Introduction
Hepatocellular carcinoma (HCC) is a primary liver
cancer that is derived from hepatocytes, accounting for 95% of all
types of primary liver cancer (1).
HCC is a malignant tumor that seriously threatens human health,
with an incidence and mortality among the highest of all tumor
types (2). The potential risk
factors for HCC include viral infections, cirrhosis, aflatoxina and
alcoholism, which are speculated to lead to fundamental genetic
mutations, causing oncogene activation and tumor suppressor gene
inactivation (3). An oncogene is a
dominant gene that can cause malignant transformation of cells
after its mutation, while a tumor suppressor gene is a recessive
gene that may lead to the transformation of normal cells into
cancerous cells due to disruption of the regulation of associated
cell division and proliferation when the tumor suppressor is
mutated or lost (4). In the
development of several tumors, the loss of tumor suppressor genes
may be more important than the activation of an oncogene since the
former exerts resistance and protection at various stages of tumor
development, including regulation of the cell cycle, inhibition of
cell adhesion and migration, suppression of proliferation of cells
with DNA damage and repair of damaged DNA (5,6). The
underlying mechanism of action behind liver cancer has been
investigated in recent years and several tumor-related genes have
been identified; however, the exact molecular mechanism of action
is yet to be elucidated (7-13).
MicroRNAs (miRNAs/miRs) are non-coding
single-stranded small RNAs that consist of ~22 nucleotides
(14). miRNAs, which are widely
found in plants, animals and human cells, exhibit a
post-transcriptional regulatory effect by degrading mRNA or
inhibiting translation of mRNA by binding to the 3' untranslated
region (3'UTR) of their target gene mRNA (15). Previous studies have shown that
miRNAs play an important regulatory role in cell proliferation,
apoptosis, insulin secretion and neurodevelopment; miRNAs are also
essential to the late stage of embryonic development (16,17).
It has also been reported that miR-196 is involved in the
regulation of mammalian limb development (5) and that miR-181 plays a role in the
regulation of mammalian blood cell differentiation (18). In mammals, miR-375 exhibits a
regulatory effect on insulin secretion (6). In addition, the development of nervous
systems of some species, such as nematodes and zebrafish, is also
regulated by lin-4, miR-142, miR-15, miR-16 and let-7 miRNAs
(19). miRNAs are widely used for
the diagnosis and prognosis of various diseases and also serve as
tumor markers for candidate cancer types (6,20,21).
Previous studies have shown that abnormal expression of miRNAs in
tumors can lead to oncogene activation and tumor suppressor gene
dysregulation, which may ultimately contribute to tumor progression
(22-24).
The dysregulated expression of miRNAs may function as an important
regulator to liver failure. The development of HCC is often
accompanied with the dysregulation of miRNAs. A number of miRNAs
have been associated with the clinicopathological features of liver
cancer (25-27).
miR-320, miR-486, miR-705 and miR-1224 were shown to be increased,
whilst the expression of miR-27B, miR-214, miR-199-3p, miR-182 and
miR-183, were shown to be decreased in hepatocellular carcinoma
tissues (27). It has been
previously demonstrated that miRNAs serve an important role in the
progression of HCC, potentially promoting the proliferation of
tumor cells, inhibiting tumor cell apoptosis and facilitating
metastasis by directly targeting their corresponding mRNAs
(28-34).
miR-129-5p regulates the development and progression
of various tumors by regulating the cell cycle, proliferation,
apoptosis, migration, invasion and angiogenesis, as well as other
physiological and pathological processes (25). In neuroblastomas, miR-129 inhibits
tumor growth by targeting Myosin X (MYO10) (35). Furthermore, miR-129-5p promotes cell
proliferation and invasion; and inhibits apoptosis through
regulating the SOX4 pathway in renal cell carcinoma (26). A previous study has reported that
miR-129-5p is an important biomarker for single ventricle heart
failure (36). In addition, the
expression of miR-129 may be an independent prognostic marker for
biochemical recurrence (BCR)-free survival in patients with
prostate cancer and the overexpression of miR-129 significantly
attenuates prostate cancer cell proliferation by regulating cell
cycle-regulated protein expression levels (27). To the best of our knowledge, no
studies exist that have investigated the correlation between
miR-129-5p and HCC. Therefore, the present study was conducted to
investigate the effect of miR-129-5p on the proliferative and
invasive capabilities of HCC cells.
Materials and methods
Clinical tissue specimens
All 45 patients with HCC involved in the present
study were enrolled from The First Affiliated Hospital of Harbin
Medical University (Harbin, China), after signing informed consent
forms from May 2015 to Sep 2017. Fresh tumor and peritumoral tissue
were frozen in liquid nitrogen immediately after dissection during
the surgery. None of the patients in the present study received any
other preoperative adjuvant therapy before surgery. The clinical
characteristics of patients with liver cancer are presented in
Table I. Among the 45 patients,
there were 29 males and 16 females were 16. All 45 patients were
clinically diagnosed with HCC. Patients with intrahepatic
metastasis, vascular invasion, or capsular invasion as found by
Computer Tomography and pathological examination were used to
classify metastatic/invasion samples. The age ranged from 27 to 71
years, of which 20 patients were ≥55 years old and 25 patients
<55 years old. There were 29 patients with intrahepatic
metastases, vascular invasion or capsular invasion
(metastatic/invasion samples, 64.4%). There were 16 patients with
metastatic/invasion. The present study was approved by the
Institutional Ethics Committee of Harbin Medical University
(approval no. KY2017059; Harbin, China).
 | Table IClinical features of the patients
with hepatocellular carcinomas. |
Table I
Clinical features of the patients
with hepatocellular carcinomas.
| Patient no. | Age (year) | Gender | Tumor size (cm x cm
x cm) | Intrahepatic
metastases | Vascular
invasion | Capsular
invasion |
|---|
| 1 | 59 | M | 3x3x2 | - | - | - |
| 2 | 61 | M | 4x4x2 | - | - | - |
| 3 | 52 | F | 5x3x3 | - | - | - |
| 4 | 50 | M | 9x7x3 | - | - | - |
| 5 | 46 | M | 10x7.5x5 | + | + | + |
| 6 | 38 | M | 3x2x2 | - | - | - |
| 7 | 70 | M | 6x4x2 | - | - | + |
| 8 | 71 | F | 7x5x3 | + | + | + |
| 9 | 51 | M | 19x12x7 | + | - | + |
| 10 | 62 | F | 12x11.5x10 | + | - | + |
| 11 | 50 | F | 6.5x6x5.5 | - | + | - |
| 12 | 57 | F | 9x8x7.5 | - | + | - |
| 13 | 43 | M | 4x3x2.5 | + | - | - |
| 14 | 70 | M | 5.5x5.5x3 | - | + | + |
| 15 | 60 | M | 18x15x7 | - | - | - |
| 16 | 66 | M | 12x9x6.5 | + | + | + |
| 17 | 67 | M | 4x3x2.5 | - | - | - |
| 18 | 60 | M | 5.5x4x4 | + | - | + |
| 19 | 49 | M | 10x8.5x5 | - | - | - |
| 20 | 47 | M | 6.5x6x3 | - | - | - |
| 21 | 68 | M | 17x12x6 | - | - | + |
| 22 | 52 | F | 12x10x8 | - | - | - |
| 23 | 39 | M | 4x3x3 | - | - | - |
| 24 | 42 | M | 7x5x3.5 | - | - | + |
| 25 | 60 | F | 9x7x4.5 | + | - | - |
| 26 | 53 | F | 14x9x8 | - | + | - |
| 27 | 50 | F | 10x7x6.5 | - | - | - |
| 28 | 27 | M | 9.5x8x7.5 | - | - | - |
| 29 | 46 | M | 4x3.5x3 | - | - | - |
| 30 | 52 | F | 8x6x5.5 | - | - | - |
| 31 | 58 | M | 6x4x4 | - | - | - |
| 32 | 46 | M | 9x6.5x5 | - | - | - |
| 33 | 37 | F | 7x2x2 | - | - | + |
| 34 | 54 | M | 3x2.7x2 | - | + | + |
| 35 | 43 | M | 4.5x1.5x1 | + | - | - |
| 36 | 69 | F | 4.5x3x3 | - | + | - |
| 37 | 57 | M | 6x5.5x4.5 | - | + | - |
| 38 | 46 | M | 4x4x1.5 | - | - | + |
| 39 | 50 | M | 7x3.5x2.5 | - | - | + |
| 40 | 67 | F | 8x6.5x4 | - | + | - |
| 41 | 70 | M | 6x3.5x3 | + | + | + |
| 42 | 55 | F | 4x2x2 | + | - | - |
| 43 | 43 | F | 5x4.5x2 | + | + | + |
| 44 | 54 | F | 3x2x2 | + | - | + |
| 45 | 62 | M | 4x3.5x3 | - | + | - |
Cell culture
The cell strain and human liver cancer cell lines
(HepG2 and Huh7) were provided by the China Center for Type Culture
Collection. Cells were seeded in DMEM (HyClone, GE Healthcare Life
Sciences), supplemented with 10% FBS (HyClone, GE Healthcare Life
Sciences), 100 U/ml penicillin and 100 µg/ml streptomycin at pH
7.2-7.4 and then cultured routinely in an incubator (37˚C, 5%
CO2, 95% air with saturated humidity).
miRNA and transfection
Transfections were performed using miRNAs analogs,
miR-129-5p mimics (5'-CUUUUUGCGGUCUGGGCUUGC-3'), miR-129-5p
inhibitor (5'-GCAAGCCCAGACCGCAAAAAGUU-3'), negative control (NC;
5'-UUCUCCGAACGUGUCACGUTT-3') and inhibitor NC
(5'-CAGUACUUUUGUGUAGUACAA-3'), which were synthesized by Shanghai
GenePharma Co., Ltd.. The NC was a synthetic scrambled double
oligonucleotide that do not target any mRNA. The transfection
reagent was Lipofectamine® 2000 (cat. no. 11668-027;
Invitrogen; Thermo Fisher Scientific, Inc.) and the operating
procedures were performed following the manufacturer's protocols.
The cells were cultured for 24 and 48 h at 37˚C and 5%
CO2 for subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
The reverse transcription of miRNA was carried out
as described previously (37).
Total RNA from tissue specimens, HepG2 cells or Huh7 cells was
isolated using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) following manufacturer's protocols.
Subsequently, the RNA was converted into cDNA using High-Capacity
cDNA Reverse Transcription kit (cat. no. 4368814; ABI; Thermo
Fisher Scientific, Inc.). The temperature of reverse transcription
is set as follows: 25˚C for 10 min, 37˚C for 120 min, 85˚C for 5
min and 4˚C for 5 min.
miRNA expression levels were quantitatively
determined using the Fast SYBR™ Green PCR Master Mix kit (cat. no.
4385610; ABI; Thermo Fisher Scientific, Inc.), with U6 snRNA as an
internal reference for miRNA. Primers were used for miRNA detection
as follows: miR-129-5p forward, 5'-CTTTTTGCGGTCTGGGCTTGC-3' and
reverse, 5'-GTGCAGGGTCCGAGGT-3' and U6 snRNA forward,
5'-TGCGGGTGCTCGCTTCGGCAGC-3' and reverse, 5'-GTGCAGGGTCCGAGGT-3'.
Primers were used for mRNA detection as follows: Bone morphogenetic
protein 2 (BMP2) forward, 5'-AAGTCTCCTCCTTCATCAGTATACGCTCG-3' and
reverse, 5'-GATATCGAATTCGATATCAAGCTGAT-3' and β-actin forward,
5'-TACCTCATGAAGATCCTCACC-3' and reverse,
5'-TTTCGTGGATGCCACAGGAC-3'. The β-actin mRNA level was used for
normalization. All primers were obtained from Invitrogen (Shanghai,
China). The annealing temperature for BMP2 and miR-129-5p was 60˚C.
The full thermocycling conditions for qPCR are as follows: Initial
denaturation at 95˚C for 2 min, followed by 40 cycles of 95˚C for
30 sec and 60˚C for 1 min. The method of ΔΔCq was used to determine
the relative quantity of mRNA expression in samples, and fold
change was determined as 2-ΔΔCq (38).
Western blotting
Cells were lysed using modified cell lysate RIPA
buffer (50 nM Tris HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium
deoxycholate and 0.5% SDS) to extract proteins. Protein
concentration was determined using a bicinchoninic acid protein
assay kit. In total, 80 µg protein was loaded into each well. After
being separated by 12% SDS-PAGE, the protein was transferred onto a
PVDF membrane. The membranes were blocked in 1% BSA (Gibco; Thermo
Fisher Scientific, Inc.) with 0.05% Tween-20 at 37˚C for 1 h. The
membranes were then incubated with rabbit monoclonal anti-BMP2
(cat. no. ab214821; 1:1,000; Abcam) and goat monoclonal
anti-β-actin (cat. no. sc-8432; 1:2,000; Santa Cruz Biotechnology,
Inc.) at 4˚C overnight. After incubation with the horseradish
peroxidase-conjugated goat anti-rabbit IgG secondary antibody (cat.
no. ab205718; 1:20,000) and rabbit anti-goat IgG (cat. no. ab6741;
1:2,000) at room temperature for 1 h, the signal was detected by
using Clarity Max™ Western ECL Substrate (Bio-Rad Laboratories,
Inc.). Tanon 1000 digital image gel analytical system (Tanon
Science & Technology Co., Ltd.) was used for photography and
quantification.
Bioinformatics analysis
Bioinformatics analysis was conducted using
TargetScan (TargetScan Release 7.2; www.targetscan.org;) to identify the target gene of
miR-129-5p. A predicted target site of miR-129-5p was identified in
the BMP2 3'UTR region.
Luciferase reporter assay
The BMP2 3'UTR and BMP2 3'UTR mutants, which were
synthesized by Shanghai GenePharma Co., Ltd., were constructed into
pmirGLO Dual-Luciferase vector (Promega Corporation) and referred
to as pGL-BMP2-3'UTR and pGL-mBMP2-3'UTR, respectively. HepG2 cells
cultured in monolayers were digested using 0.25% trypsin, mixed
into a single cell suspension with DMEM containing 10% FBS and then
seeded into 96-well culture plates at a density of 1,500-5,000
cells per well. When the cells grew to 60-70% confluence, were
co-transfected with 100 ng pGL-BMP2-3'UTR or 100 ng
pGL-mBMP2-3'UTR, miR-129-5p mimics (50 nM) and 1 ng pRL-TK. The
transfection reagent was Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) After 48 h, 200 µl
GLO-reagent (cat. no. E1960; Promega Corporation) was added to each
well. Reaction at room temperature for 5 min, the fluorescence
intensity was measured by spectrophotometer.
MTT cell cytotoxicity assay
Cells cultured in monolayers were digested using
0.25% trypsin, mixed into a single cell suspension with DMEM
containing 10% FBS and then seeded into 96-well culture plates at a
density of 1,500-5,000 cells per well. After transfection with
miR-129-5p mimics, cells were cultured at 37˚C with 5% CO2 and 95%
relative humidity for 24, 48 and 72 h. Subsequently, cells were
washed with PBS and suspended in serum-free medium. After adding 20
µl MTT solution (5 mg/ml) into each well, the cells were incubated
for 4 h at 37˚C. The culture supernatant was carefully removed from
the well, 150 µl DMSO was added into each well and the plate was
shaken for 10 min. The absorbance values at 490 nm were measured
with an enzyme-linked immunosorbent assay. Cell proliferation was
depicted with time as the horizontal axis and the absorbance value
as the vertical axis.
Invasion and migration assay
Invasion and migration of HepG2 or Huh7 cells were
detected using Transwell invasion assay and wound healing assay,
respectively. Migration of HepG2 and Huh7 cells was detected using
a scratch wound assay. Briefly, at 48 h after transfection, HepG2
or Huh7 cells, which were transfected with NC mimics, miR-129-5p
mimics, NC inhibitor or miR-129-5p inhibitor, were grown on 6-cm
dishes and to a density of 70-80%. The cell monolayer was then
scraped using a sterile cell scraper to create a cell-free zone.
The cells were cultured with serum-free DMEM medium. HepG2 or Huh7
cell migration was photographed at the time of injury and after 48
h of cultivation, using an inverted light microscope, at five
distinct positions per dish and x200 magnification.
In the invasion assay, after cell transfection in
serum-free culture, 2x104 cells were seeded into the
upper chamber of a 24-well Transwell unit with 8-µm polycarbonate
nucleopore filters (Corning, Inc.) and a 40 µl (1 mg/ml) Matrix gel
(Sigma-Aldrich; Merck KGaA), which was added prior to plating.
Before seeding the cells, the filters of the Transwell unit were
coated with the Matrix gel at 37˚C for 4 h to form a reconstructed
basement membrane. The upper compartment contained serum-free DMEM
medium whilst the lower compartment contained DMEM medium with 5%
FBS. The cells were incubated for 30 h in a humidified atmosphere
of 5% CO2 at 37˚C. The cells adhering to the lower
surface of the filter were stained with the 1% crystal violet
solution in 70% ethanol, at room temperature for 30 min and were
counted. The cells were calculated with an inverted light
microscope from ≥ five representative fields and x200
magnification.
Statistical analysis
Statistical analysis was performed using SPSS
software version 18.0 (IBM Corp). Data are presented as the mean ±
SD. Each test was repeated ≥3 times independently. Student's paired
and unpaired t-tests were used for comparisons between two groups.
One-way ANOVA analysis using GraphPad Prism 7 (GraphPad Software,
Inc.) with post hoc Tukey's multiple comparisons tests was used
when comparing multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of miR-129-5p in HCC cell
lines and tissue samples obtained from patients with liver
cancer
The clinical characteristics of patients with liver
cancer in this study are presented in Table I. The tumor size, intrahepatic
metastases, vascular invasion and capsular invasion were used to
analyze the proliferation, metastasis and invasion of tumors. To
investigate the biological role of miR-129-5p in the development of
human HCC, the expression levels of miR-129-5p in 45 pairs of fresh
liver tumoral tissues and cancerous peripheral tissue was analyzed
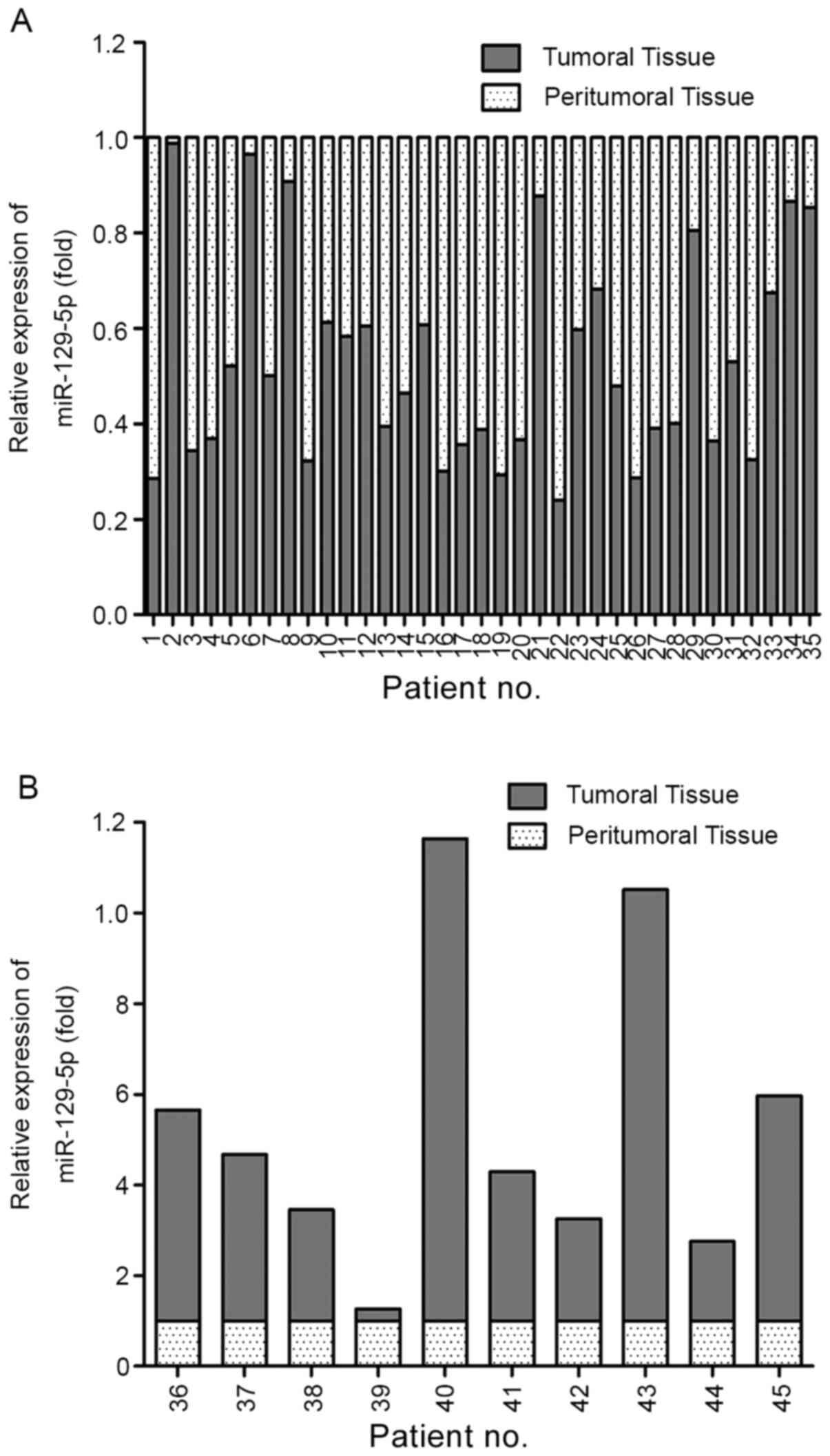
using RT-qPCR. miR-129-5p was downregulated in 77.78% of tumoral
tissues (n=35) compared with the matched peritumoral tissues
(Fig. 1A). Moreover, different
degrees of upregulation were observed in the remaining ten cases
(Fig. 1B). The results indicated
that miR-129-5p is potentially involved in the development of human
HCC.
Overexpression of miR-129-5p promotes
proliferation of HCC cells
To investigate the effect of miR-129-5p on the
cytotoxicity of HCC cells, an MTT cytotoxicity assay was performed
using HepG2 and Huh7 cells. The two types of liver cancer cells
were divided into two groups and transfected with negative control
(NC mimics) or miR-129-5p mimics and then measured for transfection
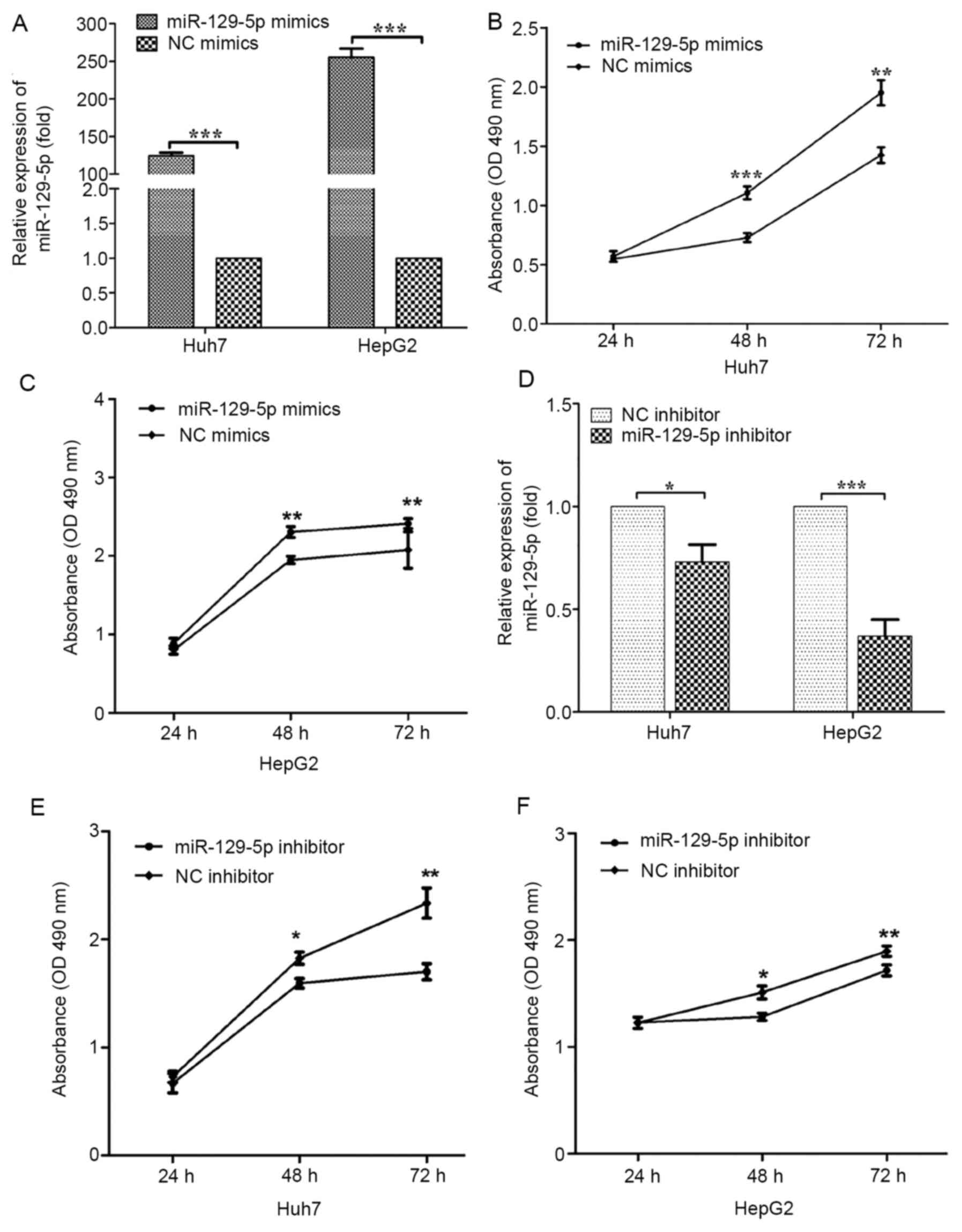
efficiency. The expression of miR-129-5p in Huh7 and HepG2 cells
was significantly higher than that in the NC mimics group (Fig. 2A). To investigate the effect of
miR-129-5p on the cytotoxicity of HCC cells, MTT assay was
performed with the same cells. The absorbance value of the cells in
the miR-129-5p overexpression group was higher compared with that
in the control group, indicating that the cytotoxicity rate was
significantly lower compared with the control group in both cell
lines (Fig. 2B and C). Therefore, miR-129-5p significantly
reduced the cytotoxicity of HepG2 and Huh7 cells in
vitro.
It was demonstrated that overexpression of
miR-129-5p can reduce the cytotoxicity of HCC, and thus, it was
investigated whether this phenotype can be reversed by knockdown of
miR-129-5p. To assess the role of miR-129-5p in HCC, cells were
transfected with miR-129-5p inhibitor NC and miR-129-5p inhibitor,
and a MTT assay was conducted after intracellular knockdown of
miR-129-5p. Transfection with the miR-129-5p inhibitor was shown to
significantly reduce the levels of miR-129-5p in both HepG2 or Huh7
cell lines (Fig. 2D). The number of
viable cells in the miR-129-5p knockdown group was significantly
lower compared with that in the control group, indicating that
knockdown of miR-129-5p significantly promoted the cytotoxicity of
both HCC cell lines (Fig. 2E and
F).
miR-129-5p promotes migration and
invasion of HCC cells
HCC is the second most deadly cancer type, with a
strong invasive and metastatic capability being the primary reasons
for the poor prognosis (2).
Therefore, the present study focused on whether the reduction of
miR-129-5p levels was related to the invasive and metastatic
capabilities of liver cancer. The expression levels of miR-129-5p
in tumor tissues of the patients (n=45) with HCC in this study were
compared between non-metastatic/invasive HCC samples (n=16) and
metastatic/invasive HCC samples, as well as those with intrahepatic
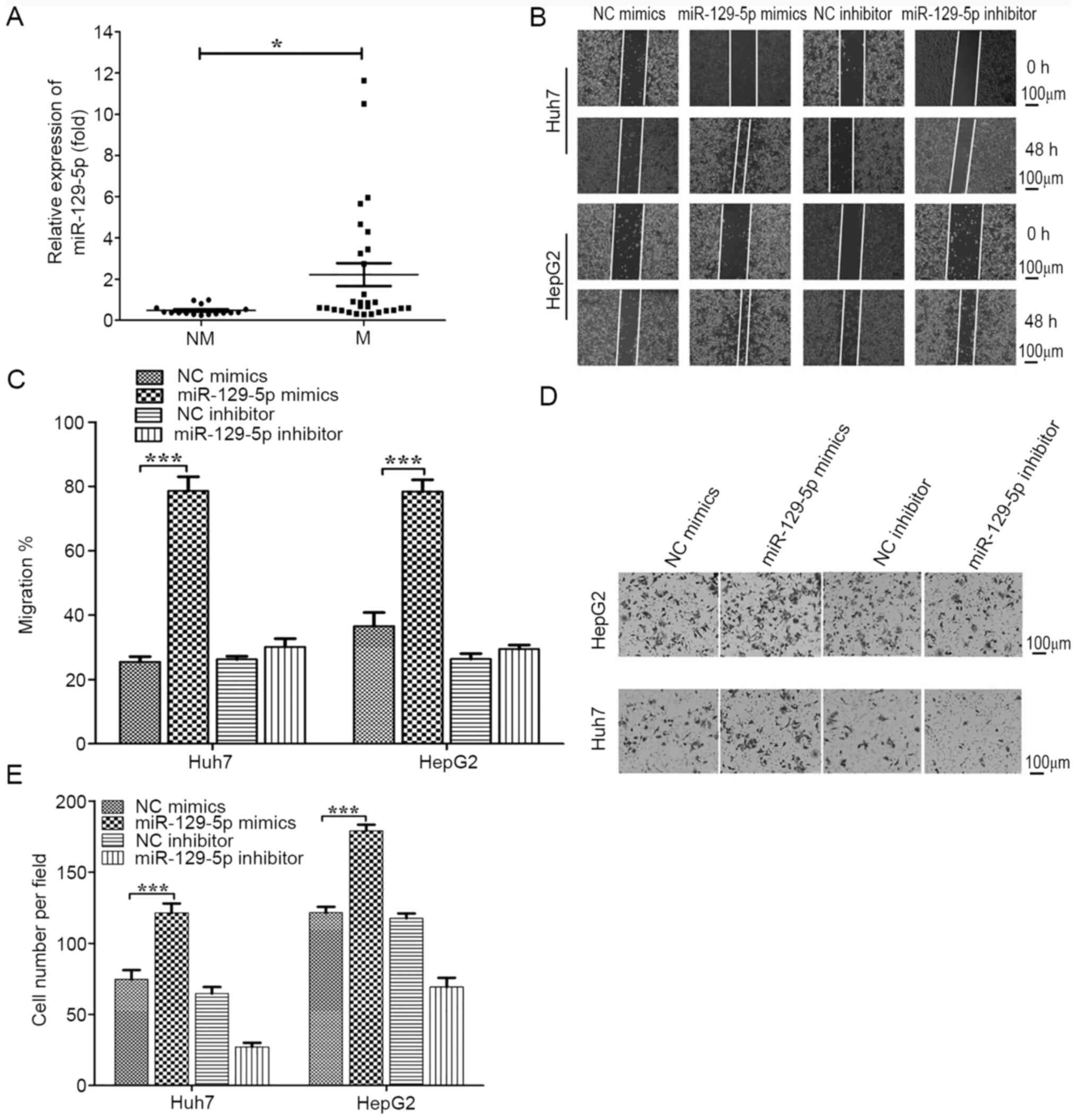
metastasis, vascular invasion or capsule invasion (n=29; Fig. 3A). It was found that miR-129-5p was
significantly downregulated in metastatic/invasive HCC samples.
To evaluate the effect of miR-129-5p on the
migration and invasion of liver cancer, wound healing and Transwell
assays were conducted using HepG2 and Huh7 cells. The results of
the migration assays suggested that the migratory capacity 48 h
after scratch in the miR-129-5p overexpression group was
significantly greater compared with that of the control group,
indicating the migration of cancer cells was significantly enhanced
(Fig. 3B and C). In the Transwell assay (Fig. 3D and E), the miR-129-5p mimics group had
significantly enhanced cell invasive capabilities compared with the
control group, whilst the migratory ability of the two cell lines
in the miR-129-5p inhibitor group was decreased, but there was no
statistical significance. These results indicated that miR-129-5p
promoted the migratory ability of liver cancer cells. Collectively,
the present results suggested that overexpression of miR-129-5p
enhanced the migration and invasion of HCC cells and thus promoted
the metastasis of HCC.
The interaction between miR-129-5p and
BMP2 3'UTR region
In order to elucidate the mechanism of miR-129-5p
action, bioinformatics analysis was conducted using www.targetscan.org to identify its target gene. A
predicted target site for miR-129-5p was identified in the BMP2
3'UTR region. To investigate whether BMP2 was a direct target of
miR-129-5p, luciferase reporter assays were performed in HepG2
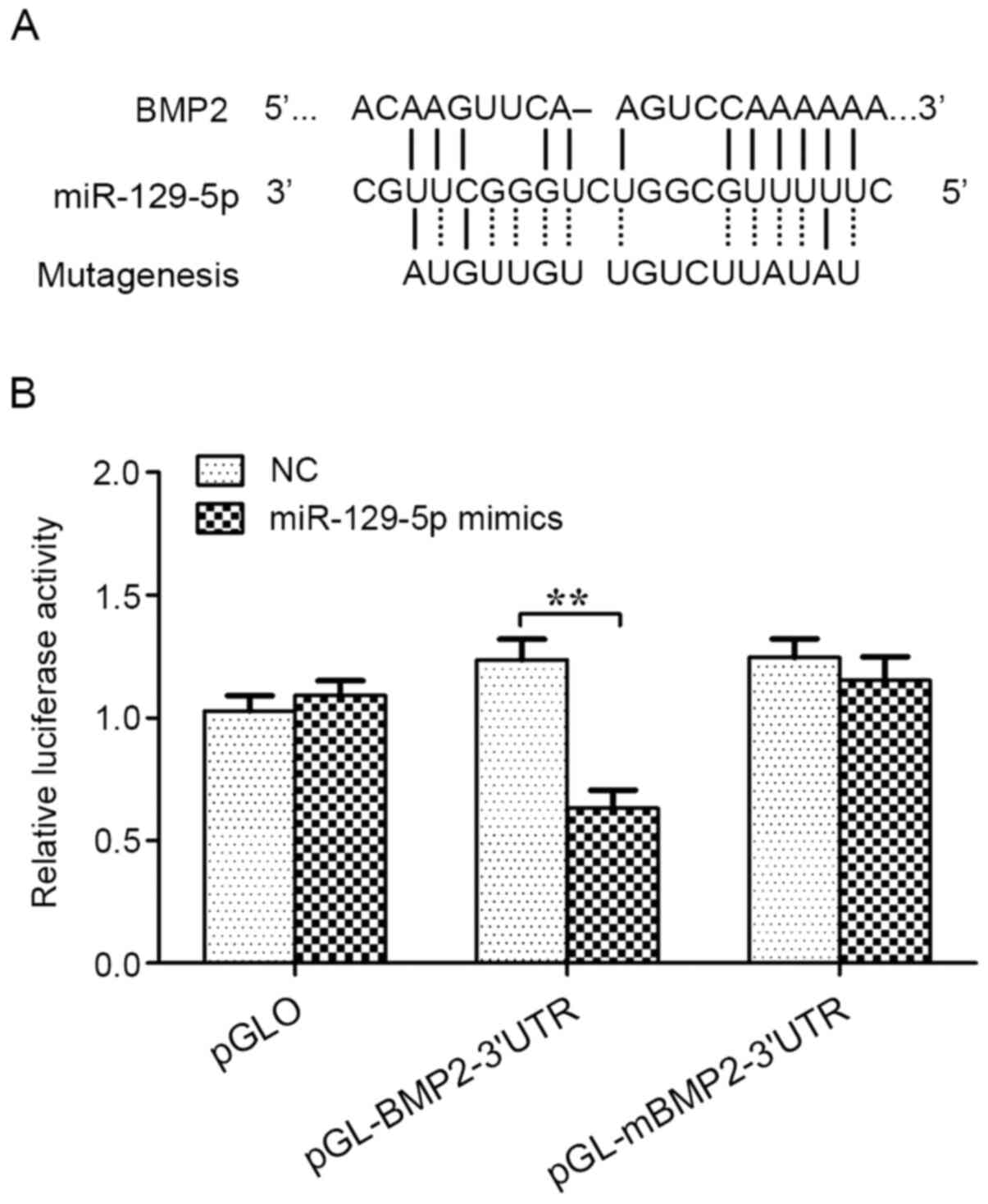
cells. The results indicated that miR-129-5p significantly reduced
the relative luciferase activity of BMP2 3'UTR (~36%). However, the
relative luciferase activity of the mutant BMP2 3'UTR was not
inhibited by miR-129-5p. Therefore, the results demonstrated that
BMP2 may be a direct target gene for miR-129-5p (Fig. 4).
miR-129-5p inhibits the expression of
BMP2
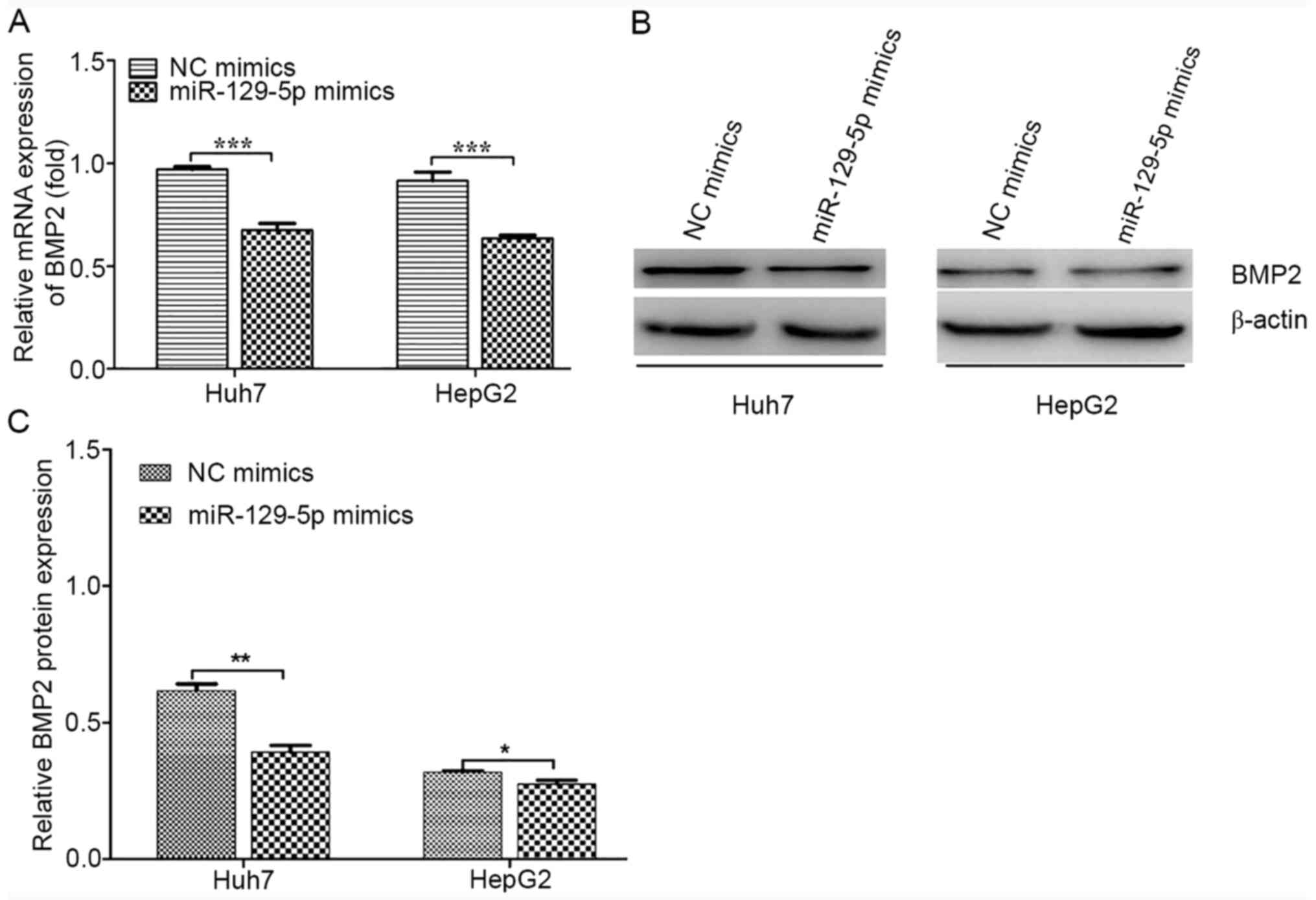
miRNAs can regulate gene expression by degrading
mRNA or by preventing translation without affecting mRNA stability
(15). To identify the regulatory
mechanism of action for miR-129-5p, BMP2 mRNA and protein
expression levels were detected following the overexpression of
miR-129-5p in HepG2 and Huh7 cells. The overexpression of
miR-129-5p appeared to reduce BMP2 mRNA and protein expression
levels in both cells (Fig. 5).
Thus, miR-129-5p may have suppressed the expression levels of BMP2
at the post-transcriptional level by degrading its mRNA.
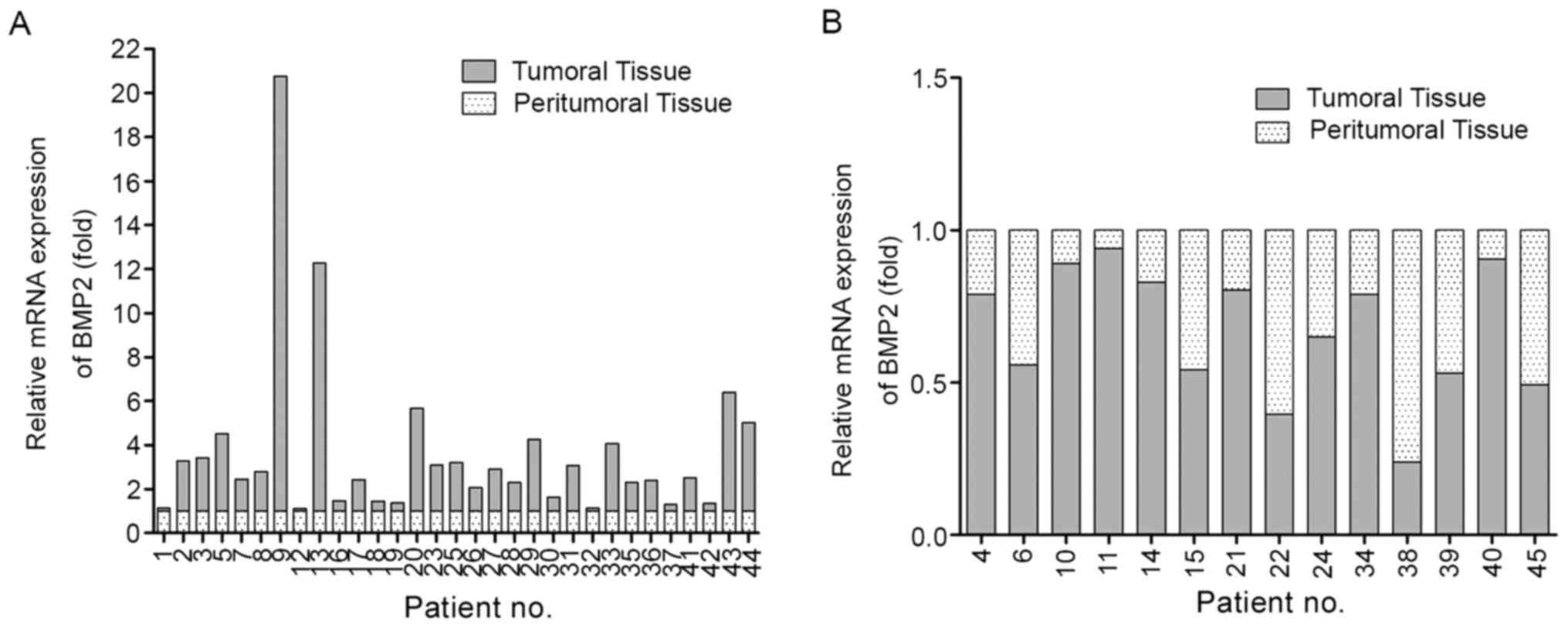
Negative correlation between
miR-129-5p and BMP2 expression levels in human HCC
The important role of BMP2 in tumor proliferation,
invasion and metastasis has been reported in previous studies
(39-44).
To determine the clinical significance of BMP2, the mRNA expression
level of BMP2 was examined in 45 pairs of liver tumoral tissues.
BMP2 was highly expressed in 31/45 cases of liver tumoral tissues,
which had different degrees of increase compared with the matched
peritumoral tissue, while the expression of BMP2 was reduced to
different degrees in the remaining 14 cases (Fig. 6A and B).
Discussion
HCC is one of the most aggressive tumor types
worldwide (1-3).
Due to the high rate of intrahepatic and extrahepatic metastasis,
patients with HCC often have a poor prognosis and high recurrence
possibility (45). Proliferation
and metastasis are the most important characteristics of cancer and
contribute to the high mortality of malignant tumors, especially
HCC (46-48).
Advancement in the research of miRNAs, as a regulatory basis behind
HCC, has provided novel insights into the development of HCC. It
has been revealed that miRNAs, such as miR-320 and miR-107, play an
important regulatory role in the proliferative and metastatic
potential of HCC (49,50). miR-129-5p exhibits a regulatory
effect on the onset and progression of various tumor types by
regulating the cell cycle, proliferation, apoptosis, migration,
invasion and angiogenesis, as well as other physiological and
pathological processes (25). In
neuroblastomas, miR-129 inhibits tumor growth by targeting
MYO10(35). Furthermore, miR-129-5p
enhances cell proliferation and invasion; and inhibits apoptosis by
regulating SOX4 in renal cell carcinoma (26). A previous study reported that
miR-129-5p is a biomarker of single ventricular heart failure
(36). miR-129 expression may be an
independent prognostic marker for BCR-free survival in patients
with prostate cancer and the overexpression of miR-129
significantly attenuates prostate cancer cell proliferation by
regulating cell cycle-regulated protein expression levels (27). However, to the best of our
knowledge, there are no reports on miR-129-5p and HCC, and thus,
the aim of the present study was to investigate the effect of
miR-129-5p on the proliferation and invasion of HCC cells.
In the present study, Huh7 was selected as a typical
HCC cell line and HepG2 was chosen as another liver cancer cell
line for the in vitro investigation into HCC. Although HepG2
has been shown to be a hepatoblastoma in recent years, >9,000
articles have used HepG2 as hepatocarcinoma or hepatoma from 1979
to March 2009. In 2019(51),
previous studies (52-55)
have used HepG2 as a HCC cell line. The aim of the present study
was to examine the effect of miR-129-5p on the proliferative and
metastatic abilities of HCC. The exact properties of HepG2 cells
was not the focus of this study and the use of HepG2 did not affect
the purpose and the results of the present study (51,56,57).
In the present study, it was found that 35/45
tumoral tissue samples had a decrease in miR-129-5p expression
levels, suggesting that miR-129-5p was involved in the progression
of liver cancer. Moreover, upregulation of miR-129-5p levels was
identified in 22.22% of tumoral tissue samples. However, the
mechanism of action responsible for the regulation of miR-129-5p in
liver cancer requires further investigation. Previous studies have
reported that miRNAs are involved in the regulation of signal
transduction pathways in HCC tumorigenesis and metastasis (49,50,58).
In the current study, miR-129-5p was found to be involved in all
stages of liver cancer and its involvement in the proliferation,
invasion and migration of liver cancer was demonstrated.
Furthermore, miR-129-5p was used to effectively distinguish 16
cases of non-metastatic/invasive HCC from 29 cases of
metastatic/invasive HCC. Collectively, the present results
suggested that miR-129-5p was involved in the invasion and
metastasis of liver cancer.
BMP, a member of the TGF-β superfamily, plays an
important role in cell proliferation, differentiation, apoptosis
and morphogenesis of various tissues and organs in the body
(59,60). BMP2 is expressed in several tumor
types, such as osteosarcoma, pancreatic cancer and breast cancer,
and has notable effects on cell proliferation and metastasis
(26,36). Although the relationship between
BMP2 and various tumors, as well as the relevant mechanism of
action, has been previously reported (26,36,59,60),
the effects of BMP2 on the development of HCC remains unknown. The
present study identified the potential target gene of miR-129-5p as
BMP2 through bioinformatics database analysis and found that
miR-129-5p can bind to a target site on the BMP2 3'UTR, using
luciferase reporter detection technology. In addition, BMP2 was
demonstrated to be the target gene of miR-129-5p by detecting the
mRNA and protein expression levels of BMP2 in cells after
transfection with miR-129-5p mimics. BMP2 expression was
post-transcriptionally regulated by miR-129-5p. Furthermore,
miR-129-5p expression levels were found to be negatively correlated
with BMP2 expression levels in liver tumoral tissue samples. It was
also found that BMP2 was highly expressed in 31/45 cases of liver
tumoral tissues, which had different degrees of increased
expression compared with the matched para-carcinoma tissue, while
14 cases had different degrees of reduced expression levels. The
last new finding of the present study is that miR-129-5p may
facilitate HCC pathogenesis through the BMP2. The newly identified
miR-129-5p/BMP2 axis provides new insight into the pathogenesis of
HCC. In conclusion, the present results demonstrated that
miR-129-5p may promote the proliferation, migration and invasion of
HCC cells, and play an important role in the cellular processes
behind the development of HCC, by targeting BMP2.
Acknowledgements
Not applicable.
Funding
The present study was supported by the fund from the
Department of Intervention, The First Affiliated Hospital of Harbin
Medical University (Harbin, Heilongjiang, China).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the study, performed the experiments and
wrote the manuscript. ZL and JS performed the experiments and
organized the images. XW and JS participated in this experiment and
made substantial contributions to the acquisition of data. ZL
analyzed and interpreted the data and organized the image. ZL and
ZC can authenticate the raw data. XW and ZC helped write the
manuscript, and were responsible for the study funding and in the
conception and design of the study, as well as providing final
approval of the version to be published. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee of Harbin Medical University (Harbin, China).
Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nakayama H and Takayama T: Role of
surgical resection for hepatocellular carcinoma based on Japanese
clinical guidelines for hepatocellular carcinoma. World J Hepatol.
7:261–269. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Braillon A: Hepatocellular carcinoma.
Lancet. 380:469–471. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Knudson AG Jr: Mutation and cancer:
Statistical study of retinoblastoma. Proc Natl Acad Sci USA.
68:820–823. 1971.PubMed/NCBI View Article : Google Scholar
|
|
5
|
He X, Yan YL, Eberhart JK, Herpin A,
Wagner TU, Schartl M and Postlethwait JH: miR-196 regulates axial
patterning and pectoral appendage initiation. Dev Biol.
357:463–477. 2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Poy MN, Hausser J, Trajkovski M, Braun M,
Collins S, Rorsman P, Zavolan M and Stoffel M: miR-375 maintains
normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci
USA. 106:5813–5818. 2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM,
Sung CO, Baek D, Haq F, Ansari AA, Lee SY, et al: Genomic portrait
of resectable hepatocellular carcinomas: Implications of RB1 and
FGF19 aberrations for patient stratification. Hepatology.
60:1972–1982. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Takai A, Dang HT and Wang XW:
Identification of drivers from cancer genome diversity in
hepatocellular carcinoma. Int J Mol Sci. 15:11142–11160.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hu TH, Huang CC, Lin PR, Chang HW, Ger LP,
Lin YW, Changchien CS, Lee CM and Tai MH: Expression and prognostic
role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular
carcinoma. Cancer. 97:1929–1940. 2003.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kawamura N, Nagai H, Bando K, Koyama M,
Matsumoto S, Tajiri T, Onda M, Fujimoto J, Ueki T, Konishi N, et
al: PTEN/MMAC1 mutations in hepatocellular carcinomas: Somatic
inactivation of both alleles in tumors. Jpn J Cancer Res.
90:413–418. 1999.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hellebrekers DM, Griffioen AW and van
Engeland M: Dual targeting of epigenetic therapy in cancer. Biochim
Biophys Acta. 1775:76–91. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nault JC and Villanueva A: Intratumor
molecular and phenotypic diversity in hepatocellular carcinoma.
Clin Cancer Res. 21:1786–1788. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Nault JC, Mallet M, Pilati C, Calderaro J,
Bioulac-Sage P, Laurent C, Laurent A, Cherqui D, Balabaud C and
Zucman-Rossi J: High frequency of telomerase reverse-transcriptase
promoter somatic mutations in hepatocellular carcinoma and
preneoplastic lesions. Nat Commun. 4(2218)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Vishnoi A and Rani S: MiRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993.PubMed/NCBI View Article : Google Scholar
|
|
16
|
van Rooij E and Olson EN: MicroRNAs:
Powerful new regulators of heart disease and provocative
therapeutic targets. J Clin Invest. 117:2369–2376. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Johnston RJ Jr, Chang S, Etchberger JF,
Ortiz CO and Hobert O: MicroRNAs acting in a double-negative
feedback loop to control a neuronal cell fate decision. Proc Natl
Acad Sci USA. 102:12449–12454. 2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kloosterman WP, Wienholds E, de Bruijn E,
Kauppinen S and Plasterk RH: In situ detection of miRNAs in animal
embryos using LNA-modified oligonucleotide probes. Nat Methods.
3:27–29. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Appolloni R, Formicola R, Scurci M,
Appolloni R and Colonna GB: Lupus anticoagulant and bilateral optic
disc edema: A case report. Ann Ophthalmol. 23:312–317.
1991.PubMed/NCBI
|
|
21
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hammond SM: MicroRNAs as tumor
suppressors. Nat Genet. 39:582–583. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Finnerty JR, Wang WX, Hébert SS, Wilfred
BR, Mao G and Nelson PT: The miR-15/107 group of microRNA genes:
Evolutionary biology, cellular functions, and roles in human
diseases. J Mol Biol. 402:491–509. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu Q, Li Y, Lv W, Zhang G, Tian X, Li X,
Cheng H and Zhu C: UCA1 promotes cell proliferation and invasion
and inhibits apoptosis through regulation of the miR129-SOX4
pathway in renal cell carcinoma. Onco Targets Ther. 11:2475–2487.
2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xu S, Yi XM, Zhang ZY, Ge JP and Zhou WQ:
miR-129 predicts prognosis and inhibits cell growth in human
prostate carcinoma. Mol Med Rep. 14:5025–5032. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fu S, Fei Q, Jiang H, Chuai S, Shi S,
Xiong W, Jiang L, Lu C, Atadja P, Li E and Shou J: Involvement of
histone acetylation of Sox17 and Foxa2 promoters during mouse
definitive endoderm differentiation revealed by microRNA profiling.
PLoS One. 6(e27965)2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ,
Zhou H and Qu LH: Liver-enriched transcription factors regulate
microRNA-122 that targets CUTL1 during liver development.
Hepatology. 52:1431–1442. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang Y, Yang P and Wang XF:
Microenvironmental regulation of cancer metastasis by miRNAs.
Trends Cell Biol. 24:153–160. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chai ZT, Kong J, Zhu XD, Zhang YY, Lu L,
Zhou JM, Wang LR, Zhang KZ, Zhang QB, Ao JY, et al: MicroRNA-26a
inhibits angiogenesis by down-regulating VEGFA through the
PIK3C2α/Akt/HIF-1α pathway in hepatocellular carcinoma. PLoS One.
8(e77957)2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Laudadio I, Manfroid I, Achouri Y, Schmidt
D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F,
et al: A feedback loop between the liver-enriched transcription
factor network and miR-122 controls hepatocyte differentiation.
Gastroenterology. 142:119–129. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato
JM and Lu SC: MicroRNAs regulate methionine adenosyltransferase 1A
expression in hepatocellular carcinoma. J Clin Invest. 123:285–298.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
35
|
Wang X, Li J, Xu X, Zheng J and Li Q:
miR-129 inhibits tumor growth and potentiates chemosensitivity of
neuroblastoma by targeting MYO10. Biomed Pharmacother.
103:1312–1318. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ramachandran S, Lowenthal A, Ritner C,
Lowenthal S and Bernstein HS: Plasma microvesicle analysis
identifies microRNA 129-5p as a biomarker of heart failure in
univentricular heart disease. PLoS One. 12(e0183624)2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lv G, Hu Z, Tie Y, Du J, Fu H, Gao X and
Zheng X: MicroRNA-451 regulates activating transcription factor 2
expression and inhibits liver cancer cell migration. Oncol Rep.
32:1021–1028. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Carragee EJ, Hurwitz EL and Weiner BK: A
critical review of recombinant human bone morphogenetic protein-2
trials in spinal surgery: Emerging safety concerns and lessons
learned. Spine J. 11:471–491. 2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Jiramongkolchai P, Owens P and Hong CC:
Emerging roles of the bone morphogenetic protein pathway in cancer:
Potential therapeutic target for kinase inhibition. Biochem Soc
Trans. 44:1117–1134. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sampath TK, Coughlin JE, Whetstone RM,
Banach D, Corbett C, Ridge RJ, Ozkaynak E, Oppermann H and Rueger
DC: Bovine osteogenic protein is composed of dimers of OP-1 and
BMP-2A, two members of the transforming growth factor-beta
superfamily. J Biol Chem. 265:13198–13205. 1990.PubMed/NCBI
|
|
42
|
Katsuno Y, Hanyu A, Kanda H, Ishikawa Y,
Akiyama F, Iwase T, Ogata E, Ehata S, Miyazono K and Imamura T:
Bone morphogenetic protein signaling enhances invasion and bone
metastasis of breast cancer cells through Smad pathway. Oncogene.
27:6322–6333. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Arnold SF, Tims E and Mcgrath BE:
Identification of bone morphogenetic proteins and their receptors
in human breast cancer cell lines: Importance of BMP2. Cytokine.
11:1031–1037. 1999.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Davies SR, Watkins G, Douglas-Jones A,
Mansel RE and Jiang WG: Bone morphogenetic proteins 1 to 7 in human
breast cancer, expression pattern and clinical/prognostic
relevance. J Exp Ther Oncol. 7:327–338. 2008.PubMed/NCBI
|
|
45
|
Budhu A, Forgues M, Ye QH, Jia HL, He P,
Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY and Wang XW:
Prediction of venous metastases, recurrence, and prognosis in
hepatocellular carcinoma based on a unique immune response
signature of the liver microenvironment. Cancer Cell. 10:99–111.
2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Aravalli RN, Cressman EN and Steer CJ:
Cellular and molecular mechanisms of hepatocellular carcinoma: An
update. Arch Toxicol. 87:227–247. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
van Zijl F, Zulehner G, Petz M, Schneller
D, Kornauth C, Hau M, Machat G, Grubinger M, Huber H and Mikulits
W: Epithelial-mesenchymal transition in hepatocellular carcinoma.
Future Oncol. 5:1169–1179. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Belghiti J and Fuks D: Liver resection and
transplantation in hepatocellular carcinoma. Liver Cancer. 1:71–82.
2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zou CD, Zhao WM, Wang XN, Li Q, Huang H,
Cheng WP, Jin JF, Zhang H, Wu MJ, Tai S, et al: MicroRNA-107: A
novel promoter of tumor progression that targets the CPEB3/EGFR
axis in human hepatocellular carcinoma. Oncotarget. 7:266–278.
2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lv G, Wu M, Wang M, Jiang X, Du J, Zhang
K, Li D, Ma N, Peng Y, Wang L, et al: miR-320a regulates high
mobility group box 1 expression and inhibits invasion and
metastasis in hepatocellular carcinoma. Liver Int. 37:1354–1364.
2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ahmed NM, Youns M, Soltan MK and Said AM:
Design, synthesis, molecular modelling, and biological evaluation
of novel substituted pyrimidine derivatives as potential anticancer
agents for hepatocellular carcinoma. J Enzyme Inhib Med Chem.
34:1110–1120. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Su CM, Hou GG, Wang CH, Zhang HQ, Yang C,
Liu M and Hou Y: Potential multifunctional agents with
anti-hepatoma and anti-inflammation properties by inhibiting NF-κB
activation. J Enzyme Inhib Med Chem. 34:1287–1297. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhao J, Wang Y, Han M, Lu H, Chen X, Liu
S, Yuan X, Han K, Liang P and Cheng J: P7TP3 inhibits tumor
development, migration, invasion and adhesion of liver cancer
through the Wnt/β-catenin signaling pathway. Cancer Sci.
111:994–1007. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Xia Y, Zhong J, Zhao M, Tang Y, Han N, Hua
L, Xu T, Wang C and Zhu B: Galactose-modified selenium
nanoparticles for targeted delivery of doxorubicin to
hepatocellular carcinoma. Drug Deliv. 26:1–11. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Aden DP, Fogel A, Plotkin S, Damjanov I
and Knowles BB: Controlled synthesis of HBsAg in a differentiated
human liver carcinoma-derived cell line. Nature. 282:615–616.
1979.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Knowles BB, Howe CC and Aden DP: Human
hepatocellular carcinoma cell lines secrete the major plasma
proteins and hepatitis B surface antigen. Science. 209:497–499.
1980.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Vasuri F, Visani M, Acquaviva G, Brand T,
Fiorentino M, Pession A, Tallini G, D'Errico A and de Biase D: Role
of microRNAs in the main molecular pathways of hepatocellular
carcinoma. World J Gastroenterol. 24:2647–2660. 2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Mukai T, Otsuka F, Otani H, Yamashita M,
Takasugi K, Inagaki K, Yamamura M and Makino H: TNF-alpha inhibits
BMP-induced osteoblast differentiation through activating SAPK/JNK
signaling. Biochem Biophys Res Commun. 356:1004–1010.
2007.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Huang RL, Yuan Y, Tu J, Zou GM and Li Q:
Opposing TNF-α/IL-1β- and BMP-2-activated MAPK signaling pathways
converge on Runx2 to regulate BMP-2-induced osteoblastic
differentiation. Cell Death Dis. 5(e1187)2014.PubMed/NCBI View Article : Google Scholar
|