Introduction
Hyperlipidemia (HLP) is a common and frequently
encountered metabolic disease characterized by abnormally elevated
total cholesterol (TC) and/or triglyceride (TG) levels in the
plasma, resulting in dormant, progressive and serious damage to
various organs, particularly the liver (1). The liver is the major site for
intermediary metabolism and the major organ for the synthesis and
metabolism of endogenous blood lipids and lipoproteins.
Extrahepatic cholesterol is mainly carried by high-density
lipoprotein (HDL) and transported to the liver via the HDL receptor
for metabolism (2). Infiltration of
liver tissues by lipids, including TG, leads to hepatic
degeneration and may cause dysregulation of lipid metabolism and
lipoprotein synthesis, particularly reduced HDL synthesis and
defective synthesis of very low-density lipoprotein (VLDL) in the
liver (3). As a result, hepatic
lipids cannot be exported via lipoproteins, leading to the
accumulation of TC and TG in the liver, ultimately resulting in
hepatic insufficiency and dysregulation of lipid metabolism
(4). Eventually, over-accumulation
of lipids caused by dysregulated lipid metabolism results in HLP.
Despite the progress in elucidating the pathogenesis of HLP, the
underlying molecular mechanisms have remained to be fully
clarified.
Although 98% of the DNA sequences of the human
genome have been transcribed, protein-coding genes account for
<2% of the genome. The remaining genomic sequences are
transcribed into non-coding RNAs (ncRNAs) (5). Based on their length, ncRNAs are
categorized into two classes: ncRNAs smaller than 200 bp are
usually referred to as small ncRNAs, including small interfering
RNA, microRNA and piWi-interacting RNA, whereas long ncRNAs
(lncRNAs) are defined as transcripts longer than 200 bp (6). lncRNAs have important roles in various
critical biological processes at the transcription level, including
genomic imprinting, chromatin inactivation, differentiation and
carcinogenesis (7). Furthermore,
accumulating evidence indicates that lncRNAs are closely related to
the development and progression of cardiovascular and myocardial
infarction (8,9). For instance, lncRNAs are involved in
the adipogenesis of white adipose tissue, energy metabolism of
brown adipocytes (10) and
accumulation of lipid droplets (11). Furthermore, myosin
heavy-chain-associated RNA, an lncRNA abundant in adult hearts, has
been suggested to protect the heart from cardiac hypertrophy and
heart failure (12). However, the
expression profile and biological function of lncRNAs in HLP remain
elusive.
As a ligand, apolipoprotein E (ApoE) is able to
contribute to the clearance of chylomicrons and VLDL and the lack
of ApoE may lead to lipoprotein metabolism disorder and predispose
an individual to lipoprotein deposition in the arterial wall. The
pathological development of atherosclerosis in ApoE knockout mice
is highly similar to that in humans and ApoE knockout mice are
prone to developing severe hyperlipidemia.
Therefore, in the present study, a microarray
analysis of the liver tissue of a model of HLP using ApoE knockout
(ApoE-/-) mice and from control mice was performed using
the Mouse lncRNA Array v1.0, 4x180K (CapitalBio Technology, Inc.,)
to identify differentially expressed lncRNAs and mRNAs.
Bioinformatics tools were used to functionally annotate the
differentially expressed genes (DEGs) and identify related
transcripts. Collectively, these results provide novel insight into
the molecular mechanisms of HLP and suggest novel therapeutic
targets for future research.
Materials and methods
Animal model
A total of 12 male 6-week-old C57BL/6J mice (weight,
23.0±0.74 g) and 12 male ApoE-/- (C57BL/6J) mice
(weight, 23.0±0.57 g) were purchased from Beijing Vital River
Laboratory Animal Technology. After 7 days of acclimation to the
specific pathogen-free environment at Henan University of Chinese
Medicine Laboratory Animal Center (Zhengzhou, China), the animals
were assigned to two groups, namely a Normal control group (n=12)
and an HLP group (n=12). The normal control group consisted of
wild-type C57BL/6J mice with a regular mouse diet (65.08%
carbohydrates, 11.85% fat, and 23.07% protein). In the HLP group
six-week-old male ApoE-/- mice were fed with a high-fat
diet (HFD) containing 2.5% cholesterol and 15% fat. HFD was
purchased from Beijing HFK Bioscience. The animals were housed in a
temperature-controlled room (22-25˚C, 45% humidity) with a 12-h
light/dark cycle for 16 weeks. Animal behavior and health were
monitored every day. On the day tissue and blood collection were to
be performed, mice were fasted beginning at 9 am for a period of 5
h and blood collection procedures were initiated at 2 pm. A total
of 600-800 µl blood was collected from each mouse. Blood from all
the 24 mice was collected via retro-orbital bleeding under
anesthesia by intraperitoneal injection of sodium pentobarbital (50
mg/kg body weight). The anesthesia state of the mice was observed;
if the mice had movement or pain, a supplementary dose of 50 mg/kg
(to reach 100 mg/kg in total) was added (13). After blood collection, the 24 mice
were euthanized with an overdose (150 mg/kg in total) of
pentobarbital. A combination of criteria was most reliable in
confirming death, including lack of pulse, breathing, corneal
reflex and response to firm toe pinch, inability to hear
respiratory sounds and heartbeat by use of a stethoscope, graying
of the mucous membranes and rigor mortis (14). Once euthanasia was performed, the
liver tissues of the mice were immediately collected and stored in
liquid nitrogen for further analysis. All of the procedures
followed the requirements of the Ethics Committee of Henan
University of Chinese Medicine (ethics review approval code no.
DWLL201704101).
Blood lipid measurement and H&E
staining
Blood was collected from mice fasted from 9 am to 2
pm and the serum was separated by centrifugation at 5,000 x g for 5
min at 22±2˚C. The mice were anesthetized for the collection of
blood and liver samples. All samples were stored in liquid nitrogen
for the subsequent analyses. An automatic biochemistry analyzer
(Siemens AG) was used for quantifying lipid concentrations,
including TG, TC and LDL cholesterol (LDL-C) in the serum.
Formalin-fixed liver tissues were embedded in paraffin and sections
(4 µm) were cut and stained with H&E (Biyuntian).
RNA extraction, labeling and
hybridization
Total RNA was extracted from homogenized liver
tissues using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. cDNA
labeled with a fluorescent dye (cyanine 3-dCTP) was produced by
Eberwine's linear RNA amplification method and subsequent enzymatic
reaction. Double-stranded cDNAs (containing the T7 RNA polymerase
promoter sequence) were synthesized from 1 µg total RNA using the
CbcScript reverse transcriptase with cDNA synthesis system
according to the manufacturer's protocol (CapitalBio Technology,
Inc.) with the T7 Oligo(dT) and T7 Oligo(dN). After completing
double-stranded cDNA (dsDNA) synthesis using DNA polymerase and
RNase H (MACHEREY-NAGEL GmbH & Co. KG), the dsDNA products were
purified using a PCR NucleoSpin Extract II Kit (MACHEREY-NAGEL GmbH
& Co. KG). 14 µl DNA was denatured in hybridization solution
(Agilent Technologies, Inc.) at 95˚C for 3 min prior to loading
onto a microarray (Mouse lncRNA Array v1.0, 4x180K; CapitalBio
Technology, Inc.). Arrays were hybridized in a hybridization oven
(Agilent Technologies, Inc.) overnight at a rotation speed of 20
rpm and a temperature of 42˚C and washed with two consecutive
solutions (0.2% SDS, 2X SSC at 42˚C for 5 min and 0.2X SSC at
23-26˚C for 5 min).
Microarray imaging and data
analysis
GeneSpring v13.0 software (Agilent Technologies,
Inc.) was used to analyze lncRNA and mRNA microarray data and
perform summarization, normalization and quality control. To
identify DEGs, the threshold for the fold-change was set at ≥2 or
≤-2 and a Benjamini-Hochberg-adjusted P-value of 0.05 was set as
the cut-off for statistically significant results. Log2
transformation of the data was performed using CLUSTER 3.0 software
(http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm)
and the genes were centered, followed by average-linkage
hierarchical clustering analysis. The data were visualized using
Java Treeview 1.1.6 (Stanford University School of Medicine).
Functional group analysis
Gene Ontology (GO) analysis was performed to explore
the functions of DEGs identified in the present study. The Database
for Visualization and Integrated Discovery (DAVID) online tool
(https://david.ncifcrf.gov/) was utilized
to perform functional enrichment analysis of DEGs, which
classified, identified and annotated the genes in terms of the GO
categories of biological process, cellular component and molecular
function. The P-values were adjusted using the false discovery rate
and the threshold for significant differences was set at
P<0.05.
Coding-non-coding gene co-expression
network analysis
The coding-non-coding gene co-expression network was
constructed based on the correlation analysis among differentially
expressed lncRNAs and mRNAs. For each pair of genes, the Pearson's
correlation coefficient was determined and pairs with a significant
correlation were selected to construct the network. The lncRNAs and
mRNAs with Pearson's correlation coefficients of no <0.99 were
incorporated into the network using the bioinformatics software
Cytoscape 3.6.1 (https://cytoscape.org/download.html).
Prediction of target genes
The cis-acting lncRNA prediction of a group of
expressed protein-coding genes was performed based on the close
association of lncRNA-mRNA pairs (minimum Pearson's correlation
coefficient of 0.99). Trans-prediction was performed using BLAT
tools (Standalone BLAT v.35x1 fast sequence search command line
tool downloaded from http://hgdownload.cse.ucsc.edu/admin/exe/) to compare
the full sequence of an lncRNA with the 3' untranslated region of
its co-expressed mRNA using default parameter settings.
Prediction of transcription
factors
The transcription factor prediction database TFactS
was downloaded from http://www.tfacts.org. Based on the co-expression data
of differentially expressed lncRNAs and mRNAs, the differentially
expressed mRNAs were compared with those in the database and the
correlations of co-expressed lncRNAs and mRNAs with the
transcription factors were obtained. The regulatory network
diagrams of co-expressed lncRNAs and mRNAs with transcription
factors were drawn using Cytoscape 3.6.1.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 5.0 Software (GraphPad Software, Inc.). Data were analyzed
for differences between the Normal control group and the HLP group
using an unpaired Student's t-test. Correlation analysis was
performed using Pearson's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Bodyweight and serum lipid level
variations in the HLP model
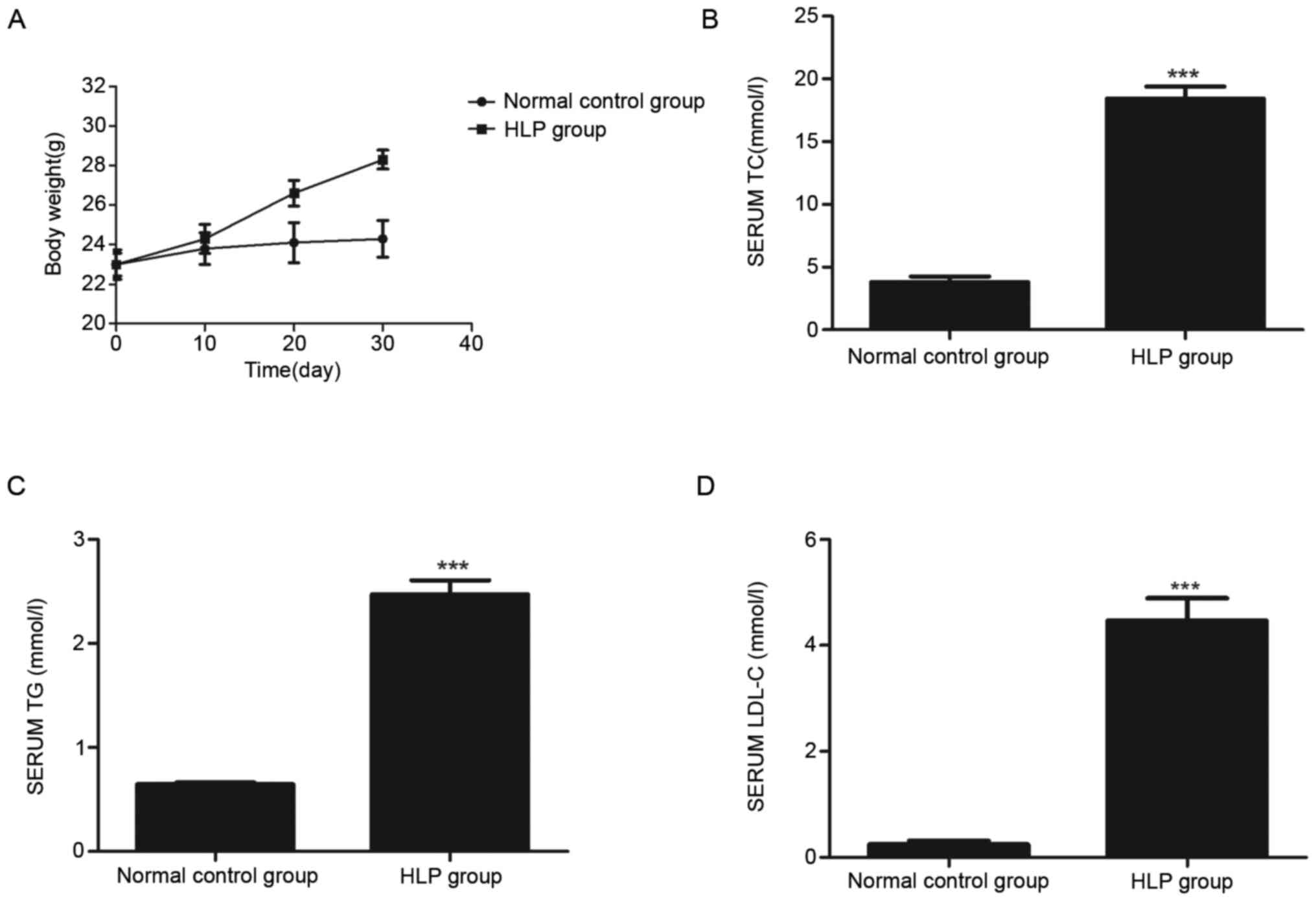
Fig. 1A presents the
measured bodyweights of mice in the Normal control group and HLP
groups at days 10, 20 and 30 during continuous feeding of the
regular and high-fat diet, respectively. Compared with the Normal
control, the bodyweight of the HLP group was significantly
increased. The plasma lipid levels suggested that the serum TC
(Fig. 1B), TG (Fig. 1C) and LDL-C (Fig. 1D) levels in the HLP group were
substantially higher than those in the Normal control group.
Histology
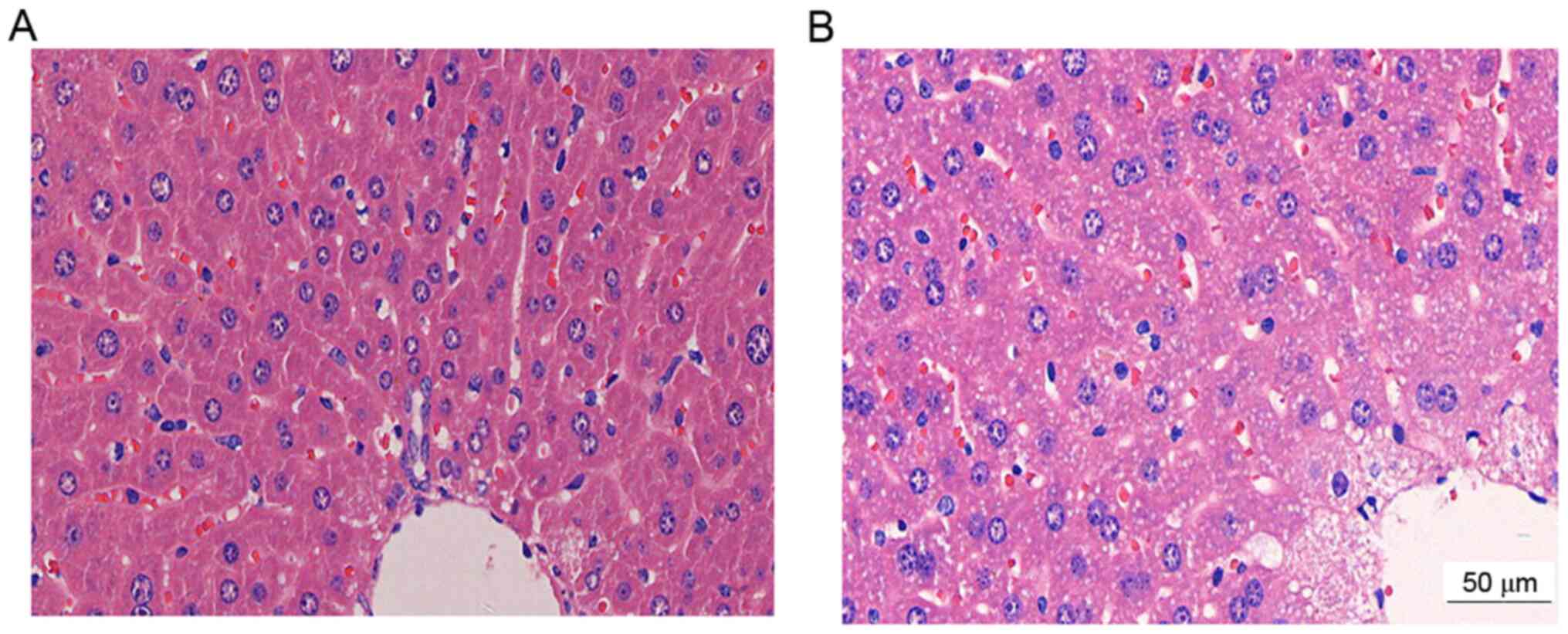
H&E staining revealed that the cell nuclei of
liver sections in the Normal control group were relatively large in
size and were polyhedral, with round nuclei deeply stained blue.
The nucleoli were clearly observed at the center of the cells. The
hepatocytes were intact and tightly packed in cords, with uniform
cytoplasm and without swelling or steatosis. The hepatic lobules
exhibited a clear edge and normal morphology. The hepatic cords
were arranged in an orderly manner displaying a radial pattern and
the central vein exhibited a normal structure (Fig. 2A). By contrast, the cell morphology
in the HLP group was clearly anomalous. The hepatocytes were
swollen and increased in size and the cytoplasm appeared cloudy. A
certain amount of inflammatory cells were observed at the periphery
and lipid vacuoles of various sizes were observed in the cytoplasm
of the hepatocytes. The lobule structure was damaged and vague
(Fig. 2B).
Differentially expressed lncRNAs and
mRNAs
A total of 96 differentially expressed mRNAs were
identified from liver samples obtained from the animals of the HLP
model and Normal control groups. Compared to those in the Normal
control, 58 mRNAs were upregulated and 38 were downregulated in the
HLP model. Upregulated genes included those encoding
carcinoembryonic antigen-related cell adhesion molecule 2
(Ceacam2), a transmembrane glycoprotein expressed in the kidney,
spleen, platelets and crypt epithelial cells, and epidermal growth
factor receptor (EGFR), an important transmembrane receptor
implicated in multiple cancer types. In addition, 104
differentially expressed lncRNAs were identified, including 46
upregulated and 58 downregulated lncRNAs in the HLP group compared
to those in the Normal control. For instance, the expression of
NONMMUT008659 was upregulated, whereas that of
ri|0610030O19|R000004O21|1299 was downregulated.
Clustering analysis of differentially
expressed lncRNAs and mRNAs
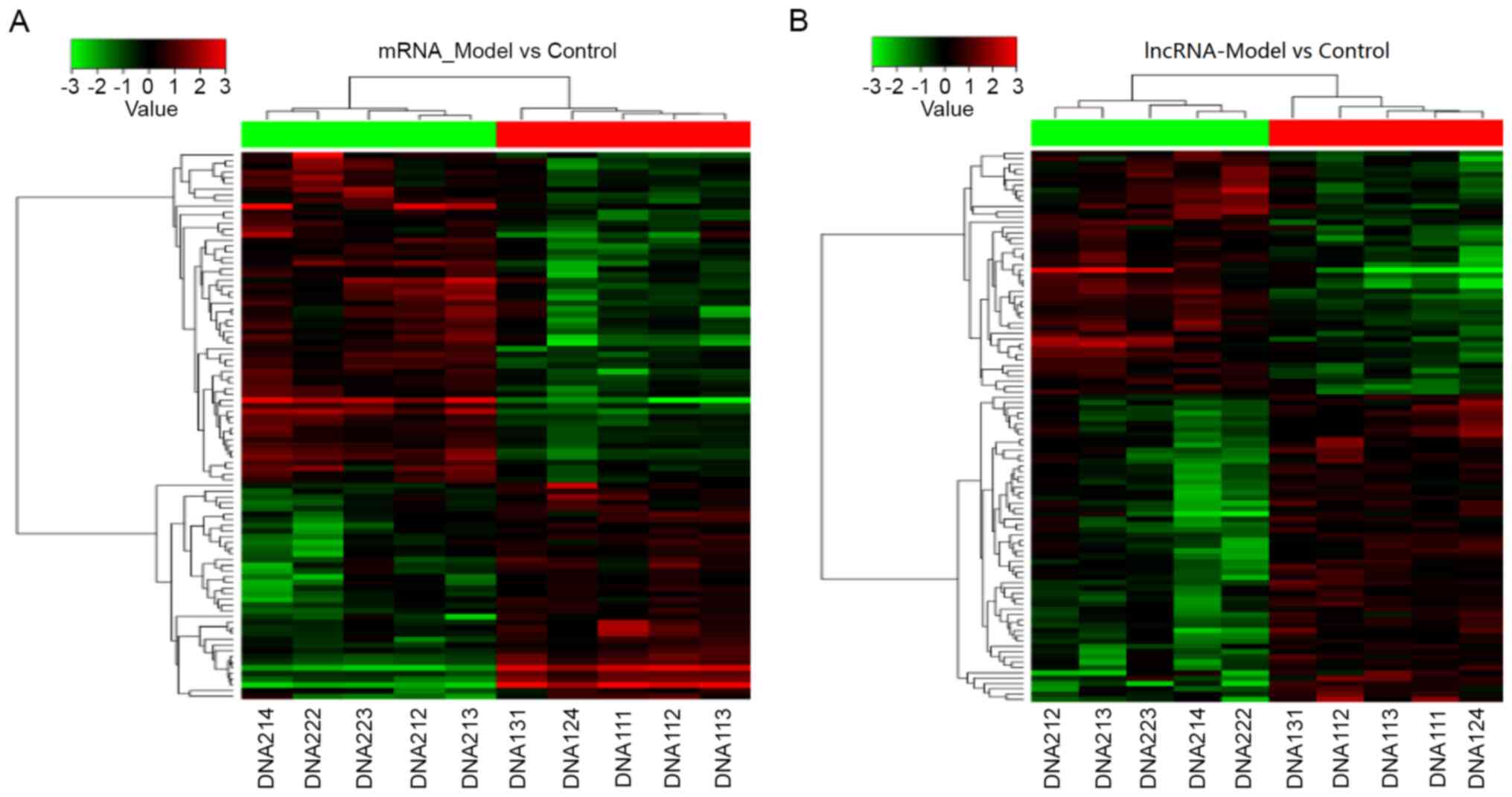
Hierarchical clustering analysis of the DEGs between
the Normal control and HLP groups was provided in a heatmap in
Fig. 3. The genes belonging to the
same group exhibited a relatively greater sequence similarity and
the classification was clear. Red and green represent the
differentially upregulated and downregulated genes in liver
tissues. This clustering result may support the functional analysis
of HLP-associated genes, suggesting that clustered genes are
regulated together via co-expression during transcription.
Functional group analysis and GO
analysis
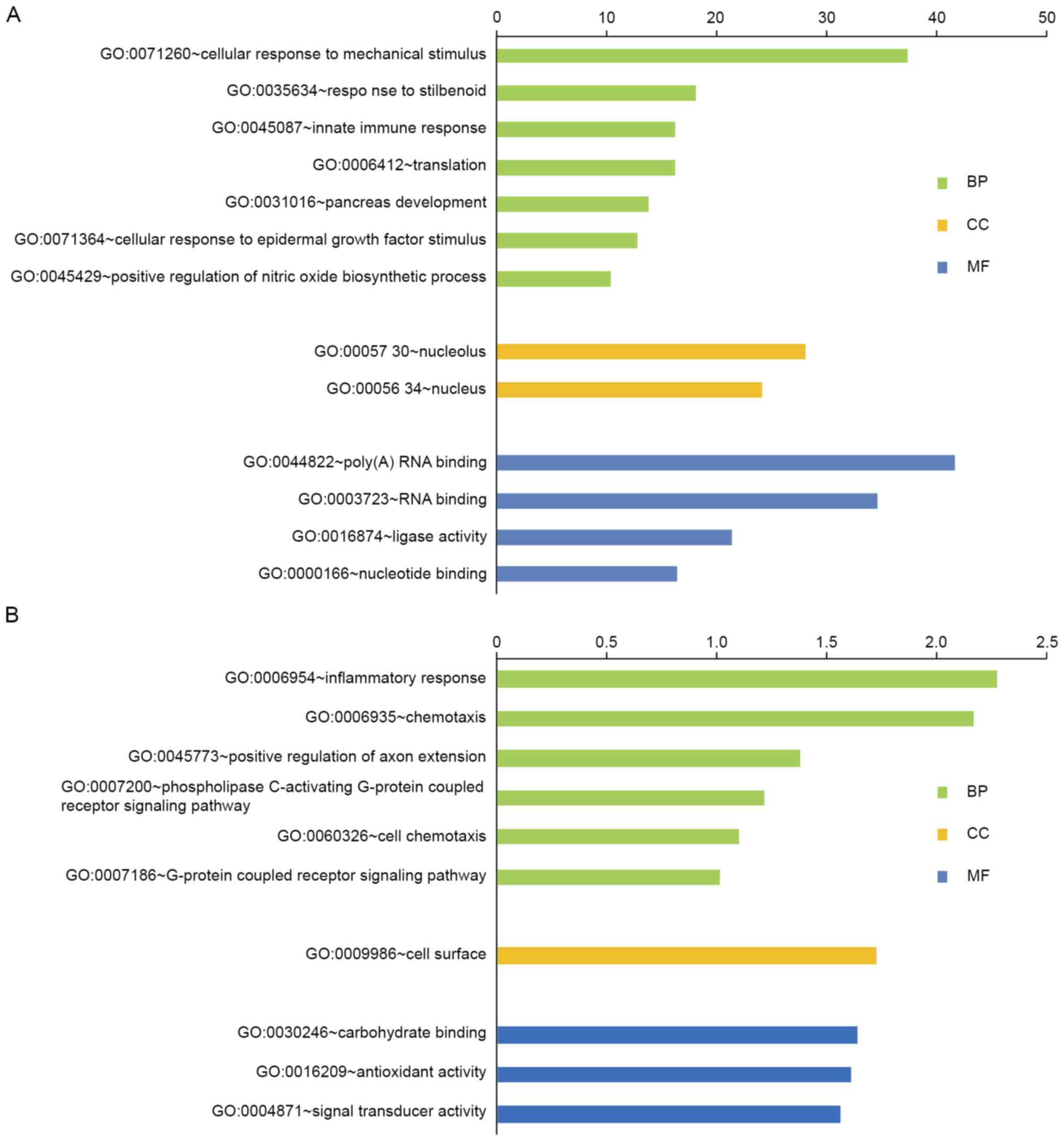
The DEGs were analyzed for their functions and
metabolic pathways using the online tool DAVID (Fig. 4). GO analysis demonstrated that the
upregulated DEGs were unambiguously enriched in biological
processes such as nitric oxide biosynthesis, innate immune response
and pancreas development. They also had a key role as a ligase in
the nucleolus and were associated with molecular functions of RNA
and nucleotide binding. The downregulated DEGs were mainly
associated with chemotaxis, inflammatory response and the G
protein-coupled receptor signaling pathway. The downregulated DEGs
were mainly associated with the cellular component of the cell
surface. Furthermore, these genes were involved in molecular
functions such as antioxidant activity, carbohydrate binding and
signal transduction.
Co-expression analysis
The co-expression network included 77 lncRNAs and
their 73 related mRNAs (Fig. 5).
This network consisted of 150 nodes and 451 connections, with
absolute values of their correlation coefficients of >0.9.
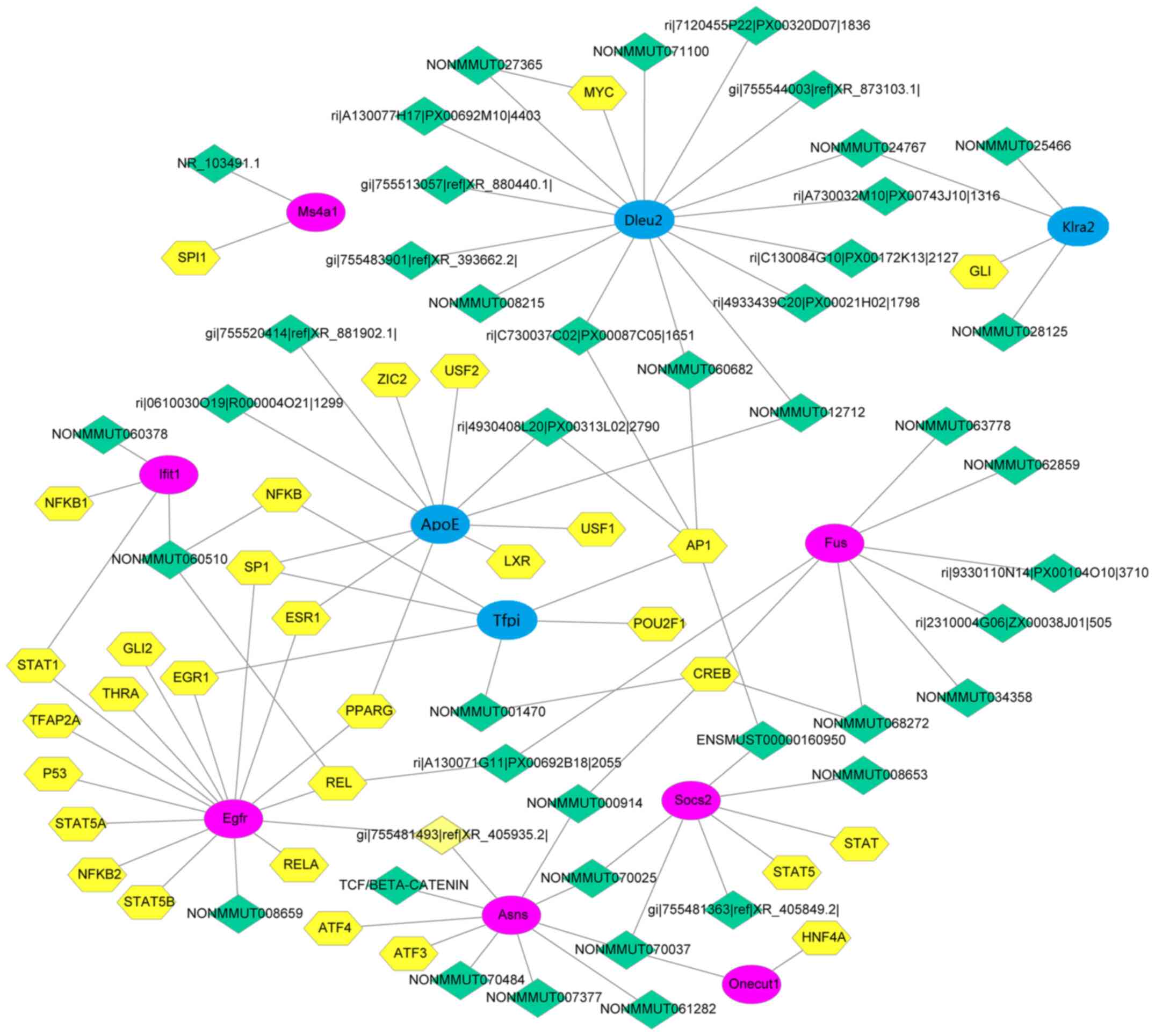
Transcription factor prediction
For transcription factor prediction, gene functional
annotation was performed using the DAVID database and they were
aligned to the database downloaded from the transcription factor
screening website http://www.tfacts.org. This led to the discovery of 7
upregulated and 4 downregulated differentially expressed mRNAs,
corresponding to 60 differentially expressed lncRNAs and 38
transcription factors. Cytoscape 3.6 was used to draw the 109 nodes
and lines (Fig. 6).
Discussion
Numerous studies have suggested that physiological
activities such as coagulation, inflammatory response, lipid
metabolism, oxidative stress and cell adhesion are closely related
to the progression of HLP (15-19).
However, the detailed underlying molecular mechanisms contributing
to the pathogenesis of HLP have so far remained elusive. During the
experiment, the body weight of the normal control group increased,
but the statistical error of C57BL/6J body weight due to the small
sample size did not increase significantly.
In the present study, a mouse lncRNA array was used
to determine the expression profile of genes in the liver of the
mouse model of HLP. Caecam2 was identified as a key upregulated
gene in HLP. Caecam2 is involved in insulin secretion by regulating
glucagon-like peptide-1, energy metabolism and energy expenditure,
as well as brown adipogenesis, and was closely associated with
metabolism in HLP. In macrophages and monocytes, the intracellular
accumulation of cholesterol upregulates the expression of several
responsive genes of liver X receptors, including ApoE, ABCA1 and
ABCG1, thereby leading to a decrease in intracellular cholesterol
pool and an increase in the extracellular lipid pool. In addition,
EGFR emerged as a significantly upregulated gene in the HLP group.
EGFR was indicated to be involved in biological processes such as
cellular response to mechanical stimulus, positive regulation of
nitric oxide biosynthesis, cellular response to EGF stimulus and
transcription. Upon ligand stimulation, EGFR initiates
intracellular signaling cascades via cytoplasmic adaptor proteins
and enzymes, and induces cell migration, adhesion, proliferation,
differentiation and apoptosis (20,21).
Several genes associated with the inflammatory
response were downregulated in the HLP group. These included C5AR2,
CYSLTR1, CXCL13 and IL1RAP (22-25).
C5AR2 and CYSLTR1 are involved in the phospholipase C-activating
GPRC signaling pathway and are strongly chemotactic (26,27).
C5AR2 is also a novel receptor essential for the mediation of human
mast cell adhesion, migration and proinflammatory mediator
production (28).
Target gene prediction revealed that AJUBA was a
target gene of ri|9430057C17. AJUBA is the receptor mediating the
internalization of secreted glucose-regulated protein 78 into
macrophages (29). In addition, it
is a critical regulator of adipocyte differentiation as it
functions as a specific co-activator of PPARγ (30). AJUBA enhances the formation of the
LXR/RXR heterodimer and is vital for activation of its target gene
LXR, which has an important role in regulating lipid and glucose
metabolism (31). In addition,
ONECUT1 was identified as the target gene of the differentially
expressed lncRNA ENSMUST00000160950. ONECUT1 is a member of the Cut
homeobox family of transcription factors and is expressed mainly in
the liver and may be activated by growth hormone. ONECUT1 regulates
the expression of genes essential for hepatic functions and is a
member of hepatic transcription factors regulating the
differentiation of liver functions by promoting the expression of
glucose kinase and glucose-6-phosphatase involved in carbohydrate
metabolism in the liver (32,33).
Transcription factor prediction identified the
suppressor of cytokine signaling 2 (SOCS2) and signal transducer
and activator of transcription 5 (STAT5). Deletion of SOCS2 in mice
was indicated to protect the liver from steatosis but aggravates
insulin resistance in high-fat diet-induced mice (34). The liver of SOCS2-knockout mice
displayed NF-κB activity and impaired glucose tolerance, and an
inflammatory response was evoked by the production of inflammatory
cytokines in the liver and adipose tissues (35). Furthermore, SOCS2 mRNA levels were
modulated in human hepatic steatosis (36). STAT5 is a member of the JAK/STAT
signaling pathway (37). The
lncRNAs NONMMUT008653, gi|755481363|ref|XR_405849.2|,
ENSMUST00000160950 and NONMMUT070025 identified in the present
study, together with SOCS2, are co-regulated by the transcription
factors STAT and STAT5.
EGFR is expressed extensively in hepatocytes,
controlling cell morphogenesis and/or homeostasis, including
proliferation, migration and transformation (38). In addition to cell proliferation and
regeneration, EGFR also exerts anti-apoptotic effects and has a
regulatory role in liver and blood lipid metabolism in adult male
mice (39). In a previous study,
the levels of EGFR or EGFR-like receptors were positively
correlated with the increase of cholesterol (40). In the present study, NONMMUT008659
and EGFR were observed to share the same transcription factors,
including NF-κB and STAT, suggesting that the pathogenesis of HLP
may be associated with the co-regulation of these transcription
factors.
ApoE is a multifunctional plasma glycoprotein (34
kDa) secreted by macrophages and other immune cells in several
tissues, including the liver, brain, kidney, adrenal gland and
adipose tissue, and is a key component that mediates the binding of
all lipoproteins. A genome-wide association study indicated that
ApoE and LDL-C levels were tightly coupled (41). ApoE participates in the
destabilization of the α-helix in the binding domain and suppresses
the degradation of LDL. Furthermore, mutant ApoE binds to LDL
receptor and disrupts efficient recycling of LDLR to the hepatocyte
surface. Thus, HLP is closely associated with ApoE protein levels.
In the present study, ApoE expression was downregulated in the
liver, with transcription factors SP1 and ESR1 co-regulating the
expression of ApoE and EGFR. These results suggested that the
lncRNAs gi|755520414|ref|XR_881902.1| and
ri|0610030O19|R000004O21|1299 may be engaged in the regulation of
ApoE expression via EGFR and SP1 to impact plasma lipid metabolism.
Hepatocyte nuclear factors (HNFs), namely HNF4A, HNF4C, ONECUT1 and
Onecut2, are expressed in a variety of organs and tissue types,
including the liver, pancreas and kidney. As mentioned above,
ONECUT1 regulates glucose metabolism and the cell cycle (42). The present results revealed that
ONECUT1 and HNF4A were regulated by NONMMUT070037, which controlled
both SOCS2 and ASNS, in the HLP group.
In summary, in the present study, 104 differentially
expressed lncRNAs were identified in the mouse model of HLP
compared to the Normal control animals. They included 46
upregulated (e.g., ENSMUST00000160950) and 58 downregulated (e.g.,
ri|9430057C17) lncRNAs. In addition, 96 differentially expressed
mRNAs were identified, including 58 upregulated genes (e.g., EGFR
and SOCS2) and 38 downregulated genes (e.g., C5AR2, APOE, CYSLTR1
and CXCL13). These results strongly indicate the significance of
DEGs in the pathogenesis of HLP. In particular, ENSMUST00000160950,
EGFR, SOCS2 and ApoE may be potential therapeutic targets for the
clinical management of HLP. However, further studies are warranted
to evaluate this hypothesis toward clinical implications.
Acknowledgements
The authors would like to thank Professor Yulong
Chen and Professor Hui Wang of the College of Traditional Chinese
Medicine, Henan University of Traditional Chinese Medicine
(Zhengzhou, China) for their help with the animal experiments and
project design.
Funding
Funding: This study was supported by a grant from the study on
the distribution rule of traditional Chinese medicine constitution
in patients with carotid atherosclerotic plaque (grant no.
20140518).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request. The microarray data of the present study were deposited in
the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/; GEO accession no.
GSE169398).
Authors' contributions
YC designed and conceived the study. BX performed
data analysis and literature collection. NW wrote the manuscript
and performed the bioinformatics analysis. XX drew the charts and
performed statistical processing of the data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Henan University of Chinese Medicine (ethics review
approval no. DWLL201704101).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ried K, Toben C and Fakler P: Effect of
garlic on serum lipids: An updated meta-analysis. Nutr Rev.
71:282–299. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tosheska Trajkovska K and Topuzovska S:
High-density lipoprotein metabolism and reverse cholesterol
transport: Strategies for raising HDL cholesterol. Anatol J
Cardiol. 18:149–154. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hassing HC, Surendran RP, Derudas B,
Verrijken A, Francque SM, Mooij HL, Bernelot Moens SJ, Hart LM,
Nijpels G, Dekker JM, et al: SULF2 strongly prediposes to fasting
and postprandial triglycerides in patients with obesity and type 2
diabetes mellitus. Obesity (Silver Spring). 22:1309–1316.
2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Walther TC and Farese RV Jr: Lipid
droplets and cellular lipid metabolism. Annu Rev Biochem.
81:687–714. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Endo H, Shiroki T, Nakagawa T, Yokoyama M,
Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et
al: Enhanced expression of long non-coding RNA HOTAIR is associated
with the development of gastric cancer. PLoS One.
8(e77070)2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hirose T, Mishima Y and Tomari Y: Elements
and machinery of non-coding RNAs: Toward their taxonomy. EMBO Rep.
15:489–507. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang Y, Sun L, Xuan L, Pan Z, Hu X, Liu
H, Bai Y, Jiao L, Li Z, Cui L, et al: Long non-coding RNA CCRR
controls cardiac conduction via regulating intercellular coupling.
Nat Commun. 9(4176)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Uchida S and Dimmeler S: Long noncoding
RNAs in cardiovascular diseases. Circ Res. 116:737–750.
2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen J, Cui X, Shi C, Chen L, Yang L, Pang
L, Zhang J, Guo X, Wang J and Ji C: Differential lncRNA expression
profiles in brown and white adipose tissues. Mol Genet Genomics.
290:699–707. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Smekalova EM, Kotelevtsev YV, Leboeuf D,
Shcherbinina EY, Fefilova AS, Zatsepin TS and Koteliansky V: lncRNA
in the liver: Prospects for fundamental research and therapy by RNA
interference. Biochimie. 131:159–172. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Han P, Li W, Lin CH, Yang J, Shang C,
Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al: A long noncoding
RNA protects the heart from pathological hypertrophy. Nature.
514:102–106. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Guo W, Zhang H, Yang A, Ma P, Sun L, Deng
M, Mao C, Xiong J, Sun J, Wang N, et al: Homocysteine accelerates
atherosclerosis by inhibiting scavenger receptor class B member1
via DNMT3b/SP1 pathway. J Mol Cell Cardiol. 138:34–48.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Leary S, Underwood W and Anthony R: AVMA
Guidelines for the Euthanasia of Animals: 2013 Edition. 2013.
American Veterinary Medical Association. Available from: avma.org/sites/default/files/2020-01/2020-Euthanasia-Final-1-17-20.pdf.
|
|
15
|
Ozay R, Uzar E, Aktas A, Uyar ME, Gürer B,
Evliyaoglu O, Cetinalp NE and Turkay C: The role of oxidative
stress and inflammatory response in high-fat diet induced
peripheral neuropathy. J Chem Neuroanat. 55:51–57. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Owens AP III, Byrnes JR and Mackman N:
Hyperlipidemia, tissue factor, coagulation, and simvastatin. Trends
Cardiovasc Med. 24:95–98. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu TT, Zeng Y, Tang K, Chen X, Zhang W
and Xu XL: Dihydromyricetin ameliorates atherosclerosis in LDL
receptor deficient mice. Atherosclerosis. 262:39–50.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xenoulis PG and Steiner JM: Lipid
metabolism and hyperlipidemia in dogs. Vet J. 183:12–21.
2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lee YJ, Seo JA, Yoon T, Seo I, Lee JH, Im
D, Lee JH, Bahn KN, Ham HS, Jeong SA, et al: Effects of low-fat
milk consumption on metabolic and atherogenic biomarkers in Korean
adults with the metabolic syndrome: A randomised controlled trial.
J Hum Nutr Diet. 29:477–486. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Singhai A, Wakefield DL, Bryant KL, Hammes
SR, Holowka D and Baird B: Spatially defined EGF receptor
activation reveals an F-actin-dependent phospho-Erk signaling
complex. Biophys J. 107:2639–2651. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zaczek A, Brandt B and Bielawski KP: The
diverse signaling network of EGFR, HER2, HER3 and HER4 tyrosine
kinase receptors and the consequences for therapeutic approaches.
Histol Histopathol. 20:1005–1015. 2005.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Coulthard LG, Hawksworth OA and Woodruff
TM: Complement: The emerging architect of the developing brain.
Trends Neurosci. 41:373–384. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Matsumoto H, Kanemitsu Y, Nagasaki T,
Tohda Y, Horiguchi T, Kita H, Kuwabara K, Tomii K, Otsuka K,
Fujimura M, et al: Staphylococcus aureus enterotoxin sensitization
involvement and its association with the CysLTR1 variant in
different asthma phenotypes. Ann Allergy Asthma Immunol.
118:197–203. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kowarik MC, Cepok S, Sellner J, Grummel V,
Weber MS, Korn T, Berthele A and Hemmer B: CXCL13 is the major
determinant for B cell recruitment to the CSF during
neuroinflammation. J Neuroinflammation. 9(93)2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Askmyr M, Ågerstam H, Hansen N, Gordon S,
Arvanitakis A, Rissler M, Juliusson G, Richter J, Järås M and
Fioretos T: Selective killing of candidate AML stem cells by
antibody targeting of IL1RAP. Blood. 121:3709–3713. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hu C, Yang J, He Q, Luo Y, Chen Z, Yang L,
Yi H, Li H, Xia H, Ran D, et al: CysLTR1 blockage ameliorates liver
injury caused by aluminum-overload via PI3K/AKT/mTOR-mediated
autophagy activation in vivo and in vitro. Mol Pharm. 15:1996–2006.
2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kistler B, Schlegel A, Grauert M, Arndt K
and Williams C: Cell type specific accessibility of C5aR2 on human
blood cells of myeloid origin. Eur Resp J. 48(PA3637)2016.
|
|
28
|
Pundir P, MacDonald CA and Kulka M: The
novel receptor C5aR2 is required for C5a-mediated human mast cell
adhesion, migration, and proinflammatory mediator production. J
Immunol. 195:2774–2787. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
La X, Zhang L, Li H, Li Z, Song G, Yang P
and Yang Y: Ajuba receptor mediates the internalization of
tumor-secreted GRP78 into macrophages through different endocytosis
pathways. Oncotarget. 9:15464–15479. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li Q, Peng H, Fan H, Zou X, Liu Q, Zhang
Y, Xu H, Chu Y, Wang C, Ayyanathan K, et al: The LIM protein Ajuba
promotes adipogenesis by enhancing PPARγ and p300/CBP interaction.
Cell Death Differ. 23:158–168. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu QY, Quinet E and Nambi P: Adipocyte
fatty acid-binding protein (aP2), a newly identified LXR target
gene, is induced by LXR agonists in human THP-1 cells. Mol Cell
Biochem. 302:203–213. 2007.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Beaudry JB, Pierreux CE, Hayhurst GP,
Plumb-Rudewiez N, Weiss MC, Rousseau GG and Lemaigre FP: Threshold
levels of hepatocyte nuclear factor 6 (HNF-6) acting in synergy
with HNF-4 and PGC-1alpha are required for time-specific gene
expression during liver development. Mol Cell Biol. 26:6037–6046.
2006.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Holterman A, Wang K, Wang M and Gannon M:
Transcriptional regulation of hepatic genes by hepatocyte nuclear
factor OC1 attenuates liver injury. J Surg Res. 158(324)2010.
|
|
34
|
Zadjali F, Santana-Farre R, Vesterlund M,
Carow B, Mirecki-Garrido M, Hernandez-Hernandez I,
Flodström-Tullberg M, Parini P, Rottenberg M, Norstedt G, et al:
SOCS2 deletion protects against hepatic steatosis but worsens
insulin resistance in high-fat-diet-fed mice. FASEB J.
26:3282–3291. 2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Leroith D and Nissley P: Knock your SOCS
off! J Clin Invest. 115:233–236. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Vesterlund M: The mechanism of action of
SOCS2 and its role in metabolism and growth. Arthritis Rheum.
60:1743–1752. 2013.
|
|
37
|
Terrell AM, Crisostomo PR, Wairiuko GM,
Wang M, Morrell ED and Meldrum DR: Jak/STAT/SOCS signaling circuits
and associated cytokine-mediated inflammation and hypertrophy in
the heart. Shock. 26:226–234. 2006.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Komposch K and Sibilia M: EGFR signaling
in liver diseases. Int J Mol Sci. 17(30)2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Scheving LA, Zhang X, Garcia OA, Wang RF,
Stevenson MC, Threadgill DW and Russell WE: Epidermal growth factor
receptor plays a role in the regulation of liver and plasma lipid
levels in adult male mice. Am J Physiol Gastrointest Liver Physiol.
306:G370–G381. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Leedman P, Giles K and Webster RJ: Method
of modulation of expression of epidermal growth factor receptor
(EGFR) involving miRNA. Patent application EP, US86738722014. Filed
28 August 2007; issued 6 March, 2008.
|
|
41
|
Deshmukh HA, Colhoun HM, Johnson T,
McKeigue PM, Betteridge DJ, Durrington PN, Fuller JH, Livingstone
S, Charlton-Menys V, Neil A, et al: Genome-wide association study
of genetic determinants of LDL-c response to atorvastatin therapy:
Importance of Lp(a). J Lipid Res. 53:1000–1011. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yamamoto K, Matsuoka T, Kawashima S,
Takebe S, Kubo F, Miyatsuka T, et al: A novel function of Onecut1
as a negative regulator of MafA. J Biol Chem. 288:21648–21658.
2013.PubMed/NCBI View Article : Google Scholar
|