Introduction
Colorectal cancer (CRC) is one of the most common
and deadliest cancers in the world, causing ~800,000 deaths in
2018(1). According to World Health
Organization data, China had 245,000 new cases and 139,000 deaths
associated with CRC in 2012, making it the fifth most common cancer
in men and the fourth most common cancer in women (2). Among cases of CRC, ~41% occur in the
proximal colon, ~22% in the distal colon and 28% involve the rectum
(3). The occurrence and
development of CRC is a complex pathological process involving
multiple signaling pathways including Wnt, Hedgehog, bone
morphogenic protein and Notch (4,5).
Therefore, identifying genes that serve an important role in CRC
may help to identify therapeutic targets for CRC.
Human prune homolog 2 with BCH domain (PRUNE2)
encodes a 340-kDa protein with a conserved BCH scaffold domain at
its C-terminus (6-8).
Proteins with the BCH domain regulate morphogenesis,
differentiation, motility and apoptosis by associating with
components of signaling networks, such as Rho, Ras and MAPK
signaling (7). Thus, PRUNE2
modulates cellular function, such as morphogenesis,
differentiation, motility, proliferation and apoptosis, by
modulating signaling networks. The function of PRUNE2 in numerous
types of tumor has been reported, for example, increased expression
of PRUNE2 is associated with favorable prognosis in neuroblastoma
(6,9); PRUNE2 also regulates the
differentiation, proliferation and invasion of neuroblastoma tumor
cells; moreover, increased PRUNE2 protein expression may serve as a
favorable prognostic marker in human leiomyosarcoma (10,11).
PRUNE2 may serve antitumor functions, however, little is known
about the effect of PRUNE2 on CRC.
The present study aimed to investigate the
biological function of PRUNE2 in CRC cell lines (SW620 and HT29) by
using Cell Counting Kit (CCK)-8, colony formation, flow cytometry
and Transwell assay. Furthermore, a mouse model was established to
investigate the effect of PRUNE2 on metastasis of CRC cells.
Materials and methods
Gene expression and survival
analysis
Gene expression RNA sequencing data from The Cancer
Genome Atlas (TCGA) were obtained for colon adenocarcinoma (COAD)
and rectal adenocarcinoma (READ) cohorts (12). mRNA expression levels were
processed as previously described (13). The association between PRUNE2
expression and relapse-free survival probability was analyzed using
the UALCAN website (ualcan.path.uab.edu/index.html). The expression levels
of PRUNE2 in COAD, READ and matched adjacent normal tissue were
analyzed using Gene Expression Profiling Interactive Analysis
(gepia.cancer-pku.cn). The expression
levels of PRUNE2 were analyzed in COAD cases with different stage
and nodal metastasis status were analyzed via UALCAN.
Tissue collection
A total of 10 CRC and adjacent healthy sections were
collected between July and September 2020 from the Department of
Pathology, First People's Hospital of Yunnan Province for western
blot and immunohistochemistry assays. No patients received any
adjuvant treatment, such as radiotherapy, chemotherapy or
immunotherapy prior to surgery. Tissue histomorphology was
confirmed by two pathologists in a blinded manner. The distance
between CRC and adjacent tissue was ≥3 cm. All patients (age,
62.6±11.82 years; 5 male and 5 female) provided written informed
consent. The present study was approved by the Ethics Committee of
the First People's Hospital of Yunnan Province.
Immunohistochemistry
Tissues were fixed with 4% paraformaldehyde at room
temperature overnight, then dehydrated, embedded in paraffin and
sliced (thickness, 5 mm). Sections were dewaxed with xylene and
ethanol (xylene for 10 and 5 min; 100% ethanol for 5 min, then 95,
80 and 70% ethanol for 2 min each). Sections were incubated with 3%
H2O2 for 10 min at room temperature to block
endogenous peroxidase/phosphatase activity. Antigen repair was
performed using 0.01 M citric acid buffer (pH 6.0) for 15 min at
100˚C and 80 kPa. The sections were washed with 1X PBS three times
for 5 min each and blocked with 5% goat serum (Beijing Solarbio
Science & Technology Co., Ltd.) in PBS for 15 min at room
temperature. Sections were incubated with PRUNE2 antibody (1:100;
cat. no. 11458-1-AP; ProteinTech Group, Inc.) overnight at 4˚C. The
sections were washed with 1X PBS three times for 5 min each and
then incubated with horseradish peroxidase (HRP)-conjugated goat
anti-rabbit IgG (1:200; cat. no. AS014; ABclonal Biotech Co., Ltd.)
for 1.5 h at room temperature. The sections were washed with 1X PBS
three times for 5 min each and visualized using a
3,3'-diaminobenzidine visualization kit (Fuzhou Maixin Biotech Co.,
Ltd.) according to the manufacturer's instructions. Sections were
counterstained with 0.5% hematoxylin for 5-10 min at room
temperature to visualize the nuclei. The sections were examined
under a light microscope (x400 magnification). Brown staining
indicated immunoreactive (positive) cells; blue staining indicated
the nuclei. Positive expression of PRUNE2 was quantified using
ImageJ 2x software (National Institutes of Health).
Cell culture
Human normal colorectal mucosa cells (FHC; CRL-1831)
and CRC cell lines [SW620 (CCL-227), SW480 (CCL-228), HT29
(HTB-38), HCT116 (CCL-247), LOVO (CCL-229), DLD-1 (CCL-221)] were
obtained from American Type Culture Collection. All cells were
authenticated using STR profiling. The cells were maintained in
high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS) and antibiotics (all Gibco;
Thermo Fisher Scientific, Inc.) at 37˚C in a humidified incubator
containing 5% CO2.
PRUNE2 overexpression and
knockdown
For overexpression of PRUNE2, PRUNE2/pcDNA3.1
overexpression plasmid was purchased from Shanghai GeneChem Co.,
Ltd. For knockdown of PRUNE2, negative control (NC) short hairpin
(sh)RNA oligo (UUCUCCGAACGUGUCACGUTT) and PRUNE2 shRNA
(GGGCCAGAATATCGATGAATT) were purchased from Shanghai GeneChem Co.,
Ltd. For cell transfection, SW620 and HT29 cells (1x105)
were plated in a 6-well plate at 37˚C overnight. Then, 100 pmol
PRUNE2 or NC shRNA or 2 µg PRUNE2/pcDNA3.1 or empty pcDNA3.1 vector
and 15 ml Lipofectamine® 2000 (cat. no. 11668-019;
Invitrogen; Thermo Fisher Scientific, Inc.) were incubated at room
temperature for 20 min according to the manufacturer's
instructions. Mixtures were transfected into cells for 48 h at
37˚C, then the subsequent experimentation was performed. For cell
experiments, SW620 and HT29 cells were divided into five groups as
follows: Normal (untransfected cells); control (ctrl; transfected
with empty pcDNA3.1 vector); PRUNE2 (transfected with
PRUNE2/pcDNA3.1 vector); shRNA-NC (transfected with NC shRNA) and
shRNA-PRUNE2 (transfected with PRUNE2 shRNA).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from cells using
TRIzol® (cat. no. 1596-026; Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was reverse-transcribed using the
RevertAid™ First Strand cDNA Synthesis kit (cat. no.
K1622; Fermentas; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, and subjected to amplification using
THERMO Maxima® SYBR Green/ROX qPCR Master Mix (cat. no.
K0223; Thermo Fisher Scientific, Inc.) as follows: 95˚C for 3 min;
followed by 40 cycles of 95˚C for 10 sec and 60˚C for 60 sec. The
relative expression levels of PRUNE2 were measured by RT-qPCR using
an ABI PRISM 7500 system (Thermo Fisher Scientific, Inc.). All
samples were normalized to β-actin and all experiments were
performed in triplicate. The mean value was used to calculate
relative mRNA expression levels using the 2-ΔΔCq method
(14). Primers (Sangon Biotech
Co., Ltd.) were as follows: PRUNE2 forward,
5'-GGGTCTTCTGGGATTATGG-3' and reverse, 5'-CTGGGCTAACAAGGTCTAC-3'
and β-actin forward, 5'-CATCGTCCACCGCAAATGCTTC-3' and reverse,
5'-ACCGACTGCTGTCACCTTCAC-3'.
Western blot assay
Total protein from cells and tissues was obtained
using RIPA lysis buffer (cat. no. R0020; Beijing Solarbio Science
& Technology Co., Ltd.), and then quantified by BCA protein
Assay. For each sample, 40 µg/lane protein was loaded onto 10%
SDS-PAGE gel, separated for 120 min at 120 V and transferred to a
PVDF membrane. Following blocking with 10% skimmed milk for 2 h at
room temperature, the membranes were incubated overnight at 4˚C
with antibodies against PRUNE2 (1:1,000; cat. no. 11458-1-AP;
ProteinTech Group, Inc.), Bcl-2 (1:1,000; cat. no. 26593-1-AP;
ProteinTech Group, Inc.), Bax (1:1,000; cat. no. 60267-1-Ig;
ProteinTech Group, Inc.), caspase-3 (1:1,000; cat. no. 19677-1-AP;
ProteinTech Group, Inc.), caspase-9 (1:1,000; cat. no. 10380-1-AP;
ProteinTech Group, Inc.), Cyclin D1 (1:1,000; cat. no. 60186-1-Ig;
ProteinTech Group, Inc.) or β-actin (1:2,000; cat. no. ab8227;
Abcam). Following washing with 1X TBST (cat. no. T1085; Beijing
Solarbio Science & Technology Co., Ltd.) three times for 10 min
each, membranes were incubated with HRP-conjugated goat anti-rabbit
or anti-mouse (both 1:2,000; cat. nos. A0208 and A0216,
respectively; both Beyotime Institute of Biotechnology) for 1.5 h
at room temperature. The blots were washed with 1X TBST three times
for 10 min each and developed with Immobilon Western
Chemiluminescent HRP Substrate (cat. no. WBKLS0100;
MilliporeSigma). The bands were imaged with a chemiluminescence
imager (Bio-Rad Laboratories, Inc.). The band intensities were
determined using ImageJ 2x software (National Institutes of Health)
and normalized to β-actin.
Cell proliferation assay
Cell proliferation was detected by CCK-8) assay. A
total of 2x104 cells/well was seeded into 96-well
plates. After incubation at 37˚C for 24 h, cells were transfected
with PRUNE2/pcDNA3.1 or PRUNE2 shRNA for 48 h. Then, 10 µl CCK-8
solution (cat. no. CP002; Signalway Antibody, LLC) was added to
each well and cells were incubated at 37˚C for 2 h. The absorbance
was measured at 450 nm using a microplate reader (cat. no.
DNM-9602; Perlong Medical Equipment Co., Ltd.).
Cell cycle assay
Following transfection for 48 h, both SW620 and HT29
cells were trypsinized, collected and centrifuged at 1,000 x g for
3 min at 4˚C. Cells were washed with 1X PBS and fixed in 70%
ethanol for 24 h at 4˚C. Cells were incubated in 100 ml RNase A
solution (1 mg/ml; cat. no. R8020-25; Beijing Solarbio Science
& Technology Co., Ltd.) in the dark at 37˚C for 30 min and
stained with 400 ml propidium iodide (PI) solution (50 µg/ml; cat.
no. C001-200; Shanghai Qihai Futai Biological Technology Co., Ltd.)
in the dark at room temperature for 10 min. Following staining,
cells were analyzed with a flow cytometer (Accuri C6; BD
Biosciences). BD Accuri C6 plus software (3.1.1.0; BD Biosciences)
was used for analysis. Red fluorescence was detected at excitation
wavelength of 488 nm, corresponding to BD Biosciences flow
cytometry FL2 detection channel.
Colony formation assay
Both SW620 and HT29 cells were trypsinized and
resuspended in DMEM. A total of 10 ml medium containing 700 cells
was added to each well of a 10 mm culture dish. After incubation at
37˚C for 24 h, the cells were transfected with PRUNE2/pcDNA3.1 or
PRUNE2 shRNA for 48 h and cultured at 37˚C in an incubator for 7
days; medium was replaced with fresh medium every 3 days. After 7
days, the cells were washed with 1X PBS three times. Then, 5 ml 4%
paraformaldehyde was used to fix cells for 15 min at room
temperature and 5 ml 0.5% crystal violet staining solution (cat.
no. C8470; Beijing Solarbio Science & Technology Co., Ltd.) was
added for 20 min at room temperature to stain the cells. The cells
were washed with 1X PBS and air dried and colonies (>50 cells)
visible to the naked eye were counted manually.
Cell invasion assay
Cell invasion assay was performed using 8-µm
Transwell chambers (cat. no. 3422; Corning, Inc.) precoated with
Matrigel (cat. no. 356234; Corning, Inc.) for 30 min at 37˚C.
Following transfection for 24 h, 200 µl cell suspension within DMEM
(4x105 cells/ml) was added to the upper chamber and 700
µl DMEM containing 10% FBS was added to the lower chamber.
Following incubation at 37˚C for 48 h, the cells were fixed with 4%
formaldehyde for 10 min at room temperature and invading cells were
stained with 0.5% crystal violet (cat. no. C8470; Beijing Solarbio
Science & Technology Co., Ltd.) for 30 min at room temperature
and air dried. The invading cells on the lower membrane surface
were imaged under a light microscope and counted.
Apoptosis assay
Following transfection for 48 h, 1x105
cells (SW620 and HT29) were collected and centrifuged at 1,000 x g
for 5 min at 4˚C. Cells were resuspended in 195 µl Annexin V-APC
binding buffer (cat. no. E-CK-A151; Elabscience Biotechnology,
Inc.) and stained with 5 µl Annexin V-APC staining solution (cat.
no. E-CK-A117; Elabscience Biotechnology, Inc.) in the dark at 4˚C
for 15 min. The cells were stained with 5 µl PI staining solution
(cat. no. E-CK-A161; Elabscience Biotechnology, Inc.) in the dark
at 4˚C for 5 min. Finally, 400 µl binding buffer was added and
samples were assessed using flow cytometer (Accuri C6; BD
Biosciences). BD Accuri C6 plus software (3.1.1.0; BD Biosciences)
was used for analysis. Tubes without Annexin V-APC or PI were used
as NC. APC (blue) was detected at an excitation wavelength of 652
nm. PI (red) was detected at an excitation wavelength of 488 nm.
The percentage of apoptotic cells was calculated as early + late
apoptotic cells.
Tumor xenografts in mice
A total of 12 female BALB/c nude mice (weight, 18±2
g; age, 6-8 weeks; Charles River Laboratories, Inc.) were house
with free food and water under specific-pathogen-free conditions at
21-25˚C with 40-70% humidity and 12/12-h light/dark cycle. The
animal experimental procedures were in accordance with Yunnan
Administration Guidelines for Laboratory Animals and approved by
the Animal Ethics Committee of Kunming University of Science and
Technology. pGV492-gcGFP and pGV492-gcGFP-PRUNE2 overexpression
vector (Shanghai GeneChem Co., Ltd.) were transfected into SW620
cells as aforementioned. Following 48 h transfection, cells were
trypsinized with 0.25% trypsin (Gibco) and sub-cultured. Following
cell adherence, puromycin (cat. no. P8230; Beijing Solarbio Science
& Technology Co., Ltd.) was added at 5 µg/ml for screening
cells and solution was replaced with complete DMED with 5 µg/ml
puromycin every 2-3 days. After 1 week, cells were cultured in
complete medium with 2 µg/ml puromycin. RT-qPCR was performed to
verify the expression levels of PRUNE2 in the screened
PRUNE2-GFP-SW620 and the NC-GFP-SW620 cells. Cell lines
successfully screened were named NC-GFP-SW620 and PRUNE2-GFP-SW620.
A total of 2x106 NC-GFP- or PRUNE2-GFP-SW620 cells in
0.1 ml PBS were subcutaneously injected into the axilla of mice. A
total of 12 mice were randomly divided into two groups (n=6/group)
as follows: NC-GFP-SW620 and PRUNE2-GFP-SW620. All mice were
anesthetized with pentobarbital sodium (50 mg/kg) by
intraperitoneal injection at 5, 10, 15, 20 and 25 days after
inoculation, then fluorescence of cell inoculation sites in mice
was observed using an IVIS® Spectrum In Vivo
Bioluminescence imaging system (PerkinElmer, Inc.) at 5, 10, 15, 20
and 25 days after inoculation. Tumor diameter was measured with
Vernier calipers every 3 days after tumor emergence (14 days after
inoculation) to measure tumor volume [(volume=(4/3) x π x
radius3] for 18 days before mice were sacrificed. All
mice were euthanized with pentobarbital sodium (150 mg/kg) by
intraperitoneal injection and cervical dislocation. Subcutaneous
tumors were collected at 32 days after cell injection.
Statistical analysis
Data are presented as the mean ± SD. All experiments
were performed in triplicate. All data were analyzed using GraphPad
Prism 5.0 software (GraphPad Software, Inc.). The log-rank test
statistical analysis was used for the curves of relapse-free
survival probability. Differences between two groups were analyzed
using paired Student's t-test. Differences between >2 groups
were compared using one-way ANOVA followed by Tukey's-post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
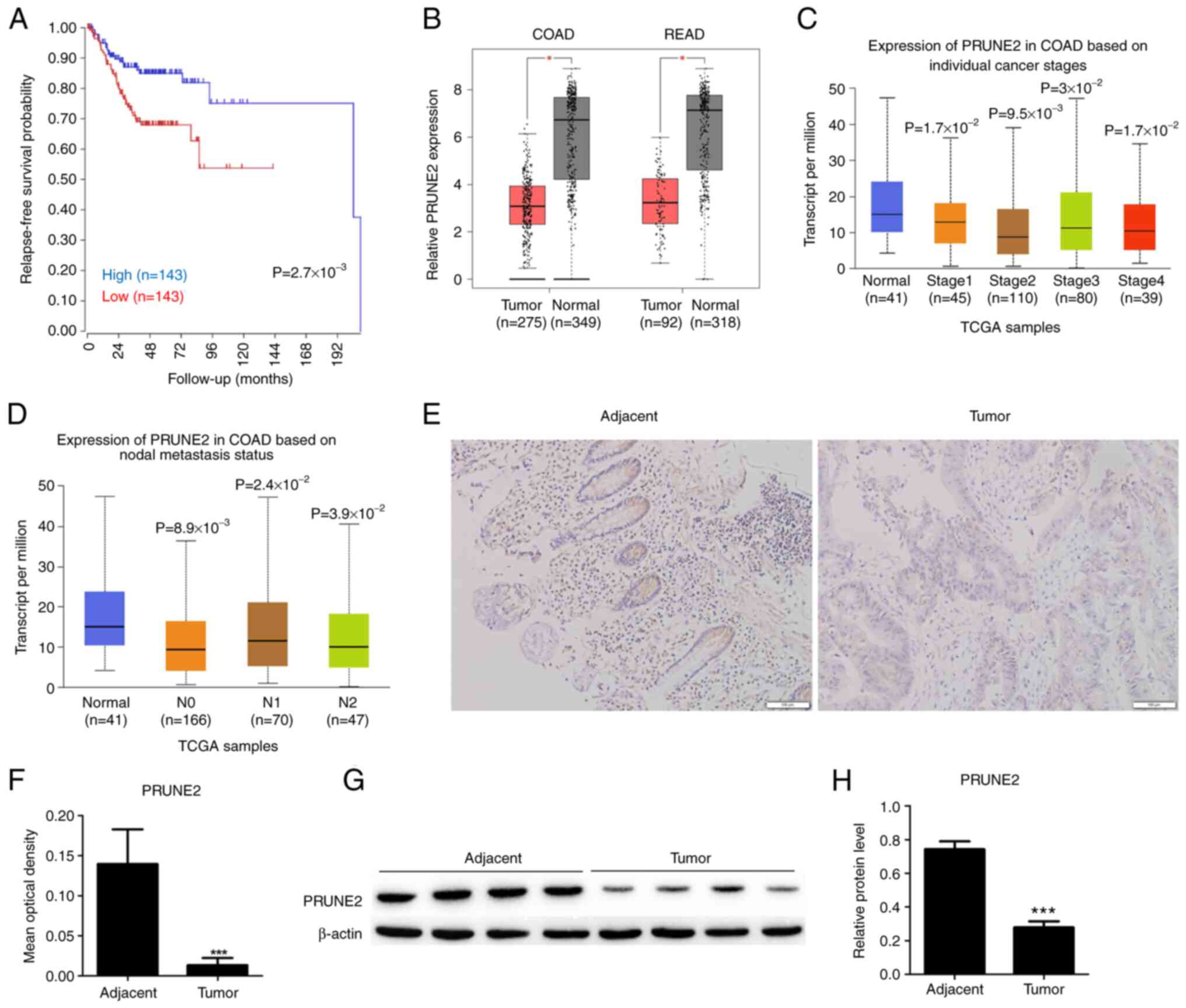
PRUNE2 downregulation is associated
with poor CRC patient survival
A total of 286 patients were included for
relapse-free survival probability analysis using clinical and
PRUNE2 expression data from TCGA. There was a significant
difference in relapse-free survival probability between patients
with low and high PRUNE2 levels, suggesting that low PRUNE2
expression was associated with lower relapse-free survival
probability (Fig. 1A). Statistical
analysis of mRNA expression data from TCGA showed that PRUNE2 was
underexpressed in COAD and READ compared with adjacent normal
tissue (Fig. 1B). Moreover,
expression of PRUNE2 was significantly downregulated in CRC tissue
compared with adjacent normal tissue at all tumor stages (Fig. 1C) and metastasis statuses (Fig. 1D). Immunohistochemistry of paired
CRC and normal tissue revealed that PRUNE2 was primarily localized
in the cytoplasm and its expression levels were lower in CRC
compared with adjacent tissue (Fig.
1E and F). The western
blotting results were consistent with the immunohistochemistry
results, the expression levels of PRUNE2 was lower in tumor tissues
compared with adjacent tissue (Fig.
1G and H). These data
indicated that PRUNE2 was downregulated in CRC and associated with
poor patient survival.
Effect of PRUNE2 on viability of CRC
cell lines
PRUNE2 expression was detected in human normal
colorectal mucosa cells (FHC) and six CRC cell lines (SW620, SW480,
HT29, HCT116, LOVO, DLD-1) using RT-qPCR and western blotting.
PRUNE2 mRNA levels were lower in CRC cell lines compared with FHC
cells (Fig. 2A). Western blotting
results were consistent with RT-qPCR results, PRUNE2 protein levels
were lower in CRC cell lines compared with FHC cells (Fig. 2B). Two CRC cell lines (SW620 and
HT29) were randomly selected from several cells with low expression
and used in subsequent experiments. PRUNE2 was overexpressed or
knocked down via transient transfection of the pcDNA3.1/PRUNE2
vector and shRNA PRUNE2 into the SW620 and HT29 CRC cell lines;
empty pcDNA3.1 vector and shRNA NC were used as controls. Western
blotting was performed to evaluate protein levels of PRUNE2; PRUNE2
was significantly upregulated by pcDNA3.1/PRUNE2 and downregulated
by PRUNE2 shRNA in SW620 and HT29 cells (Fig. 2C). CCK-8 assay revealed that PRUNE2
overexpression significantly decreased proliferation and PRUNE2
knockdown significantly increased proliferation of CRC cells
(Fig. 2D). These data indicated
that PRUNE2 was expressed at low levels and decreased viability in
CRC cell lines.
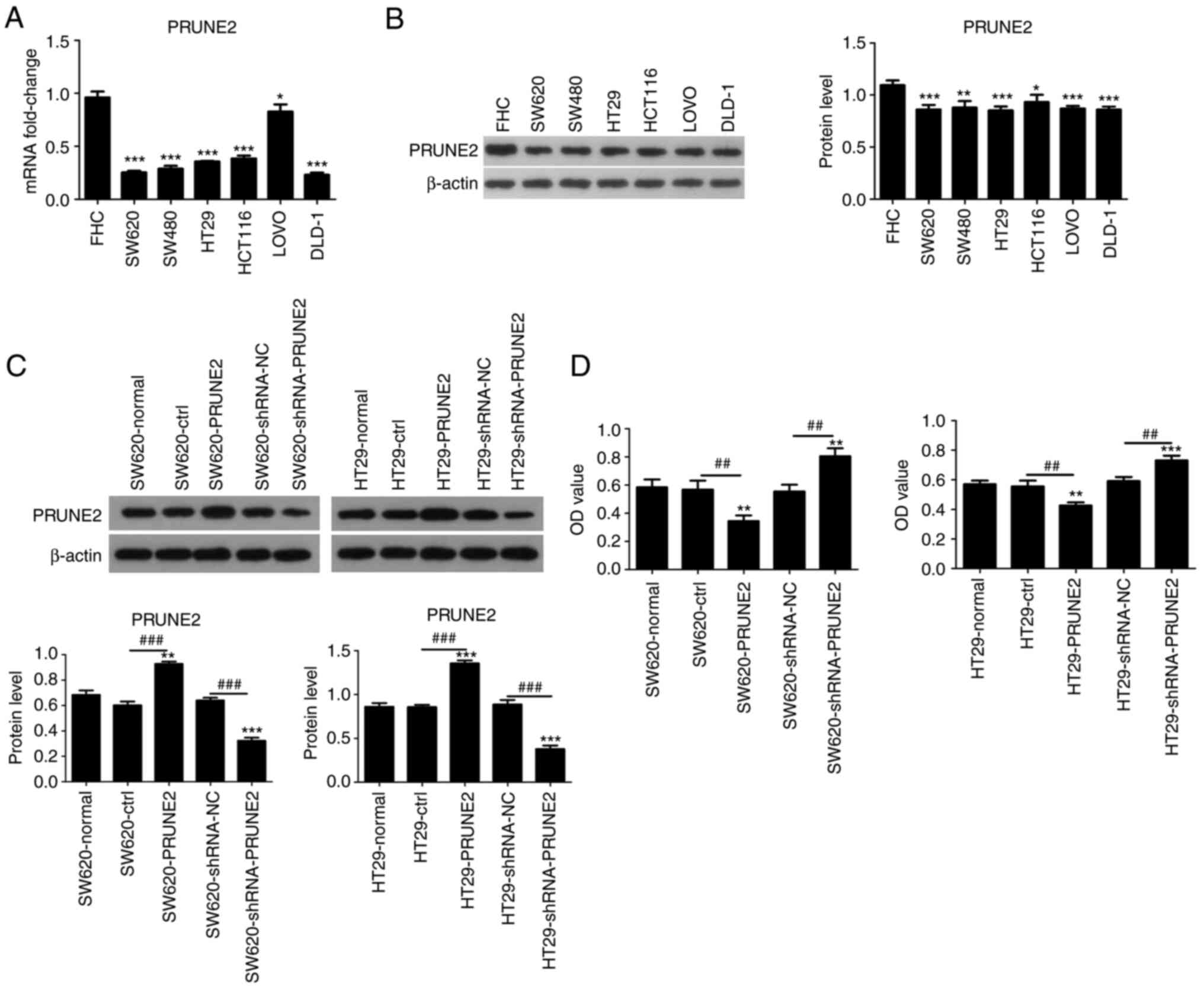 | Figure 2PRUNE2 expression levels in cell
lines and effect on cell viability. (A) mRNA and (B) protein levels
in human normal colorectal mucosa cells (FHC) and CRC cell lines
(SW620, SW480, HT29, HCT116, LOVO, DLD-1) were determined by
reverse transcription-quantitative PCR and western blotting,
respectively. *P<0.05, **P<0.01,
***P<0.001 vs. FHC. (C) Transfection efficiency of
the pCDNA3.1/PRUNE2 plasmid and shRNA PRUNE2 were detected by
western blot assay. (D) Viability of CRC cells was detected by Cell
Counting Kit-8 assay. Data are presented as the mean ± SD (n=3).
Data were analyzed using one-way ANOVA. **P<0.01,
***P<0.001 vs. normal; ##P<0.01,
###P<0.001. PRUNE2, prune homolog 2 with BCH domain;
sh, short hairpin; CRC, colorectal cancer; OD, optical density;
ctrl, control; NC, negative control. |
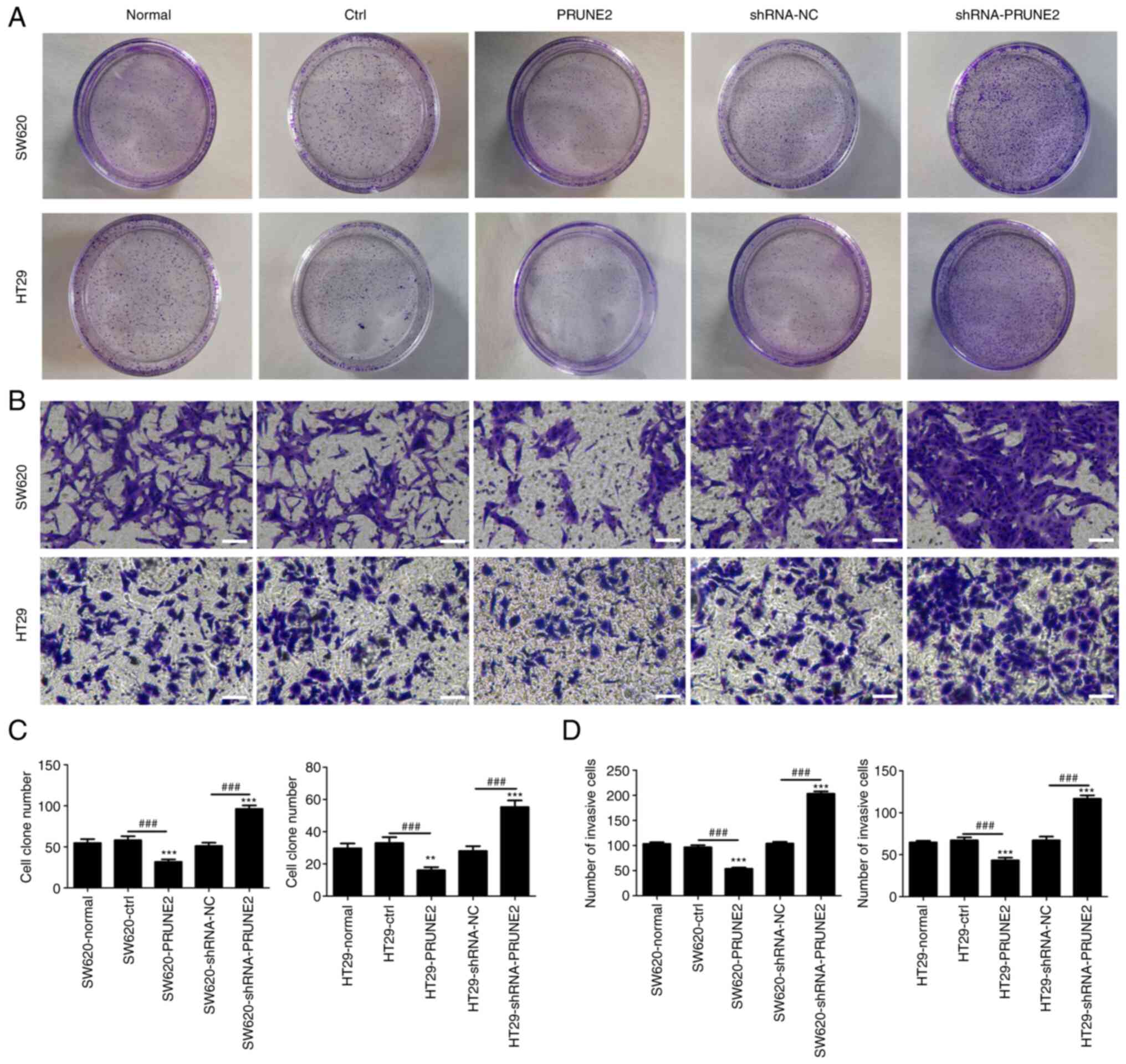
Effect of PRUNE2 on colony formation
and invasion of CRC cell lines
To determine the effect of PRUNE2 on colony
formation and invasion, colony formation and Transwell assays were
performed on two CRC cell lines. PRUNE2 overexpression
significantly decreased the number of cell colonies, whereas PRUNE2
knockdown significantly increased the number of cell colonies
(Fig. 3A and C), suggesting that PRUNE2 may be involved
in regulating clonogenic ability of CRC cells. Transwell assay
demonstrated that the number of invading cells in the PRUNE2 group
was significantly decreased and that in the shRNA-PRUNE2 group was
significantly increased compared with the ctrl and shRNA-NC groups,
respectively (Fig. 3B and D). These results indicated that PRUNE2
inhibited proliferation and invasion of CRC cell lines.
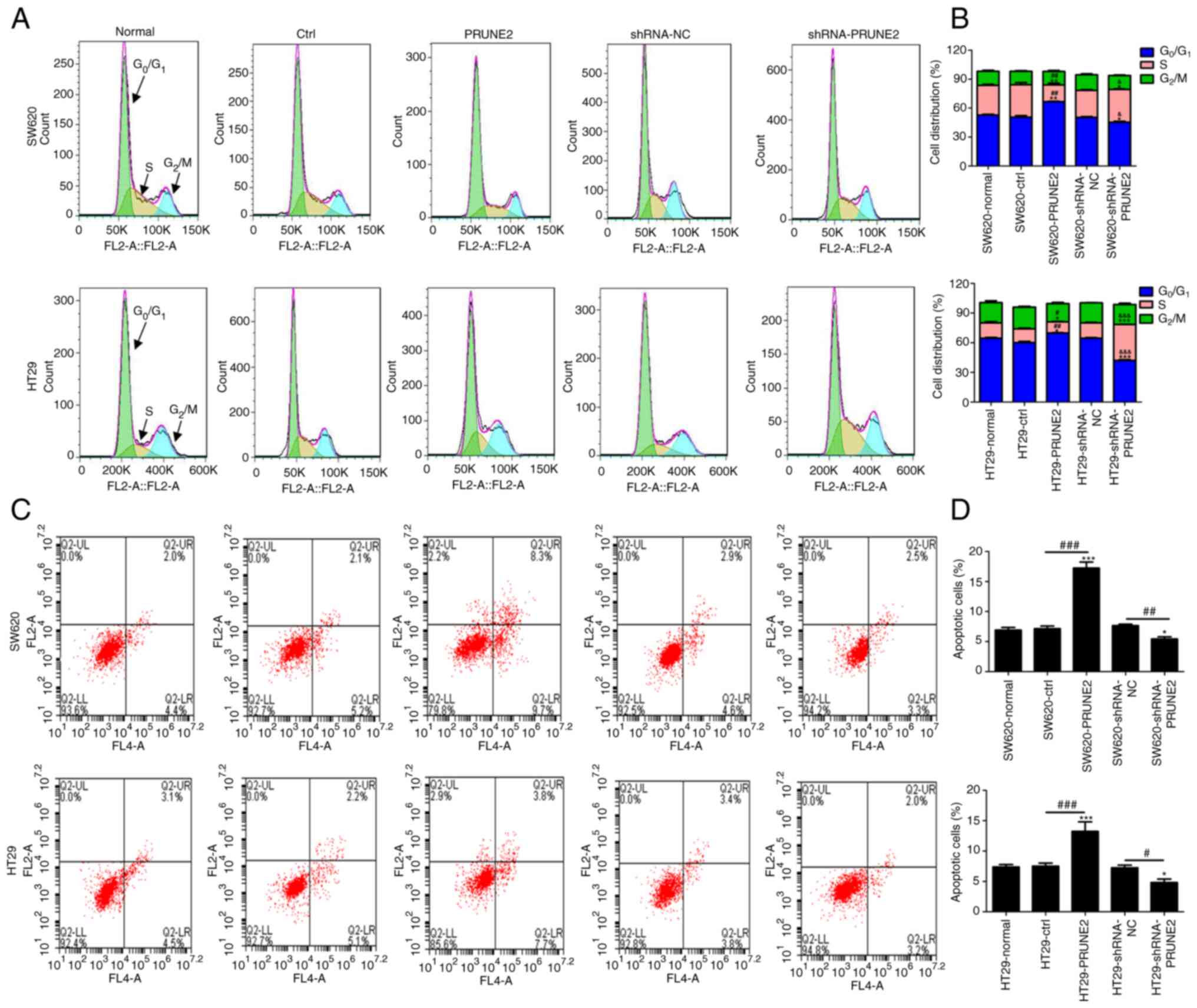
PRUNE2 overexpression arrests CRC
cells at G0/G1 stage and induces
apoptosis
The effect of PRUNE2 on CRC cell cycle progression
was detected by flow cytometry. The results indicated increased
accumulation of CRC cells in G0/G1 phase in
the PRUNE2 group compared with the ctrl (Fig. 4A and B). This was accompanied by a significant
decrease in the percentage of cells in S phase. PRUNE2 knockdown
decreased the percentage of cells in G0/G1
phase, and increased the percentage of cells in S phase. To
determine the effect of PRUNE2 on cell apoptosis, annexin/PI double
staining kit and flow cytometry were used. The results indicated
that apoptosis was significantly decreased in CRC cells transfected
with PRUNE2 shRNA and significantly increased in CRC cells
transfected with PRUNE2 overexpressing cells compared with NC and
ctrl, respectively (Fig. 4C and
D). These data indicated that
PRUNE2 overexpression prevented G0/G1 to S
stage transition and promoted cell apoptosis.
PRUNE2 expression in CRC cells
decreases protein expression of apoptosis markers
As aforementioned, PRUNE2 induced cell apoptosis.
Hence, western blotting was used to detect protein levels of Bcl-2,
Bax, caspase-3, caspase-9 and Cyclin D1. PRUNE2 overexpression
significantly increased expression of proapoptotic proteins Bax,
caspase-3 and caspase-9 and significantly decreased expression of
antiapoptotic proteins Bcl-2 and Cyclin D1 in CRC cell lines
(Fig. 5A and B). These results indicated PRUNE2
overexpression promoted the expression of apoptotic markers.
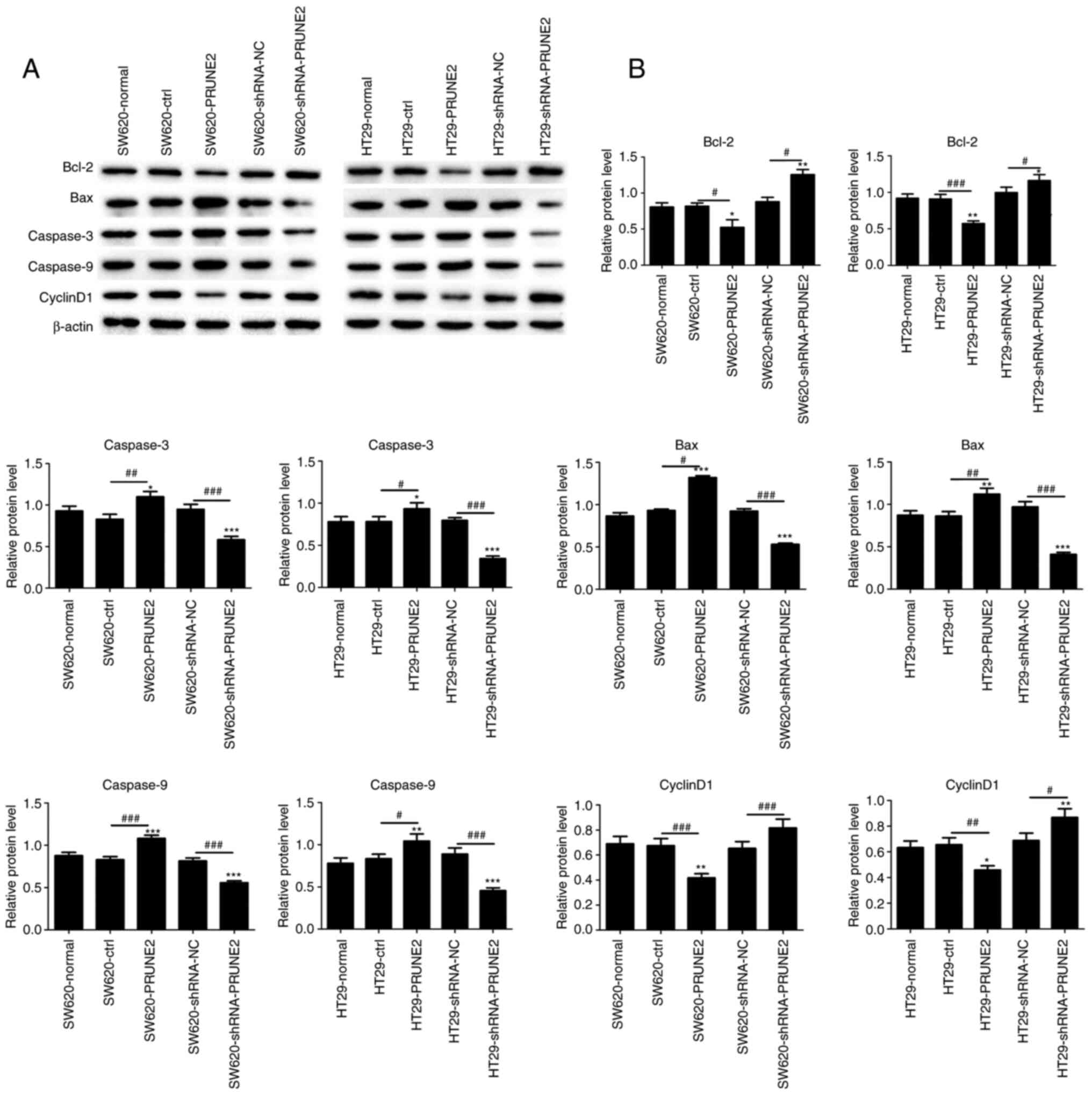 | Figure 5Effect of PRUNE2 on expression of
apoptotic markers in colorectal cells. (A) Western blot analysis of
expression levels of apoptotic (Bax, caspase-3 and caspase-9) and
antiapoptotic proteins (Bcl-2 and Cyclin D1) with β-actin as a
loading control. (B) Quantitative results of Bcl-2, Bax, caspase-3,
caspase-9 and cyclin D1 protein levels relative to β-actin. Data
are presented as the mean ± SD (n=3). Data were analyzed using
one-way ANOVA. *P<0.05, **P<0.01 and
***P<0.001 vs. normal; #P<0.05,
##P<0.01 and ###P<0.001. PRUNE2, prune
homolog 2 with BCH domain; ctrl, control; sh, short hairpin; NC,
negative control. |
Tumor growth is inhibited by PRUNE2
overexpression in vivo
To investigate the putative role of PRUNE2 in CRC
cells in vivo, tumorigenic potential of PRUNE2-GFP-SW620
cells was assessed in a mouse tumor xenograft model. Expression
levels of PRUNE2 in screened PRUNE2-GFP-SW620 and NC-GFP-SW620
cells were detected by RT-qPCR, showed significant high expression
in the PRUNE2-GFP-SW620 group compared with that of the
NC-GFP-SW620 group (Fig. 6A). The
size, weight and growth curves of xenograft tumors were
significantly decreased in the PRUNE2 overexpression compared with
the NC-GFP group (Fig. 6B-D).
Fluorescence in vivo bioluminescence imaging demonstrated
that PRUNE2 overexpression markedly inhibited tumorigenic ability
of CRC cells and tumor growth in mice at 5, 10, 15, 20 and 25 days
after inoculation (Fig. 6E). These
results indicated that PRUNE2 overexpression suppressed the
proliferation of CRC cells in vivo and confirmed results
obtained in vitro.
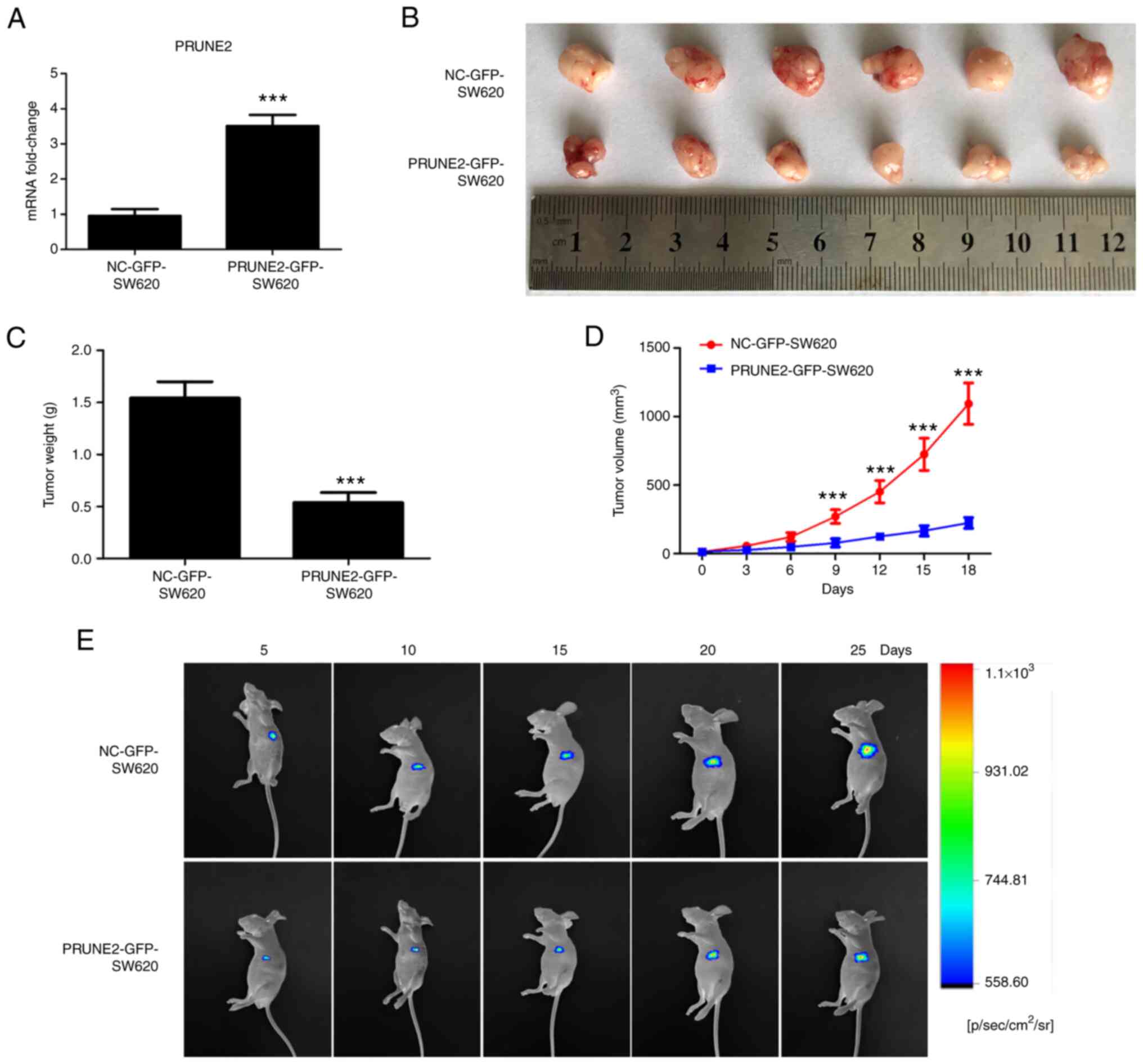 | Figure 6PRUNE2 overexpression inhibits
tumorigenic ability of SW620 cells in vivo. (A) Expression
levels of PRUNE2 in NC-GFP-SW620 and PRUNE2-GFP-SW620 cells were
measured by reverse transcription-quantitative PCR. (B)
Representative images of mouse xenograft tumors derived from
NC-GFP-SW620 and PRUNE2-GFP-SW620 groups. Subcutaneous tumors were
collected 32 days after inoculation. (C) Tumor weight was measured.
Data were analyzed using Student's t-test. (D) Growth curves
(volume) of xenograft tumors in NC-GFP-SW620 and PRUNE2-GFP-SW620
groups at 0, 3, 6, 9, 12, 15 and 18 days after emergence of the
subcutaneous tumors. Data were analyzed using two-way ANOVA. (E)
Fluorescence of cell inoculation sites in mice were observed using
an In Vivo Bioluminescence imaging system at 5, 10, 15, 20
and 25 days after inoculation. Data are presented as the mean ± SD
(n=6). ***P<0.001 vs. NC-GFP-SW620. PRUNE2, prune
homolog 2 with BCH domain; NC, negative control; p, pico; sr,
steradian. |
Discussion
Due to its high incidence, late diagnosis and poorly
understood molecular mechanisms, there is a lack of effective
treatments for CRC, especially for patients with advanced disease
(15). There is therefore a need
to characterize the disease and identify novel promising
treatments. PRUNE2, a human homolog of the Drosophila prune
gene, is regulated by long non-coding RNAs in human prostate cancer
and serves as a tumor suppressor (16). PRUNE2 homolog 2 is a susceptibility
gene for Alzheimer's disease and an important regulator of Rho
signaling (9,17). PRUNE2 is constitutively expressed
in adult nerve (18) and prostate
tissue (19), which indicates
that, in addition to promoting apoptosis in neuroblastoma, PRUNE2
may serve a physiological role in these tissues. PRUNE2 is
downregulated in epithelial-derived skin, prostate and colon cancer
tissue (8), which suggests that
inhibition of PRUNE2 expression may be associated with tumor
progression. However, the function of PRUNE2 in CRC is still
unknown. To the best of our knowledge, the present study is the
first to demonstrate that PRUNE2 serves a key role in CRC cells
both in vivo and in vitro.
Using clinical data from TCGA, PRUNE2 was shown to
be downregulated in colorectal tumor samples compared with normal
colorectal tissue. Although most prior studies have reported that
PRUNE2 expression is decreased in various types of cancer (6,10,11,16),
including prostate cancer, neuroblastoma and leiomyosarcoma, few
studies have investigated PRUNE2 expression in CRC. Low PRUNE2
expression is associated with poor long-term clinical outcomes in
patients with CRC (20).
Meta-analysis of patients treated with bevacizumab identified
PRUNE2 as a prognostic biomarker for CRC (20). This supports the hypothesis that
PRUNE2 serves as tumor suppressor gene of CRC. The present
immunohistochemistry and western blot assays also exhibited low
expression of PRUNE2 in CRC tissue. These results suggested that
PRUNE2 may be a human CRC suppressor. Numerous studies have
revealed that PRUNE2 expression is associated with various types of
cancer (6,10,11,16).
PRUNE2 may contribute to the maintenance of mature nervous systems
(18). In addition, PRUNE2 mRNA
expression is associated with the survival of patients with
leiomyosarcoma (21). DNA damage
induces programmed expression of PRUNE2 during apoptosis in
neuroblastoma cells (22). PRUNE2
also serves a role in suppressing prostate cancer (16). To the best of our knowledge,
however, few studies have reported PRUNE2 expression in CRC. The
functional role of PRUNE2 in the proliferation, invasion and
apoptosis of CRC cells remains to be reported.
PRUNE2 was previously reported to have low
expression in colon cancer (8),
but its function in CRC has not previously been reported. Here,
PRUNE2 overexpression suppressed the proliferation, invasion and
colony formation of CRC cells and induced cell cycle arrest in
G0/G1 stage, which suggested that PRUNE2 may
be associated with proliferative capacity of CRC cells; by
contrast, PRUNE2 silencing increased cell proliferation and
invasion. PRUNE2 regulates cell cycle transition in neuroblastoma
cells (23). The present study
observed arrest in G0/G1 phase following
overexpression of PRUNE2; these results indicated a potential role
of PRUNE2 in DNA replication. The present study demonstrated that
upregulation of PRUNE2 decreased cell proliferation. Salameh et
al (16) reported that PRUNE2
overexpression decreases proliferation of prostate cancer cells.
Flow cytometric analysis revealed that PRUNE2 overexpression
promoted apoptosis of CRC cells, suggesting that PRUNE2 may inhibit
CRC development via apoptosis inhibition. This supports the
hypothesis that PRUNE2 promotes cell death triggered by apoptotic
stimuli (8). In neuroblastoma and
prostate cancer, PRUNE2 protein is highly expressed and serves a
prognostic role (6,24). However, the association between
PRUNE2 expression and its prognostic role in CRC requires further
study. Increased methylation of PRUNE2 promoter is associated with
nodal metastasis and inversely associated with PRUNE2 expression in
head and neck cancer (25).
Further studies are required to identify potential genes that
regulate expression of PRUNE2 by regulating the promoter or
methylation of PRUNE2 and thus influence the development of CRC.
PRUNE2 regulates cell processes (26) and signaling (7), such as Rho, Ras and MAPK signaling;
it may have a similar regulation mode to other tumors or exhibit a
CRC-specific regulation pattern; further studies should investigate
this.
PRUNE2 inhibits expression of Bcl-2 and other
antiapoptotic Bcl-2 family proteins to promote mitochondrial
apoptosis in DNA-damaged cells and caspase-3 and caspase-9
activation are mediated by PRUNE2 expression (8). The present study detected expression
of apoptosis-associated proteins Bcl-2, Bax, caspase-3, caspase-9
and Cyclin D1 using western blot assay. PRUNE2 overexpression
increased expression of proapoptotic proteins and decreased
expression of antiapoptotic proteins in CRC cell lines. Cyclin D1
is a G1 phase cyclin and protooncogene that binds to and
activates cyclin-dependent kinases in G1 phase to
facilitate entry into S phase and promote cell cycle progression
(27,28).
To confirm the effect of PRUNE2 on CRC, a nude mouse
tumor xenograft model was generated. PRUNE2 overexpression markedly
inhibited the weight and growth of xenograft tumors. These results
indicated that PRUNE2 overexpression suppressed the tumorigenic
ability of CRC cells in vivo and confirmed results obtained
in vitro. Limitations exist in the present study; the
mechanism by which PRUNE2 promotes apoptosis in CRC is unknown and
the association between PRUNE2 expression and CRC development was
not analyzed. Investigation of the association between PRUNE2 level
and CRC grade and degree of malignancy is required to understand
the potential biomarker role of PRUNE2 in CRC. Future studies
should investigate the mechanism by which PRUNE2 promotes apoptosis
in CRC.
The functional role of PRUNE2 in proliferation,
invasion and apoptosis of CRC cells remains to be reported. To the
best of our knowledge, the present study is the first to report
that expression levels of PRUNE2 are associated with development of
CRC by promoting proliferation and invasion and inhibiting
apoptosis. CRC cells with higher expression of PRUNE2 showed
decreased proliferation and invasion and lower tumorigenic ability,
suggesting that PRUNE2 may be involved in CRC development and may
be a potential tumor suppressor in CRC. Further investigation of
the association between PRUNE2 and CRC is required to understand
the role of PRUNE2 in CRC. The present study suggested that PRUNE2
may function as a tumor-suppressive gene in CRC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81860522), Yunnan Health
Training Project of High Level Talents (grant no. H-2018039), Joint
Foundation of Kunming Medical University and Yunnan Provincial
Science and Technology Department (grant nos. 202001AY070001-114
and 2019FE001-121), Internal Division of Yunnan Provincial Health
Commission (grant no. 2018NS0263) and Clinical Medical Center of
Yunnan Provincial Health Commission (grant no.
2020LCZXKF-XH03).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ and QG conceived and supervised the study. TL, YZ
and QG designed the study. TL, SH and WY performed the experiments
and analyzed the data. TL, YZ and QG confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Affiliated Hospital of Kunming University of
Science and Technology (approval no. KHLL2021-YJ097) and the Animal
Ethics Committee of Kunming University of Science and Technology
(approval no. 202060446). Written informed consent was obtained
from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gu MJ, Huang QC, Bao CZ, Li YJ, Li XQ, Ye
D, Ye ZH, Chen K and Wang JB: Attributable causes of colorectal
cancer in China. BMC Cancer. 18(38)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cheng L, Eng C, Nieman LZ, Kapadia AS and
Du XL: Trends in colorectal cancer incidence by anatomic site and
disease stage in the United States from 1976 to 2005. Am J Clin
Oncol. 34:573–580. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vinson KE, George DC, Fender AW, Bertrand
FE and Sigounas G: The notch pathway in colorectal cancer. Int J
Cancer. 138:1835–1842. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Geissler K and Zach O: Pathways involved
in Drosophila and human cancer development: The notch,
hedgehog, wingless, runt, and trithorax pathway. Ann Hematol.
91:645–669. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Machida T, Fujita T, Ooo ML, Ohira M,
Isogai E, Mihara M, Hirato J, Tomotsune D, Hirata T, Fujimori M, et
al: Increased expression of proapoptotic BMCC1, a novel gene with
the BNIP2 and Cdc42GAP homology (BCH) domain, is associated with
favorable prognosis in human neuroblastomas. Oncogene.
25:1931–1942. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pan CQ and Low BC: Functional plasticity
of the BNIP-2 and Cdc42GAP Homology (BCH) domain in cell signaling
and cell dynamics. FEBS Lett. 586:2674–2691. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tatsumi Y, Takano R, Islam MS, Yokochi T,
Itami M, Nakamura Y and Nakagawara A: BMCC1, which is an
interacting partner of BCL2, attenuates AKT activity, accompanied
by apoptosis. Cell Death Dis. 6(e1607)2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Soh UJ and Low BC: BNIP2 extra long
inhibits RhoA and cellular transformation by Lbc RhoGEF via its BCH
domain. J Cell Sci. 121:1739–1749. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhao LR, Tian W, Wang GW, Chen KX and Yang
JL: The prognostic role of PRUNE2 in leiomyosarcoma. Chin J Cancer.
32:648–652. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Price ND, Trent J, El-Naggar AK, Cogdell
D, Taylor E, Hunt KK, Pollock RE, Hood L, Shmulevich I and Zhang W:
Highly accurate two-gene classifier for differentiating
gastrointestinal stromal tumors and leiomyosarcomas. Proc Natl Acad
Sci USA. 104:3414–3419. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cancer Genome Atlas Network. Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vaidyanathan S, Cato K, Tang L, Pavey S,
Haass NK, Gabrielli BG and Duijf PH: In vivo overexpression of Emi1
promotes chromosome instability and tumorigenesis. Oncogene.
35:5446–5455. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kazemi T, Younesi V, Jadidi-Niaragh F and
Yousefi M: Immunotherapeutic approaches for cancer therapy: An
updated review. Artif Cells Nanomed Biotechnol. 44:769–779.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Salameh A, Lee A, Cardó-Vila M, Nunes DN,
Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM,
Hosoya H, et al: PRUNE2 is a human prostate cancer suppressor
regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad
Sci USA. 112:8403–8408. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Potkin SG, Guffanti G, Lakatos A, Turner
JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, et
al: Hippocampal atrophy as a quantitative trait in a genome-wide
association study identifying novel susceptibility genes for
Alzheimer's disease. PLoS One. 4(e6501)2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Iwama E, Tsuchimoto D, Iyama T, Sakumi K,
Nakagawara A, Takayama K, Nakanishi Y and Nakabeppu Y:
Cancer-related PRUNE2 protein is associated with nucleotides and is
highly expressed in mature nerve tissues. J Mol Neurosci.
44:103–114. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Harris JL, Richards RS, Chow CW, Lee S,
Kim M, Buck M, Teng L, Clarke R, Gardiner RA and Lavin MF: BMCC1 is
an AP-2 associated endosomal protein in prostate cancer cells. PLoS
One. 8(e73880)2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Quintanilha JCF, Wang J, Sibley AB, Xu W,
Espin-Garcia O, Jiang C, Etheridge AS, Ratain MJ, Lenz HJ,
Bertagnolli M, et al: Genome-wide association studies of survival
in 1520 cancer patients treated with bevacizumab-containing
regimens. Int J Cancer. 150:279–289. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yang JL, Cogdell D, Eddy J, Trent J, Price
N and Zhang W: Expression of PRUNE2 mRNA and its positive
correlation with non-coding RNA PCA3 in leiomyosarcoma. Zhonghua
Zhong Liu Za Zhi. 34:497–500. 2012.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
22
|
Islam MR, Takano R, Yokochi T, Akter J,
Nakamura Y, Nakagawara A and Tatsumi Y: Programmed expression of
pro-apoptotic BMCC1 during apoptosis, triggered by DNA damage in
neuroblastoma cells. BMC Cancer. 19(542)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Islam MS, Tatsumi Y, Takano R, Yokochi T,
Akter J, Ozaki T, Nakamura Y, Ohira M and Nakagawara A:
Transcriptional regulation of BMCC1 mediated by E2F1 in
neuroblastoma cells. Biochem Biophys Res Commun. 478:81–86.
2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Clarke RA, Zhao Z, Guo AY, Roper K, Teng
L, Fang ZM, Samaratunga H, Lavin MF and Gardiner RA: New genomic
structure for prostate cancer specific gene PCA3 within BMCC1:
Implications for prostate cancer detection and progression. PLoS
One. 4(e4995)2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang
CY, Tang CH, Lee YC and Yang SF: A novel melatonin-regulated lncRNA
suppresses TPA-induced oral cancer cell motility through
replenishing PRUNE2 expression. J Pineal Res.
71(e12760)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bozgeyik E, Kocahan S, Temiz E and Bagis
H: miR-19a and miR-421 target PCA3 long non-coding RNA and restore
PRUNE2 tumor suppressor activity in prostate cancer. Mol Biol Rep:
Nov 27, 2021 (Epub ahead of print). doi:
10.1007/s11033-021-06996-5.
|
|
27
|
Masuda M, Hirakawa N, Nakashima T,
Kuratomi Y and Komiyama S: Cyclin D1 overexpression in primary
hypopharyngeal carcinomas. Cancer. 78:390–395. 1996.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bayat Z, Ghaemi Z, Behmanesh M and Soltani
BM: Hsa-miR-186-5p regulates TGFβ signaling pathway through
expression suppression of SMAD6 and SMAD7 genes in colorectal
cancer. Biol Chem. 402:469–480. 2021.PubMed/NCBI View Article : Google Scholar
|