Introduction
Crohn's disease (CD) is a chronic inflammatory
disease of the gastrointestinal tract, with an evolution marked by
alternating episodes of exacerbation and remission (1). The initial phase of CD is
characterized by chronic digestive symptoms, such as diarrhea and
weight loss, while longstanding CD is often complicated by the
formation of strictures or fistulas (1). According to the Montreal
classification, phenotypically, CD can be classified into three
categories: B1-inflammatory (non-stricturing, non-penetrating),
B2-stricturing and B3-penetrating (2).
Previous research has suggested that these
phenotypic differences in CD are not due to intrinsic differences
in the disease itself but rather due to the different stages of
intestinal injury, manifesting a more or less protracted disease
evolution (3-5).
Clinically, the behavioral classification of CD is difficult and
may not truly reflect the natural history of the disease, as the
incidence of stricture formation may differ substantially, not only
among different patients but also in the same individual (3).
The frequency of disease-associated fibrostenotic
complications increases with time in the evolution of CD (4,5).
Indeed, intestinal strictures represent a common complication of
CD, with as many as 40% of patients with ileal CD developing
clinical symptoms of obstruction (6,7).
Intestinal obstruction secondary to stricture formation is a
frequent indication for surgical management. Progressive intestinal
resection further predisposes to postoperative morbidity, as these
patients are more likely to develop short bowel syndrome (8).
Although immunosuppressive and biological treatments
are expected to lower the CD-related incidence of complications in
patients with high-risk CD, the rate of complications does not
significantly decrease despite the early initiation of
immunosuppressive therapy (9).
To the best of our knowledge, the pathophysiological
mechanisms underpinning the stricturing process have not been
clarified. However, some clinical, environmental and endoscopic
parameters have been postulated as risk factors. An age at onset
below 40 years, perianal disease at diagnosis and the need for
steroids during the first flare, together with a history of
smoking, have a positive predictive clinical value for disabling CD
(6). In particular, smoking is a
risk factor for a faster rate of progression from diagnosis to the
first stricture (10). The
endoscopic parameter associated with a higher risk of surgical
intervention is the presence of deep mucosal ulcerations (11). Furthermore, the location of
inflammation in the small bowel rather than the colon has also been
indicated to be predictive of stricturing disease (12).
Some genetic variants have been associated with a
higher risk of progression from an inflammatory, structuring or
penetrating phenotype: The rs4263839 variant in the TNF superfamily
member 15 gene and rs2066847 in nucleotide binding oligomerization
domain containing 2 (NOD2) have been associated with progression to
the B2 or B3 phenotype during 10 years of follow-up (13). The effects of NOD2 and its genetic
variants have been extensively studied in CD, with the results
highlighting their importance in disease progression (14,15).
Rs2066844, rs2066847 and rs2066847 have been associated with
stricturing and penetrating phenotypes, whereas rs2066847 has been
most strongly associated with a severe CD course (14-17).
However, a large genetic association study that included >19,000
patients with CD from 16 countries in Europe, North America and
Australasia did not indicate an association with NOD2 and
stricturing disease after accounting for disease location (18).
The inability to determine which patients are more
prone to develop strictures and which could have a rapidly
progressive disease remains an important knowledge gap. The present
study aimed to identify the gene expression profiles of tissue
samples from stricturing and inflammatory CD to improve the
understanding of the different molecular events implicated in the
two phenotypes.
For this purpose, the present study evaluated the
gene expression profile of a panel of genes previously associated
with inflammatory bowel disease (IBD) in paired mucosa samples of
12 patients with inflammatory CD and 9 patients with stricturing
CD.
Materials and methods
Patients
A total of 21 patients with CD (12 patients with
inflammatory CD and 9 patients with stricturing CD) were enrolled
at the Department of Gastroenterology and Hepatology of Elias
Emergency University Hospital (Bucharest, Romania) and at Fundeni
Clinical Institute (Bucharest, Romania) between May 2016 and
September 2018. The diagnosis was made using the available
guidelines from the European Crohn's and Colitis Organization
(19). For each patient with CD,
paired colonic inflamed mucosa (IM) and non-inflamed mucosa (NIM)
samples were obtained during colonoscopy, based on the endoscopic
mucosal appearance of the same colonic segment. Recorded
demographical data of the patients consisted of age, sex, disease
location based on Montreal classification (2), disease evolution in months and class
of drugs used as treatment at biopsy sampling. The inclusion
criteria were: Diagnosis of CD, age >18 years and active lesions
seen at colonoscopy. The exclusion criteria were: Lack of
inflammatory lesions at colonoscopy and penetrating disease
pattern. Samples for gene expression analysis were collected in
RNAlater and were then processed at the laboratory for dry storage
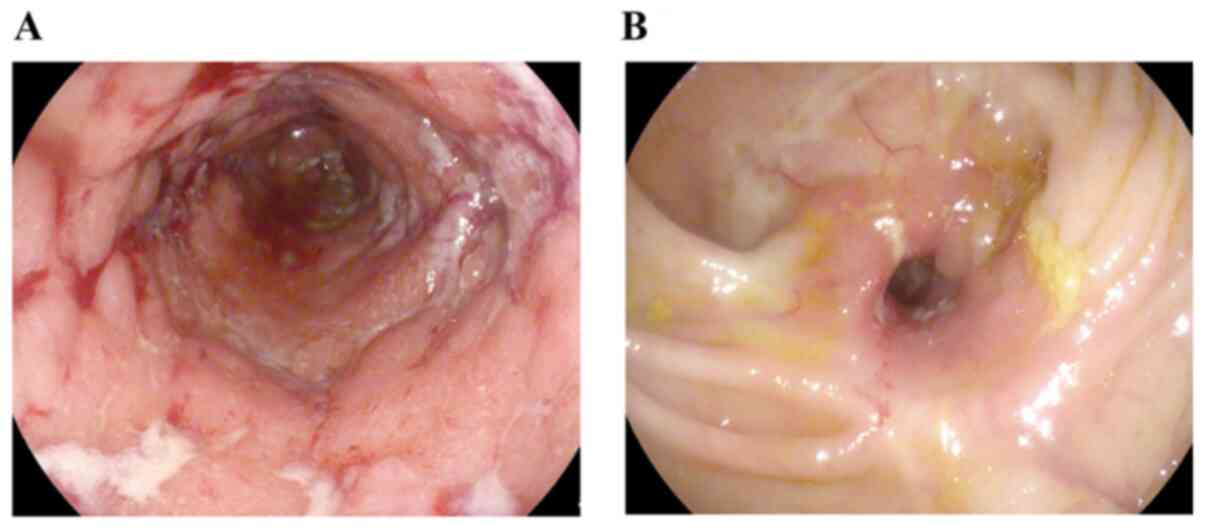
at -80˚C. Endoscopically, IM was characterized by the presence of
erythema, aphthous lesions, cobblestoning or deep ulcers. Stenosis
was defined as the endoscopic appearance of the narrowing of the
luminal caliber (Fig. 1) and was
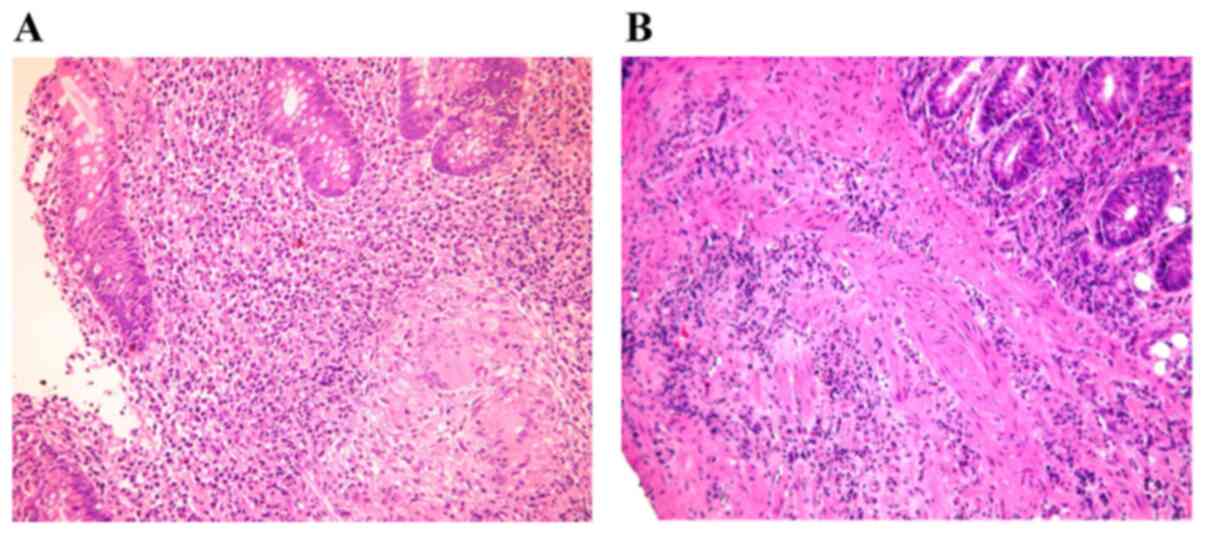
confirmed by computed tomography. Histological evaluation of the
biopsies from the two groups was performed (Fig. 2). The tissue was fixed in formalin
(10%) for 24 h at room temperature using an Excelsior AS Tissue
Processor (Thermo Scientific, Inc.) and embedded in paraffin at
60˚C using the Modular Tissue Embedding center Myr EC350
(Especialidades Médicas Myr, S.L.). Sections with a thickness of 4
µm were stained with Hematoxylin 7211 (Thermo Fisher Scientific,
Inc.) and Eosin-Y (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol (staining at room temperature for 4 and 2.5
min, respectively), using the automatic stainer Gemini AS slide
stainer (Thermo Scientific, Inc.) and examined by a pathologist
using a light microscope DM750 (Leica Microsystems GmbH).
Ethical considerations
All patients included in the present study were of
Romanian origin. The present study was presented to and evaluated
and approved by the Elias Emergency University Hospital Ethics
Committee (registration number 6598; May 11, 2015; Bucharest,
Romania) and by the Fundeni Clinical Institute Ethics Committee
(registration number 8007; February 23, 2018; Bucharest, Romania).
All patients included in the present study signed a written
informed consent form prior to biopsy sampling. The study protocol
conformed to the ethical guidelines of the 1975 Declaration of
Helsinki and its later amendments.
Total RNA isolation and quantitative
PCR
Fresh-frozen tissues were preserved in RNAlater
(Thermo Fisher Scientific, Inc.). Total RNA isolation was performed
using the RNeasy Mini Kit (Qiagen GmbH) according to the
manufacturer's protocols. RNA quality and quantity were assessed
using a NanoDrop 2000 instrument (Thermo Fisher Scientific, Inc.).
RNA (600 ng) was reverse transcribed to cDNA using the RT2 First
Strand Kit (Qiagen GmbH) according to the manufacturer's protocols.
The expression of 84 key genes (Table
SI) was evaluated using the Human CD RT2 Profiler
PCR Array (PAHS-169Z; Qiagen GmbH), as previously reported
(20), using SYBR Green chemistry
(RT2 SYBR Green ROX qPCR Mastermix; Qiagen GmbH) according to the
manufacturer's protocols using the ABI-7500 fast instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.). In the
present study, the 2-ΔΔCq method was used (21). Each ΔCt value indicates the Ct value
of the target gene relative to the geometric mean of the two
housekeeping genes (GAPDH and HPRT1). The two housekeeping genes
were selected as reported by Milanesi et al (22). The ΔΔCt value represents the ΔCt
value of IM compared with NIM. The 2-ΔΔCt values were
calculated for the stricturing and inflammatory CD samples:
ΔΔCt=(Ct, target gene - Ct, HKs)IM-(Ct, target
gene - Ct, HKs)NIM
The fold change (FC) between the two groups (B2 vs.
B1) was further identified as follows: FC=2-ΔΔCt
(B2)/2-ΔΔCt (B1).
Statistical analysis
Statistical analysis was performed using the
Statistical Package for the Social Sciences software (SPSS version
17.0; SPSS, Inc.). The categorical variable sex was tested using
the means of the χ2 test, and the continuous variable
age (mean age in years ± SD) was evaluated using an unpaired
t-test. The non-parametric Mann-Whitney U test was used to assess
the difference in gene expression levels between stricturing and
inflammatory CD. The analysis of gene expression data has been
performed comparing the mean ± SD of the 2-ΔΔCq values.
Since the gene expression array is experimentally validated and
includes many internal controls (see Table SI), no experimental replicates have
been evaluated. Comparisons were considered significant with
P<0.05 and a FC>|3|. The graphs were generated using GraphPad
Prism 8 (GraphPad Software, Inc.).
Results
In the two groups of patients, no statistically
significant difference was found in age (P=0.238) or sex
(χ2=1.311; P=0.252). Notably, more patients in the
stricturing CD group were treated with biologic agents and the
patients in the structuring CD group had a longer disease evolution
from diagnosis; however, this was not statistically significant.
The characteristics of the patients are presented in Table I.
 | Table IClinical and demographic features of
the patients enrolled in the present study. |
Table I
Clinical and demographic features of
the patients enrolled in the present study.
| Parameters | Patients with
stricturing CD (B2; n=9) | Patients with
inflammatory CD (B1; n=12) | P-value |
|---|
| Age, years (mean ±
SD) | 50.2±20.2 | 40.4±12.4 | P=0.238 |
| Age at onset, years
(mean ± SD) | 50.6±21.0 | 35.6±12.2 | P=0.053 |
| Male, % | 80 | 50 | P=0.252;
χ2=1.311 |
| Perianal disease,
% | 0 | 16.7 (n=2) | - |
| Extension (Montreal
score), % | | | |
|
L1 | 44.4 (n=4) | 0 (n=0) | - |
|
L2 | 11.2 (n=1) | 41.7 (n=5) | - |
|
L3 | 44.4 (n=4) | 58.3 (n=7) | - |
| Months of
evolution | 62.2±99.6 | 36.4±45.1 | P=0.434 |
| Medications (at
biopsy acquisition), % | | | |
|
Biological | 33.4 (n=3) | 8.3 (n=1) | - |
|
5-ASA | 22.2 (n=2) | 33.3 (n=4) | - |
|
5-ASA+cortisone | 22.2 (n=2) | 33.3 (n=4) | - |
|
None | 22.2 (n=2) | 25.0 (n=3) | - |
Differential gene expression analysis between
patients with stricturing and inflammatory CD revealed that, in the
mucosa from patients with a stricturing phenotype (B2), 27 genes
were upregulated and six genes were downregulated with an FC>|3|
compared with the inflammatory phenotype (B1; Table II), and many of these were not
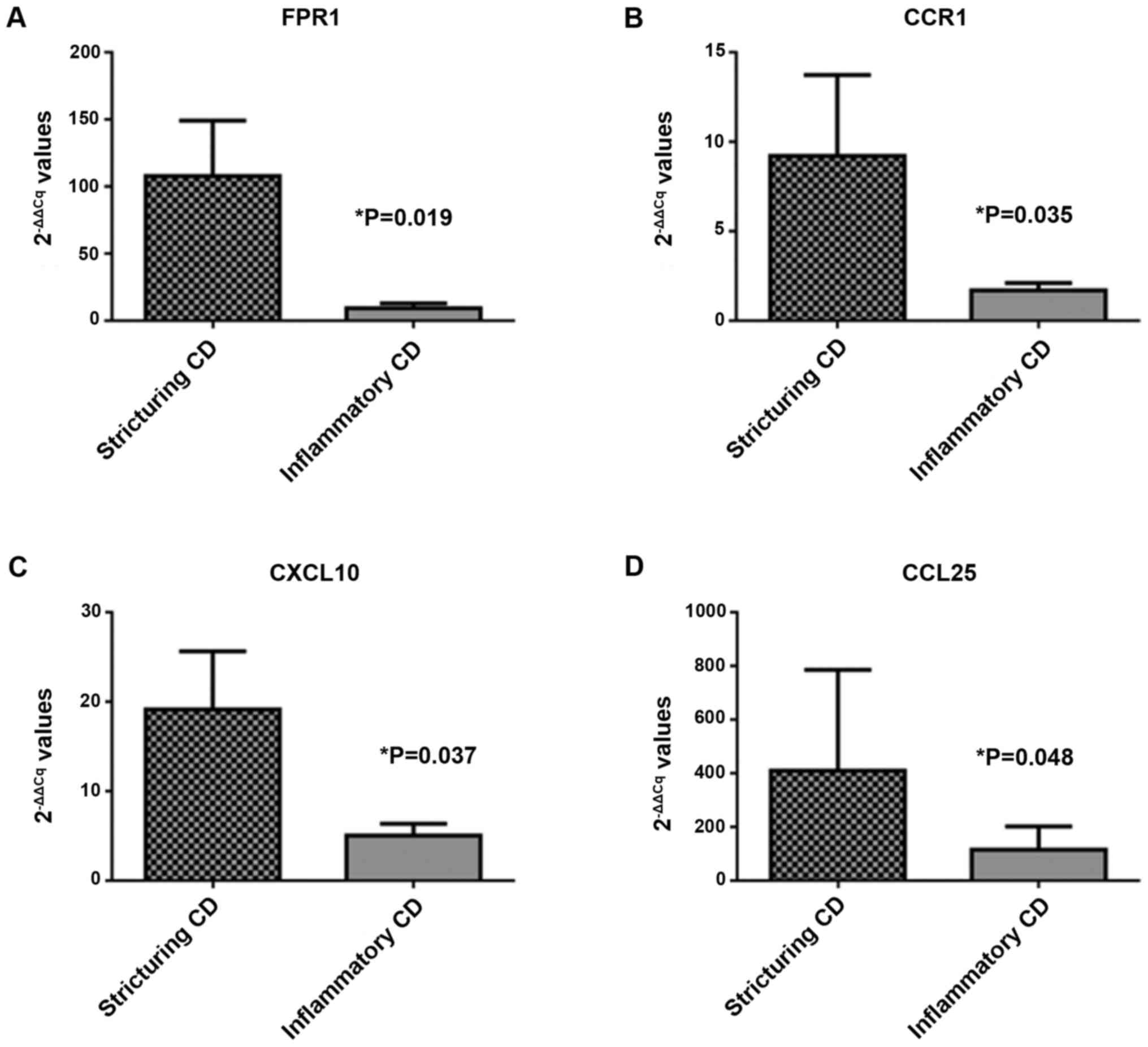
statistically significant (P>0.05). Four transcripts reached
statistical significance (P<0.05): C-C chemokine receptor type 1
(CCR1; P=0.035; FC=5.44), IFN-γ-inducible protein 10 (CXCL10;
P=0.037; FC=3.81), C-C chemokine ligand 25 (CCL25; P=0.048;
FC=3.56) and formyl-peptide receptor 1 (FPR1; P=0.019; FC=11.61;
Fig. 3).
 | Table IIDifferential expression of genes in
patients with stricturing (B2) and inflammatory (B1) Crohn's
disease. |
Table II
Differential expression of genes in
patients with stricturing (B2) and inflammatory (B1) Crohn's
disease.
| Gene | Fold change (B2 vs.
B1) | P-value |
|---|
| FPR1 | 11.61 | 0.019 |
| CCR1 | 5.44 | 0.035 |
| CXCL10 | 3.81 | 0.037 |
| CCL25 | 3.56 | 0.048 |
| MUC1 | -7.68 | 0.082 |
| VWF | 3.25 | 0.082 |
| IFNG | 3.61 | 0.104 |
| S100A9 | 5.84 | 0.104 |
| EGR3 | 7.71 | 0.130 |
| IL2RA | 4.98 | 0.130 |
| MMP1 | 116.34 | 0.195 |
| MMP3 | 35.51 | 0.195 |
| TNF | 3.39 | 0.195 |
| ALDOB | 21.26 | 0.234 |
| CXCR1 | 7.18 | 0.234 |
| CXCL8 | 17.47 | 0.234 |
| S100A8 | 9.67 | 0.234 |
| IL1RN | 4.67 | 0.279 |
| CHI3L1 | 4.35 | 0.328 |
| TDO2 | 13.83 | 0.328 |
| CXCL12 | 3.83 | 0.383 |
| SELL | 19.66 | 0.383 |
| CSTA | 3.19 | 0.442 |
| SELE | 13.67 | 0.442 |
| MMP7 | 6.59 | 0.506 |
| IL6 | 12.65 | 0.560 |
| REG1B | -4.80 | 0.574 |
| IL1B | 9.70 | 0.646 |
| PCK1 | -5.36 | 0.646 |
| CCL2 | 5.01 | 0.721 |
| DEFA5 | -5.80 | 0.721 |
| REG1A | -13.30 | 0.799 |
| SAA1 | -7.08 | 0.879 |
Discussion
The present study aimed to identify the differences
in gene expression profiles between inflammatory and stricturing CD
phenotypes. For this purpose, the present study analyzed paired
mucosal samples from 9 patients with B2 CD and 12 patients with
B1/B1p CD. Four transcripts, namely CCR1, CXCL10, CCL25 and FPR1,
with significantly higher expression levels in stricturing than in
inflammatory CD mucosa samples were identified.
The main limitation of the present study was the low
number of patients with the stricturing phenotype. Furthermore,
disease duration may also contribute to a change in mucosal
inflammatory pathways, affecting gene expression profiles.
Regarding the possible effect of biological treatment (more
prominent in the stricturing phenotype group) on the gene
expression levels, according to our previous results (23), the anti-TNF medications did not
affect the levels of the differentially expressed genes identified
in the present study. Nonetheless, these findings may be important
for further research in the field of IBD, and each transcript will
be addressed further.
CXCL10, CCL25 and CCR1 belong to the chemokine
signaling pathway and serve as chemokines and chemokine receptors.
Chemokines are key actors in directing the balance between
physiological and pathophysiological inflammation in the
gastrointestinal mucosa (24). In
IBD, chemokines attract immune cells to inflamed and
epithelial-damaged sites, and a majority of chemokines have been
reported to be elevated in the mucosa of IBD (24).
The CCR1 gene encodes CCR1, which belongs to the G
protein-coupled receptors. These receptors interact with C-C motif
chemokine ligand 3 (also referred to as macrophage inflammatory
protein-1α), C-C motif chemokine ligand 5 or regulated on
activation normal T expressed and secreted protein, C-C motif
chemokine ligand 7 or monocyte chemoattractant protein-3, and C-C
motif chemokine ligand 23 or myeloid progenitor inhibitory
factor-1(25). The first
association between CCR1 and chronic bowel inflammation was
revealed in animal models and was described by Ajuebor et al
(24) in 2001. This study
demonstrated increased mRNA levels in colonic tissue during the
chronic phase of colitis (>7 days after induced colitis)
(24).
Recently, a Slovakian study revealed an increase in
CCR1 mRNA expression in IM samples from patients with ulcerative
colitis (UC) and CD compared with non-inflamed mucosal samples
(26). Our previous results
suggested that CCR1 may be a marker of molecular activity in CD
(27). In the present study,
increased CCR1 gene expression was observed in stricturing CD
compared with inflammatory CD. This suggested that CCR1 may be
implicated in disease progression and tissue destruction.
CXCL10 and CCL25 are two chemokines involved in
T-cell recruitment, and thus, serve an important role in regulating
the gastrointestinal immune response and enabling mucosal
inflammation (28).
CXCL10 is secreted by immune cells, such as T-cells,
B-cells, NK cells and myeloid cells (28). It targets the C-X-C motif chemokine
receptor 3 (CXCR3) receptor (28).
It can be detected in blood, normal colonic mucosa and tissues with
active inflammation, including IBD (28). The activation of CXCR3 upregulates
CXCL10, which is expressed by non-immune cells, such as epithelial
cells and fibroblasts (29).
Inflammation induced by interferon γ, tumor necrosis factor α and
interleukin-1β can further increase CXCL10 expression (30). CCL25, which is the only C-C motif
chemokine receptor 9 ligand, was considered to be exclusively
expressed in the lamina propria by mononuclear cells of the small
intestine and the thymus for years (31). However, recent data have extended
these observations, revealing its expression in the colon and liver
(32).
Studies examining the roles of CXCL10 and CXCR3 in
IBD continue to find novel advancements. Plasmatic levels of CXCL10
are markedly increased in patients with IBD compared with non-IBD
controls (33). In patients with
CD, CXCL10 serum levels remain upregulated during flares and
remission (34). Furthermore,
higher expression levels of CXCL10 and CXCR3 have been observed in
multiple studies comparing active IBD mucosa with mucosa from
non-IBD controls (34,35).
CCL25 gene expression is positively associated with
both the Mayo endoscopic subscore and mucosal TNFα levels (36). The elevated serum levels of this
chemokine have also been reported in patients with UC (28). To the best of our knowledge, the
present study was the first to report higher gene expression levels
of CXCL10 and CCL25 in stricturing CD, suggesting the presence of
more prominent inflammatory stimuli in these patients compared with
in patients with purely inflammatory CD.
FPR1 was the most significantly upregulated gene in
stricturing CD found in the present study. It encodes a G-coupled
protein receptor and serves a crucial role in inflammation, as it
is a prominent catalyst for neutrophil sensing and chemoattraction
(37). FPR1 was the first member
described in this family (38).
Upon activation, it regulates multiple functions, such as
chemotaxis, degranulation, reactive oxygen species synthesis and
phagocytosis (39). The main
ligands for FPR1 are formylated peptides of both mitochondrial and
bacterial origin secreted by invading pathogens or released from
apoptotic cells (40).
Data regarding FPR families in murine models has
revealed their role in rapid neutrophil mobilization, in which
neutrophil infiltration augmented wound-healing capacities in
sterile skin lesions (41). In the
intestinal lumen, in which the number of N-formyl peptides is
elevated due to the bacterial burden, the polymorphonuclear
cell-mediated response is important (39). Increased intramucosal levels of
these peptides, possibly as a consequence of epithelial barrier
damage, can contribute to the activation of FPR1 and the
development of intestinal crypt abscesses in acute and chronic IBD
(34). In the present study,
increased FPR1 gene expression was noted in stricturing CD compared
with the inflammatory phenotype. Consequently, this puts emphasis
on data that implicate FPR1 in wound healing. To the best of our
knowledge, the present study was the first to report a link between
FPR1 and CD in humans.
In conclusion, the present study revealed four
transcripts that were differentially expressed between inflammatory
and stricturing CD. CCR1, CXCL10, CCL25 and FPR1 were all
significantly upregulated in stricturing CD. Despite the low number
of patients analyzed, the present preliminary results are
promising. Future studies on the transcriptional profiling of CD
may offer vital insights into the mechanisms of stricture formation
in CD and aid in predicting disease evolution and stricturing
complications.
Supplementary Material
Gene table of the Human Crohn's
Disease RT2 Profiler PCR Array (96-well plates).
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Ministry of
Education and Research in Romania (grant nos. 7PFE/16.10.2018 and
PN 1N/2019_19.29.01.05).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CGT and AOO confirm the authenticity of all the raw
data. CGT and AOO contributed to conception and design, data
acquisition, drafting and revising of the manuscript. CMP, NB, EM
and MD contributed to data acquisition, analysis and
interpretation. TEM, GB and IT contributed to data acquisition and
drafting of the manuscript. AK and EMI have been involved in
conception and design of the study, drafting the manuscript and
revising it critically for important intellectual content. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Elias
Emergency University Hospital Ethics Committee (registration number
6598; May 11, 2015; Bucharest, Romania) and the Fundeni Clinical
Institute Ethics Committee (registration number 8007; February 23,
2018; Bucharest, Romania). All patients included in the present
study signed a written informed consent form on enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bettenworth D, Nowacki TM, Cordes F,
Buerke B and Lenze F: Assessment of stricturing Crohn's disease:
Current clinical practice and future avenues. World J
Gastroenterol. 22:1008–1016. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Satsangi J, Silverberg MS, Vermeire S and
Colombel JF: The Montreal classification of inflammatory bowel
disease: Controversies, consensus, and implications. Gut.
55:749–753. 2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Freeman HJ: Natural history and long-term
clinical course of Crohn's disease. World J Gastroenterol.
20:31–36. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cosnes J, Gower-Rousseau C, Seksik P and
Cortot A: Epidemiology and natural history of inflammatory bowel
diseases. Gastroenterology. 140:1785–1794. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Cosnes J, Cattan S, Blain A, Beaugerie L,
Carbonnel F, Parc R and Gendre JP: Long-term evolution of disease
behavior of Crohn's disease. Inflamm Bowel Dis. 8:244–250.
2002.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Solberg IC, Vatn MH, Høie O, Stray N,
Sauar J, Jahnsen J, Moum B and Lygren I: IBSEN Study Group.
Clinical course in Crohn's disease: Results of a norwegian
population-based ten-year follow-up study. Clin Gastroenterol
Hepatol. 5:1430–1438. 2007.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Louis E, Collard A, Oger AF, Degroote E,
Aboul Nasr El Yafi F and Belaiche J: Behaviour of Crohn's disease
according to the Vienna classification: Changing pattern over the
course of the disease. Gut. 49:777–782. 2001.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Frolkis AD, Lipton DS, Fiest KM, Negrón
ME, Dykeman J, deBruyn J, Jette N, Frolkis T, Rezaie A, Seow CH, et
al: Cumulative incidence of second intestinal resection in Crohn's
disease: A systematic review and meta-analysis of population-based
studies. Am J Gastroenterol. 109:1739–1748. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Cosnes J, Nion-Larmurier I, Beaugerie L,
Afchain P, Tiret E and Gendre JP: Impact of the increasing use of
immunosuppressants in Crohn's disease on the need for intestinal
surgery. Gut. 54:237–241. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Louis E, Michel V, Hugot JP, Reenaers C,
Fontaine F, Delforge M, El Yafi F, Colombel JF and Belaiche J:
Early development of stricturing or penetrating pattern in Crohn's
disease is influenced by disease location, number of flares, and
smoking but not by NOD2/CARD15 genotype. Gut. 52:552–557.
2003.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Allez M and Lémann M: Role of endoscopy in
predicting the disease course in inflammatory bowel disease. World
J Gastroenterol. 16:2626–32. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Pernat Drobež C, Ferkolj I, Potočnik U and
Repnik K: Crohn's disease candidate gene alleles predict time to
progression from inflammatory B1 to stricturing B2, or penetrating
B3 phenotype. Genet Test Mol Biomarkers. 22:143–151.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Adler J, Rangwalla SC, Dwamena BA and
Higgins PD: The prognostic power of the NOD2 genotype for
complicated Crohn's disease: A meta-analysis. Am J Gastroenterol.
106:699–712. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tolentino YF, Elia PP, Fogaça HS, Carneiro
AJ, Zaltman C, Moura-Neto R, Luiz RR, Carvalho Mda G and de Souza
HS: Common NOD2/CARD15 and TLR4 polymorphisms are associated with
Crohn's disease phenotypes in southeastern brazilians. Dig Dis Sci.
61:2636–2647. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cleynen I, González JR, Figueroa C, Franke
A, McGovern D, Bortlík M, Crusius BJ, Vecchi M, Artieda M,
Szczypiorska M, et al: Genetic factors conferring an increased
susceptibility to develop Crohn's disease also influence disease
phenotype: Results from the IBDchip European project. Gut.
62:1556–1565. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Schnitzler F, Friedrich M, Wolf C,
Angelberger M, Diegelmann J, Olszak T, Beigel F, Tillack C,
Stallhofer J, Göke B, et al: The NOD2 p.Leu1007fsX1008 mutation
(rs2066847) is a stronger predictor of the clinical course of
Crohn's disease than the FOXO3A intron variant rs12212067. PLoS
One. 9(e108503)2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cleynen I, Boucher G, Jostins L, Schumm
LP, Zeissig S, Ahmad T, Andersen V, Andrews JM, Annese V, Brand S,
et al: Inherited determinants of Crohn's disease and ulcerative
colitis phenotypes: A genetic association study. Lancet.
387:156–167. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Magro F, Gionchetti P, Eliakim R,
Ardizzone S, Armuzzi A, Barreiro-de Acosta M, Burisch J, Gecse KB,
Hart AL, Hindryckx P, et al: Third European evidence-based
consensus on diagnosis and management of ulcerative colitis. Part
1: Definitions, diagnosis, extra-intestinal manifestations,
pregnancy, cancer surveillance, surgery, and ileo-anal pouch
disorders. J Crohns Colitis. 11:649–670. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ţieranu CG, Dobre M, Mănuc TE, Milanesi E,
Pleşea IE, Popa C, Mănuc M, Ţieranu I, Preda CM, Diculescu MM, et
al: Gene expression profile of endoscopically active and inactive
ulcerative colitis: Preliminary data. Rom J Morphol Embryol.
58:1301–1307. 2017.PubMed/NCBI
|
|
20
|
Dobre M, Milanesi E, Mănuc TE, Arsene DE,
Ţieranu CG, Maj C, Becheanu G and Mănuc M: Differential intestinal
mucosa transcriptomic biomarkers for Crohn's disease and ulcerative
colitis. J Immunol Res. 2018(9208274)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Milanesi E, Dobre M, Manuc T, Becheanu G,
Tieranu CG, Ionescu EM and Manuc M: Mucosal gene expression changes
induced by anti-TNF treatment in inflammatory bowel disease
patients. Drug Dev Res. 80:831–836. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Grimm MC and Doe WF: Chemokines in
inflammatory bowel disease Mucosa: Expression of RANTES, Macrophage
Inflammatory Protein (MIP)-1α, MIP-1β, and γ-Interferon-Inducible
Protein-10 by Macrophages, Lymphocytes, Endothelial Cells, and
Granulomas. Inflamm Bowel Dis. 2:88–96. 1996.PubMed/NCBI
|
|
24
|
Ajuebor MN, Hogaboam CM, Kunkel SL,
Proudfoot AE and Wallace JL: The chemokine RANTES is a crucial
mediator of the progression from acute to chronic colitis in the
rat. J Immunol. 166:552–558. 2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kristensen NN, Olsen J, Gad M and Claesson
MH: Genome-wide expression profiling during protection from colitis
by regulatory T cells. Inflamm Bowel Dis. 14:75–87. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mayer L, Sandborn WJ, Stepanov Y, Geboes
K, Hardi R, Yellin M, Tao X, Xu LA, Salter-Cid L, Gujrathi S, et
al: Anti-IP-10 antibody (BMS-936557) for ulcerative colitis: A
phase II randomised study. Gut. 63:442–450. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dobre M, Manuc T, Milanesi E, Pleşea IE,
Ţieranu EN, Popa C, Mănuc M, Preda CM, Ţieranu I, Diculescu MM, et
al: Mucosal CCR1 gene expression as a marker of molecular activity
in Crohn's disease: Preliminary data. Rom J Morphol Embriol.
58:1263–1268. 2017.PubMed/NCBI
|
|
28
|
Trivedi PJ, Bruns T, Ward S, Mai M,
Schmidt C, Hirschfield GM, Weston CJ and Adams DH: Intestinal CCL25
expression is increased in colitis and correlates with inflammatory
activity. J Autoimmun. 68:98–104. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Singh UP, Venkataraman C, Singh R and
Lillard JW Jr: CXCR3 Axis: Role in inflammatory bowel disease and
its therapeutic implication. Endocrine Metab Immune Disord Targets.
7:111–123. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Singh UP, Singh NP, Murphy EA, Price RL,
Fayad R, Nagarkatti M and Nagarkatti PS: Chemokine and cytokine
levels in inflammatory bowel disease patients. Cytokine. 77:44–49.
2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Svensson M and Agace WW: Role of
CCL25/CCR9 in immune homeostasis and disease. Expert Rev Clin
Immunol. 2:759–773. 2006.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Vasilyeva E, Abdulkhakov S, Cherepnev G,
Martynova E, Mayanskaya I, Valeeva A, Abdulkhakov R, Safina D,
Khaiboullina S and Rizvanov A: Serum cytokine profiles in children
with Crohn's disease. Mediators Inflamm.
2016(7420127)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hosomi S, Oshitani N, Kamata N, Sogawa M,
Okazaki H, Tanigawa T, Yamagami H, Watanabe K, Tominaga K, Watanabe
T, et al: Increased numbers of immature plasma cells in peripheral
blood specifically overexpress chemokine receptor CXCR3 and CXCR4
in patients with ulcerative colitis. Clin Exp Immunol. 163:215–224.
2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Østvik AE, Granlund AV, Torp SH, Flatberg
A, Beisvåg V, Waldum HL, Flo TH, Espevik T, Damås JK and Sandvik
AK: Expression of Toll-like receptor-3 is enhanced in active
inflammatory bowel disease and mediates the excessive release of
lipocalin 2. Clin Exp Immunol. 173:502–511. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Uguccioni M, Gionchetti P, Robbiani DF,
Rizzello F, Peruzzo S, Campieri M and Baggiolini M: Increased
expression of IP-10, IL-8, MCP-1, and MCP-3 in Ulcerative Colitis.
Am J Pathol. 155:331–336. 1999.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kolaczkowska E and Kubes P: Neutrophil
recruitment and function in health and inflammation. Nat Rev
Immunol. 13:159–175. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Le Y, Ye RD, Gong W, Li J, Iribarren P and
Wang JM: Identification of functional domains in the formyl peptide
receptor-like 1 for agonist-induced cell chemotaxis. FEBS J.
272:769–778. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Boulay F, Tardif M, Brouchon L and Vignais
P: Synthesis and use of a novel N-formyl peptide derivative to
isolate a human N-formyl peptide receptor cDNA. Biochem Biophys Res
Commun. 168:1103–1109. 1990.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Dorward DA, Lucas CD, Chapman GB, Haslett
C, Dhaliwal K and Rossi AG: The role of formylated peptides and
formyl peptide receptor 1 in governing neutrophil function during
acute inflammation. Am J Pathol. 185:1172–1184. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Leoni G, Gripentrog J, Lord C, Riesselman
M, Sumagin R, Parkos CA, Nusrat A and Jesaitis AJ: Human neutrophil
formyl peptide receptor phosphorylation and the mucosal
inflammatory response. J Leukoc Biol. 97:87–101. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
VanCompernolle SE, Clark KL, Rummel KA and
Todd SC: Expression and function of formyl peptide receptors on
human fibroblast cells. J Immunol. 171:2050–2056. 2003.PubMed/NCBI View Article : Google Scholar
|