Introduction
Lysine (K) methylation of histone (H) is a type of
epigenetic modification. There are numerous sites on the lysine
residue of histone 3 (H3) that may be methylated, including H3K4,
H3K9 and H3K27. Methylation of histones have multiple effects on
gene expression. For example, H3K4 dimethylation (H3K4me2) may
trigger gene transcription; however, H3K9 dimethylation (H3K9me2)
will inhibit gene transcription (1-3).
Histone methylation status is controlled by a balance of histone
lysine methyltransferases (HKMTs) and histone lysine demethylases
(HKDMs).
Proliferation and activation of hepatic stellate
cells (HSC), and balancing extracellular matrix (ECM) synthesis and
metabolism have become research hotspots of hepatic fibrosis
treatment (4,5). There have been several reports that
modification of aberrant histone methylation status may inhibit the
proliferation and activation of HSCs to prevent the development of
hepatic fibrosis (6,7). Yang et al (6) found that the histone H3K27
methyltransferase inhibitor, DZNep, displays anti-hepatic fibrosis
activity through alteration of the aberrant H3K27 methyl group in
HSCs. Dong et al (7)
demonstrated that knockout of lysine demethylase 4D (KDM4D)
protein, a demethylase of H3K9 and H3K36, increased methylation
levels of H3K9 and H3K36 in HSCs to decrease α-smooth muscle actin
(SMA) and Col I expression, thereby inhibiting the proliferation
and activation of HSC. The results of these studies indicated that
reduction of histone methylation in activated HSCs by corresponding
inhibitors or siRNA methods may inhibit HSC proliferation and
activation. It has been demonstrated that the H3K9 methylation
level was decreased during HSC activation (7); however, it is unknown whether
modification of H3K9 methylation by histone H3K9 demethylase
inhibitors affects HSC proliferation, activation and ECM synthesis,
and metabolism in the activated HSCs.
Although numerous histone demethylase inhibitors
have been developed, including GSK-J1 for H3K27, and CP2 for H3K9
and H3K36 (8,9), there are still no inhibitors of H3K9
demethylase available. 5-carboxy-8-hydroxyquinoline (IOX1) is a
potent 8-hydroxyquinoline-based histone demethylase inhibitor that
is capable of chelation with Fe (II) in histone H3K9 demethylase
active center to impede single electron transfer to block hydroxyl
methylation, resulting in an increase of H3K9 methylation in cells
(10). So far, few studies have
reported the application of IOX1 to prevent and treat disease. Hu
et al (11) have reported
that following pre-treatment with IOX1, the percentage of cells in
the G0/G1 phase was increased and the percentage of cells in the S
phase was decreased in cells, indicating that IOX1 significantly
inhibited the proliferation of vascular smooth muscle cells by
slowing down the progression of the cell cycle from the G0/G1 to
the S phase. IOX1 stimulation on angiotensin II (Ang II)-pretreated
vascular smooth muscle cells resulted in the elevation of H3K9
methylation enrichment in the cyclin D1 gene promoter region,
therefore inhibiting cyclin D1 gene expression. Since cyclin D1 is
involved in cell proliferation, downregulation of cyclin D1 will
lead to cellular cycle arrest at the G0/G1 phase. Therefore, it was
hypothesized that IOX1 has anti-atherosclerotic activity based on
the findings that IOX1 decreases proliferation of vascular smooth
muscle cells via inhibition of cyclin D1 expression.
Based on these findings, the present study measured
the activity of IOX1 on cellular proliferation, apoptosis and
production of α-SMA, Col I, and its ECM metabolism-related enzymes,
effects of matrix metalloproteinase-1 (MMP-1) and tissue inhibitor
of metalloproteinases 1 (TIMP-1) protein expression via H3K9me2
modification in HSC cells. In addition, chromatin
immunoprecipitation (ChIP) was performed to investigate the effects
of IOX1 on H3K9me2 methylation of promoters and downregulation of
α-SMA, Col I, MMP-1 and TIMP-1 gene in HSC-LX-2 cells.
Materials and methods
Cell line and materials
The LX-2 cell line was purchased from the Cell Bank
of Culture of the Chinese Academy of Sciences. IOX1 and dimethyl
sulfoxide were obtained from MilliporeSigma. Dulbecco's modified
Eagle's medium (DMEM) and fetal bovine serum (FBS) were from Gibco
(Thermo Fisher Scientific, Inc.). Trypsin and rainbow marker were
purchased from Beijing Solabao Information Technology Co., Ltd.
Cell lysis buffer, protease inhibitors and bicinchoninic acid assay
(BCA) protein quantification kit were from Shanghai Biyuntian
Biotechnology Co., Ltd. Annexin V-FITC/PI apoptosis detection kit
was purchased from Jiangsu Kaiji Biotechnology Co., Ltd.
Anti-β-actin, anti-H3, goat anti-rabbit IgG + H, anti-H3K9
dimethylation, anti-α-SMA, anti-Col I, anti-MMP-1 and anti-TIMP-1
antibodies were purchased from Abcam. The chromatin
immunoprecipitation (ChIP) kit was purchased from MilliporeSigma
(cat. no. 17-371).
Cell culture and treatment
LX-2 cells were grown in DMEM supplemented with 10%
FBS, 100 U/ml penicillin and 100 mg/ml streptomycin. The cells were
pretreated with 5 ng/ml TGF-β (PeproTech China) for 48 h, and then
the cells were harvested for subsequent experiments.
Cellular proliferation assay
The effect of different IOX1 concentrations on LX-2
cell proliferation was detected by Real-time Cell Analysis (ACEA
Biosciences, Inc.). The RTCA cellular functional analysis system
detects the electrical impedance of adherent cells through the
electrode at the bottom of the EPlate, which reflects the number,
viability, morphology and adherence of the cells by using the cell
index (CI) as an index. Cells in the logarithmic growth phase were
seeded at a density of 5x103 cells per well. Following a
10 h synchronization with DMEM (Gibco; Thermo Fisher Scientific,
Inc.), the cells were treated with DMSO (MilliporeSigma) or IOX1 at
a final concentration of 50, 100, 200 and 400 µM at 37˚C,
respectively. Untreated cells were used as controls. The effect of
IOX1 on LX-2 cell proliferation was observed and recorded once
every 15 min for 72 h. Each assay condition was performed in
triplicate. RTCA software (xCELLigence DP System; Agilent
Technologies, Inc.) calculated corresponding CI values. The
concentration of IOX1 administered in the subsequent experiments
was determined based on the results of the RTCA experiment.
Morphological alteration of the LX-2
cells observed by inverted microscopy
LX-2 cells in the logarithmic growth phase were
randomly divided into the control group (not treated with TGF-β)
and IOX1 treatment groups (0, 100, 200 and 300 µM, respectively).
After 48 h, the morphological changes of the IOX1-treated cells
were observed under a conventional light microscopy (x100).
Flow cytometry
LX-2 cells (1x106 in an uncoated 10-cm
culture dish) were treated with 0, 100, 200 and 300 µM IOX1 at
37˚C, respectively. Untreated cells were used as negative controls.
After 48 h, the cells were collected by trypsin digestion without
EDTA. A total of 1x108 cells in each condition were
prepared as a suspension in 200 µl Annexin-VFITC binding solution
and propidium iodide staining solution. The cellular suspension was
kept on ice, away from light for 20 min and was then analyzed using
a flow cytometer.
Western blotting
LX-2 cells were collected, lysed with lysis buffer,
kept on ice for 10 min, sonicated and centrifuged at 2,000 x g for
20 min at 4˚C. The protein concentration of the supernatant was
assayed using a BCA protein quantification kit. A total of 40 µg
the sample was loaded onto an SDS-PAGE gel (10%), electrophoresed,
transferred onto a PVDF membrane, blocked with 5% skimmed milk in
TBS-0.1% Tween-20 buffer (TBST) at room temperature for 90 min, and
incubated with the primary antibodies at 4˚C overnight: Anti-α-SMA
(cat. no. 5694; 1:1,500), Col I (cat. no. 34710; 1:1,500), MMP-1
(cat. no. 134184; 1:1,500), TIMP-1 (cat. no. 211926; 1:1,500),
anti-H3K9 (cat. no. 1220; 1:500), anti-β-actin (cat. no. 8226;
1:1,500) and anti-H3 (cat. no. 1791; 1:1,000; all from Abcam). The
membrane was washed with TBST buffer three times, reacted with
HRP-labeled anti-rabbit antibody (cat. no. ab191866; Abcam;
1:4,000) at room temperature for 90 min, washed with TBST buffer
three times, and visualized by enhanced chemiluminescence
(MilliporeSigma). The probed protein bands were quantitatively
analyzed using Image Lab image analysis software (Image Lab
Software; Bio-Rad Laboratories, Inc.).
ChIP
ChIP assay was performed using a ChIP kit
(MilliporeSigma), according to the manufacturer's instructions.
LX-2 cells in a 10-cm culture dish were fixed in 3 ml 4%
formaldehyde at room temperature for 10 min, collected in a tube,
washed twice with PBS, suspended in 1 ml PBS containing 5 µl
protease inhibitors, and sonicated to break the DNA at 200-1,000 bp
(the DNA size was checked by agarose gel electrophoresis).
Subsequently, antibodies (anti-H3K9me2) were added and the mixtures
were shaken mildly overnight at 4˚C, followed by addition of 60 µl
Protein G Agarose, washing with ChIP buffer, addition of 8 µl 5M
NaCl, mixing and cross-linking overnight at 65˚C. The DNA of the IP
sample was isolated. ChIP-enriched samples were analyzed in
triplicate by quantitative PCR (qPCR) using promoter primers of
α-SMA, Col I, MMP-1 and TIMP-1. The primer sequences were as
follows: Col I forward, 5'-GGCGAGAGAGGTGAACAAGG-3' and reverse,
5'-GCCAAGGTCTCCAGGAACAC-3'; α-SMA forward,
5'-CCGAATGCAGAAGGAGATCA-3' and reverse, 5'-GTGGACAGAGAGGCCAGGAT-3';
TIMP-1 forward, 5'-TGCAGGATGGACTCTTGCAC-3' and reverse,
5'-GCATTCCTCACAGCCAACAG-3'; and MMP-1 forward,
5'-GAGTGCCTGATGTGGCTCAG-3 and reverse, 5'-TTCTCAATGGCATGGTCCAC-3'.
qPCR was performed using SYBR® Green mix from Thermo
Fisher Scientific, Inc. The reaction conditions included an initial
pre-denaturation step at 94˚C for 10 min followed by 50 cycles at
94˚C for 20 sec and 60˚C for 1 min. Data were analyzed using the
2-ΔΔCq method and normalized to input samples (12). ΔCq (normalized
ChIP)=Ct(ChIP)-[Ct(Input)-6.64].
Statistical analysis
All data were analyzed using SPSS statistical
software (version 20.0; IBM Corp.) and expressed as the mean ±
standard deviation. One-way analysis of variance was used for
comparison between groups. Half maximal inhibitory concentrations
(IC50) were automatically calculated using the
xCELLigence system of RTCA. The least significant difference test
was used for comparison between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Cytotoxicity of IOX1 on LX-2
cells
The cytotoxicity of IOX1 on LX-2 cells was analyzed
using RTCA. LX-2 cells were treated with different concentrations
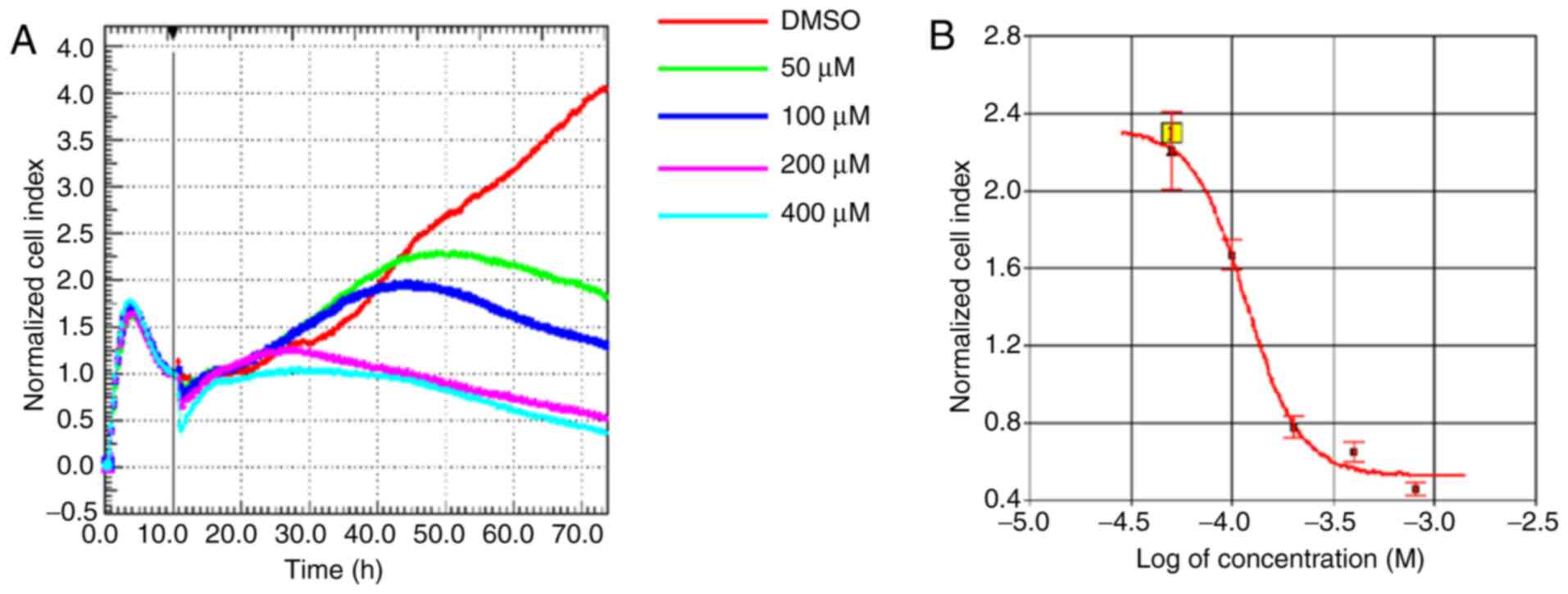
of IOX1. Live cells were counted by RTCA for 72 h. Compared with
the normal control group, cytotoxic effects of IOX1 were observed
after 20 h, which was more notable in cells treated with higher
concentrations of IOX1. A cellular viability curve of LX-2 cells
and the IOX1 concentration at 48 h was prepared based on the RTCA
results, which indicated that the IC50 of IOX1 at 48 h
was 100 µM (Fig. 1). According to
the dose-response curve, 100, 200, and 300 µM of IOX1 were selected
for subsequent experimental tests.
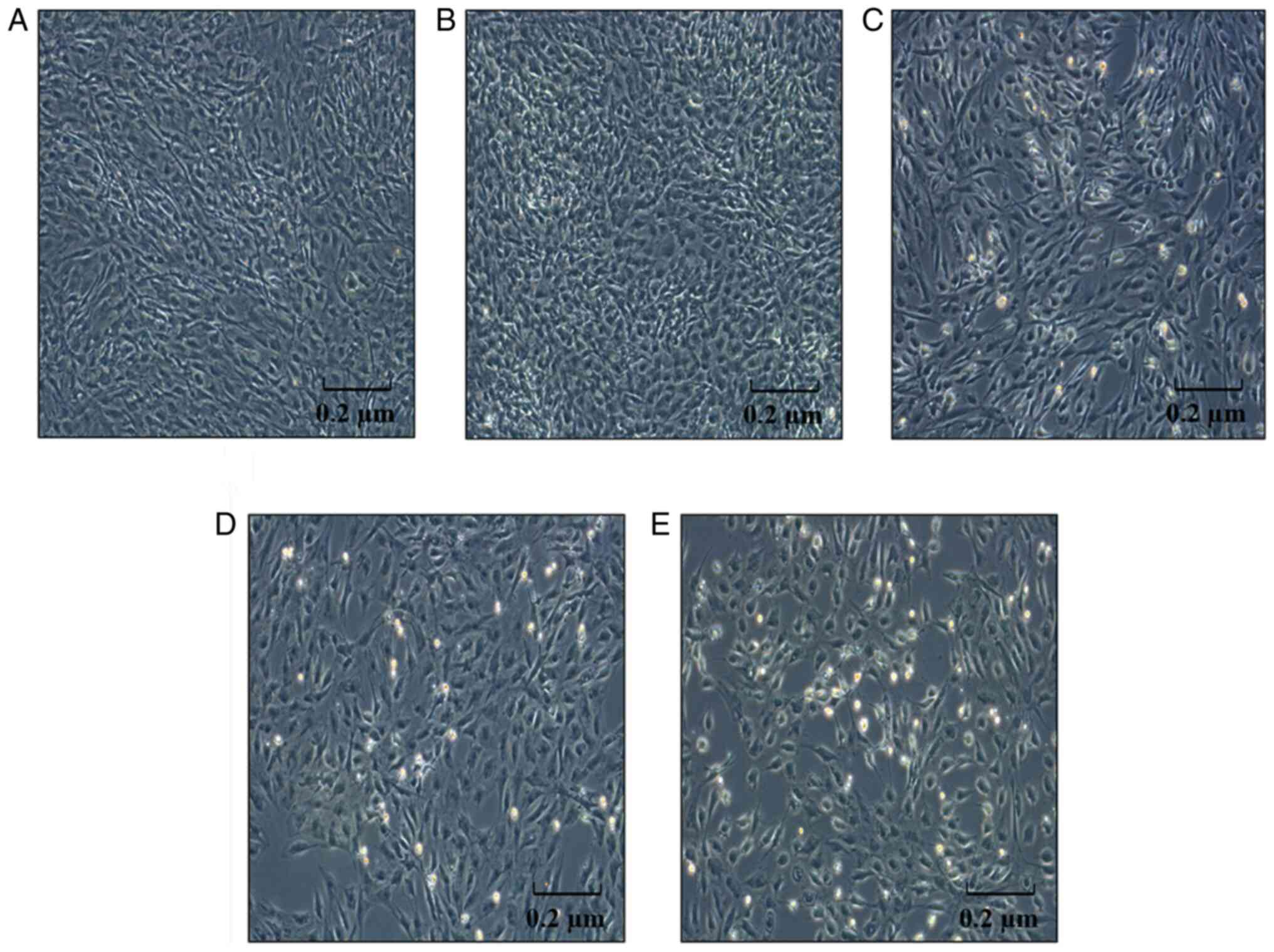
IOX1 treatment induces alterations in
LX-2 cellular morphology at 48 h
In parallel with increasing concentrations and
prolonged treatment duration of IOX1, LX-2 cells became brighter
and some were necrosed, demonstrating a concentration- and
time-dependent cytotoxicity (Fig.
2).
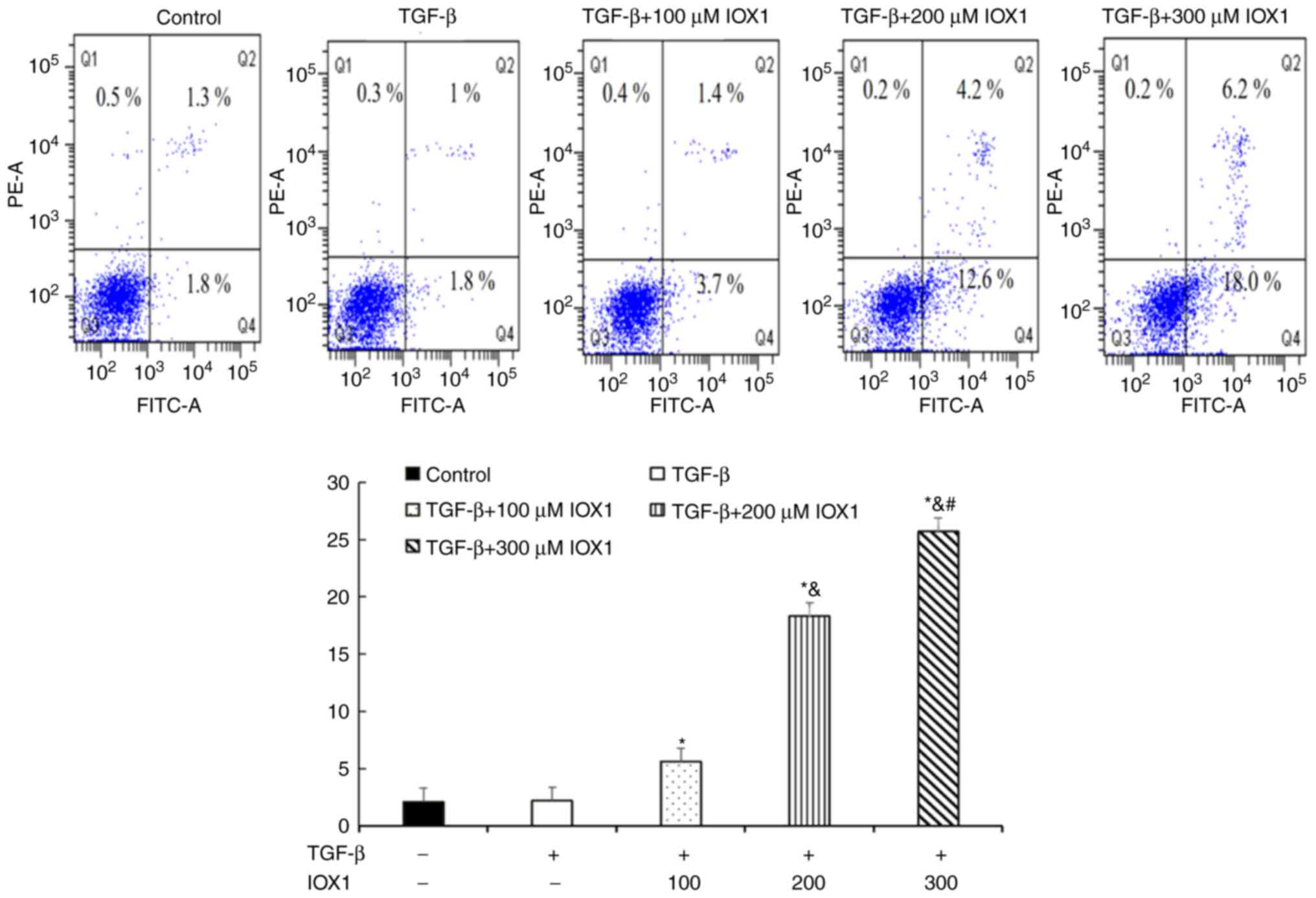
IOX1 treatment induces apoptosis of
the LX-2 cells
Using flow cytometry, cellular apoptosis of LX-2
cells treated with different concentrations of IOX1 was observed
(Fig. 3). The apoptotic rate of
cells treated with 100, 200 or 300 µM IOX1 was significantly higher
than that in the control group (P<0.05).
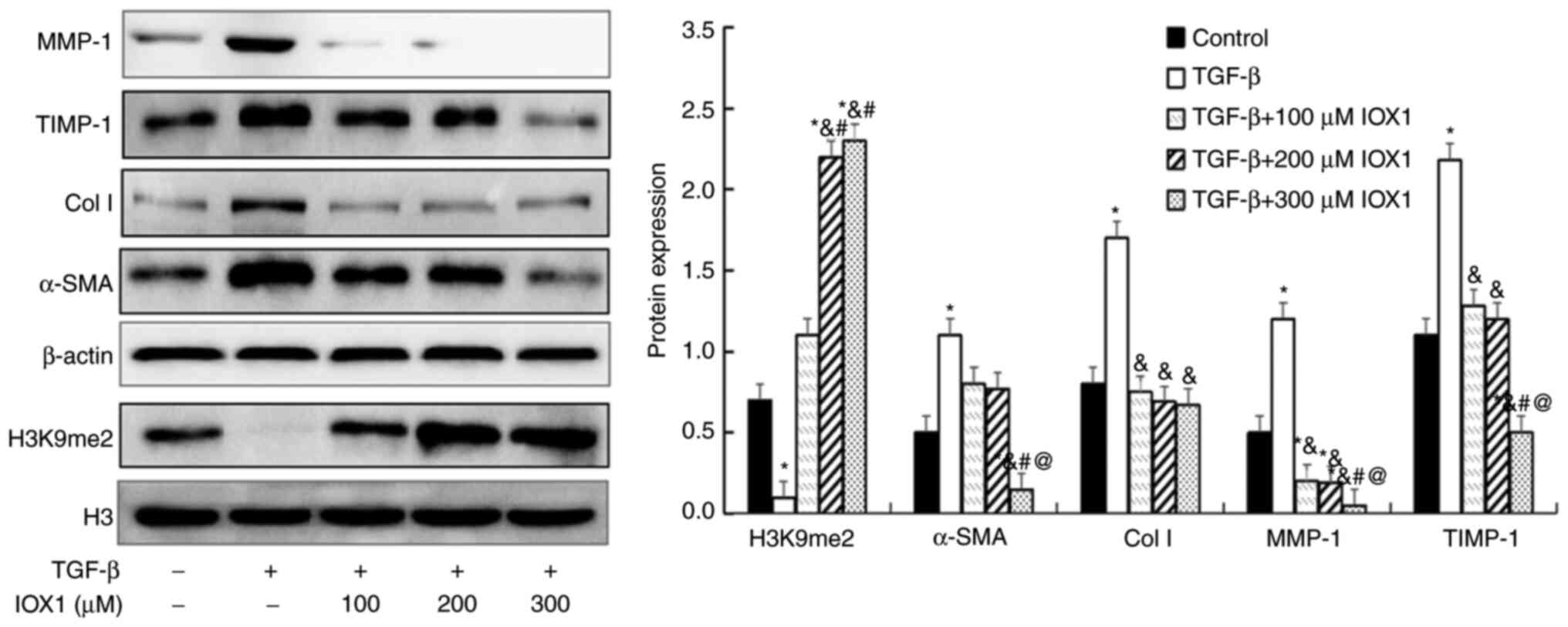
IOX1 treatment increases H3K9me2, but
decreases α-SMA, Col I, MMP-1 and TIMP-1 protein levels in
TGF-β-induced LX-2 cells
To inspect the anti-fibrotic activity and mechanism
of IOX1, H3K9me2 and protein expression of fibrosis-related factors
(α-SMA, Col I, MMP-1 and TIMP1) were measured by western blotting
in a cellular fibrosis model and in IOX1 treated cells. Compared
with controls, H3K9me2 levels were significantly increased in the
100, 200 or 300 µM IOX1-treated groups. By contrast, protein
expression of α-SMA, Col I, MMP-1 and TIMP1 were significantly
decreased in the TGF-β-induced LX-2 cells treated with 300 µM IOX1
compared with controls (P<0.05). The levels of Col I, MMP-1 and
TIMP1 were decreased in cells treated with 100-200 µM IOX1 compared
with controls (P<0.05; Fig.
4).
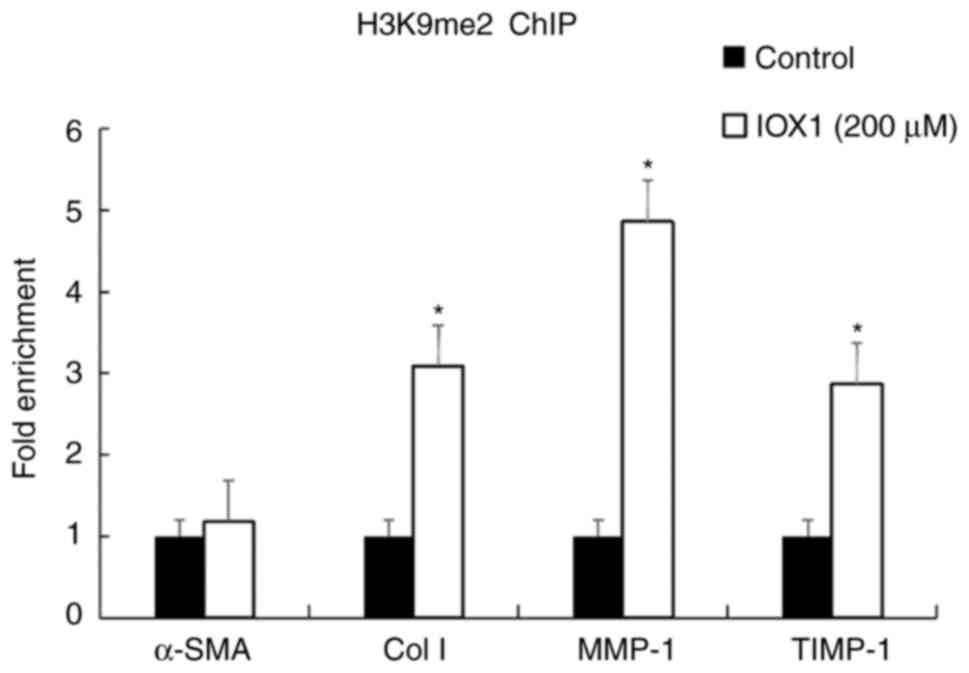
IOX1 induces H3K9 methylation at
promotor regions of fibrotic factors
To investigate the molecular mechanism of the
anti-fibrotic activity of IOX1, ChIP was performed to observe
whether IOX1 treatment may increase H3K9me2 levels at the promotor
region of the fibrotic factors (α-SMA, Col I, MMP-1 and TIMP1).
Cell lysates of LX-2 cells treated with 200 µM IOX1 for 48 h was
immunoprecipitated by H3K9me2 antibody. Total RNA of the
immunoprecipitates was then extracted, reversed transcribed into
cDNA, and then underwent qPCR with corresponding promotor region
primers of α-SMA, Col I, MMP-1 or TIMP1, respectively. Compared
with controls, H3K9me2 levels at promotor regions of MMP-1, TIMP-1
and Col I were significantly increased in IOX1-treated cells
(P<0.05). However, the H3K9me2 level at the promotor region of
α-SMA were not significantly different from the control group
(P>0.05; Fig. 5). Combined with
our previous findings in Fig. 4,
this suggested that IOX1 increases H3K9me2 at the promotor regions
of MMP-1, TIMP-1 and Col I, and may attenuate cellular fibrosis of
LX-2 induced by TGF-β.
Discussion
HSCs located in the space of Disse between
hepatocytes and sinusoidal endothelial cells, the principal
function of which is to store and metabolize vitamin A in its
physiological state. Therefore, they are referred to as lipid
storage cells. Under pathological conditions, HSCs lose their lipid
phenotype and begin to synthesize ECM. The synthetic speed of ECM
is more than that of the degradation, resulting in a large
deposition of ECM in the cells. This process is known as activation
of the HSC. Therefore, activation of HSC is a key process during
the development of hepatic fibrosis (13,14).
TGF-β is currently known as the strongest cytokine that promotes
hepatic fibrosis, which activates HSC through the TGF-β-Smad
signaling pathway to increase ECM synthesis in HSCs and alters ECM
metabolism, thereby promoting liver fibrosis development (15,16).
In the present study, the human activated HSC LX-2 cell line was
used as the research object. To mimic the activation of HSC, LX-2
cells were stimulated with TGF-β. With this cellular model, the
effect of IOX1 on H3K9 methylation and ECM metabolism was
investigated. Histone demethylase inhibitors include IOX1, JIB-04,
GSK and ML324. Among these, GSK specifically inhibits the activity
of KDM6, ML324 specifically inhibits the activity of KDM4, while
IOX1 and JIB-04 may inhibit the activity of KDM3 in addition to the
activity of KDM4 and KDM6. The only substrate of KDM3 is H3K9me,
and KDM3 may demethylate H3K9me. In the present study, IOX1 was
selected as an inhibitor of KDM3 as IOX1 treatment may inhibit the
proliferation of cardiomyocytes in angiotensin-pretreated
cardiomyocytes and IOX1 may inhibit cell cycle-associated proteins
through upregulation of H3K9me levels (11).
A 48-h intervention with IOX1 demonstrated that the
IC50 of IOX1 on the LX-2 cells was 100 µM. These data
suggested that the IC50 of IOX1 is higher than that of
other histone demethylase inhibitors. The IC50 of the
H3K27 demethylase inhibitor, GSK, and the ML324 inhibitor, JMJD2,
are 9.9 nM and 0.2 µM, respectively (17). We hypothesized that IOX1 contains a
hydrophilic hydroxyl group (18),
which may have a different polarity to the other histone
demethylase inhibitors, resulting in lower cellular permeability
and thus a relatively large IC50. Based on these data,
100, 200, 300 µM IOX1 were selected as the conditions for the
subsequent experiments. Further observations revealed that at 100
µM or more IOX1 may significantly improve H3K9me2 levels in LX-2
cells. The level of H3K9me2 was significantly increased with
increasing doses of IOX1, suggesting that 100 µM or more IOX1 may
upregulate the H3K9 dimethyl group through inhibition of the
activity or expression of H3K9 demethylase.
Cellular apoptosis analysis by flow cytometry
demonstrated that IOX1 treatment over 100 µM for 48 h significantly
promoted cellular apoptosis of the TGF-β-induced LX-2 cells and
enhanced the apoptotic rate of the cells in a dose-dependent
manner. This finding suggested that IOX1 induced cellular
morphological alterations and that death of the LX-2 cells may
occur through an apoptotic mechanism. Indeed, a large number of
studies have confirmed that decreasing activated HSCs or promoting
the apoptosis of the activated HSCs may effectively restrain the
development of liver fibrosis (19). It has been demonstrated that drugs
that promote HSC apoptosis reveal anti-fibrosis activity (20). Liu et al (20) demonstrated that tanshinone extracted
from Salvia miltiorrhiza Bunge revealed a potential anti-fibrotic
activity through regulation of the ERK-Bax-Caspase pathway to
promote HSC apoptosis. Therefore, based on these results and those
of the present study, we hypothesized that IOX1 may exert
anti-hepatic fibrosis via promoting the apoptosis of HSC cells
through the regulation of H3K9 methylation.
Morphological and behavioral alterations (including
cellular mobility and contractility) of HSCs during fibrosis
recruit cytoskeletal proteins, including α-SMA, which serve an
important role in this morphological change (21,22).
Therefore, the expression of α-SMA is considered a marker of HSC
activation (23,24). In addition, activated HSCs express a
large number of ECMs, the most abundant being Col I (25,26).
Therefore, repression of HSC activation through a decrease in ECM
synthesis, particularly Col I, is an effective strategy for
treating liver fibrosis. In the present study, western blot
analysis indicated that IOX1 above 100 µM significantly
downregulated the expression of Col I in LX-2 cells, but 100-200 µM
IOX1 did not alter α-SMA levels. However, 300 µM IOX1 significantly
decreased α-SMA protein expression, suggesting that IOX1 may
inhibit the synthesis of Col I and α-SMA. Further analysis using
ChIP assays demonstrated that H3K9me2 expression in the promoter
region of the Col I was significantly increased in LX-2 cells
treated with 200 µM IOX1 for 48 h. This indicated that IOX1 may
decrease Col I gene expression via elevation of the H3K9me2 level
at promotor region. However, no notable alteration of H3K9me2 was
observed in the α-SMA promotor region in this experiment.
Perugorria et al (27)
reported that the expression of α-SMA gene in activated HSCs was
controlled by H3K4me3. Therefore, we hypothesized that IOX1 has no
direct regulatory effect on the expression of α-SMA gene.
The increase in ECM accumulation during HSC
activation is not the only factor in ECM deposition. Indeed, ECM
degradation serves a more critical role in the development of liver
fibrosis (28,29). MMP-1 is a metalloproteinase that
degrades collagen-based ECM, whose activity is inhibited by TIMP-1
(30,31). During hepatic fibrosis, the
expression of MMP-1 is downregulated, resulting in decreased ECM
degradation. An increase in TIMP-1 expression inhibits the activity
of MMP-1, leading to an imbalance of ECM synthesis and metabolism,
which ultimately leads to the development of hepatic fibrosis
(32). Therefore, a correction of
this imbalance between MMP-1 and TIMP-1 to promote degradation of
ECM may effectively reverse the occurrence and development of
hepatic fibrosis. The results of the present study demonstrated
that the MMP-1 protein expression level was higher in the TGF-β
stimulated cells, but quickly declined in the TGF-β-induced LX-2
cells stimulated with 100-300 µM IOX1 for 48 h. In our previous
experiments, it was found that MMP-1, TIMP-1 and the ratio of
TIMP-1/MMP-1 were upregulated during hepatic fibrosis, resulting in
increased ECM deposition, thereby promoting the occurrence and
development of hepatic fibrosis (33). Based on these results and those of
our previous study, we hypothesized that the increase of MMP-1 in
the TGF-β-stimulated cells was due to the upregulation of Col I
expression, while at the concentration of 100-300 µM IOX1, the
expression of Col I in the cells is downregulated, at which time
the cells no longer need to produce a large amount of MMP-1 to
degrade ECM, resulting in the decreased expression of MMP-1.
Wang et al (34) reported that expression of histone
H3K4 demethylase and retinal-binding protein (RBP2) were increased
in the livers of patients with cirrhosis, compared with normal
healthy controls. Treatment with RBP2 siRNA in the TGF-β-induced
HSC LX-2 cell line increased the H3K4 methylation level and
downregulated α-SMA and vimentin, thereby inhibiting the
proliferation and activation of HSCs. The results of the present
study demonstrated that the TIMP-1 protein expression level was
markedly downregulated with IOX1 treatment. Further analysis with
H3K9me2 ChIP suggested that the level of H3K9me2 enrichment in the
promoter region of MMP-1 and TIMP-1 genes was increased after 48-h
stimulation with 300 µM IOX1 in the LX-2 cells. As a demethylase
inhibitor, IOX1 may upregulate H3K9 methylation, thereby inhibiting
gene transcription; therefore, the expression level of MMP-1 is
lower than the baseline. However, protein expression of MMP-1 does
not equate to its activity. TIMP-1, as an MMP-1 inhibitory protein,
may inhibit MMP-1 activity. A total of 300 µM IOX1 treatment
downregulated TIMP-1 levels and the inhibitory effect on MMP-1
activity was weakened. These data demonstrated that IOX1 may
inhibit the expression of TIMP-1 protein through H3K9me2, thereby
decreasing the inhibition of MMP-1 activity by TIMP-1 to improve
the degradation of extracellular matrix proteins by MMP-1.
The histone demethylase inhibitor IOX1 not only
inhibited LX-2 cell proliferation and activation, and promoted LX-2
apoptosis, but also upregulated H3K9me2 levels in the promoter
region of Col I, MMP-1 and TIMP-1 genes. These alterations may
decrease the synthesis of Col I and increase the degradation of ECM
through activation of ECM metabolic enzymes, thereby exerting its
anti-hepatic fibrotic activity.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Guizhou
Medical University 2018 Academic Seedling Cultivation and
Innovation Exploration Special Project [(2018)5779-19], Guizhou
Platform [(20185101)] and Guizhou Support [(20204Y126)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QY and XY conceived and designed the experiments. TT
performed the experiments. RJX and BH analyzed the data. KZD was
responsible for data acquisition. TT and QY confirm the
authenticity of the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study protocol was reviewed and approved by the
Ethics Committee of Guizhou medical university (Guiyang, China;
approval no. 1900005).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lei W, Long Y, Li S, Liu Z, Zhu F, Hou FF
and Nie J: Homocysteine induces collagen I expression by
downregulating histone methyltransferase G9a. PLoS One.
10(e0130421)2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sowa Y and Sakai T: Development of novel
epigenetic molecular-targeting agents. Nihon Rinsho. 73:1263–1267.
2015.PubMed/NCBI(In Japanese).
|
|
3
|
Füllgrabe J, Heldring N, Hermanson O and
Joseph B: Cracking the survival code: Autophagy-related histone
modifications. Autophagy. 10:556–561. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Stanković V, Mihailović V, Mitrović S and
Jurišić V: Protective and therapeutic possibility of medical herbs
for liver cirrhosis. Rom J Morphol Embryol. 58:723–729.
2017.PubMed/NCBI
|
|
5
|
Walker C, Mojares E and Del Río Hernández
A: Role of extracellular matrix in development and cancer
progression. Int J Mol Sci. 19(3028)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yang Y, Chen XX, Li WX, Wu XQ, Huang C,
Xie J, Zhao YX, Meng XM and Li J: EZH2-mediated repression of Dkk1
promotes hepatic stellate cell activation and hepatic fibrosis. J
Cell Mol Med. 21:2317–2328. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dong F, Jiang S, Li J, Zhu L, Huang Y,
Jiang X, Hu X, Zhou Q, Zhang Z and Bao Z: The histone demethylase
KDM4D promotes hepatic fibrogenesis by modulating Toll-like
receptor 4 signaling pathway. EBioMedicine. 39:472–483.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Raeisossadati R, Móvio MI, Walter LT,
Takada SH, Del Debbio CB and Kihara AH: Small molecule GSK-J1
affects differentiation of specific neuronal subtypes in developing
rat retina. Mol Neurobiol. 56:1972–1983. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Rotili D, Tomassi S, Conte M, Benedetti R,
Tortorici M, Ciossani G, Valente S, Marrocco B, Labella D,
Novellino E, et al: Pan-histone demethylase inhibitors
simultaneously targeting Jumonji C and lysine-specific demethylases
display high anticancer activities. J Med Chem. 57:42–55.
2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mettananda S, Fisher CA, Sloane-Stanley
JA, Taylor S, Oppermann U, Gibbons RJ and Higgs DR: Selective
silencing of α-globin by the histone demethylase inhibitor IOX1: A
potentially new pathway for treatment of β-thalassemia.
Haematologica. 102:e80–e84. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hu Q, Chen J, Zhang J, Xu C, Yang S and
Jiang H: IOX1, a JMJD2A inhibitor, suppresses the proliferation and
migration of vascular smooth muscle cells induced by angiotensin II
by regulating the expression of cell cycle-related proteins. Int J
Mol Med. 37:189–196. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hernández-Aquino E and Muriel P:
Beneficial effects of naringenin in liver diseases: Molecular
mechanisms. World J Gastroenterol. 24:1679–1707. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ezhilarasan D, Sokal E and Najimi M:
Hepatic fibrosis: It is time to go with hepatic stellate
cell-specific therapeutic targets. Hepatobiliary Pancreat Dis Int.
17:192–197. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tsuchida T and Friedman SL: Mechanisms of
hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol.
14:397–411. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yoshida K, Matsuzaki K, Murata M,
Yamaguchi T, Suwa K and Okazaki K: Clinico-pathological importance
of TGF-β/phospho-smad signaling during human hepatic
fibrocarcinogenesis. Cancers (Basel). 10(183)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kim MS, Cho HI, Yoon HJ, Ahn YH, Park EJ,
Jin YH and Jang YK: JIB-04, a small molecule histone demethylase
inhibitor, selectively targets colorectal cancer stem cells by
inhibiting the Wnt/β-catenin signaling pathway. Sci Rep.
8(6611)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hopkinson RJ, Tumber A, Yapp C, Chowdhury
R, Aik W, Che KH, Li XS, Kristensen JBL, King ONF, Chan MC, et al:
5-Carboxy-8-hydroxyquinoline is a broad spectrum 2-oxoglutarate
oxygenase inhibitor which causes Iron translocation. Chem Sci.
4:3110–3117. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Schwabe RF and Luedde T: Apoptosis and
necroptosis in the liver: A matter of life and death. Nat Rev
Gastroenterol Hepatol. 15:738–752. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu K, Li X, Cao Y, Ge Y, Wang J and Shi
B: miR-132 inhibits cell proliferation, invasion and migration of
hepatocellular carcinoma by targeting PIK3R3. Int J Oncol.
47:1585–1593. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sun YY, Li XF, Meng XM, Huang C, Zhang L
and Li J: Macrophage phenotype in liver injury and repair. Scand J
Immunol. 85:166–174. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Koyama Y, Xu J, Liu X and Brenner DA: New
developments on the treatment of liver fibrosis. Dig Dis.
34:589–596. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Marrone G, Shah VH and Gracia-Sancho J:
Sinusoidal communication in liver fibrosis and regeneration. J
Hepatol. 65:608–617. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Fung E and Tsukamoto H: Morphogen-related
therapeutic targets for liver fibrosis. Clin Res Hepatol
Gastroenterol. 39 (Suppl 1):S69–S74. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Omar R, Yang J, Liu H, Davies NM and Gong
Y: Hepatic stellate cells in liver fibrosis and siRNA-based
therapy. Rev Physiol Biochem Pharmacol. 172:1–37. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen PJ, Huang C, Meng XM and Li J:
Epigenetic modifications by histone deacetylases: Biological
implications and therapeutic potential in liver fibrosis.
Biochimie. 116:61–69. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Perugorria MJ, Wilson CL, Zeybel M, Walsh
M, Amin S, Robinson S, White SA, Burt AD, Oakley F, Tsukamoto H, et
al: Histone methyltransferase ASH1 orchestrates fibrogenic gene
transcription during myofibroblast transdifferentiation.
Hepatology. 56:1129–1139. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Irvine KM, Wockner LF, Hoffmann I,
Horsfall LU, Fagan KJ, Bijin V, Lee B, Clouston AD, Lampe G,
Connolly JE and Powell EE: Multiplex serum protein analysis
identifies novel biomarkers of advanced fibrosis in patients with
chronic liver disease with the potential to improve diagnostic
accuracy of established biomarkers. PLoS One.
11(e0167001)2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Magdaleno F, Arriazu E, Ruiz de Galarreta
M, Chen Y, Ge X, Conde de la Rosa L and Nieto N: Cartilage
oligomeric matrix protein participates in the pathogenesis of liver
fibrosis. J Hepatol. 65:963–971. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang H, Yang LM and Huang L: Clinical
effects of qianggan capsule on the liver tissue pathology and
PDGF-BB, TGF-beta1, TIMP-1, and MMP-1 factors in patients with
chronic hepatitis B. Zhongguo Zhong Xi Yi Jie He Za Zhi.
31:1337–1340. 2011.PubMed/NCBI(In Chinese).
|
|
31
|
Yu Y, Li ZQ, Chen K, Zhan P, Liao J and
Ruan QY: Significance and expressions of MMP-1, TIMP-1 and TGF-β1
in valve tissue of rheumatic heart disease. Sichuan Da Xue Xue Bao
Yi Xue Ban. 48:52–56. 2017.PubMed/NCBI(In Chinese).
|
|
32
|
Agrinier N, Thilly N, Boivin JM, Dousset
B, Alla F and Zannad F: Prognostic value of serum PIIINP, MMP1 and
TIMP1 levels in hypertensive patients: A community-based
prospective cohort study. Fundam Clin Pharmacol. 27:572–580.
2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tian T, Zhan W, Xie R, Yu L, Zheng L, Tang
L, Liu X, Guo X, Zhang J, Han B, et al: Epigenetic histone
methylation regulates MMP-1 expression in myofibroblastichepatic
stellate cell. Int J Clin Exp Med. 10:15187–15195. 2017.
|
|
34
|
Wang Q, Wang LX, Zeng JP, Liu XJ, Liang XM
and Zhou YB: Histone demethylase retinoblastoma binding protein 2
regulates the expression of α-smooth muscle actin and vimentin
incirrhotic livers. Brazilian J Med Biol Res. 46:739–745.
2013.PubMed/NCBI View Article : Google Scholar
|