of a
96-well PCR plate in the array. GAPDH, β-2-microglobulin (B2M),
ACTB, hypoxanthine-guanine phosphoribosyltransferase (HPRT1) and
ornithine decarboxylase antizyme 1 (OAZ1) were used as housekeeping
genes for normalization. The primers used were as follows: C-ros
oncogene 1 receptor tyrosine kinase (ROS) forward, 5'-GCAAATA
ATCTAGGGTTTGGTGA-3' and reverse, 5'-TCAGTGGGAT TGTAACAACCAG-3'; NLR
family pyrin domain-containing (NLRP)11 forward,
5'-TGTTCAGCGCATCTTTCAA-3' and reverse,
5'-CTCCAGTAGACAAGGCTCTTCA-3'; Dectin-1 forward,
5'-AGCCTACCTGTAGGTCGACAA-3' and reverse,
5'-CTGAGGTCAAGATAAATGCAGAAA-3'; Dectin-3 forward,
5'-CCAGCTGATACCTTCGGTTA-3' and reverse, 5'-TGCCTCTCTTACAGCGTGAA-3';
NLRP3 forward, 5'-TGAAGTGCTGAAACAGCAGAG-3' and reverse, 5'-AAA
GACGACGGTCAGCTCAG-3'; TNF-α forward, 5'-GCCCG ACTATCTCGACTTTG-3'
and reverse, 5'-ATGTTCGTCCT CCTCACAGG-3'; IL-8 forward,
5'-AAGACATACTCCAAAC CTTTCCAC-3' and reverse, 5'-AATTTCTGTGTTGGCGC
AGT-3'; B2M forward, 5'-TGTCTTTCAGCAAGGACTGG-3' and reverse,
5'-AACTATCTTGGGCTGTGACAAA-3'; ACTB forward,
5'-AGTCCGCCTAGAAGCATTTG-3' and reverse, 5'-CTGTCCACCTTCCAGCAGAT-3';
HPRT1 forward, 5'-ACGTCTTGCTCGAGATGTGA-3' and reverse, 5'-AATCC
AGCAGGTCAGCAAAG-3'; OAZ1 forward, 5'-GGAACCGT AGACTCGCTCAT-3' and
reverse, 5'-TGAGCGTTTATTTGC ACGAT-3'; and GAPDH forward,
5'-GGGAGCCAAAAGGGT CATCA-3' and reverse,
5'-TGGTTCACACCCATGACGAA-3'.
Lentiviral transfection of THP-1
cells
The design of the Dectin-1 shRNA (interference
sequence, 5'-CAATTACAC TTCGACTCTCAA-3'), a scrambled shRNA
(5'-TTCTCCGA ACGTGTCACGT-3') and the packaging of the lentivirus
particles were performed by Shanghai GeneChem Co., Ltd. A total of
4x104 THP-1 cells were cultured in supplemented RPMI
medium in 96-well plates for 24 h. Subsequently, 4 µl HitransG P
(25X; Shanghai GeneChem Co., Ltd.) enhanced infection solution was
added prior to cell transduction with lentiviral particles at a
multiplicity of infection (MOI) of 50 (virus number/cell number
=50). Cells were incubated for 12 h, and then the culture medium
was replaced with fresh medium. GFP expression was observed by
fluorescence microscopy 72 h after transduction. After 72 h of
infection, fresh medium containing 2 µg/ml puromycin was added for
72 h to select positively stably transduced cells. Stably
transduced cells were maintained in 1 µg/ml puromycin. The third
passage of stable clones was collected at 9 days after transduction
for reverse transcription (RT)-qPCR analysis.
RT-qPCR
THP-1 macrophages were incubated with heat-killed
T. marneffei spores or hyphae (25:1, fungi to cell) cultured
at 37˚C in 5% CO2 for 0, 1, 2, 4, 8 and 12 h. Following
these treatments, cells were harvested, and total RNA was extracted
using TRIzol® reagent. RT-qPCR was performed using a One
Step TB Green™ Prime Script™ RT-PCR kit II on an Applied Biosystems
7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). GAPDH was used as the internal control. The
thermocycling conditions were as follows: Reverse transcription at
42˚C for 5 min; pre-denaturation for 10 sec at 95˚C; followed by 40
cycles of 5 sec at 94˚C and 34 sec at 60˚C. The specific primers
used were Dectin-1 forward, 5'-CAACTGGGCTCTAATCTCC-3' and reverse,
5'-GCACACTACACAGTTGGTC-3'; and GAPDH forward,
5'-GACCTGACCTGCCGTCTA-3' and reverse 5'-AGGAGTGGGTGTCGCTGT-3'.
Finally, the Cq values for each reaction were collected and the
changes in the expression of the target gene were normalized to
GAPDH. Relative mRNA expression levels were calculated by fold
changes using the 2-ΔΔCq formula, where ΔCq is the
difference between the target gene and GAPDH, and ΔΔCq for the
sample = ΔCq of treated condition - ΔCq of control condition
(17).
Western blotting
THP-1 macrophages were incubated with heat-killed
T. marneffei spores or hyphae (25:1, fungi to cell) cultured
at 37˚C in 5% CO2 for 0, 1, 2, 4, 8 and 12 h. After
treatment, cells were lysed in RIPA lysis buffer containing
protease inhibitor cocktail and the phosphatase inhibitor PhosSTOP
(cat. no. 78440; Thermo Fisher Scientific, Inc.). The protein
concentration was determined using a BCA assay. Total protein (20
µg/lane) was separated by 10% SDS-PAGE and transferred to a PVDF
membrane (MilliporeSigma). After blocking with 5% skimmed milk for
1.5 h at room temperature, the membranes were incubated with the
following primary antibodies at 4˚C overnight: Anti-Dectin-1
(1:1,000), anti-NF-κB p65 (1:1,000), anti-p-NF-κB p65 (1:1,000),
anti-IκBα (1:1,000), anti-p-IκBα (1:1,000), anti-Syk (1:1,000),
anti-p-Syk (1:1,000) and anti-β-actin (1:5,000). The membranes were
then incubated with the appropriate HRP-conjugated secondary
antibodies (1:5,000) at 37˚C for 1 h. The immunoreactive bands were
visualized with ECL reagents.
Immunofluorescence assay for detecting
the nuclear translocation of NF-κB
Cells were cultured on glass dishes. THP-1
macrophages were incubated with heat-killed T. marneffei
spores for 4 h, and THP-1 macrophages with Dectin-1 shRNA or
scrambled shRNA were incubated with heat-killed T. marneffei
spores for 4 h. Stimulated and unstimulated THP-1 macrophages were
washed three times with PBS at various time points, fixed with 4%
paraformaldehyde at 37˚C for 30 min, and permeabilized with 0.2%
Triton X-100 for 15 min, and then washed with PBS. After blocking
with 5% BSA in PBS for 30 min at 37˚C, the cells were incubated
with rabbit anti-NF-κB-p65 antibodies at 4˚C overnight. The cells
were washed with PBST, and then incubated with Alexa 555-conjugated
anti-rabbit IgG at 37˚C for 1 h. Finally, using a drop of ProLong™
Diamond antifade mountant medium with DAPI (cat. no. P36962;
Invitrogen; Thermo Fisher Scientific, Inc.) nuclei were stained for
10 min at room temperature and sealed. The slides were carefully
observed under a confocal microscope.
Cytokine quantification using
ELISA
After stimulation of THP-1 macrophages
(5x105 cells/ml) with heat-killed T. marneffei
spores for 8 or 18 h, specific commercial ELISA kits (R&D
Systems, Inc.) were used to measure the quantity of TNF-α (cat. no.
DTA00D) and IL-8 (cat. no. D8000C) in the cell-free culture
supernatants. The experimental procedure was performed according to
the manufacturer's protocols. All experiments were performed in
triplicate. Data are presented as the mean ± standard
deviation.
Statistical analysis
Experiments were conducted at least three times.
Data are presented as the mean ± standard deviation. Differences
among the groups were evaluated using one-way ANOVA followed by
Tukey's post hoc test. Intragroup (time) and intergroup (fluences)
comparisons of the Cytokine quantification were analyzed by two-way
ANOVA followed by Tukey's post hoc test using SPSS version 22.0
(IBM Corp.). P<0.05 was considered to indicate a statistically
significant difference.
Results
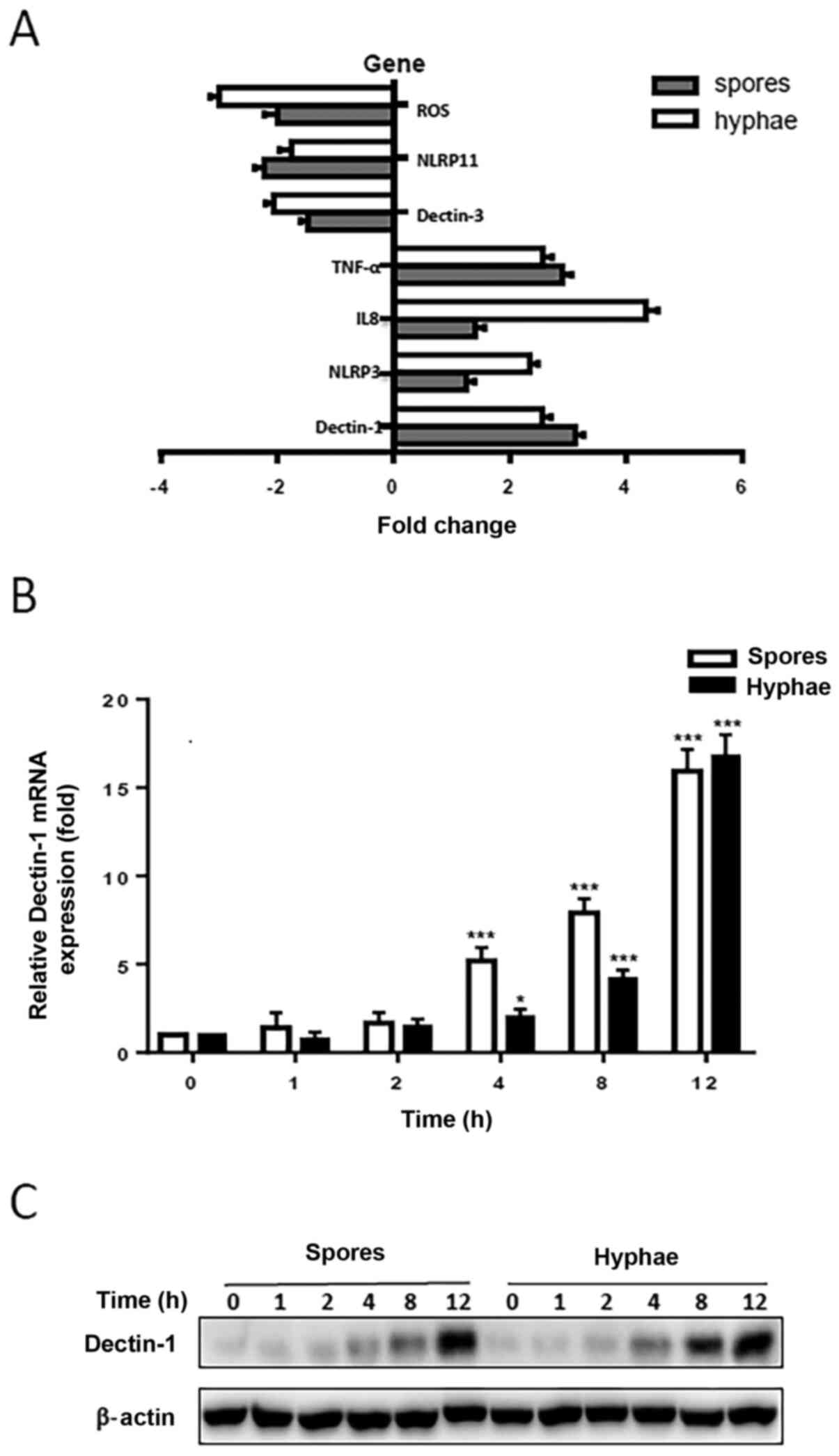
Dectin-1 expression is increased
following T
marneffei infection in vitro. To identify the
potential genes that may be involved in T. marneffei
infection, an in vitro model of THP-1 macrophages infected
with heat-killed T. marneffei spores and hyphae was
established, and a PCR-array was used to screen differential gene
expression in the THP-1 macrophages together with or without T.
marneffei spores or hyphae. It was revealed that the expression
levels of ROS, Dectin-3 and NLRP11 were downregulated, whereas the
levels of Dectin-1, NLRP3, TNF-α and IL-8 were upregulated compared
with untreated THP-1 macrophages (Fig.
1A). Since Dectin-1 had previously been suggested to serve
important roles in the antifungal response, a focus was placed on
the study of Dectin-1 and its regulatory roles in the antifungal
immune response to T. marneffei infection. To investigate
whether Dectin-1 expression was affected by T. marneffei
infection in vitro, THP-1 macrophages were infected with
heat-killed T. marneffei spores or hyphae for various
durations. The expression levels of Dectin-1 in THP-1 macrophages
were examined using RT-qPCR and western blotting. The relative mRNA
expression levels of Dectin-1 in THP-1 macrophages were
significantly increased in response to T. marneffei
infection. Compared with in the uninfected control cells, Dectin-1
mRNA expression levels were significantly increased 4, 8 and 12 h
after infection with heat-killed T. marneffei spores or
hyphae (Fig. 1B). Western blotting
results revealed that Dectin-1 protein expression levels were
elevated 4, 8 and 12 h following infection with heat-killed T.
marneffei spores and hyphae (Fig.
1C). Western blotting and RT-qPCR results confirmed that the
expression levels of Dectin-1 were gradually increased in the cells
with the prolongation of stimulation time. These results suggested
that T. marneffei infection may promote the expression
levels of Dectin-1 in vitro.
T. marneffei triggers the activation
of Syk/NF-κB signaling pathways
To study the Dectin-1 expression patterns in the
antifungal immune response, THP-1 macrophages were exposed to
heat-killed T. marneffei spores or hyphae for 1, 2, 4, 8 or
12 h. Western blotting was used to analyze Dectin-1 protein
expression levels in THP-1 macrophages. It was observed that
Dectin-1 protein expression levels were upregulated in the THP-1
macrophages stimulated with spores and hyphae compared with in the
control cells (Fig. 1C). The
phosphorylation levels of Syk, p65 and IκBα were detected using
western blotting, and were increased in a time-dependent manner;
however, IκBα phosphorylation began to decrease slightly at 4 h
poststimulation (Fig. 2A). After
Dectin-1 expression was knocked down, the phosphorylation levels of
Syk, p65 and IκBα were inhibited (Fig.
2B). These results indicated that activation of Syk/NF-κB may
be involved in the T. marneffei-induced inflammatory
response.
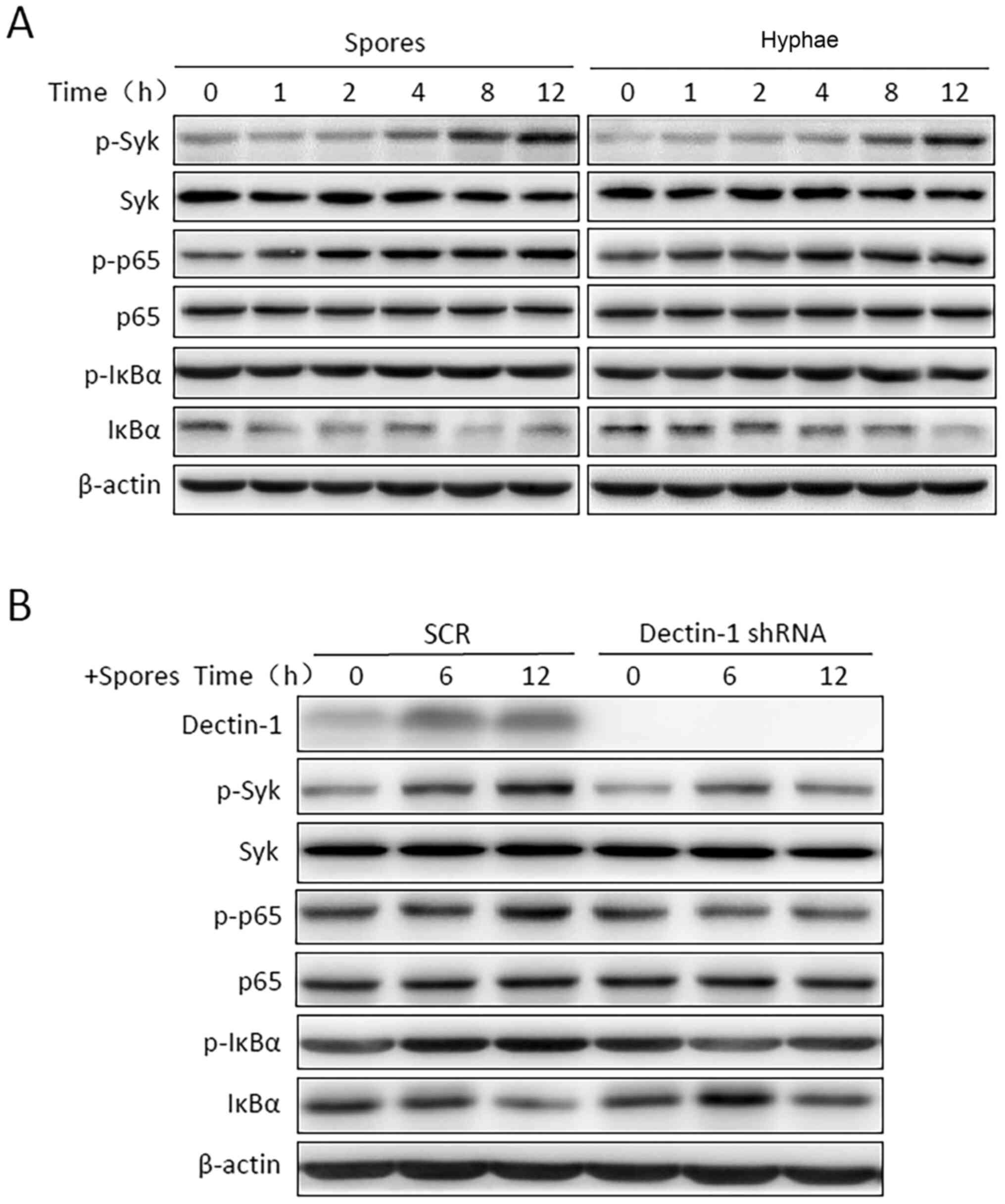 | Figure 2Talaromyces marneffei triggers
the activation of the Syk/NF-κB signaling pathway. (A) THP-1
macrophages were incubated with heat-killed T. marneffei
spores or hyphae for the indicated time periods. (B) THP-1
macrophages were transfected with Dectin-1 shRNA or SCR shRNA.
Cells were then incubated with T. marneffei spores for the
indicated time. Syk, p65 and IκBα activation were determined using
immunoblotting with anti-p-Syk, anti-p-p65 and anti-p-IκBα
antibodies. Immunoblotting was performed with antibodies against
Syk, p-Syk, p65, p-p65, IκBα and p-IκBα, and β-actin was used to
confirm equal protein loading. p, phosphorylated; Syk, spleen
tyrosine kinase; IκBα, nuclear factor of κ light polypeptide gene
enhancer in B-cells inhibitor, α; SCR, scrambled; shRNA, short
hairpin RNA; Dectin-1, dendritic cell-associated C-type
lectin-1. |
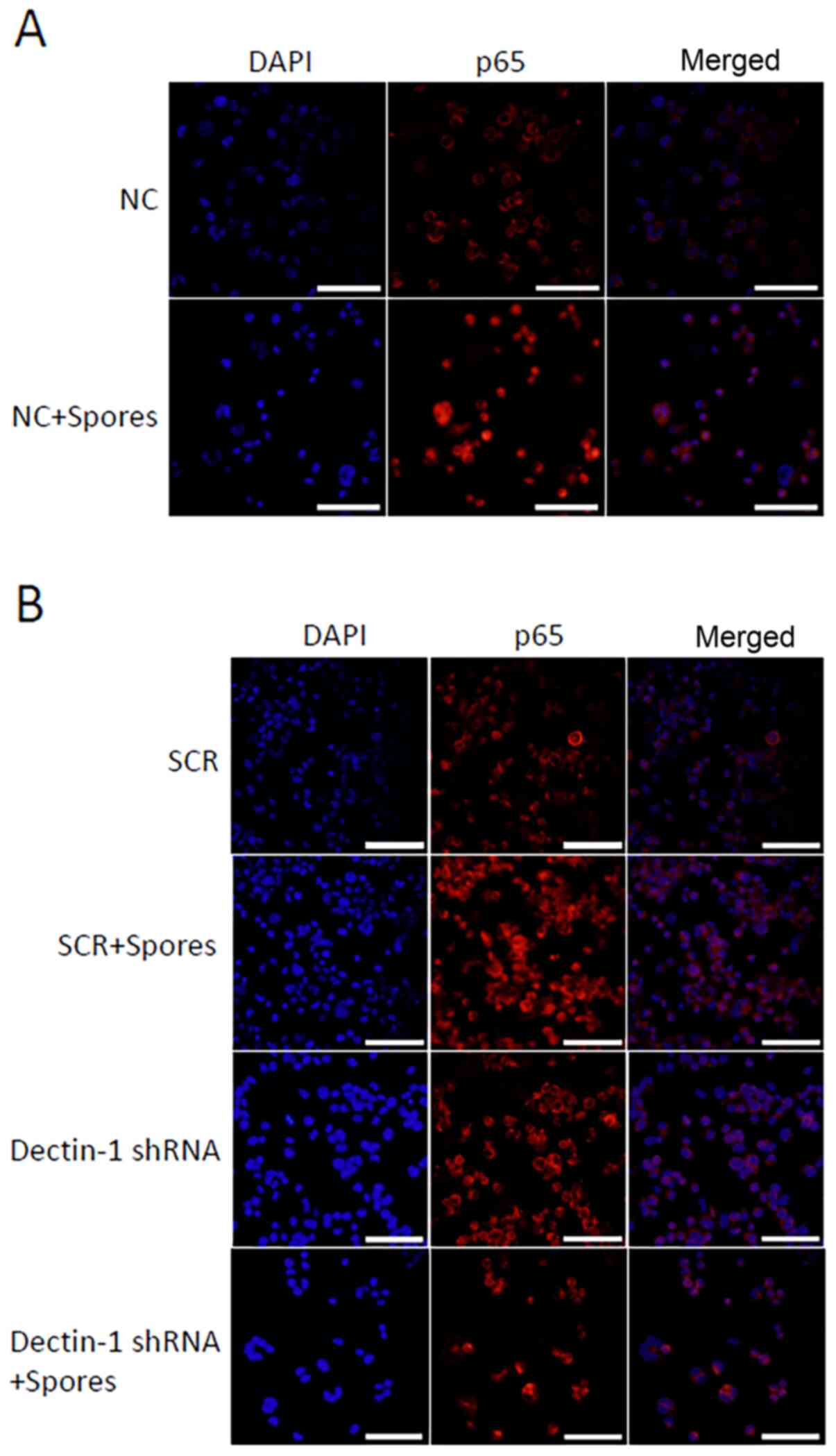
T. marneffei induces the translocation
of NF-κB in THP-1 macrophages
To further confirm the effect of T. marneffei
on NF-κB activation in THP-1 macrophages, immunofluorescence
analysis was used to assess the cellular localization of NF-κB in
THP-1 macrophages. The stimulated or unstimulated THP-1
macrophages, fixed 4 h after stimulation, were stained with diluted
rabbit anti-NF-κB-p65 antibodies. As revealed in Fig. 3, T. marneffei induced the
translocation of NF-κB in THP-1 macrophages. As shown in Fig. 3A, NF-κB-p65 was primarily localized
in the cytoplasm of normal untreated cells, whereas it was
predominately located in the nuclei of the T.
marneffei-stimulated cells. Following knockdown of Dectin-1
expression, the nuclear translocation of p65 protein was inhibited
(Fig. 3B). Compared with in the
SCR + spores group, the fluorescence intensity of nuclear p65 in
THP-1 macrophages was reduced in the shDectin-1 + spores group.
These results demonstrated that T. marneffei elicited
inflammatory activity in THP-1 macrophages by modulating
subcellular localization of the transcriptional factor NF-κB.
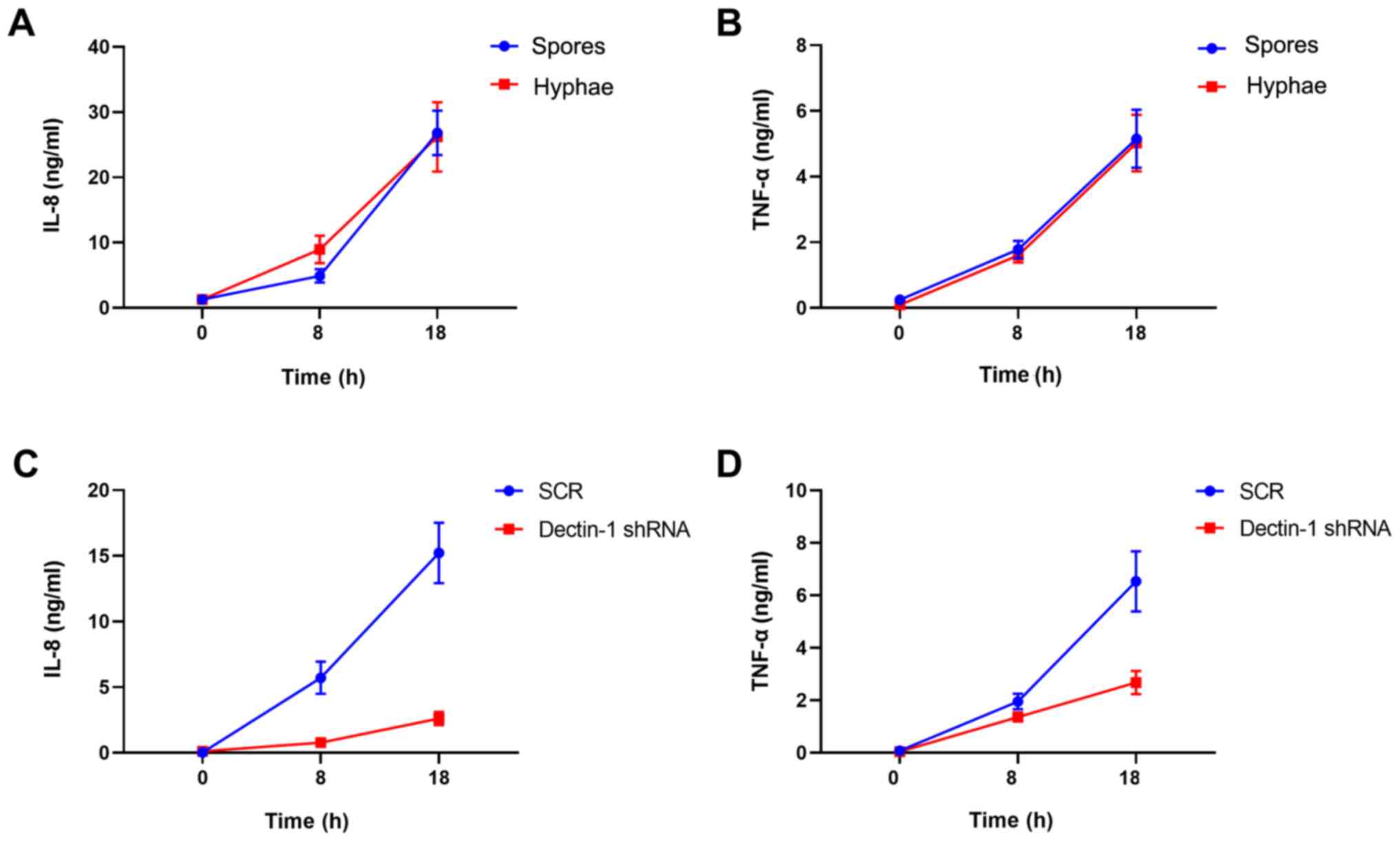
Knockdown of Dectin-1 inhibits
cytokine release from macrophages
Since the expression of inflammatory cytokines is
regulated by NF-κB, the levels of TNF-α and IL-8 were finally
detected using an ELISA. The interaction between T.
marneffei and THP-1 macrophages resulted in increased secretion
of TNF-α and IL-8 (Fig. 4A and
B). THP-1 macrophages were
transfected with Dectin-1 shRNA or scrambled shRNA. Two-way ANOVA
analysis demonstrated that knockdown of Dectin-1 had statistical
significance at the IL-8 level and TNF-α level (data not shown).
The Dectin-1 shRNA group exhibited lower IL-8 and TNF-α levels than
in the SCR group. Knockdown of Dectin-1 in THP-1 macrophages
decreased the production of TNF-α and IL-8. Dynamic changes of IL-8
and TNF-α levels are presented in Fig.
4C and D. These results
demonstrated that Dectin-1 may be involved in the release of
cytokines from macrophages following fungal stimulation, and
supported the hypothesis that Dectin-1 may be pivotal in the
recognition of T. marneffei and subsequent macrophage
activation.
Proposed mechanism of Dectin-1
involvement in the recognition of T
marneffei on human macrophages and contribution
to their immunomodulatory capacity. The results of the present
study indicated that T. marneffei bind and activate the
Dectin-1 receptor, leading to activation of the NF-κB signaling
pathway, including activation of IKK complexes, release of p50/p65
complexes from the inhibitor complex with IκBα and translocation of
the phosphorylated p50/p65 heterocomplex to the nucleus, where the
transcription of proinflammatory genes, including those for
cytokines, such as TNF-α and IL-8, was promoted (Fig. 5).
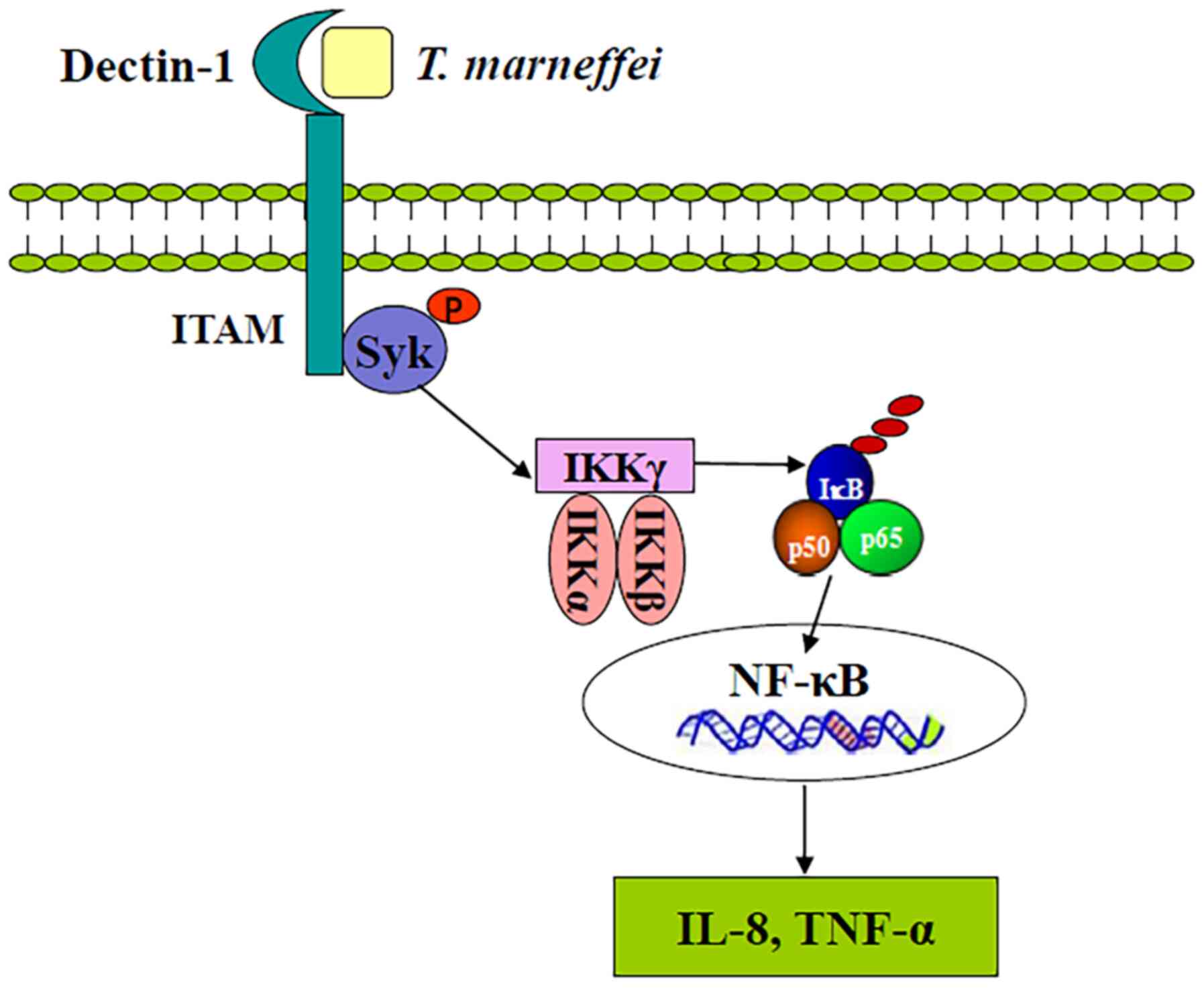 | Figure 5Schematic representation of Dectin-1
and its roles in the immune response to Talaromyces
marneffei infection addressed in the present study. T.
marneffei binds and activates Dectin-1 receptor, leading to
activation of the NF-κB signaling pathway, including activation of
IKK complexes, release of p50/p65 complexes from the inhibitor
complex with IκBα, and translocation of the phosphorylated p50/p65
heterocomplex to the nucleus, where the transcription of
proinflammatory genes, such as the cytokines TNF-α and IL-8, is
induced. Syk, spleen tyrosine kinase; Dectin-1, dendritic
cell-associated C-type lectin-1; IκBα, nuclear factor of κ light
polypeptide gene enhancer in B-cells inhibitor, α; p,
phosphorylated. |
Discussion
Due to the lack of effective antifungal agents,
talaromycosis is well known as a severe disease that can cause
disseminated infection, and treatment of this disease remains a
challenge (18). Nakamura et
al (19) demonstrated that
Dectin-1 was essential in sensing T. marneffei for the
activation of bone marrow-derived DCs. However, the interaction
between macrophages and T. marneffei remains largely
unknown. Innate immunity acting as the front-line defense plays an
essential role in resisting fungal infections. Dectin-1 is a member
of the C-type lectin family and functions as an innate PRR involved
in antifungal immunity. Although the role of Dectin-1 in mediating
talaromycosis remains poorly understood, several studies have
indicated its role in antifungal immunity. Gantner et al
(20) reported that, via
β-glucan-containing particles, Dectin-1 expression enhanced
TLR-mediated activation of NF-κB. In addition, in macrophages and
DCs, Dectin-1 and TLRs were revealed to be synergistic in mediating
the production of cytokines, such as IL-12 and TNF. Sun et
al (21) demonstrated that
Dectin-1 may serve an important role in Aspergillus-induced
innate immune responses in human bronchial epithelial cells. In
addition, Cohen-Kedar et al (22) presented evidence for β-glucan- and
fungal-induced activation of human intestinal epithelial cells and
the potential underlying mechanism, which involved the Dectin-1/Syk
pathway. These findings indicated that Dectin-1 may be a critical
component of the antifungal immune response in macrophages.
Therefore, in the present study, the role and mechanism of Dectin-1
in T. marneffei infection of macrophages was
investigated.
To investigate whether Dectin-1 recognized the
spores and hyphae of T. marneffei, PMA-induced THP-1
macrophages stimulated with T. marneffei spores or hyphae
were used, and the expression levels of Dectin-1 mRNA and protein
were determined. In the present study, it was revealed that THP-1
macrophages interacted with distinct T. marneffei
morphotypes, and increased expression levels of Dectin-1 were
observed in macrophages in response to T. marneffei
infection. These findings suggested that Dectin-1 was involved in
the recognition of T. marneffei by macrophages.
NF-κB is an important transcriptional regulator,
which controls the expression of various pro-inflammatory
mediators. The NF-κB family of transcription factors consists of
five members, p50, p52, p65 (RelA), c-Rel and RelB. The underlying
mechanism of NF-κB signaling consists of a series of positive and
negative regulatory elements. Firstly, inducing stimuli initiate
IKK activation leading to phosphorylation, ubiquitination and
degradation of IκB proteins. IκB is an inhibitory protein that acts
to prevent NF-κB migration into the nucleus, and the degradation of
IκB leads to the release of NF-κB p65/p50 dimers into the nucleus
(23,24). Zhu et al (25) demonstrated that stimulation of
RAW264.7 cells with Candida albicans hyphae triggered Syk
phosphorylation and IκBα degradation. Sun et al (26) identified that Aspergillus
fumigatus infection induced IκBα phosphorylation and
NF-κB-mediated activation of THP-1 macrophages. Furthermore, Rogers
et al (27) revealed the
classical Syk-dependent NF-κB pathway, and showed that following
zymosan binding to Dectin-1, tyrosine phosphorylation of an
ITAM-like sequence by the activated Src family of kinases occurred,
which resulted in the expression of docking sites for the Syk
protein. Duan et al (28)
revealed that Candida parapsilosis could stimulate the
inflammatory response, increase the expression of Dectin-1, and
activate NF-κB and MAPK signaling pathways in macrophages.
Gringhuis et al (29)
reported that Dectin-1 expressed on human DCs activated the
Syk-dependent canonical NF-κB subunits p65 and c-Rel. The results
of the present study are consistent with these findings in which
Dectin-1 in THP-1 macrophages recognized pathogens and induced the
activation of Syk, in turn triggering the downstream molecules,
IκBα and NF-κB. In the present study, it was observed that the
phosphorylation levels of Syk, p65 and IκBα protein were increased,
and p65 nuclear translocation was induced following stimulation of
THP-1 macrophages with T. marneffei. These results suggested
that T. marneffei could induce Syk-mediated activation of
NF-κB in THP-1 macrophages. In addition, it was revealed that
silencing of Dectin-1 inhibited the phosphorylation of Syk, p65 and
IκBα in THP-1 macrophages induced by T. marneffei spores. It
was hypothesized that Dectin-1 may participate in the immunological
defense against T. marneffei, and the Dectin-1/Syk/NF-κB
signaling pathway may serve an indispensable role in T.
marneffei infection.
Inflammatory cytokines play a critical role in the
development of fungal infectious diseases. Monocytes/macrophages
are an important part of innate immunity against fungi, and they
primarily produce cytokines, such as TNF-α, IL-1, IL-6, IL-8 and
granulocyte-colony stimulating factor (30,31).
TNF-α is a pro-inflammatory cytokine produced by immune cells,
primarily T lymphocytes. TNF-α belongs to a family of both soluble
and cell-bound cytokines that have a wide range of functions, such
as host defense, inflammation and apoptosis (32,33).
IL-8, also known as CXCL8, is a proinflammatory CXC chemokine
involved in inflammatory reactions. The biological effects of IL-8
are mediated by two highly related chemokine receptors, CXCR1
(IL-8RA) and CXCR2 (IL-8RB). IL-8 exerts its function alongside
other cytokines and chemokines, thus causing chemoattraction of
leukocytes to sites of inflammation, recruitment and activation of
neutrophils to phagocytosis, and bacterial clearance (34,35).
Li et al (36) demonstrated
that Candida albicans could induce NF-κB activation and
cytokine (TNF-α and IL-8) production. Furthermore, Hohl et
al (37) identified that
antibody-mediated blockade of Dectin-1 partially inhibited
TNF-α/macrophage inflammatory protein-2 induction by metabolically
active conidia of Aspergillus fumigatus. The present study
revealed that T. marneffei treatment in THP-1 macrophages
resulted in increased levels of two pro-inflammatory cytokines
(TNF-α and IL-8). Furthermore, Dectin-1 silencing in THP-1
macrophages with T. marneffei stimulation resulted in
significantly decreased TNF-α and IL-8 cytokine production. These
results indicated that Dectin-1 is important in inducing the
secretion of proinflammatory cytokines. Consistently, knockdown of
Dectin-1 reduced inflammation by decreasing the levels of
pro-inflammatory cytokines. Taken together, the results
demonstrated the essential pro-inflammatory role of Dectin-1 in
talaromycosis.
In conclusion, the present study confirmed that
Dectin-1 was involved in the recognition of T. marneffei on
human macrophages, and contributed to their immunomodulatory
capacity (Fig. 5). It was verified
that T. marneffei induced immune responses by activation of
Dectin-1 as well as the NF-κB signaling pathway in THP-1
macrophages. Dectin-1 was shown to be an important receptor for
T. marneffei on THP-1 macrophages and it was revealed to be
involved in the induction of a pro-inflammatory cytokine response.
Therefore, it was hypothesized that regulating the expression of
Dectin-1 receptor could interfere with the innate immune state of
the host, and thus regulate the defense ability of the host against
T. marneffei infection. Although the underlying molecular
mechanisms remain to be elucidated, the present findings may partly
explain the immune response associated with T. marneffei
infection, and contributed to an improved understanding of the
immune response against T. marneffei. By studying the
precise responses to T. marneffei infection in macrophages,
these findings may enable the further exploration and development
of novel antifungal strategies. However, there are limitations of
the present study. Dectin-1 expression was only investigated at the
cellular level, and the mechanisms underlying the effects of
Dectin-1 on T. marneffei infection were not evaluated in
vivo. Therefore, the effects of Dectin-1 on T. marneffei
infection must be further verified using in vivo
experiments.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81572052), the Natural
Science Foundation of Jiangsu province, China (grant no.
BK20151178), the Natural Science Foundation for Yang Scholars of
Jiangsu province (grant no. BK20180184), the Health and Family
Planning Commission for Yang Technology talents of Changzhou (grant
no. QN201710) and the Young Talent Development Plan of Changzhou
Health Commission (grant no. 2020-233).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YP, YC and WS performed the experiments. YP and YC
prepared the manuscript. YaW, JM, WZ, HZ and WS analyzed the data.
HZ, YuW, WZ and WS were responsible for study conception and design
of the study. JM and YaW contributed to literature searching and
processing. WZ and WS confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Capponi M, Segretain G and Sureau P:
Penicillosis from Rhizomys sinensis. Bull Soc Pathol Exot
Filiales. 49:418–421. 1956.PubMed/NCBI(In French).
|
|
2
|
Segretain G: Penicillium marneffei
n.sp., agent of a mycosis of the reticuloendothelial system.
Mycopathol Mycol Appl. 11:327–353. 1959.PubMed/NCBI View Article : Google Scholar : (In French).
|
|
3
|
Boyce KJ and Andrianopoulos A: Fungal
dimorphism: The switch from hyphae to yeast is a specialized
morphogenetic adaptation allowing colonization of a host. FEMS
Microbiol Rev. 39:797–811. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cogliati M, Roverselli A, Boelaert JR,
Taramelli D, Lombardi L and Viviani MA: Development of an in vitro
macrophage system to assess Penicillium marneffei growth and
susceptibility to nitric oxide. Infect Immun. 65:279–284.
1997.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tsang CC, Lau SKP and Woo PCY: Sixty Years
from Segretain's Description: What Have We Learned and Should Learn
About the Basic Mycology of Talaromyces marneffei?
Mycopathologia. 184:721–729. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hu Y, Zhang J, Li X, Yang Y, Zhang Y, Ma J
and Xi L: Penicillium marneffei infection: An emerging
disease in mainland China. Mycopathologia. 175:57–67.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Le T, Huu Chi N, Kim Cuc NT, Manh Sieu TP,
Shikuma CM, Farrar J and Day JN: AIDS-associated Penicillium
marneffei infection of the central nervous system. Clin Infect
Dis. 51:1458–1462. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Sellge G and Kufer TA: PRR-signaling
pathways: Learning from microbial tactics. Semin Immunol. 27:75–84.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Akira S, Uematsu S and Takeuchi O:
Pathogen recognition and innate immunity. Cell. 124:783–801.
2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Salazar F and Brown GD: Antifungal Innate
Immunity: A Perspective from the Last 10 Years. J Innate Immun.
10:373–397. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Goyal S, Castrillón-Betancur JC, Klaile E
and Slevogt H: The Interaction of Human Pathogenic Fungi With
C-Type Lectin Receptors. Front Immunol. 9(1261)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sancho D and Reis e Sousa C: Signaling by
myeloid C-type lectin receptors in immunity and homeostasis. Annu
Rev Immunol. 30:491–529. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Saijo S and Iwakura Y: Dectin-1 and
Dectin-2 in innate immunity against fungi. Int Immunol. 23:467–472.
2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wevers BA, Kaptein TM, Zijlstra-Willems
EM, Theelen B, Boekhout T, Geijtenbeek TB and Gringhuis SI: Fungal
engagement of the C-type lectin mincle suppresses dectin-1-induced
antifungal immunity. Cell Host Microbe. 15:494–505. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Brown GD: Dectin-1: A signalling non-TLR
pattern-recognition receptor. Nat Rev Immunol. 6:33–43.
2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Saijo S, Ikeda S, Yamabe K, Kakuta S,
Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et
al: Dectin-2 recognition of alpha-mannans and induction of Th17
cell differentiation is essential for host defense against
Candida albicans. Immunity. 32:681–691. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Limper AH, Adenis A, Le T and Harrison TS:
Fungal infections in HIV/AIDS. Lancet Infect Dis. 17:e334–e343.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nakamura K, Miyazato A, Koguchi Y, Adachi
Y, Ohno N, Saijo S, Iwakura Y, Takeda K, Akira S and Fujita J:
Toll-like receptor 2 (TLR2) and dectin-1 contribute to the
production of IL-12p40 by bone marrow-derived dendritic cells
infected with Penicillium marneffei. Microbes Infect.
10:1223–1227. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gantner BN, Simmons RM, Canavera SJ, Akira
S and Underhill DM: Collaborative induction of inflammatory
responses by dectin-1 and Toll-like receptor 2. J Exp Med.
197:1107–1117. 2003.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sun WK, Lu X, Li X, Sun QY, Su X, Song Y,
Sun HM and Shi Y: Dectin-1 is inducible and plays a crucial role in
Aspergillus-induced innate immune responses in human
bronchial epithelial cells. Eur J Clin Microbiol Infect Dis.
31:2755–2764. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cohen-Kedar S, Baram L, Elad H, Brazowski
E, Guzner-Gur H and Dotan I: Human intestinal epithelial cells
respond to β-glucans via Dectin-1 and Syk. Eur J Immunol.
44:3729–3740. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hayden MS and Ghosh S: Shared principles
in NF-kappaB signaling. Cell. 132:344–362. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hinz M and Scheidereit C: The IκB kinase
complex in NF-κB regulation and beyond. EMBO Rep. 15:46–61.
2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhu LL, Zhao XQ, Jiang C, You Y, Chen XP,
Jiang YY, Jia XM and Lin X: C-type lectin receptors Dectin-3 and
Dectin-2 form a heterodimeric pattern-recognition receptor for host
defense against fungal infection. Immunity. 39:324–334.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sun H, Xu XY, Tian XL, Shao HT, Wu XD,
Wang Q, Su X and Shi Y: Activation of NF-κB and respiratory burst
following Aspergillus fumigatus stimulation of macrophages.
Immunobiology. 219:25–36. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rogers NC, Slack EC, Edwards AD, Nolte MA,
Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL,
Brown GD, et al: Syk-dependent cytokine induction by Dectin-1
reveals a novel pattern recognition pathway for C type lectins.
Immunity. 22:507–517. 2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Duan Z, Chen X, Du L, Liu C, Zeng R, Chen
Q and Li M: Inflammation Induced by Candida parapsilosis in
THP-1 Cells and Human Peripheral Blood Mononuclear Cells (PBMCs).
Mycopathologia. 182:1015–1023. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gringhuis SI, den Dunnen J, Litjens M, van
der Vlist M, Wevers B, Bruijns SC and Geijtenbeek TB: Dectin-1
directs T helper cell differentiation by controlling noncanonical
NF-kappaB activation through Raf-1 and Syk. Nat Immunol.
10:203–213. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Koh TJ and DiPietro LA: Inflammation and
wound healing: The role of the macrophage. Expert Rev Mol Med.
13(e23)2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Idriss HT and Naismith JH: TNF alpha and
the TNF receptor superfamily: Structure-function relationship(s).
Microsc Res Tech. 50:184–195. 2000.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Aggarwal BB, Gupta SC and Kim JH:
Historical perspectives on tumor necrosis factor and its
superfamily: 25 years later, a golden journey. Blood. 119:651–665.
2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Akdis M, Aab A, Altunbulakli C, Azkur K,
Costa RA, Crameri R, Duan S, Eiwegger T, Eljaszewicz A, Ferstl R,
et al: Interleukins (from IL-1 to IL-38), interferons, transforming
growth factor β, and TNF-α: Receptors, functions, and roles in
diseases. J Allergy Clin Immunol. 138:984–1010. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang W and Chen H: The study on the
interleukin-8 (IL-8)]. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi.
19:697–702. 2002.PubMed/NCBI(In Chinese).
|
|
36
|
Li M, Liu ZH, Chen Q, Zhou WQ, Yu MW, Lü
GX, Lü XL, Shen YN, Liu WD and Wu SX: Insoluble beta-glucan from
the cell wall of Candida albicans induces immune responses
of human THP-1 monocytes through Dectin-1. Chin Med J (Engl).
122:496–501. 2009.PubMed/NCBI
|
|
37
|
Hohl TM, Van Epps HL, Rivera A, Morgan LA,
Chen PL, Feldmesser M and Pamer EG: Aspergillus fumigatus
triggers inflammatory responses by stage-specific beta-glucan
display. PLoS Pathog. 1(e30)2005.PubMed/NCBI View Article : Google Scholar
|