Introduction
Knee osteoarthritis is the most common chronic
degenerative bone and joint disease in middle-aged and elderly
individuals worldwide (1). Knee
osteoarthritis is a heterogeneous disease with an incidence of ~3%
(2,3), and results from a multifactorial
imbalance in the synthesis and degradation of knee chondrocytes
subchondral bone and extracellular matrix (4). Abnormal joint metabolism leads to
degeneration of knee articular cartilage and subchondral bone,
joint fiber hyperplasia, synovial non-specific inflammation and
other lesions (5). Due to the
continuous damage caused by chronic inflammation and the
progressive structural changes of the knee joint tissue, the
disease progresses continuously (6). An irreversible loss of knee function
and pain are among the common causes of decreased quality of life
and disability associated with knee osteoarthritis (7).
The main clinical manifestations of knee
osteoarthritis are slow-developing knee pain, tenderness,
stiffness, joint swelling, limited mobility and knee deformity
(8). The occurrence and development
of knee osteoarthritis are the result of the interaction between
biomechanical and biological factors, including the mechanical
environment around the knee joint, the metabolic rate of the
individual, the apoptosis of chondrocytes and the levels of related
cytokines, such as interleukin (IL) IL-4, IL-10 and tumor necrosis
factor (TNF)-α (9-11).
A number of risk factors for knee osteoarthritis have been
identified, including age, sex, obesity, inflammation, strain,
trauma and genetics (12-14).
The development and progression of knee osteoarthritis is complex
and at present, effective treatment strategies and clear
therapeutic targets have not been identified (15).
Long non-coding RNAs (lncRNAs) are RNA molecules
>200 nucleotides in length that are the by-product of RNA
polymerase II transcription, and were previously thought to have no
biological function (16). lncRNAs
do not encode proteins, but play a critical role in gene regulation
and epigenetic modifications (17-19).
lncRNA affect cartilage matrix production and metabolism,
synovitis, autophagy and apoptosis of chondrocytes (20,21).
Gaballah et al (18)
reported that ZNF1 antisense RNA 1 can promote chondrocyte
proliferation and inhibit apoptosis by regulating the expression
level of Wnt family member 3A. A previous study by Wang et
al (22) suggested that nuclear
paraspeckle assembly transcript 1 can act on microRNA-181c to
promote the proliferation of human synoviocytes and accelerate the
progression of osteoarthritis (22). Although the specific function of
lncRNAs has not been elucidated, previous studies have reported
that the abnormal expression of lncRNAs is closely related to
osteoarthritis (23,24).
The aim of the present study was to examine the
lncRNA expression profiles of cartilage from young patients with
amputations without joint lesions, and from patients with knee
osteoarthritis who underwent total knee arthroplasty. The lncRNA
expression profiles of 13 patients were compared to identify new
potential regulatory targets involved in the pathogenesis of the
disease. The results of the present study may aid in identifying
the role of lncRNAs in the progression of knee osteoarthritis.
Materials and methods
Subjects
The present study recruited 13 patients from the
Department of Orthopedic Surgery, The Second Affiliated Hospital of
Anhui Medical University, between February 2016 and December 2018,
including six males and seven females. The 13 patients were divided
into two groups: Six patients aged 29-49 years in the normal group
(NC), and seven patients aged 67-82 years in the knee
osteoarthritis group (OA; average age 38.17±7.70 vs. 71.71±6.02
years). The NC group was made up of young patients with amputations
without joint disease. Inclusion criteria for the NC group were as
follows: i) No history of joint lesions; and ii) lower limb injury
treated with amputation. All patients in the OA group were patients
with knee osteoarthritis who underwent total knee arthroplasty. The
inclusion criteria for the OA group were as follows: i) Diagnosed
with knee osteoarthritis based on clinical manifestations, combined
with X-ray examination, knee pain and severely affected quality of
life, with failure of conventional oral drug therapy; ii) patients
with primary total knee arthroplasty; iii) knee flexion
malformation <15˚, varus deformity <15˚ or valgus deformity
<15˚; and iv) Kellgren Lawrence III-IV level (25). Exclusion criteria for both groups
were as follows: i) Severe internal and external valgus and flexion
contracture deformity (flaw angle >15˚); ii) obese patients
(body mass index >35); iii) patients with heart and lung
dysfunction, cerebrovascular disease or diabetes; iv) history of
open joint surgery; and v) active infection. The present study was
approved by The Second Affiliated Hospital of Anhui Medical
University Ethics Committee. Written informed consent was obtained
from each patient or their relatives prior to surgery.
Sample collection and total RNA
extraction
Approximately 5 g of cartilage tissue was collected
from each patient and stored at -80˚C until further analysis. Total
RNA was extracted from the tissue suspension using the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The tissue was
homogenized prior to RNA extraction. The amount and purity of the
total RNA were assessed using a NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.), and RNA integrity was analyzed
using an RNA Nano 6000 kit from the Agilent Bioanalyzer 2100 system
(Agilent Technologies, Inc.).
Preparation for lncRNA-sequencing
library
A lncRNA library was constructed using 3 µg of total
RNA per sample. The Ribo-Zero™ Gold kit (Illumina, Inc.) was used
to remove the rRNA from the sample and select the different index
tags to build the library according to the manufacturer's
instructions. The NEB Next Ultra™ Directional RNA Library Prep kit
for Illumina (New England BioLabs, Inc.) was used to construct a
sequencing library using rRNA-depleted RNA, according to the
manufacturer's instructions.
High-throughput sequencing
Sequencing libraries were generated using the
NEBNext Ultra™ Directional RNA LibraryPrep kit for Illumina (New
England BioLabs, Inc.) according to the manufacturer's protocol.
Raw reads were obtained by Illumina sequencing and high-quality
sequences (clean reads) were obtained through a series of data
processing procedures such as removing low-quality sequences,
de-linking contamination and rRNA including removing reads of joint
contamination (the base number of joint contamination in reads was
greater than 5 bp, removing low-quality reads (reads intermediate
value Q ≤19 accounted for >50% of the total base), removing
reads with N ratio >5% and removing reads matched with rRNA.
All subsequent analyses were based on clean reads.
Library sequencing was carried out on a HiSeq 4000 platform
(Illumina, Inc.) according to the commercially available protocols
from Chongqing Western Biotechnology, Inc.
Identification of differentially
expressed lncRNAs
The analysis of differences in lncRNA expression
between the two groups (NC and OA group) was performed using the
DEGseq (2010) R package (http://dblp.uni-trier.de/db/journals/bioinformatics/bioinformatics26.html#WangFWWZ10).
The P-value was adjusted using the q-value. P<0.05 and |log2
(fold-change)|>1 were set as the threshold for significant
differential expression.
Reverse transcription-quantitative PCR
(RT-qPCR)
To validate the reliability of the RNA-sequencing
data, 6 differentially expressed lncRNAs were randomly selected and
RT-qPCR was used to examine the expression level of the lncRNAs.
Total RNA was extracted from the cartilage using the TRIzol reagent
(Invitrogen, Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. RT-qPCR reactions were performed using
the Luna Universal One-Step RT-qPCR kit (New England BioLabs, Inc.)
according to the manufacturer's instructions. The following
thermocycling conditions were used: 15 sec at 55˚C and 1 min at
95˚C, followed by 40 cycles of 10 sec at 95˚C and 30 sec at 60˚C,
and 30 sec at 50˚C. GAPDH was used as an internal control and the
relative expression levels of candidate lncRNAs were calculated
using the 2-ΔΔCq method (26). GAPDH forwards primer,
AATGGGCAGCCGTTAGGAAA and reverse primer, GCCCAATACGACCAAATCAGAG.
Experiments were conducted in at least triplicate. The primer
sequences that were used in the present study are listed in
Table SI.
GO and KEGG enrichment analyses
Target mRNAs of the differentially expressed lncRNAs
were classified according to the principles of GO classification.
GO collects information from GO and NCBI databases, annotating and
classifying genes based on biological processes, molecular
functions and cellular components (http://www.geneontology.org/). KEGGis a comprehensive
database for the systematic analysis of gene function, which is
based on manually drawn metabolic pathways and is divided into
metabolism, genetic information processing, cellular processes,
environmental information processing, biological systems and human
diseases (http://www.genome.jp/kegg/).
Statistical analysis
Data analyses were performed using SPSS software
(version 19.0; IBM Corp.) and data are expressed as the mean ± SD.
Comparisons of data between groups were performed by the Student's
t-test. Categorical data were analyzed by the χ2 test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical characteristics
The clinical characteristics of the 13 patients are
presented in Table I. The NC group
consisted of six patients including three men and three women with
a mean age of 38.17±7.70 years. The OA group consisted of seven
patients including three women and three men with a mean age of
71.71±6.02. A significant difference was identified between the
ages of the two groups (P=0.001). There were no significant
differences between the two groups regarding sex, BMI and limb side
(P=0.817, 0.389 and 0.751, respectively).
 | Table IBaseline clinical
characteristics. |
Table I
Baseline clinical
characteristics.
| Variable | NC, n=6 | OA, n=7 | P-value |
|---|
| Age, years (mean ±
SD) | 38.17±7.70 | 71.71±6.02 | 0.001 |
| Sex
(male/female) | 3/3 | 3/4 | 0.817 |
| BMI (mean ±
SD) | 20.82±2.11 | 22.16±3.09 | 0.389 |
| Side
(left/right) | 2/4 | 4/3 | 0.751 |
Differentially expressed lncRNAs
A total of 23,583 lncRNAs were identified in
osteoarthritis cartilage, including 5,255 upregulated and 5,690
downregulated lncRNAs, compared with normal cartilage (log2
fold-change >1; P<0.05; Fig.
1). The most significantly upregulated lncRNA was MSTRG.95856
(log2 fold-change = 11.6488). The most significantly downregulated
lncRNA was ENSG00000279725 (log2 fold-change = -9.6546). According
to the microarray data, the number of downregulated lncRNAs was
larger than that of the upregulated lncRNAs. However, among
significantly differentially expressed lncRNAs (log2 fold-change
>6), there were more upregulated than downregulated lncRNAs.
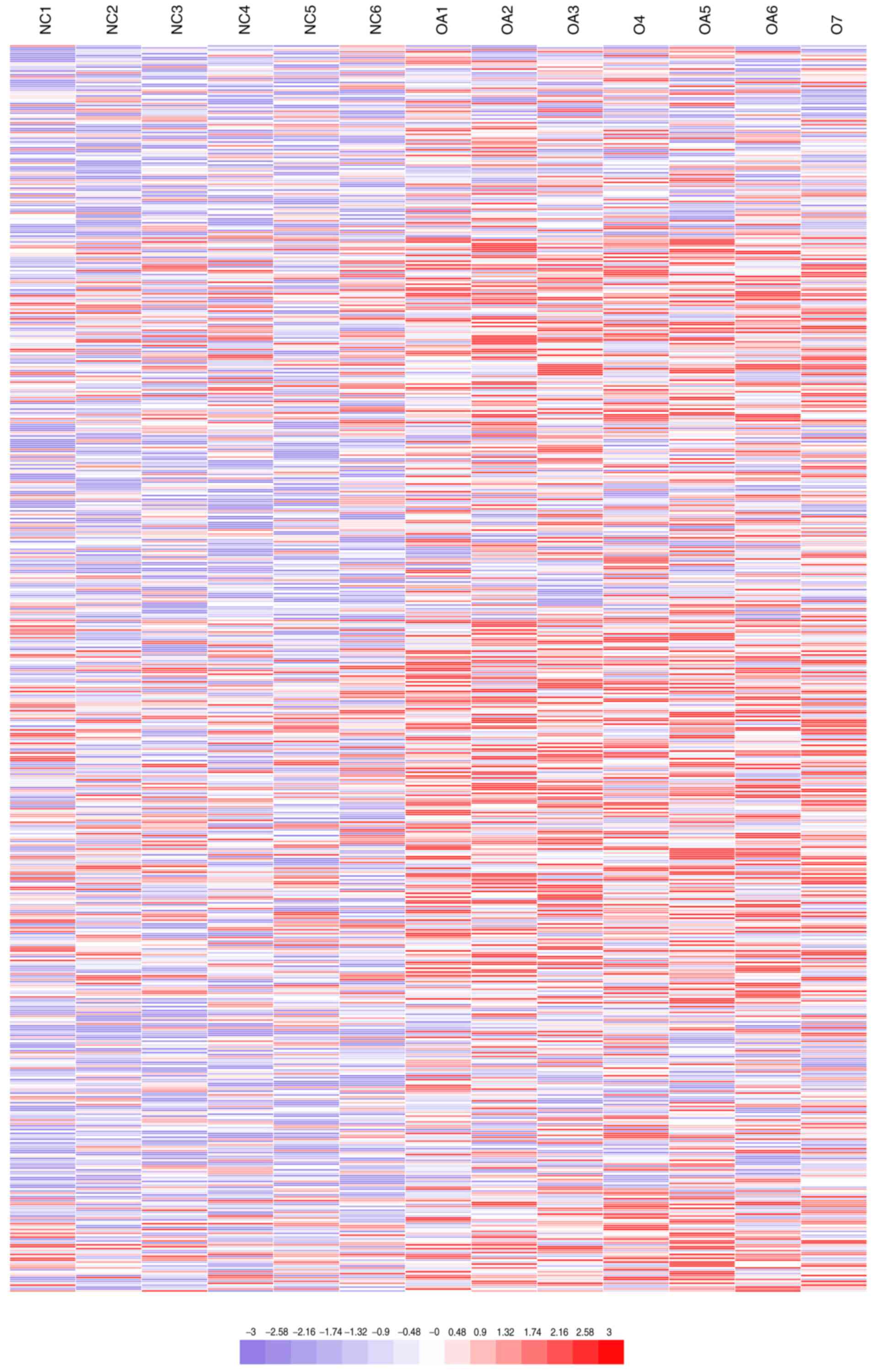
Hierarchical clustering analysis displayed the differential
expression of lncRNAs (Fig. 2),
where red and blue represent expression values higher and lower
than the median expression value, respectively. The results
indicated that differences between osteoarthritis and normal
cartilage could be identified by differences in the expression
profiles of lncRNAs associated with osteoarthritis (Table II).
 | Table IIMost significantly expressed top 15
entries in upregulated and downregulated lncRNAs. |
Table II
Most significantly expressed top 15
entries in upregulated and downregulated lncRNAs.
| lncRNA ID | Position | Log2
(fold-change) | Regulation | P-value |
|---|
| MSTRG.95856 |
chr17:64319415-64413776 | 11.64880695 | Up |
8.83x10-245 |
|
ENSG00000246430 |
chr8:56436674-56446734 | 11.12541347 | Up |
3.69x10-122 |
|
ENSG00000253730 |
chr17:50183289-50201632 | 9.970804581 | Up |
4.75x10-116 |
|
ENSG00000261959 |
chr17:50183289-50201632 | 9.970804581 | Up |
4.35x10-241 |
|
ENSG00000214145 |
chr3:194355247-194369743 | 9.961630563 | Up |
2.60x10-221 |
|
ENSG00000254855 |
chr11:66314487-66317044 | 9.259743264 | Up |
4.07x10-232 |
|
ENSG00000255468 |
chr11:66314487-66317044 | 9.259743264 | Up |
2.53x10-206 |
|
ENSG00000269053 |
chr19:17351448-17377350 | 8.885696373 | Up |
2.93x10-190 |
|
ENSG00000282851 |
chr19:17351448-17377350 | 8.885696373 | Up |
7.99x10-111 |
| MSTRG.106332 |
chr19:17351448-17377350 | 8.885696373 | Up |
3.09x10-214 |
| MSTRG.143488 |
chr22:39223359-39244751 | 8.774787060 | Up |
4.01x10-172 |
| MSTRG.92410 |
chr17:31303766-31314112 | 8.727920455 | Up |
6.30x10-233 |
|
ENSG00000227502 |
chr6:113857362-113863471 | 8.715226609 | Up |
3.50x10-94 |
|
ENSG00000244161 |
chr3:58192257-58214697 | 8.640244936 | Up |
7.63x10-237 |
|
ENSG00000258810 |
chr14:20801228-20803278 | 8.628141719 | Up |
5.65x10-160 |
|
ENSG00000279725 |
chr10:127737235-127741186 | -9.654636029 | Down |
1.26x10-23 |
|
ENSG00000280302 |
chr18:12254319-12277595 | -8.214319121 | Down |
1.22x10-79 |
|
ENSG00000250012 |
chr3:126103562-126197994 | -7.740118043 | Down |
6.01x10-116 |
| MSTRG.155259 |
chr3:126103562-126197994 | -7.740118043 | Down |
7.25x10-14 |
|
ENSG00000231966 |
chr1:179743163-179816198 | -6.693486957 | Down |
1.35x10-23 |
|
ENSG00000203288 |
chr1:151806071-151831872 | -6.524023102 | Down |
4.52x10-102 |
|
ENSG00000234614 |
chr1:151806071-151831872 | -6.524023102 | Down |
3.04x10-170 |
| MSTRG.13443 |
chr1:151806071-151831872 | -6.524023102 | Down |
8.04x10-48 |
|
ENSG00000229178 |
chr3:195614947-195620233 | -6.438791853 | Down |
6.39x10-61 |
|
ENSG00000242086 |
chr3:195614947-195620233 | -6.438791853 | Down |
8.84x10-211 |
| MSTRG.162045 |
chr3:195614947-195620233 | -6.438791853 | Down |
4.62x10-19 |
| MSTRG.162046 |
chr3:195614947-195620233 | -6.438791853 | Down |
5.26x10-151 |
|
ENSG00000227388 |
chr9:35752990-35756613 | -6.384897330 | Down |
3.85x10-29 |
|
ENSG00000226133 |
chr1:48102068-48104707 | -6.266786541 | Down |
2.40x10-70 |
|
ENSG00000261026 |
chr8:22713251-23000000 | -6.200762405 | Down |
1.36x10-271 |
GO and KEGG pathway analyses
lncRNAs do not encode proteins and they function
through cis- or trans-regulation of protein-coding genes (27). For the target genes of
differentially expressed lncRNAs, the functions of these lncRNAs
were indirectly predicted according to the target genes. GO and
KEGG pathway analyses of the target genes of the differential
lncRNAs were then performed.
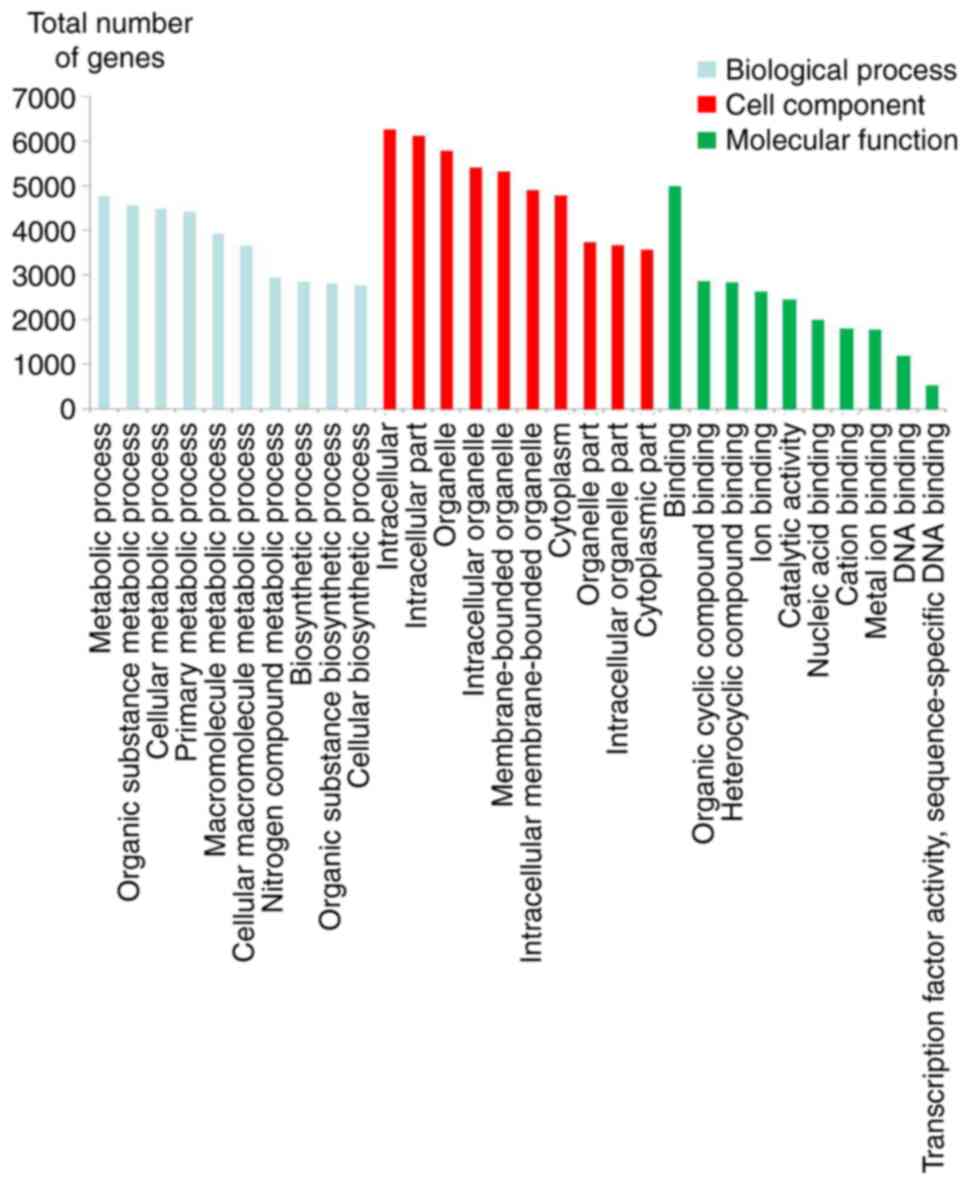
Table III and
Fig. 3 display the top 10 enriched
GO entries with the most significant difference in target genes of
lncRNAs in each group. In biological processes, target genes were
mainly enriched in the metabolic process of substances, including
‘organic substance metabolic process’. In cellular components,
target genes were mainly enriched in ‘intracellular part’. In
molecular functions, target genes were mainly enriched in
‘binding’. However, target genes do not appear to be mainly
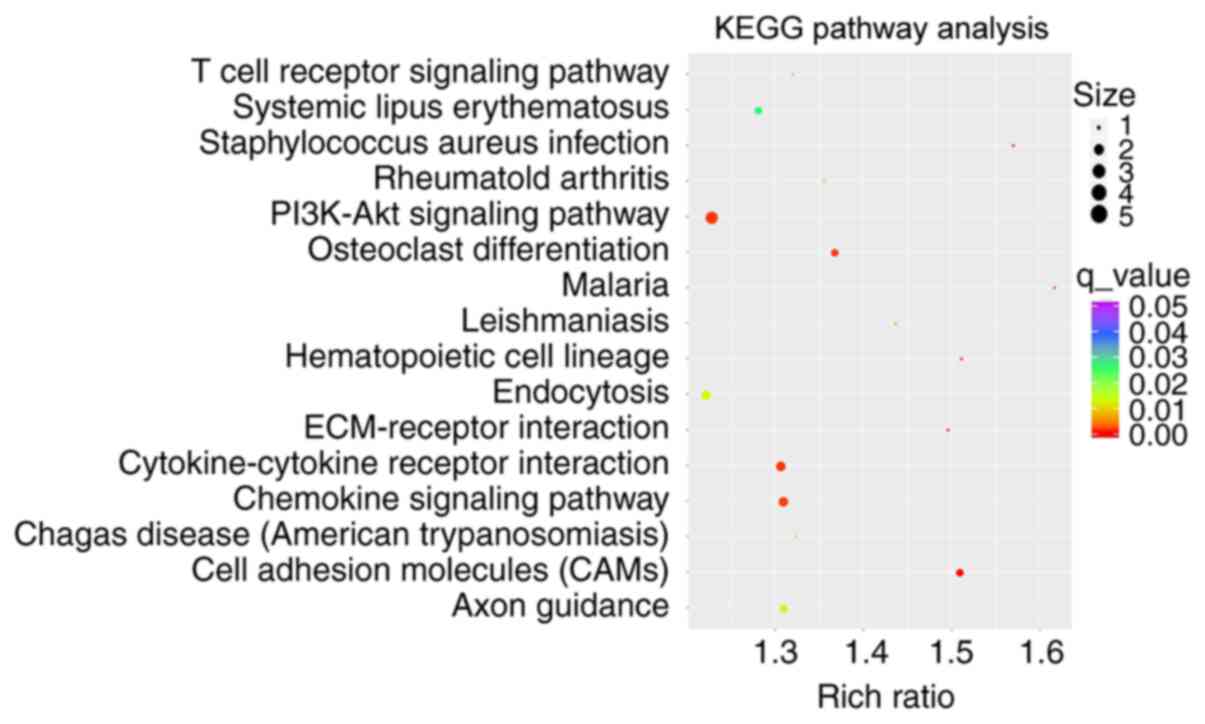
enriched in all of the terms listed here. Furthermore, Table IV and Fig. 4 display the KEGG pathways with the
most significant differences in target genes for lncRNAs between
the two groups. In the KEGG pathway analysis, the enrichment
pathways associated with significant expression of lncRNAs were
‘PI3K-Akt signaling pathway’, ‘endocytosis’, ‘cytokine-cytokine
receptor interaction’ and ‘chemokine signaling pathway’.
 | Table IIITop 15 significantly enriched GO
terms in biological processes, cellular components and molecular
functions of the differentially expressed lncRNAs. |
Table III
Top 15 significantly enriched GO
terms in biological processes, cellular components and molecular
functions of the differentially expressed lncRNAs.
| A, Upregulated |
|---|
| Category | GO accession
no. | Description | Corrected
P-value | Total number of
genes |
|---|
| GOTERM_BP | GO:0008152 | Metabolic
process |
7.1x10-11 | 4,779 |
| GOTERM_BP | GO:0071704 | Organic substance
metabolic process |
4.2x10-11 | 4,573 |
| GOTERM_BP | GO:0044237 | Cellular metabolic
process |
2.7x10-14 | 4,486 |
| GOTERM_BP | GO:0009058 | Biosynthetic
process |
2.6x10-09 | 2,858 |
| GOTERM_BP | GO:1901576 | Organic substance
biosynthetic process |
2.7x10-09 | 2,820 |
| GOTERM_CC | GO:0043226 | Organelle |
2.5x10-17 | 5,787 |
| GOTERM_CC | GO:0043229 | Intracellular
organelle |
6.0x10-20 | 5,414 |
| GOTERM_CC | GO:0043227 | Membrane-bounded
organelle |
1.4x10-18 | 5,316 |
| GOTERM_CC | GO:0043231 | Intracellular
membrane-bounded organelle |
3.9x10-22 | 4,902 |
| GOTERM_CC | GO:0005737 | Cytoplasm |
2.2x10-11 | 4,784 |
| GOTERM_MF | GO:0097159 | Organic cyclic
compound binding |
2.9x10-6 | 2,861 |
| GOTERM_MF | GO:1901363 | Heterocyclic
compound binding |
2.3x10-6 | 2,836 |
| GOTERM_MF | GO:0043167 | Ion binding |
3.4x10-6 | 2,639 |
| GOTERM_MF | GO:0043169 | Cation binding |
6.6x10-5 | 1,804 |
| GOTERM_MF | GO:0046872 | Metal ion
binding |
2.5x10-5 | 1,779 |
| B,
Downregulated |
| Category | GO accession
no. | Description | Corrected
P-value | Total number of
genes |
| GOTERM_BP | GO:0044238 | Primary metabolic
process |
6.6x10-11 | 4,424 |
| GOTERM_BP | GO:0043170 | Macromolecule
metabolic process |
7.5x10-15 | 3,935 |
| GOTERM_BP | GO:0044260 | Cellular
macromolecule metabolic process |
4.4x10-17 | 3,660 |
| GOTERM_BP | GO:0006807 | Nitrogen compound
metabolic process |
7.8x10-09 | 2,947 |
| GOTERM_BP | GO:0044249 | Cellular
biosynthetic process |
1.6x10-10 | 2,781 |
| GOTERM_CC | GO:0005622 | Intracellular |
1.0x10-29 | 6,263 |
| GOTERM_CC | GO:0044424 | Intracellular
part |
3.0x10-27 | 6,127 |
| GOTERM_CC | GO:0044422 | Organelle part |
1.6x10-7 | 3,740 |
| GOTERM_CC | GO:0044446 | Intracellular
organelle part |
8.1x10-8 | 3,675 |
| GOTERM_CC | GO:0044444 | Cytoplasmic
part |
4.1x10-5 | 3,572 |
| GOTERM_MF | GO:0005488 | Binding |
2.6x10-6 | 4,991 |
| GOTERM_MF | GO:0003824 | Catalytic
activity |
5.0x10-5 | 2,445 |
| GOTERM_MF | GO:0003676 | Nucleic acid
binding |
1.9x10-4 | 1,994 |
| GOTERM_MF | GO:0003677 | DNA binding |
8.9x10-10 | 1,185 |
| GOTERM_MF | GO:0003700 | Transcription
factor activity, sequence-specific DNA binding |
1.1x10-5 | 526 |
 | Table IVKEGG pathway analysis of the
differentially expressed lncRNAs. |
Table IV
KEGG pathway analysis of the
differentially expressed lncRNAs.
| A, Upregulated |
|---|
| KEGG pathway | Term | P-value | Total number of
genes |
|---|
| hsa04320 | PI3K-Akt signaling
pathway |
1.4458x10-5 | 214 |
| hsa05630 | Hematopoietic cell
lineage |
6.3426x10-6 | 60 |
| hsa04722 | Osteoclast
differentiation |
2.0712x10-5 | 92 |
| hsa05803 | Staphylococcus
aureus infection |
2.3805x10-5 | 44 |
| hsa03638 | Chemokine signaling
pathway |
3.8041x10-5 | 115 |
| hsa04327 | Malaria |
3.6349x10-5 | 37 |
| hsa02874 | Leishmaniasis |
2.4737x10-4 | 51 |
| hsa00381 | Axon guidance |
4.6875x10-4 | 82 |
| B,
Downregulated |
| KEGG pathway | Term | P-value | Total number of
genes |
| hsa00273 | Cell adhesion
molecules (CAMs) |
8.5728x10-09 | 98 |
| hsa03256 | Cytokine-cytokine
receptor interaction |
1.416x10-5 | 130 |
| hsa04438 | ECM-receptor
interaction |
1.484x10-5 | 58 |
| hsa02931 | Endocytosis |
5.5678x10-4 | 141 |
| hsa01375 | Chagas disease
(American trypanosomias) |
9.3938x10-4 | 68 |
| hsa02306 | Rheumatoid
arthritis |
1.03056x10-3 | 57 |
| hsa00561 | T cell receptor
signaling pathway |
1.20597x10-3 | 66 |
| hsa01934 | Systemic lupus
erythematosus |
1.49979x10-3 | 79 |
RT-qPCR validation of lncRNA
expression
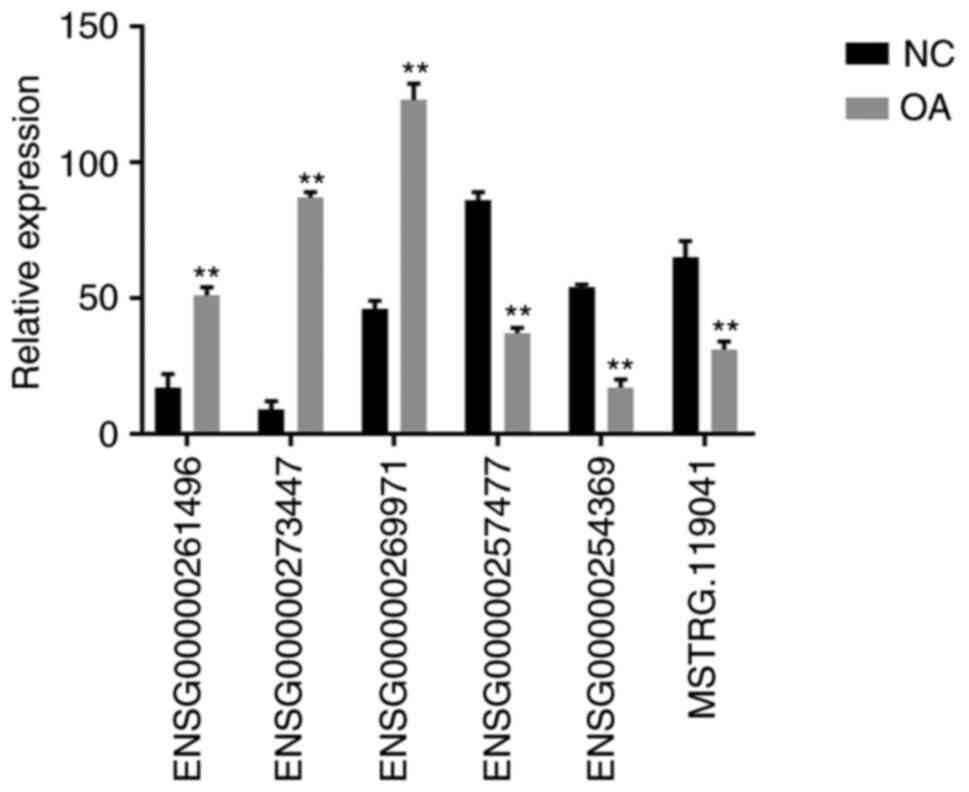
To verify the sequencing results of the lncRNAs,
three upregulated lncRNAs (ENSG00000261496, ENSG00000273447 and
ENSG00000269971) and three downregulated lncRNAs (ENSG00000257477,
ENSG00000254369 and MSTRG.119041) were randomly selected. The
expression levels of the six lncRNAs were detected by RT-qPCR in 13
cartilage samples (six NC and seven OA). The results of the RT-qPCR
analysis were consistent with the microarray data (Fig. 5). Each lncRNA displayed a similar
trend of upregulation or downregulation.
Discussion
lncRNAs do not encode proteins, but function at the
RNA level, regulating gene expression and epigenetic modifications
(28). Abnormal expression of
lncRNAs is involved in the pathogenesis of a number of diseases,
such as glioma, allergic dermatitis and rheumatism (29-31).
Previous studies have reported that the abnormal expression of
lncRNAs in osteoarthritis cartilage in vitro is related to
the degradation of the extracellular matrix of chondrocytes,
suggesting that lncRNAs are involved in the pathogenesis of
osteoarthritis (32,33). However, the potential targets and
functions of lncRNAs associated with skeletal development and
osteoarthritis are not fully understood. The present study
systematically screened the lncRNA expression profiles of cartilage
from normal control patients and patients with knee osteoarthritis
to identify novel lncRNA targets involved in the mechanisms
underlying osteoarthritis. The new lncRNA targets might aid in
identifying the role of lncRNAs in the progression of knee
osteoarthritis.
The results displayed differences in the expression
levels of a number of lncRNAs between normal and osteoarthritis
cartilage, ranging from a few to several thousands. The variable
differences may be attributed to the small sample size. However,
RT-qPCR was performed to verify the functions of randomly selected
lncRNAs. The results suggested that the trend was similar to that
of the microarray data, which provided evidence of differential
lncRNA expression levels between normal and osteoarthritis
cartilage.
Bioinformatics methods were used to perform cis- and
trans-target analyses to identify the target genes of
differentially expressed lncRNAs, as it is difficult to predict the
function of lncRNA based solely on nucleotide sequences (34,35).
Then, the target genes were subjected to GO and KEGG enrichment
analyses to explore the regulation of lncRNAs according to target
gene function.
Target analysis identified a number of potential
lncRNA regulatory targets. Certain targets were previously reported
to play an important role in the pathogenesis of osteoarthritis,
although their potential function as regulatory targets for lncRNA
shad not previously been suggested (36,37).
For example, in the present study, integrin-α5 (ITGA5) was found to
be a potential target of lncRNA ENSG00000257477, which is expressed
at a 2.2787-fold higher level in normal cartilage than in
osteoarthritis chondrocytes. ITGA5 can mediate mutual adhesion and
two-way signal transduction between chondrocytes and the
extracellular matrix (38). ITGA5
is also involved in physiological processes including inflammation,
differentiation and migration (39), and the dedifferentiation of
chondrocytes leads to a reduction in ITGA5(40). Another example is JNK, a potential
target of lncRNA ENSG00000261496, which was found in the present
study to be expressed at a 4.4304-fold higher level in
osteoarthritis than in normal chondrocytes. Elevated levels of JNK
and activation of the JNK signaling pathway may aggravate
chondrocyte inflammation and cause cartilage degradation (41). Mice overexpressing JNK are more
susceptible to osteoarthritis compared to normal mice (42). In the present study, certain targets
that have not previously been reported to play a role in
osteoarthritis were identified as having a potential regulatory
role in osteoarthritis. For example, high mobility group protein B1
(HMGB1), a late inflammatory regulator, has been studied for its
role in coronary atherosclerosis and diabetes (43,44).
However, results from the present study suggested that the
expression level of HMGB1 was higher in articular cartilage tissue
than in normal cartilage (~42.0320-fold), indicating that it may be
a regulatory target of lncRNA ENSG00000272825, which plays a
proinflammatory role (45).
Interferon induced transmembrane protein 3 (IFITM3), a member of
the IFITM gene family, is involved in cell adhesion, immune cell
regulation and stem cell differentiation and maturation (46). IFITM3also plays a role in tumor
proliferation and viral infection (47). Microarray data suggested that IFITM3
may be the regulatory target of lncRNA ENSG00000277310. The
expression level of IFITM3 was lower in arthritic cartilage than in
normal cartilage, which may be related to the continuous
dedifferentiation of chondrocytes [(48), (Fig
S1)].
The PI3K-Akt signaling pathway is involved in the
release of the proinflammatory cytokines IL-1α, IL-1β, TNF-α and
IL-6 and the regulation of the joint inflammatory microenvironment,
based on significantly abundant KEGG signaling pathways. The
present study suggested that the PI3K-Akt signaling pathway is
related to the occurrence and progression of knee osteoarthritis.
In addition, the present study identified several chemokine
signaling pathways that are involved in the differentiation and
metabolism of osteoclasts, which may also be related to the
development of joint inflammation (49).
The present study had several limitations. Firstly,
the age of the patients in the NC group was lower than that of the
patients in the OA group, therefore, a number of the detected
transcripts may be age-related rather than disease-specific. Future
studies should include a larger number of age-matched control
cartilage samples. Secondly, as only a few patients agreed to
donate knee cartilage, the present study consisted of a small
sample size. In order to provide support for the conclusions made
in the present study, further investigations should include larger
cohorts. Thirdly, only indirect experimental evidence of the
functional link between lncRNAs and their predicted target genes
was included in the present study. Therefore, this also requires
further investigation.
In summary, the present study used RNA microarray
data to describe the expression profiles of lncRNAs in
osteoarthritis and normal cartilage. Bioinformatics methods were
used to predict the target genes and potential functions of
differentially expressed lncRNAs. The results obtained by
microarray analysis may contribute to the diagnosis of
osteoarthritis and predict the development of osteoarthritis.
Differentially expressed lncRNAs may also help to identify novel
potential candidate targets for the treatment of knee
osteoarthritis.
Supplementary Material
Interaction networks of significantly
enriched metabolic pathways for certain regulatory targets. Red
boxes represent downregulated genes and green boxes represent
upregulated genes, compared with the NC group. NC, normal
control.
Primers for reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: The present study received funding from the Major
Research and Development Project of Anhui Department of Science and
Technology (grant no. 1804h08020269).
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL and WC analyzed and interpreted the sequencing
data regarding osteoarthritis and contributed to writing the
manuscript. JJ designed the study. HY, JZ and QC performed the
surgeries and collected tissue. XZ, JL and SZ performed secondary
analysis and production of partial sequencing data and charts. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Second
Affiliated Hospital of Anhui Medical University Ethics Committee
(approval no. SL-YX2019-019). Written informed consent was obtained
from each patient or their relatives prior to surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Health Quality Ontario. Structured
education and neuromuscular exercise program for Hip and/or knee
osteoarthritis: A health technology assessment. Ont Health Technol
Assess Ser. 18:1–110. 2018.PubMed/NCBI
|
|
2
|
Zheng S, Tu L, Cicuttini F, Han W, Zhu Z,
Antony B, Wluka A, Winzenberg T, Meng T, Aitken D, et al: Effect of
Vitamin D supplementation on depressive symptoms in patients with
knee osteoarthritis. J Am Med Dir Assoc. 20:1634–1640.e1.
2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Musumeci G, Loreto C, Imbesi R, Trovato
FM, Di Giunta A, Lombardo C, Castorina S and Castrogiovanni P:
Advantages of exercise in rehabilitation, treatment and prevention
of altered morphological features in knee osteoarthritis. A
narrative review. Histol Histopathol. 29:707–719. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Petersen KK, Olesen AE, Simonsen O and
Arendt-Nielsen L: Mechanistic pain profiling as a tool to predict
the efficacy of 3-weeks non-steroidal anti-inflammatory drugs
(NSAIDs) plus paracetamol in patients with painful knee
osteoarthritis. Pain. 160:486–492. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liang Y, Idrees E, Szojka A, Andrews S,
Kunze M, Mulet-Sierra A, Jomha N and Adesida AB: Chondrogenic
differentiation of synovial fluid mesenchymal stem cells on human
meniscus-derived decellularized matrix requires exogenous growth
factors. Acta Biomater. 80:131–143. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Driban JB, Eaton CB, Amin M, Stout AC,
Price LL, Lu B, Lo GH, McAlindon TE and Barbe MF: Glucose
homeostasis influences the risk of incident knee osteoarthritis:
Data from the osteoarthritis initiative. J Orthop Res.
35:2282–2287. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Thorson C, Galicia K, Burleson A, Bouchard
O, Hoppensteadt D, Fareed J and Hopkinson W: Matrix
metalloproteinases and their inhibitors and proteoglycan 4 in
patients undergoing total joint arthroplasty. Clin Appl Thromb
Hemost. 25(1076029619828113)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Davis HC, Luc-Harkey BA, Seeley MK, Troy
Blackburn J and Pietrosimone B: Sagittal plane walking biomechanics
in individuals with knee osteoarthritis after quadriceps
strengthening. Osteoarthritis Cartilage. 27:717–780.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhou X, Jiang L, Zhang Y, Zhang J, Zhou D,
Wu L, Huang Y and Xu N: Genetic variation of aggrecanase-2
(ADAMTS5) in susceptibility to osteoarthritis. Braz J Med Biol Res.
52(e8109)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bar-Or D, Thomas G, Rael LT, Frederick E,
Hausburg M, Bar-Or R and Brody E: On the mechanisms of action of
the low molecular weight fraction of commercial human serum albumin
in osteoarthritis. Curr Rheumatol Rev. 15:189–200. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Giunta S, Castorina A, Marzagalli R,
Szychlinska MA, Pichler K, Mobasheri A and Musumeci G: Ameliorative
effects of PACAP against cartilage degeneration. morphological,
immunohistochemical and biochemical evidence from in vivo and in
vitro models of rat osteoarthritis. Int J Mol Sci. 16:5922–5944.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gauffin H, Sonesson S, Meunier A,
Magnusson H and Kvist J: Knee arthroscopic surgery in middle-aged
patients with meniscal symptoms: A 3-year follow-up of a
prospective, randomized study. Am J Sports Med. 45:2077–2084.
2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tasci Bozbas G, Sendur OF and Aydemir AH:
Primary knee osteoarthritis increases the risk of falling. J Back
Musculoskelet Rehabil. 30:785–789. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Castrogiovanni P, Di Rosa M, Ravalli S,
Castorina A, Guglielmino C, Imbesi R, Vecchio M, Drago F,
Szychlinska MA and Musumeci G: Moderate physical activity as a
prevention method for knee osteoarthritis and the role of
synoviocytes as biological key. Int J Mol Sci.
20(511)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sharma L: Osteoarthritis year in review
2015: Clinical. Osteoarthritis Cartilage. 24:36–48. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Huynh NPT, Anderson BA, Guilak F and
McAlinden A: Emerging roles for long noncoding RNAs in skeletal
biology and disease. Connect Tissue Res. 58:116–141.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tian W, Jiang C, Huang Z, Xu D and Zheng
S: Comprehensive analysis of dysregulated lncRNAs, miRNAs and mRNAs
with associated ceRNA network in esophageal squamous cell
carcinoma. Gene. 696:206–218. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gaballah HH, Gaber RA, Elrashidy MA,
Elshahat DA, Hablus MA and Ebeid AM: Expression of long non-coding
RNA CCHE1 in colorectal carcinoma: Correlations with
clinicopathological features and ERK/COX-2 pathway. Mol Biol Rep.
46:657–667. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Szychlinska MA, Trovato FM, Di Rosa M,
Malaguarnera L, Puzzo L, Leonardi R, Castrogiovanni P and Musumeci
G: Co-expression and Co-localization of cartilage glycoproteins
CHI3L1 and lubricin in osteoarthritic cartilage: Morphological,
immunohistochemical and gene expression profiles. Int J Mol Sci.
17(359)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao X, Li J, Lian B, Gu H, Li Y and Qi Y:
Global identification of Arabidopsis lncRNAs reveals the regulation
of MAF4 by a natural antisense RNA. Nat Commun.
9(5056)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gardner OF, Musumeci G, Neumann AJ, Eglin
D, Archer CW, Alini M and Stoddart MJ: Asymmetrical seeding of MSCs
into fibrin-poly(ester-urethane) scaffolds and its effect on
mechanically induced chondrogenesis. J Tissue Eng Regen Med.
11:2912–2921. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang Q, Wang W, Zhang F, Deng Y and Long
Z: NEAT1/miR-181c regulates osteopontin (OPN)-mediated synoviocyte
proliferation in osteoarthritis. J Cell Biochem. 118:3775–3784.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cen X, Huang XQ, Sun WT, Liu Q and Liu J:
Long noncoding RNAs: A new regulatory code in osteoarthritis. Am J
Transl Res. 9:4747–4755. 2017.PubMed/NCBI
|
|
24
|
Huang H, Zheng J, Shen N, Wang G, Zhou G,
Fang Y, Lin J and Zhao J: Identification of pathways and genes
associated with synovitis in osteoarthritis using bioinformatics
analyses. Sci Rep. 8(10050)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hayes B, Kittelson A, Loyd B, Wellsandt E,
Flug J and Stevens-Lapsley J: Assessing radiographic knee
osteoarthritis: An online training tutorial for the
kellgren-lawrence grading scale. MedEdPORTAL.
12(10503)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yuan J, Yue H, Zhang M, Luo J, Liu L, Wu
W, Xiao T, Chen X, Chen X, Zhang D, et al: Transcriptional
profiling analysis and functional prediction of long noncoding RNAs
in cancer. Oncotarget. 7:8131–8142. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sun X, Jia B, Qiu XL, Chu HX, Zhang ZQ,
Wang ZP and Zhao JJ: Potential functions of long non-coding RNAs in
the osteogenic differentiation of human bone marrow mesenchymal
stem cells. Mol Med Rep. 19:113–114. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bian EB, Chen EF, Xu YD, Yang ZH, Tang F,
Ma CC, Wang HL and Zhao B: Exosomal lncRNA-ATB activates astrocytes
that promote glioma cell invasion. Int J Oncol. 54:713–721.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang X, Bao K, Wu P, Yu X, Wang C, Ji L
and Hong M: Integrative analysis of lncRNAs, miRNAs, and
mRNA-Associated ceRNA network in an atopic dermatitis recurrence
model. Int J Mol Sci. 19(3263)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang Y, Xu YZ, Sun N, Liu JH, Chen FF,
Guan XL, Li A, Wang F, Zhao QF, Wang HY, et al: Long noncoding RNA
expression profile in fibroblast-like synoviocytes from patients
with rheumatoid arthritis. Arthritis Res Ther.
18(227)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chen L, Zhang Y, Rao Z, Zhang J and Sun Y:
Integrated analysis of key mRNAs and lncRNAs in osteoarthritis. Exp
Ther Med. 16:1841–1849. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Boehme KA and Rolauffs B: Onset and
progression of human osteoarthritis-can growth factors,
inflammatory cytokines, or differential miRNA expression
concomitantly induce proliferation, ECM degradation, and
inflammation in articular cartilage? Int J Mol Sci.
19(2282)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Dolcino M, Pelosi A, Fiore PF, Patuzzo G,
Tinazzi E, Lunardi C and Puccetti A: Long non-coding RNAs play a
role in the pathogenesis of psoriatic arthritis by regulating
MicroRNAs and genes involved in inflammation and metabolic
syndrome. Front Immunol. 9(1533)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yan ZF, Zhao XY, Liu W and Liu XP: UCA1
impacts progress of rheumatoid arthritis by inducing the apoptosis
of fibroblast-like synoviocyte. Eur Rev Med Pharmacol Sci.
22:914–920. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pearson MJ, Philp AM, Heward JA, Roux BT,
Walsh DA, Davis ET, Lindsay MA and Jones SW: Long intergenic
noncoding RNAs mediate the human chondrocyte inflammatory response
and are differentially expressed in osteoarthritis cartilage.
Arthritis Rheumatol. 68:845–856. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu Y, Yau MS, Yerges-Armstrong LM, Duggan
DJ, Renner JB, Hochberg MC, Mitchell BD, Jackson RD and Jordan JM:
Genetic determinants of radiographic knee osteoarthritis in African
Americans. J Rheumatol. 44:1652–1658. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Xu D, Jiang Y, Yang L, Hou X, Wang J, Gu
W, Wang X, Liu L, Zhang J and Lu H: Long noncoding RNAs expression
profile and functional networks in rheumatoid arthritis.
Oncotarget. 8:95280–95292. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tanaka N, Ikeda Y, Yamaguchi T, Furukawa
H, Mitomi H, Nakagawa T, Tohma S and Fukui N: α5β1 integrin induces
the expression of noncartilaginous procollagen gene expression in
articular chondrocytes cultured in monolayers. Arthritis Res Ther.
15(R127)2013.PubMed/NCBI View
Article : Google Scholar
|
|
40
|
Zhang Y, Ding J, Xu C, Yang H, Xia P, Ma S
and Chen H: rBMSCs/ITGA5B1 promotes human vascular smooth muscle
cell differentiation via enhancing nitric oxide production. Int J
Stem Cells. 11:168–176. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Choi HJ and Zhu BT: Upregulated cyclin
B1/CDK1 mediates apoptosis following 2-methoxyestradiol-induced
mitotic catastrophe: Role of Bcl-XL phosphorylation. Steroids.
150(108381)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Collins JA, Arbeeva L, Chubinskaya S and
Loeser RF: Articular chondrocytes isolated from the knee and ankle
joints of human tissue donors demonstrate similar redox-regulated
MAP kinase and akt signaling. Osteoarthritis Cartilage. 27:703–711.
2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhou B, Li Q, Wang J, Chen P and Jiang S:
Ellagic acid attenuates streptozocin induced diabetic nephropathy
via the regulation of oxidative stress and inflammatory signaling.
Food Chem Toxicol. 123:16–27. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Monte E, Rosa-Garrido M, Karbassi E, Chen
H, Lopez R, Rau CD, Wang J, Nelson SF, Wu Y, Stefani E, et al:
Reciprocal regulation of the cardiac epigenome by chromatin
structural proteins hmgb and Ctcf: Implications for transcriptional
regulation. J Biol Chem. 291:15428–15446. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yang H, Antoine DJ, Andersson U and Tracey
KJ: The many faces of HMGB1: Molecular structure-functional
activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol.
93:865–873. 2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Min J, Feng Q, Liao W, Liang Y, Gong C, Li
E, He W, Yuan R and Wu L: IFITM3 promotes hepatocellular carcinoma
invasion and metastasis by regulating MMP9 through p38/MAPK
signaling. FEBS Open Bio. 8:1299–1311. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Luo X, Huang S, Luo S, Liao H, Wang Y,
Deng X, Ma F, Ma CW and Zhou L: Identification of genes underlying
the enhancement of immunity by a formula of lentinan, pachymaran
and tremelia polysaccharides in immunosuppressive mice. Sci Rep.
8(10082)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lee HM, Sugino H, Aoki C, Shimaoka Y,
Suzuki R, Ochi K, Ochi T and Nishimoto N: Abnormal networks of
immune response-related molecules in bone marrow cells from
patients with rheumatoid arthritis as revealed by DNA microarray
analysis. Arthritis Res Ther. 13(R89)2011.PubMed/NCBI View
Article : Google Scholar
|
|
49
|
Sucur A, Jajic Z, Artukovic M, Matijasevic
MI, Anic B, Flegar D, Markotic A, Kelava T, Ivcevic S, Kovacic N,
et al: Chemokine signals are crucial for enhanced homing and
differentiation of circulating osteoclast progenitor cells.
Arthritis Res Ther. 19(142)2017.PubMed/NCBI View Article : Google Scholar
|