Introduction
Glucocorticoids (GCs) are used clinically to treat a
number of diseases, and it is estimated that 1-2% of the worldwide
population are undergoing long-term GC therapy (1-4).
However, long-term administration with GCs has been shown to result
in multiple complications, common among which is GC-induced
osteoporosis (GIO), which is characterized by systemic damage of
the bone size and microarchitecture (5). A growing number of studies have
revealed that osteoblast dysfunction disrupts the balance between
bone formation and bone resorption in terms of the role that is
exerted by bone mass in GIO, for example, by inhibiting osteoblast
proliferation and differentiation, as well as enhancing the rate of
osteoblast apoptosis (5-7).
Cell senescence is considered to be a state of cell
cycle arrest, as well as physiological and metabolic dysfunction,
which occurs under normal environmental conditions, with a gradual
increase in its prevalence towards death (8). A previous study showed that cell
senescence serves an important role in the development of
osteoporosis (9). However, whether
osteoblast senescence contributes to the development of GIO has yet
to be elucidated.
Hydrogen sulfide (H2S) is an endogenously
generated secondary messenger that is involved in regulating a wide
range of physiological processes, including having cardioprotective
and neuroprotective effects, as well exerting a beneficial role
following acute lung injury (10-12).
Furthermore, H2S also has a critical role in the
occurrence and development of osteoporosis (13,14).
Xu et al (13) demonstrated
that H2S attenuates oxidative stress-induced
osteoblastic cell damage and inhibits cell proliferation, as well
as differentiation, through a MAPK-dependent signaling pathway.
Additionally, Yang et al (14) found that H2S mitigates
dexamethasone (Dex)-induced osteoblast injury by activating the
AMP-activated protein kinase signaling (14); however, whether H2S
inhibits Dex-induced osteoblast dysfunction through repressing cell
senescence has yet to be elucidated.
MicroRNAs (miRs), as a class of highly conserved
noncoding RNAs, which are able to post-transcriptionally modulate
the expression of target genes and serve a critical role in various
diseases (15-17).
Numerous studies have shown that miRs contribute to the
pathogenesis and development of osteoporosis (18-20).
Notably, previous research showed that miR-22 was negatively
correlated with the function of various cells (17,21,22).
In the present study, alterations in the expression profile of
biomarkers, such as p53 and p21, which are associated with cell
senescence in Dex-treated MC3T3-E1 cells, and the effects of sodium
hydrosulfide (NaHS) on Dex-induced osteoblast damage and
senescence, were examined. Subsequently, the expression levels of
miR-22 in Dex-treated osteoblast cells with or without NaHS
administration were investigated to determine whether miR-22 is
involved with the protective role that NaHS has in Dex-induced
osteoblast dysfunction and cell senescence.
Materials and methods
Cell culture and drug
administration
Osteoblastic murine MC3T3-E1 cells were cultured in
α-MEM (α-MEM, Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS
(Thermo Fisher Scientific, Inc.) at a temperature of 37˚C and in an
atmosphere containing 5% CO2/95% air. Absolute ethanol
was used to dissolve the Dex (Sigma-Aldrich; Merck KGaA) and the
cells were treated with Dex at a final concentration of 1 µM for 72
h, as previously described (23).
The p53 inhibitor, pifithrin-α (Sigma-Aldrich; Merck KGaA) was
dissolved in DMSO and the cells were treated with pifithrin-α at a
concentration of 20 µM for 48 h (23). NaHS (Sigma-Aldrich; Merck KGaA) was
dissolved in saline solution and used at 20 µM for 72 h. Dex and
NaHS after 24 h of cell culture, and pifithrin-α was used after 48
h. The cell number used in these experiments was 2x105
cells/ml (200 µl each well in 48-well plate and 2 ml each well in
6-well plate).
Small interfering (si)RNA transfection
assay
Xfect™ RNA transfection reagent (Takara
Biotechnology Co., Ltd.) was used to perform sirtuin 1 (sirt1)
siRNA transfections (200 nM; cat. no. sc-40987; Santa Cruz
Biotechnology, Inc.) into osteoblastic MC3T3-E1 cells, with a
non-targeting siRNA (Santa Cruz Biotechnology, Inc.) served as
negative control. The sequences of control siRNA are as follows:
5'-UUCUUCGAACGUGUCACGUTT-3' and 5'-ACGUGACACGUUCGGAGAATT-3'.
Osteoblasts were pretreated with control or sirt1 siRNA for 24 h
and then immediately subjected to subsequent experimentation.
Cell viability analysis
MTT detection assays were performed to measure the
viability of the osteoblasts. Briefly, osteoblastic cells were
cultivated in a 48-well plate at a density of 2x105
cells/ml (200 µl each well) at 37˚C and the cells were subjected to
Dex and NaHS treatment for 72 h, and pifithrin-α. was added for the
last 48h. Cells were then exposed to MTT for 2-4 h at 37˚C in the
dark and a microplate reader was used to detect absorbance of
formazan crystals dissolved in DMSO at 550 nm.
Alkaline phosphatase (ALP) activity
detection
The method described by Bowers and McComb was
performed to measure ALP activity (24). Briefly, Triton X-100 was added into
1x105 osteoblasts at a final concentration of 1%. The
supernatant was collected by centrifugation at 14,000 x g at 4˚C
for 20 min. A total of 25 µl supernatant was added into the
reaction mixture, which contained assay buffer and para-nitrophenyl
phosphate (cat. no. P0321S; Alkaline Phosphatase Assay kit;
Beyotime Institute of Biotechnology). Subsequently, the mixture was
incubated for 10 min at 37˚C and a microplate reader was used to
measure the absorbance of p-nitrophenol at 405 nm after termination
of the reaction.
Dual luciferase assay
The psiCHECK2 luciferase reporter plasmid (Promega
Corporation) was used to clone the synthesized wild-type (WT) and
mutant 3'-untranslated region (UTR) of sirt1. The psiCHECK2 plasmid
with WT or mutant derivatives and miRNA control or miR-22 mimic
were co-transfected into the osteoblasts. Xfect™ RNA transfection
reagent (Takara Biotechnology Co., Ltd.) was used to perform
plasmid and miRNA control, as well as miR-22 mimic transfection.
Cell lysates were collected 24 h post-transfection and the
dual-luciferase reporter system (Dual-Luciferase®
Reporter Assay System; cat. no. E1910; Promega Corporation) was
used to measure firefly and Renilla luciferase activity. The
ratio of luminescence between the experimental reporter (firefly)
to the control reporter (Renilla) was used to reflect the
relative luciferase activity of sirt1.
Reverse transcription-quantitative PCR
(qPCR)
Total RNA was extracted from the osteoblasts using
TRIzol® (Takara Biotechnology Co., Ltd.) and reverse
transcribed into cDNA using SuperScript™ II reverse transcriptase
(Thermo Fisher Scientific, Inc.) with a special stem-loop primer
for miR-22 and oligodeoxythymidine for mRNAs with the following
temperature protocol: 25˚C for 10 min; 42˚C for 1 h and 72˚C for 10
min). A MiniOpticon real-time PCR detection system (Bio-Rad
Laboratories, Inc.) was used to perform qPCR. The reaction solution
consisted of 5.0 µl diluted cDNA, 0.2 µM/l of each paired primer,
1x Sybr Green qPCR Mix buffer (Toyobo Biotech Co., Ltd.) and 4.9 µl
DEPC-treated water. The annealing temperature was set at 58-62˚C
and amplification was set at 40 cycles. The temperature range to
detect the melting temperature of the PCR product was set from
60-95˚C. To determine the relative quantitation of gene expression,
the comparative Cq (threshold cycle) method with arithmetic
formulae (2-ΔΔCq) was used (25). mRNA levels of sirt1, p53 and p21
were normalized relative to the house-keeping gene β-actin. The
primers used were as follows: Sirt1 (accession number
NM_001159589.2) sense strand, 5'-CTGTTTCCTGTGGGATACCTGACT-3' and
antisense strand, 5'-ATCGAACATGGCTTGAGGATCT-3' (26); p53 (accession number NM_001127233.1)
sense strand, 5'-AGAGACCGCCGTACAGAAGA-3' and antisense strand,
5'-CTGTAGCATGGGCATCCTTT-3' (27);
p21 (accession number NM_001111099.2) sense strand,
5'-AGCAAAGTGTGCCGTTGT CT-3' and antisense strand,
5'-AGAAATCTGTCAGGCTGGTC-3'; β-actin (accession number NM_007393.5)
sense strand, 5'-AGCCATGTACGTAGCCATCC-3' and antisense strand,
5'-CTCTCAGCTGTGGTGGTGAA-3' (28).
The sequence of miR-22 was AAGCUGCCAGUUGAAGAACUGU (29).
Western blot analysis
Osteoblast proteins were extracted using RIPA buffer
including 1% protease inhibitor cocktail (Thermo Fisher Scientific,
Inc.) and BCA assay was used to calculate the protein
concentration. Then 30 µg proteins per lane was subjected to
SDS-PAGE (10% gels) to separate, prior to their transfer onto PVDF
membranes. After blocking with nonfat dry milk dissolved in TBS
containing 0.05% Tween-20 for 1-2 h at room temperature, the
membranes were incubated with antibodies against sirt1 (Santa Cruz
Biotechnology, Inc.; cat. no. sc-74665); p53 (ProteinTech Group,
Inc.; cat. no. 10442-1-AP) and p21 (ProteinTech Group, Inc.; cat.
no. 28248-1-AP) and GAPDH (Santa Cruz Biotechnology, Inc.; cat. no.
sc-137179) overnight at 4˚C at a dilution of 1:1,000. Subsequently,
the membranes were incubated with a secondary horseradish
peroxidase-conjugated antibody for 1-2 h at room temperature at a
dilution of 1:2,000, including goat anti-rabbit IgG-HRP (Santa Cruz
Biotechnology, Inc.; cat. no. sc-2004) and goat anti-mouse IgG-HRP
(Santa Cruz Biotechnology, Inc.; cat. no. sc-2005). An enhanced
chemiluminescence western blotting detection system (Santa Cruz
Biotechnology, Inc.) and a GeneGnome HR scanner (Syngene) were used
to visualize the immunoreactive proteins and chemiluminescent
signal from the membranes. The expression levels of the proteins of
interest were normalized to GAPDH.
Bioinformatics analysis
The TargetScan (http://www.targetscan.org/) and miRanda, (http://www.microrna.org/microrna/home.do) tools were
used to perform target prediction. The target genes predicted by
TargetScan and miRanda were screened based on their scoring
criteria. In the TargetScan algorithm, target genes with a context
percentile <50 were excluded. In the miRanda algorithm, target
genes with Max-Energy >-10 were excluded. The overlapping
prediction results were selected as the candidate target genes of
miR-22 (30,31).
Statistical analysis
SPSS 16.0 (SPSS, Inc.) was used to perform the data
analysis. Data are presented as the mean ± SEM. Statistical
comparisons between two groups were determined using two-tailed
paired Student's t-test. One-way or two-way ANOVAs, with
Bonferroni's post hoc tests were performed for comparisons among
multiple groups. P<0.05 was considered to indicate a
statistically significant value.
Results
Osteoblast senescence contributes to
Dex-induced osteoblast dysfunction
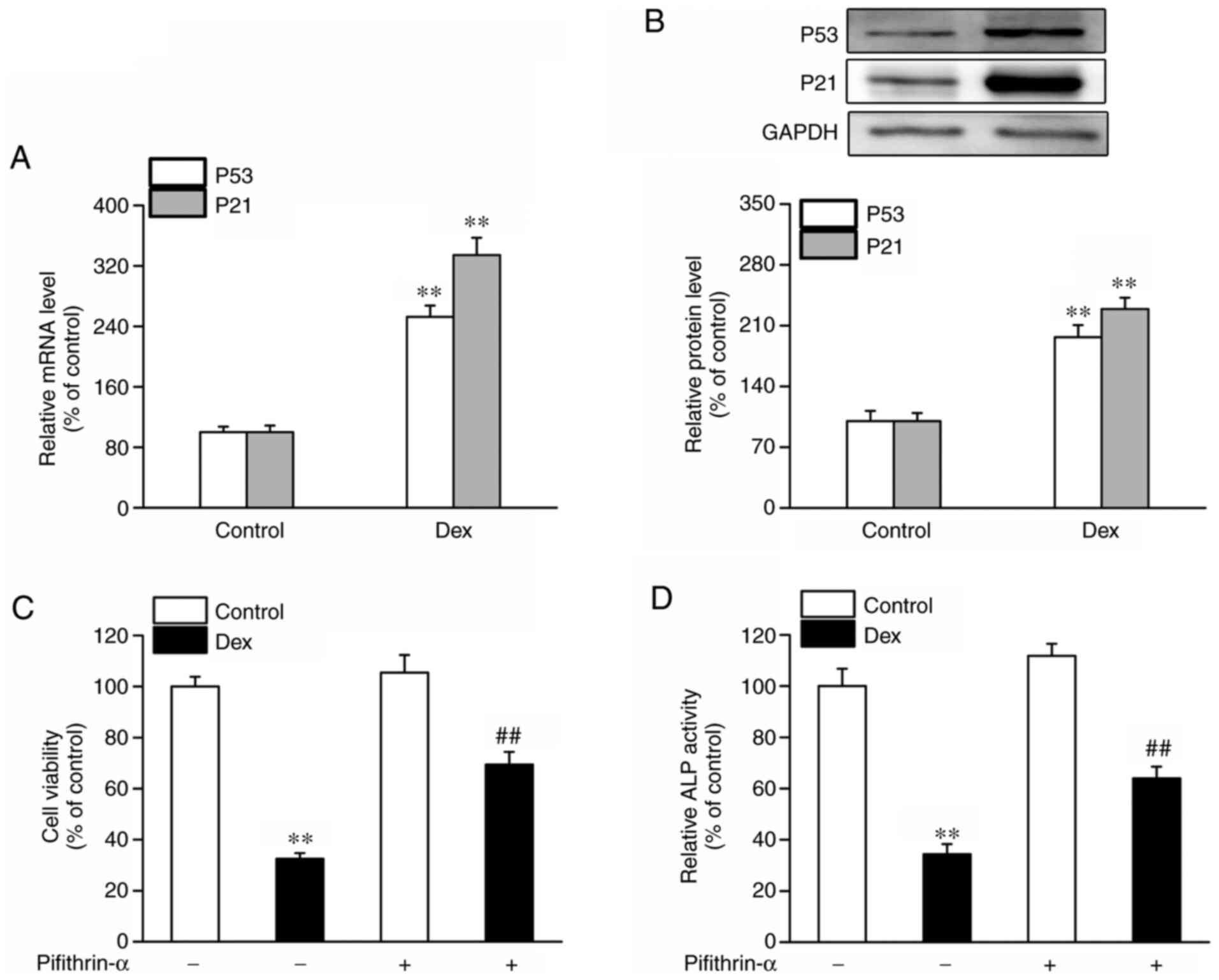
Firstly, alterations in the levels of the
senescence-associated biomarkers, p53 and p21, in osteoblastic
MC3T3-E1 cells exposed to Dex, were measured in the present study.
As shown in Fig. 1A and B, the mRNA and protein expression levels
of p53 and p21 were significantly increased in the Dex-treated
osteoblasts, suggesting that osteoblast senescence was involved in
Dex-induced cell damage. Subsequently, a p53 inhibitor
(pifithrin-α) was used to measure the impact of Dex-induced
osteoblast damage. It was found that Dex treatment resulted in a
decreased cell viability and ALP activity, with these results being
partly reversed by pifithrin-α, as shown by the increased cell
viability and ALP activity (Fig. 1C
and D).
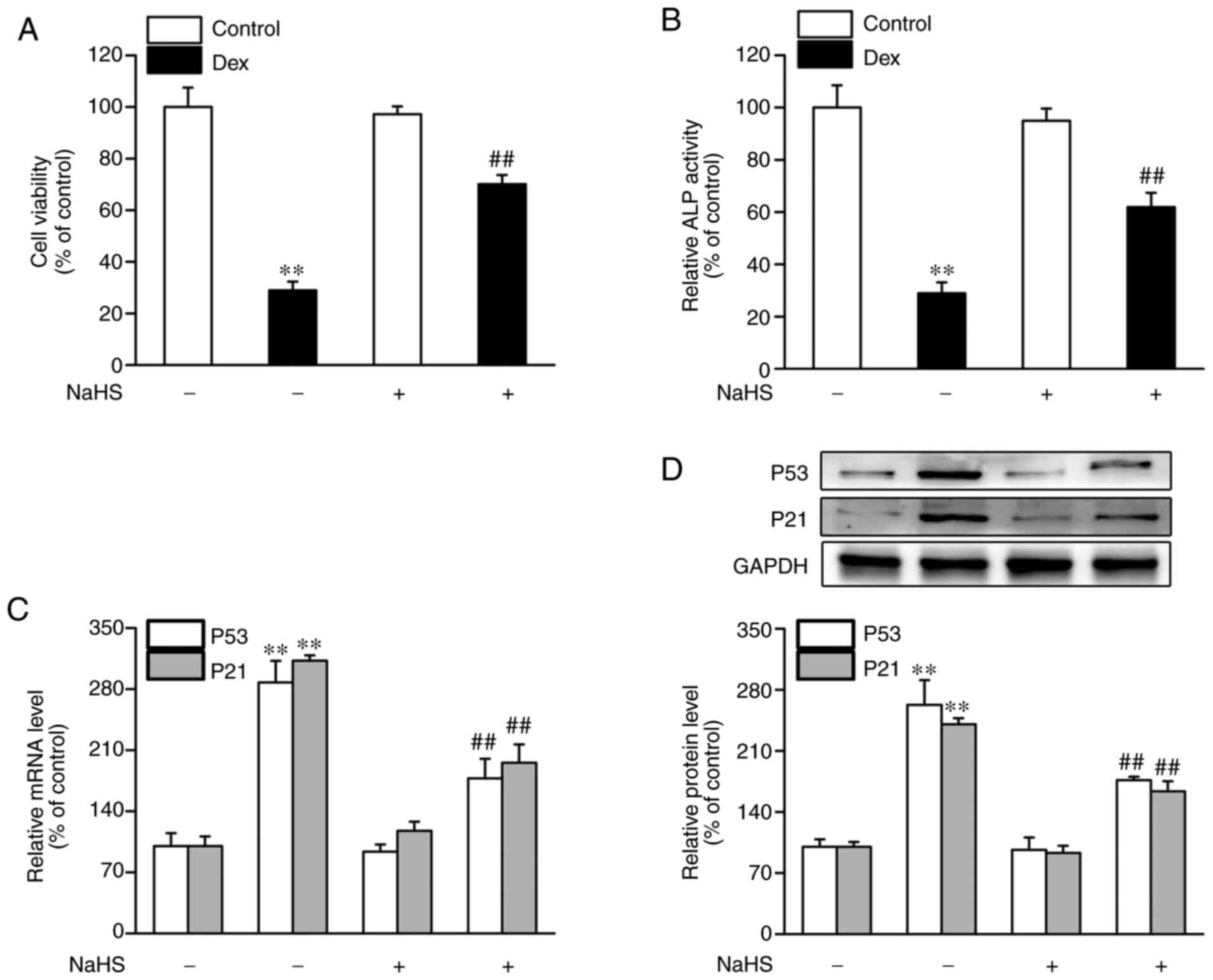
NaHS mitigates Dex-induced osteoblast
dysfunction and senescence
Subsequently, the effects of NaHS on osteoblast
dysfunction and senescence exposed to Dex were measured. As shown
in Fig. 2, compared with control
group, cell viability and ALP activity in the Dex-treated group
were markedly decreased, whereas the expression of p53 and p21 at
the mRNA and protein levels was significantly increased. However,
NaHS attenuated Dex-induced osteoblast damage and senescence, as
shown by the increased cell viability (Fig. 2A) and the increase in ALP activity
(Fig. 2B) in the Dex and NaHS
treated group. Additionally, NaHS treatment caused an increase in
the mRNA and protein expression levels of p53 and p21 (Fig. 2C and D).
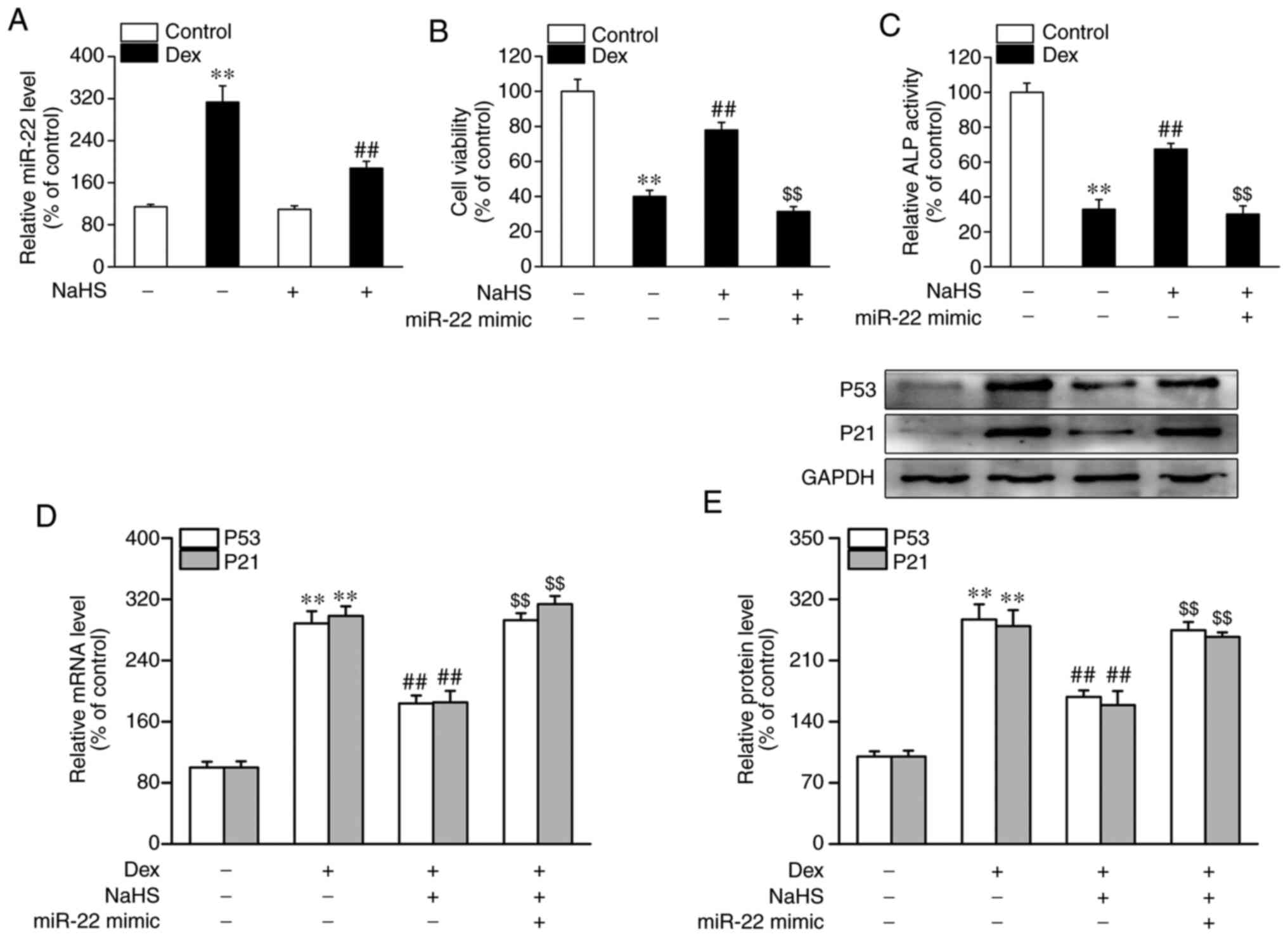
miR-22 mimic blocks the beneficial
effects of NaHS on Dex-induced osteoblast dysfunction and
senescence
In the present set of experiments, it was found that
the miR-22 levels in MC3T3-E1 cells that Dex-treated were
significantly increased (Fig. 3A).
Subsequently, the present study investigated whether alterations in
the miR-22 levels were also associated with the protective effect
of NaHS against Dex-induced osteoblast damage and senescence. As
shown in Fig. 3A, the increased
miR-22 levels in Dex-treated MC3T3-E1 cells was reversed upon
administration of NaHS. Moreover, as shown in Fig. S1A, miR-22 mimic transfection
resulted in a markable increase expression of miR-22. It was found
that the miR-22 mimic blocked the NaHS-induced increases in cell
viability and ALP activity that were observed in MC3T3-E1 cells
exposed to Dex (Fig. 3B and
C). Furthermore, the miR-22 mimic
also reversed the NaHS-induced downregulation of p53 and p21 at the
mRNA and protein levels in MC3T3-E1 cells subjected to Dex
(Fig. 3D and E).
sirt1 is the target of miR-22 in
osteoblastic MC3T3-E1 cells
As previously mentioned, miRs are able to
post-transcriptionally and negatively regulate the expression of
target genes (15-17).
TargetScan and miRanda were used to perform target prediction
analysis of miR-22, and sirt1 was found to be an overlapping gene.
As shown in Fig. 4A, one
evolutionarily conserved sequence of the sirt1 gene was
complementary to miR-22. As shown in Fig. 4B and C, the miR-22 mimic was able to
significantly suppress the mRNA and protein expression levels of
sirt1. Sirt1 3'-UTR luciferase reporter constructs and miR-22
mimics were transfected into osteoblastic MC3T3-E1 cells to confirm
the direct regulatory effect of miR-22 on sirt1 expression. A
significant decrease in luciferase activity in the miR-22 mimic
group was found compared with the miR control group (Fig. 4D). No decrease in luciferase
activity was observed upon co-transfection of miR-22 mimic with the
sirt1 3'UTR luciferase reporter construct containing a mutated
target sequence. These results suggested that miR-22 could suppress
sirt1 expression in osteoblastic MC3T3-E1 cells through binding to
response elements in its 3'-UTR.
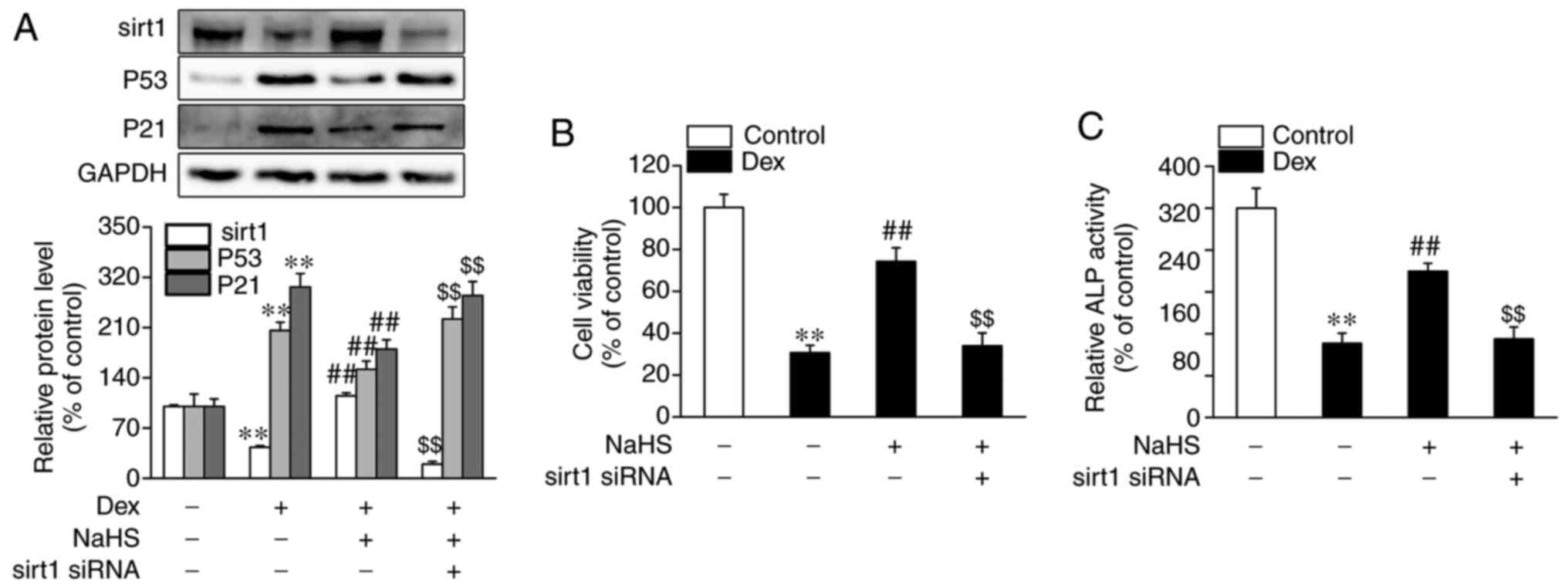
Silencing of sirt1 blocks the
protective role of NaHS against Dex-induced osteoblast damage and
senescence
As shown in Fig. 5A,
the protein expression level of sirt1 was significantly decreased
in Dex-treated osteoblasts, although this was reversed upon
administration of NaHS. Furthermore, the addition of sirt1 siRNA
led to a marked decrease in mRNA and protein expression levels of
sirt1 (Fig. S1B and C). Consequently, whether sirt1 is
associated with the beneficial effects of NaHS against Dex-induced
osteoblast senescence was investigated. As shown in Fig. 5A, sirt1 siRNA blocked the inhibitory
effect of NaHS against Dex-induced osteoblast senescence, as shown
by the increased protein levels of p53 and p21. In addition, sirt1
siRNA also abolished the beneficial effects of NaHS against
Dex-induced osteoblast damage, as evidenced by decreased cell
viability and ALP activity (Fig. 5B
and C).
Discussion
In the present study, it has been shown that
osteoblast senescence may contribute to Dex-induced dysfunction and
that a p53 inhibitor (pifithrin-α) could partly reverse the
osteoblast damage induced by Dex, as revealed by increased levels
of cell proliferation and ALP activity. Moreover, NaHS mitigated
the Dex-induced damage and senescence through targeting the
miR-22/sirt1 pathway, as miR-22 mimic and sirt1 siRNA blocked the
beneficial effects of NaHS on Dex-induced osteoblast dysfunction
and senescence, and sirt1 is a target of miR-22.
An accumulating number of studies have suggested
that cell senescence exerts a dominant role in a wide variety of
diseases, including hepatocarcinogenesis (32), intervertebral disc degeneration
(33), diabetic nephropathy
(34), diabetic cardiomyopathy
(35), Parkinson's disease
(36) and Alzheimer's disease
(37). Additionally, the crucial
role of cell senescence in the development of osteoporosis has also
been widely recognized (38,39).
Zhang et al (38) revealed
that osteoblastic cell senescence may contribute to osteoporosis
resulting from estrogen deficiency, and this process is accelerated
upon treatment with serum from ovariectomized rats. Khosla et
al (39) determined that this
may be a novel therapeutic paradigm for restraining, or even
reversing, age-associated osteoporosis via targeting cellular
senescence.
It is widely accepted that sirt1 protects
osteoblastic cells against multiple unfavorable factors, including
hypoxia (40); TiAl6V4 particles
and CoCrMo particles (41);
hydrogen peroxide (42); and sodium
fluoride (43). Interestingly, it
is also recognized that sirt1 prevents cell senescence, with sirt1
knockdown inducing cell senescence (44-48).
Ota et al (44) demonstrated
that overexpression of sirt1 could provide protection against
stress-caused endothelial damage through repressing premature
senescence of human endothelial cells. Zu et al (46) also showed that sirt1 prevents
primary porcine aortic endothelial cell senescence by targeting
serine/threonine kinase 11. In the present study, senescence and
dysfunction in osteoblastic MC3T3-E1 cells were phenotypically
shown to be associated with decreased sirt1 expression levels.
Among many factors that negatively regulate gene expression, an
accumulating number of studies have provided confirmatory evidence
in support of an essential role for miRs (15-17).
miR-22 is a widely expressed miR, and bioinformatics analysis and
previous studies (21,22,49)
have shown that sirt1 is the target of miR-22 in osteoblastic
MC3T3-E1 cells. Chen et al (21) demonstrated that miR-22 induces human
glioblastoma cell dysfunction through the direct targeting of
sirt1, as revealed by suppressed levels of cell proliferation,
motility and invasion. Zou et al (22) also revealed that miR-22 could
restrict cell growth and metastasis in breast cancer through
targeting sirt1 and the miR-22/sirt1 signaling pathway, also
proposing that this potentially forms the basis of a novel
therapeutic strategy for breast cancer. Another study demonstrated
that upregulation of miR-22 contributes to
ischemia-reperfusion-induced myocardial damage by directly
targeting sirt1, which exerts a critical role in the regulation of
mitochondrial function (17). In
the present study, it was also shown that upregulation of miR-22 is
involved in Dex-induced osteoblastic cell senescence and injury
through targeting the regulation of sirt1 expression. However,
further research is required to determine whether alteration of the
mitochondrial function in Dex-treated osteoblast cells is involved
in the underlying mechanism of action.
Previous studies have demonstrated that NaHS, an
H2S donor, exerts a protective role in multiple
diseases, including chronic restrain stress (CRS)-induced cognitive
damage (50), cerebral IR injury
(51), chronic kidney disease
(52,53) and high pulmonary blood flow-induced
pulmonary artery collagen remodeling (54). Interestingly, Li et al
(50) showed that H2S
mitigates CRS-induced cognitive damage and hippocampal impairment
through the upregulation of sirt1. A growing number of studies have
shown that H2S may protect a variety of cell types from
adverse factors by delaying cell senescence (55-57).
Zheng et al (57)
demonstrated that NaHS delays cellular senescence of human
umbilical vascular endothelial cells through upregulating sirt1
expression. In agreement with these results, the results of the
present study have also demonstrated that NaHS mitigated
Dex-induced osteoblastic cell senescence and impairment through the
upregulation of sirt1 expression.
A limitation of the present study was that the
results were gained in vitro. Further in vivo studies
would be beneficial to confirm the present findings. Interestingly,
Ma et al (58) demonstrated
that exogenous H2S attenuates Dex-induced inhibition of
osteoblast proliferation and osteogenic differentiation by
activating the Wnt/β-catenin signaling pathway. Additionally, Xia
et al (59) indicated that
activation of the Wnt/β-catenin signaling pathway is negatively
related to cell senescence. As such, it was speculated that
exogenous H2S alleviates Dex-induced osteoblast
dysfunction through inhibition of senescence. Furthermore, future
studies should investigate the mechanism of action behind how Dex
treatment leads to increased expression levels of miR-22, as well
as the other targets of miR-22 in Dex-treated osteoblasts.
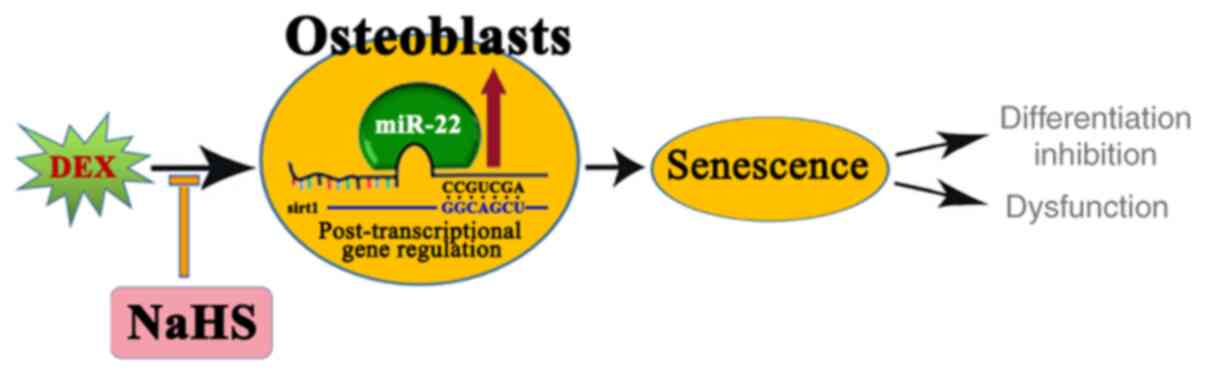
As summarized in Fig.
6, the present study demonstrated that NaHS prevented
Dex-induced osteoblastic MC3T3-E1 cell senescence and damage
through targeting the miR-22/sirt1 pathway.
Supplementary Material
Impact of miR-22 mimic transfection
and sirt1 siRNA on the expression of miR-22 and sirt1 at the mRNA
and protein expression levels in osteoblastic MC3T3E1 cells.
Osteoblasts were transfected with miR control or miR22 mimic (200
nM) for 24 h. RT-qPCR was performed to measure the expression
levels of miR22 (A). Osteoblasts were transfected with control
siRNA or sirt1 siRNA for 24 h. RT-qPCR and western blot analyses
were performed to determine sirt1 mRNA (B) and protein (C)
expression levels, respectively, in osteoblasts. Data are shown as
the mean ± SEM (n=4). **P<0.01 vs. the control. miR,
microRNA; RT-qPCR; reverse transcription PCR; siRNA, small
interfering RNA; sirt1, sirtuin 1.
Acknowledgements
Not applicable.
Funding
The present study was supported by the key
incubation project of Li Peng from the Science and Technology
Department of Ningxia Medical University, Key Incubation
Project.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
ZDL was responsible for study design and manuscript
preparation. PL performed manuscript preparation, data collection
and statistical analysis. WWM contributed to data interpretation.
The literature search and dual luciferase assay were performed by
SZ, LZ and ZRC. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
van Staa TP, Leufkens HG, Abenhaim L,
Begaud B, Zhang B and Cooper C: Use of oral corticosteroids in the
United Kingdom. QMJ. 93:105–111. 2000.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fardet L, Petersen I and Nazareth I:
Prevalence of long-term oral glucocorticoid prescriptions in the UK
over the past 20 years. Rheumatology (Oxford). 50:1982–1990.
2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Overman RA, Yeh JY and Deal CL: Prevalence
of oral glucocorticoid usage in the United States: A general
population perspective. Arthritis Care Res (Hoboken). 65:294–298.
2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Silverman S, Curtis J, Saag K, Flahive J,
Adachi J, Anderson F, Chapurlat R, Cooper C, Diez-Perez A,
Greenspan S, et al: International management of bone health in
glucocorticoidexposed individuals in the observational GLOW study.
Osteoporos Int. 26:419–420. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ventura A, Brunetti G, Colucci S, Oranger
A, Ladisa F, Cavallo L, Grano M and Faienza MF:
Glucocorticoid-induced osteoporosis in children with 21-hydroxylase
deficiency. Biomed Res Int. 2013(250462)2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Delany AM, Dong Y and Canalis E:
Mechanisms of glucocorticoid action in bone cells. J Cell Biochem.
56:295–302. 1994.PubMed/NCBI View Article : Google Scholar
|
|
7
|
O'Brien CA, Jia D, Plotkin LI, Bellido T,
Powers CC, Stewart SA, Manolagas SC and Weinstein RS:
Glucocorticoids act directly on osteoblasts and osteocytes to
induce their apoptosis and reduce bone formation and strength.
Endocrinology. 145:1835–1841. 2004.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Burton DG and Krizhanovsky V:
Physiological and pathological consequences of cellular senescence.
Cell Mol Life Sci. 71:4373–4386. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wu G, Xu R, Zhang P, Xiao T, Fu Y, Zhang
Y, Du Y, Ye J, Cheng J and Jiang H: Estrogen regulates stemness and
senescence of bone marrow stromal cells to prevent osteoporosis via
ERβ-SATB2 pathway. J Cell Physiol. 233:4194–4204. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Elrod JW, Calvert JW, Morrison J, Doeller
JE, Kraus DW, Tao L, Jiao X, Scalia R, Kiss L, Szabo C, et al:
Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury
by preservation of mitochondrial function. Proc Natl Acad Sci USA.
104:15560–15565. 2007.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kimura Y and Kimura H: Hydrogen sulfide
protects neurons from oxidative stress. FASEB J. 18:1165–1167.
2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Esechie A, Kiss L, Olah G, Horváth EM,
Hawkins H, Szabo C and Traber DL: Protective effect of hydrogen
sulfide in a murine model of acute lung injury induced by combined
burn and smoke inhalation. Clin Sci (Lond). 115:91–97.
2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xu ZS, Wang XY, Xiao DM, Hu LF, Lu M, Wu
ZY and Bian JS: Hydrogen sulfide protects MC3T3-E1 osteoblastic
cells against H2O2-induced oxidative damage-implications for the
treatment of osteoporosis. Free Radic Biol Med. 50:1314–1323.
2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang M, Huang Y, Chen J, Chen YL, Ma JJ
and Shi PH: Activation of AMPK participates hydrogen
sulfide-induced cyto-protective effect against dexamethasone in
osteoblastic MC3T3-E1 cells. Biochem Biophys Res Commun. 454:42–47.
2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Banzhaf-Strathmann J, Benito E, May S,
Arzberger T, Tahirovic S, Kretzschmar H, Fischer A and Edbauer D:
MicroRNA-125b induces tau hyperphosphorylation and cognitive
deficits in Alzheimer's disease. EMBO J. 33:1667–1680.
2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Harada M, Luo X, Murohara T, Yang B,
Dobrev D and Nattel S: MicroRNA regulation and cardiac calcium
signaling: Role in cardiac disease and therapeutic potential. Circ
Res. 114:689–705. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Du JK, Cong BH, Yu Q, Wang H, Wang L, Wang
CN, Tang XL, Lu JQ, Zhu XY and Ni X: Upregulation of microRNA-22
contributes to myocardial ischemia-reperfusion injury by
interfering with the mitochondrial function. Free Radic Biol Med.
96:406–417. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shi C, Qi J, Huang P, Jiang M, Zhou Q,
Zhou H, Kang H, Qian N, Yang Q, Guo L and Deng L: MicroRNA-17/20a
inhibits glucocorticoid-induced osteoclast differentiation and
function through targeting RANKL expression in osteoblast cells.
Bone. 68:67–75. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang Y, Gao Y, Cai L, Li F, Lou Y, Xu N,
Kang Y and Yang H: MicroRNA-221 is involved in the regulation of
osteoporosis through regulates RUNX2 protein expression and
osteoblast differentiation. Am J Transl Res. 9:126–135.
2017.PubMed/NCBI
|
|
20
|
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL,
Wong CW, Chan KM, Li G, Waye MM and Zhang JF: H19 activates Wnt
signaling and promotes osteoblast differentiation by functioning as
a competing endogenous RNA. Sci Rep. 6(20121)2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen H, Lu Q, Fei X, Shen L, Jiang D and
Dai D: miR-22 inhibits the proliferation, motility, and invasion of
human glioblastoma cells by directly targeting SIRT1. Tumour Biol.
37:6761–6768. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zou Q, Tang Q, Pan Y, Wang X, Dong X,
Liang Z and Huang D: MicroRNA-22 inhibits cell growth and
metastasis in breast cancer via targeting of SIRT1. Exp Ther Med.
14:1009–1016. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tang YH, Yue ZS, Li GS, Zeng LR, Xin DW,
Hu ZQ and Xu CD: Effect of β-ecdysterone on glucocorticoid-induced
apoptosis and autophagy in osteoblasts. Mol Med Rep. 17:158–164.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bowers GN Jr and McComb RB: A continuous
spectrophotometric method for measuring the activity of serum
alkaline phosphatase. Clin Chem. 12:70–89. 1966.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang HH, Ma XJ, Wu LN, Zhao YY, Zhang PY,
Zhang YH, Shao MW, Liu F, Li F and Qin GJ: SIRT1 attenuates high
glucose-induced insulin resistance via reducing mitochondrial
dysfunction in skeletal muscle cells. Exp Biol Med (Maywood).
240:557–565. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cho SJ, Rossi A, Jung YS, Yan W, Liu G,
Zhang J, Zhang M and Chen X: Ninjurin1, a target of p53, regulates
p53 expression and p53-dependent cell survival, senescence, and
radiation-induced mortality. Proc Natl Acad Sci USA. 110:9362–9367.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lee CJ, Kim HT, Song KW, Kim SS, Park HH
and Yoon YD: Ovarian expression of p53 and p21 apoptosis regulators
in gamma-irradiated mice. Mol Reprod Dev. 75:383–391.
2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739.
2002.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Meng SS, Wang H, Xue DB and Zhang WH:
Screening and validation of differentially expressed extracellular
miRNAs in acute pancreatitis. Mol Med Rep. 16:6412–6418.
2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Sun F, Yang X, Jin Y, Chen L, Wang L, Shi
M, Zhan C, Shi Y and Wang Q: Bioinformatics analyses of the
differences between lung adenocarcinoma and squamous cell carcinoma
using The Cancer Genome Atlas expression data. Mol Med Rep.
16:609–616. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mossanen JC, Kohlhepp M, Wehr A, Krenkel
O, Liepelt A, Roeth AA, Möckel D, Heymann F, Lammers T, Gassler N,
et al: CXCR6 inhibits hepatocarcinogenesis by promoting natural
killer T- and CD4+ T-cell-dependent control of
senescence. Gastroenterology. 156:1877–1889.e4. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chen J, Xie JJ, Jin MY, Gu YT, Wu CC, Guo
WJ, Yan YZ, Zhang ZJ, Wang JL, Zhang XL, et al: Sirt6
overexpression suppresses senescence and apoptosis of nucleus
pulposus cells by inducing autophagy in a model of intervertebral
disc degeneration. Cell Death Dis. 9(56)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen K, Dai H, Yuan J, Chen J, Lin L,
Zhang W, Wang L, Zhang J, Li K and He Y: Optineurin-mediated
mitophagy protects renal tubular epithelial cells against
accelerated senescence in diabetic nephropathy. Cell Death Dis.
9(105)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gu J, Wang S, Guo H, Tan Y, Liang Y, Feng
A, Liu Q, Damodaran C, Zhang Z, Keller BB, et al: Inhibition of p53
prevents diabetic cardiomyopathy by preventing early-stage
apoptosis and cell senescence, reduced glycolysis, and impaired
angiogenesis. Cell Death Dis. 9(82)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Williams-Gray CH, Wijeyekoon RS, Scott KM,
Hayat S, Barker RA and Jones JL: Abnormalities of age-related T
cell senescence in Parkinson's disease. J Neuroinflammation.
15(166)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Virgili J, Lebbadi M, Tremblay C, St-Amour
I, Pierrisnard C, Faucher-Genest A, Emond V, Julien C and Calon F:
Characterization of a 3xTg-AD mouse model of Alzheimer's disease
with the senescence accelerated mouse prone 8 (SAMP8) background.
Synapse. 72:2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang J, Lazarenko OP, Blackburn ML,
Badger TM, Ronis MJ and Chen JR: Blueberry consumption prevents
loss of collagen in bone matrix and inhibits senescence pathways in
osteoblastic cells. Age (Dordr). 35:807–820. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Khosla S, Farr JN and Kirkland JL:
Inhibiting cellular senescence: A new therapeutic paradigm for
age-related osteoporosis. J Clin Endocrinol Metab. 103:1282–1290.
2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhou L, Wang SI, Moon YJ, Kim KM, Lee KB,
Park BH, Jang KY and Kim JR: Overexpression of SIRT1 prevents
hypoxia-induced apoptosis in osteoblast cells. Mol Med Rep.
16:2969–2975. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Deng Z, Wang Z, Jin J, Wang Y, Bao N, Gao
Q and Zhao J: SIRT1 protects osteoblasts against particle-induced
inflammatory responses and apoptosis in aseptic prosthesis
loosening. Acta Biomater. 49:541–554. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
He N, Zhu X, He W, Zhao S, Zhao W and Zhu
C: Resveratrol inhibits the hydrogen dioxide-induced apoptosis via
Sirt 1 activation in osteoblast cells. Biosci Biotechnol Biochem.
79:1779–1786. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Gu X, Han D, Chen W, Zhang L, Lin Q, Gao
J, Fanning S and Han B: SIRT1-mediated FoxOs pathways protect
against apoptosis by promoting autophagy in osteoblast-like
MC3T3-E1 cells exposed to sodium fluoride. Oncotarget.
7:65218–65230. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ota H, Akishita M, Eto M, Iijima K, Kaneki
M and Ouchi Y: Sirt1 modulates premature senescence-like phenotype
in human endothelial cells. J Mol Cell Cardiol. 43:571–579.
2007.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ota H, Tokunaga E, Chang K, Hikasa M,
Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y and Kaneki M: Sirt1
inhibitor, Sirtinol, induces senescence-like growth arrest with
attenuated Ras-MAPK signaling in human cancer cells. Oncogene.
25:176–185. 2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zu Y, Liu L, Lee MY, Xu C, Liang Y, Man
RY, Vanhoutte PM and Wang Y: SIRT1 promotes proliferation and
prevents senescence through targeting LKB1 in primary porcine
aortic endothelial cells. Circ Res. 106:1384–1393. 2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Huang J, Gan Q, Han L, Li J, Zhang H, Sun
Y, Zhang Z and Tong T: SIRT1 overexpression antagonizes cellular
senescence with activated ERK/S6k1 signaling in human diploid
fibroblasts. PLoS One. 3(e1710)2008.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhao G, Cui J, Zhang JG, Qin Q, Chen Q,
Yin T, Deng SC, Liu Y, Liu L, Wang B, et al: SIRT1 RNAi knockdown
induces apoptosis and senescence, inhibits invasion and enhances
chemosensitivity in pancreatic cancer cells. Gene Ther. 18:920–928.
2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ming GF, Wu K, Hu K, Chen Y and Xiao J:
NAMPT regulates senescence, proliferation, and migration of
endothelial progenitor cells through the SIRT1 AS
lncRNA/miR-22/SIRT1 pathway. Biochem Biophys Res Commun.
478:1382–1388. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li XN, Chen L, Luo B, Li X, Wang CY, Zou
W, Zhang P, You Y and Tang XQ: Hydrogen sulfide attenuates chronic
restrain stress-induced cognitive impairment by upreglulation of
Sirt1 in hippocampus. Oncotarget. 8:100396–100410. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jiang WW, Huang BS, Han Y, Deng LH and Wu
LX: Sodium hydrosulfide attenuates cerebral ischemia/reperfusion
injury by suppressing overactivated autophagy in rats. FEBS Open
Bio. 7:1686–1695. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Askari H, Seifi B, Kadkhodaee M, Sanadgol
N, Elshiekh M, Ranjbaran M and Ahghari P: Protective effects of
hydrogen sulfide on chronic kidney disease by reducing oxidative
stress, inflammation and apoptosis. EXCLI J. 17:14–23.
2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wu D, Luo N, Wang L, Zhao Z, Bu H, Xu G,
Yan Y, Che X, Jiao Z, Zhao T, et al: Hydrogen sulfide ameliorates
chronic renal failure in rats by inhibiting apoptosis and
inflammation through ROS/MAPK and NF-κB signaling pathways. Sci
Rep. 7(455)2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Li X, Du J, Jin H, Geng B and Tang C:
Sodium hydrosulfide alleviates pulmonary artery collagen remodeling
in rats with high pulmonary blood flow. Heart Vessels. 23:409–419.
2008.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yang G, Zhao K, Ju Y, Mani S, Cao Q,
Puukila S, Khaper N, Wu L and Wang R: Hydrogen sulfide protects
against cellular senescence via S-sulfhydration of Keap1 and
activation of Nrf2. Antioxid Redox Signal. 18:1906–1919.
2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Wang WJ, Cai GY, Ning YC, Cui J, Hong Q,
Bai XY, Xu XM, Bu R, Sun XF and Chen XM: Hydrogen sulfide mediates
the protection of dietary restriction against renal senescence in
aged F344 rats. Sci Rep. 6(30292)2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zheng M, Qiao W, Cui J, Liu L, Liu H, Wang
Z and Yan C: Hydrogen sulfide delays nicotinamide-induced premature
senescence via upregulation of SIRT1 in human umbilical vein
endothelial cells. Mol Cell Biochem. 393:59–67. 2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ma J, Shi C, Liu Z, Han B, Guo L, Zhu L
and Ye T: Hydrogen sulfide is a novel regulator implicated in
glucocorticoids-inhibited bone formation. Aging (Albany NY).
11:7537–7552. 2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Xia W, Zhuang L, Deng X and Hou M: Long
noncoding RNA-p21 modulates cellular senescence via the
Wnt/β-catenin signaling pathway in mesenchymal stem cells. Mol Med
Rep. 16:7039–7047. 2017.PubMed/NCBI View Article : Google Scholar
|