Introduction
Oral cavity cancer is an intractable malignancy that
has become a highly relevant public health issue worldwide
(1). According to global cancer
statistics from 2018, there were ~354,864 newly diagnosed cases and
177,384 mortalities of oral cavity cancer (2). Furthermore, >90% of oral cavity
cancer types are identified as oral squamous cell carcinoma (OSCC)
(3). Although great progress has
been made in diagnosis, surgery and chemotherapy strategies, the
overall prognosis of OSCC remains unfavorable owing to local
recurrence and metastasis (4).
Cisplatin (DDP) has been reported as an effective first-line
chemotherapy drug for the treatment of OSCC, but the therapeutic
effect of DDP often fails as a result of the rapid development of
drug resistance (5). Thus, it is
important to identify the underlying molecular mechanisms of
chemoresistance in OSCC to develop a novel target for improving DDP
sensitivity.
Long non-coding RNAs (lncRNAs) are >200
nucleotides and have been identified as crucial regulatory
transcripts lacking protein-coding functions (6). Previous studies have reported that
dysregulation of lncRNAs is implicated in the initiation and
development of various tumors, including OSCC (7,8).
LncRNA opa-interacting protein 5 antisense RNA 1 (OIP5-AS1) is
derived from the antisense of OIP5 gene; it acts as a carcinogenic
factor by promoting cell proliferation, migration and invasion in
OSCC progression (9). Moreover, a
recent study observed that OIP5-AS1 could induce DDP resistance by
regulating microRNA (miRNA/miR)-340-5p in osteosarcoma (10). ever, the function and mechanism of
OIP5-AS1 in DDP-resistant OSCC cells are yet to be fully
elucidated.
In recent decades, miRNAs (small non-coding RNAs ~22
nucleotides in length) have been reported to negatively regulate
gene expression in part by repressing translation of target mRNAs
(11). Numerous miRNAs have been
found to be abnormally expressed and closely associated with
physiological activities, including proliferation, metastasis and
development in OSCC (12-14).
miR-27b-3p, a form of mature miR-27b, has been revealed to exert a
tumor-suppressive effect by regulating its target genes, including
MET and frizzled class receptor 7in OSCC (15,16).
Previous studies have suggested that miR-27b-3p could reduce
resistance of some drugs in breast cancer and prostate cancer
(17,18). In addition, a recent study reported
that miR-27b could improve the chemotherapy sensitivity of OSCC
cells to DDP (19), indicating the
involvement of miR-27b-3p in DDP resistance of OSCC.
Tripartite motif-containing 14 (TRIM14), located on
chromosome 9q22, was first identified in HIV-infected human and
simian lymphomas (20). It has been
shown that TRIM14 is upregulated in OSCC, and the overexpression of
TRIM14 could facilitate the progression of OSCC by interacting with
miR-195-5p (21). Furthermore,
previous studies have demonstrated that TRIM14 contributes to drug
resistance in gliomas and oral tongue squamous cell carcinoma
(22,23). However, the involvement of TRIM14 in
DDP-resistant in OSCC remains unknown.
Therefore, the aim of the present study was to
identify the function ofOIP5-AS1, and to explore whether the
involvement of OIP5-AS1 in the DDP resistance of OSCC was mediated
via the miR-27b-3p/TRIM14 axis.
Materials and methods
Clinical samples and cell culture
Samples of OSCC tumor tissues and normal adjacent
tissues (the distance from the tumor margin was >5 cm) were
collected from 30 patients (17 male and 13 females; 12 patients
aged >60 and 18 patients aged <60; age range, 27-78 years)
who underwent oral surgical operation at The First Hospital of
Qiqihar (Qiqihar, China) from January 2015 to June 2017. Tissues
excised during the surgery were instantly frozen in liquid nitrogen
and stored at -80˚C until subsequent use. The study was performed
with the approval of the Ethical Committee of The First Hospital of
Qiqihar. Written informed consent was obtained from all
participating patients.
A normal human oral keratinocyte cell line (NHOK)
was obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China) and incubated in
Keratinocyte-SFM medium (Invitrogen; Thermo Fisher Scientific,
Inc.) containing 10% FBS (Invitrogen; Thermo Fisher Scientific,
Inc.) and 1% antibiotics (100 U/ml penicillin and 100 µg/ml
streptomycin; Invitrogen; Thermo Fisher Scientific, Inc.) in a 5%
CO2 incubator at 37˚C. Human OSCC cell lines (SCC-15,
SCC-9 and Cal-27) were purchased from the American Type Culture
Collection, and the human OSCC cell line HSC-3 was acquired from
Cell Bank of Japanese Collection of Research Bioresources. All OSCC
cells were cultured at 37˚C with 5% CO2 under a humid
atmosphere in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.).
A gradually increasing dose of DDP (from 1.5 to 25
µg/ml over a 10-month period; Sigma-Aldrich; Merck KGaA) at 37˚C
was added to the culture medium of SCC-9/DDP and HSC-3/DDP cells
for maintaining the DDP-resistant phenotype, as previously
described (24).
Cell transfection
OIP5-AS1 or TRIM14 overexpression vectors were
established by inserting OIP5-AS1 or TRIM14 cDNA sequence into
pcDNA3.1 (pcDNA; Invitrogen; Thermo Fisher Scientific, Inc.),
obtaining pcDNA-OIP5-AS1 or pcDNA-TRIM14. And the pcDNA3.1 empty
vector (pcDNA; Invitrogen; Thermo Fisher Scientific, Inc.) acted as
a negative control. Small interfering (si)RNA againstOIP5-AS1
(si-OIP5-AS1: 5'-GGCAGTAGAATCACTTAAA-3') and its scrambled negative
control (si-NC: 5'-TACCGACTGGCAATTCATG-3'), miR-27b-3p mimic
(miR-27b-3p: 5'-TTCACAGTGGCTAAGTTCTGC-3') and miR-27b-3p inhibitor
(anti-miR-27b-3p: 5'-GCAGAACTTAGCCACTGTGAA-3'), as well as their
scrambled NC (miR-NC: 5'-GGTTCCATCGTACACTGTTCA-3' or anti-miR-NC:
5'-CCATCAGTCCCCATCGCCA-3') were obtained from Shanghai GenePharma
Co., Ltd. Transfection of all aforementioned plasmids or
oligonucleotides was performed in OSCC cells (2x105
cells/well)using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. After 48 h incubation at 37˚C,
transfected cells were harvested and utilized for further
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Extraction of RNA from OSCC tissues and cells was
conducted using TRIzol® reagent (Gibco; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. cDNA
was synthesized using PrimeScript™ RT Master mix kit (Takara Bio,
Inc.), followed by incubation at 37˚C for 15 min and 85˚C for 5
sec. Relative expression levels of OIP5-AS1 and TRIM14 were
analyzed using the SYBR® Premix Ex Taq™ kit (Takara Bio,
Inc.), and the amplification parameters were: Denaturation at 95˚C
for 10 min, followed by 40 cycles of denaturation at 95˚C for 30
sec, annealing at 60˚C for 30 sec and extension at 72˚C for 1 min.
The quantitative analysis of miR-27b-3p was performed using an
All-in-One™ miRNA RT-qPCR Detection kit (GeneCopoeia, Inc.), and
the PCR cycling profile was as follows: Denaturation at 95˚C for 2
min; followed by 40 cycles of annealing at 95˚C for 5 sec; and
extension at 60˚C for 35 sec. Subsequently, the expression levels
of OIP5-AS1, miR-27b-3p and TRIM14 were calculated using
2-ΔΔCq method (25),
normalizing to GAPDH or U6 small nuclear RNA. The specific primer
sequences used were as follows: OIP5-AS1 forward,
5'-TGCGAAGATGGCGGAGTAAG-3' and reverse, 5'-TAGTTCCTCTCCTCTGGCCG-3';
miR-27b-3p forward, 5'-ACACTCCAGCTGGGTTTCACAGTGGCTAAG-3' and
reverse, 5'-TGGTGTCGTGGAGTCG-3'; TRIM14 forward,
5'-GCAGAGACAGAGCTAGACTGTAAAGGT-3' and reverse,
5'-CCTGGTCACACAATTGATATGGA-3'; GAPDH forward,
5'-AGAAGGCTGGGGCTCATTTG-3' and reverse, 5'-AGGGGCCATCCACAGTCTTC-3';
and U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse,
5'-AACGCTTCACGAATTTGCGT-3'.
Drug resistance assay and cell
proliferation assay
An MTT assay (Sigma-Aldrich; Merck KGaA) was
performed to measure DDP resistance and cell proliferation,
according to the manufacturer's instructions. Transfected
DDP-resistant OSCC cells were cultured at 37˚C for 48 h prior to
exposure to different doses of DDP (1, 2, 4, 6, 8, 16, 32 and 64
µg/ml), and then the MTT assay was performed. The IC50
was calculated using a viability curve.
For the proliferation assay, transfected
DDP-resistant OSCC cells (6x103 cells/well) were
incubated for 48 h, and then 20 µl MTT solution (5 mg/ml) was added
to each well at the different time points (0, 24, 48 and 72 h),
followed by incubation for another 4 h at 37˚C. After removing the
cell culture medium, 100 µl DMSO (Sigma-Aldrich; Merck KGaA) was
added into each well. The optical density (OD) was detected with a
microplate reader at 490 nm (ELX808; BioTek Instruments, Inc.).
Cell migration and invasion assay
The cell migratory and invasive abilities were
detected using Transwell chambers (24-well; Sigma-Aldrich; Merck
KGaA) according to the manufacturer's instructions. Transfected
SCC-9/DDP and HSC-3/DDP cells (1x106) were inoculated
into the upper chamber with serum-free medium for migration assay.
In total, 5x104 transfected DDP-resistant OSCC cells
were added into the upper chamber coated with Matrigel at 37˚C for
4-5 h (BD Biosciences) for the invasion assay. The lower chamber
contained complete medium with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). After 24 h of incubation, the cells on the
surface of the upper chamber were scraped with cotton swabs,
whereas cells that migrated or invaded to the lower chamber were
fixed with methanol for 30 min at 4˚C and stained with 0.1% crystal
violet solution for 20 min at 37˚C. Cells were analyzed with an
inverted fluorescent microscope (magnification, x100).
Western blot analysis
Western blotting was conducted according to previous
description (26). Total protein
from tissues and cells was isolated using pre-cold RIPA buffer
(Beyotime Institute of Biotechnology) including protease inhibitor.
Total protein was quantified using a BCA protein assay kit
(Invitrogen; Thermo Fisher Scientific, Inc.). Separated proteins
(30 µg) using 10% SDS-PAGE were transferred onto nitrocellulose
membranes (EMD Millipore). The membranes were probed with primary
antibodies againstTRIM14 (1:800; cat. no. ab185349; Abcam),
E-cadherin (1:1,000; cat. no. ab1416; Abcam), N-cadherin (1:1,000;
cat. no. ab76011; Abcam), Vimentin (1:200; cat. no. ab8978; Abcam)
and GAPDH (1:5,000; cat. no. ab9485; Abcam) at 4˚C overnight.
Subsequently, the corresponding horseradish peroxidase conjugated
goat-anti-rabbit secondary antibody (1:10,000; cat. no. ab205178,
Abcam) was probed in the membranes to bind these primary
antibodies. Protein bands were detected with an ECL detection
system (Cytiva) and analyzed using Quantity One v4.6.2software
(Bio-Rad Laboratories, Inc.). And the expression levels of protein
were normalized to GAPDH.
Dual-luciferase reporter assay
Using the bioinformatics website starBase v2.0
(http://starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA),
the binding sites between the miR-27b-3p and OIP5-AS1 or TRIM14
3'-untranslated regions (UTR) were predicted and analyzed. A
luciferase activity reporter assay was conducted to further assess
the binding relationship between miR-27b-3p and OIP5-AS1 or TRIM14
3'-UTR. Partial sequences of OIP5-AS1 and TRIM143'-UTR containing
the putative (wild-type; WT) or mutated putative binding sites for
miR-27b-3p were amplified and cloned into psiCHECK-2 vector
(Promega Corporation), resulting inOIP5-AS1 WT or MUT
andTRIM143'-UTRWT or MUT reporter plasmids. Then, SCC-9/DDP and
HSC-3/DDP cells (2x105 cells/well) were co-transfected
with 100 ng of the constructed reporter plasmids and 100 nM miR-NC
or miR-27b-3p and were incubated for 48 h at 37˚C. Luciferase
activities were measured with the LD400 luminometer (Beckman
Coulter, Inc.) at 48 h post-transfection. Firefly luciferase
activity was normalized to that of Renilla luciferase.
Tumor xenograft assay
Male BALB/C nude mice (n=6 per group; age, 4 weeks,
18-20 g weight) were obtained from the Shanghai Experimental Animal
Center. A total of 12 mice were kept in an environmental room
equipped with a constant temperature of 20˚C, a humidity of 60% and
a programmed 12 h light/dark cycle for circadian control, and were
randomly divided into 2 groups (the sh-NC+cisplatin group, and the
sh-OIP5-AS1+cisplatin). All mice were allowed free access to
drinking water and sterilized standard diet. The animal experiment
was performed as per the protocol approved by the Institutional
Committee for Animal Research of The First Hospital of Qiqihar. The
short hairpin (sh)-OIP5-AS1 lentivirus was obtained from Shanghai
GenePharma Co., Ltd, and a lentivirus empty vector was used as the
sh-NC. Subsequently, these obtained lentivirus vectors were
transfected into 293T cells (Invitrogen; Thermo Fisher Scientific,
Inc.) along with lentivirus packaging vectors (psPAX2 and pMD2. G,
Addgene, Inc.), followed by incubation for 72 h at 37˚C. After
collection with cell supernatants including sh-OIP5-AS1 or sh-NC
lentivirus, SCC-9 cells were infected with sh-OIP5-AS1 or sh-NC
lentivirus, followed by screening with puromycin (Sigma-Aldrich;
Merck KGaA). One week later, stable lentiviro-transfected SCC-9
cells were established. Subsequently, transfected cells
(5x106) were subcutaneously injected into the left flank
of the nude mice. At 7 days after injection, 3 mg/kg DDP (dissolved
in PBS buffer; Sigma-Aldrich; Merck KGaA) was intraperitoneally
injected once every 4 days. Tumor volume was measured every 4 days
after the first injection (the largest tumor diameter was 128 mm).
After 31 days, mice were euthanized by the administration of 5%
isoflurane followed by cervical dislocation. Tumors were excised,
weighed, and were stored at -80˚C for subsequent experiments.
Statistical analysis
GraphPad Prism 7.0 software (GraphPad Software,
Inc.) was used for statistical analysis. Paired Student's t-test or
one-way ANOVA with Tukey's tests were used to analyze the
differences in the data between two groups or among multiple
groups, respectively. The correlation between OIP5-AS1, miR-27b-3p
and TRIM14 was detected using Pearson's correlation analysis. Data
are presented as the mean ± SD. P<0.05 was considered to
indicate a statistically significant difference.
Results
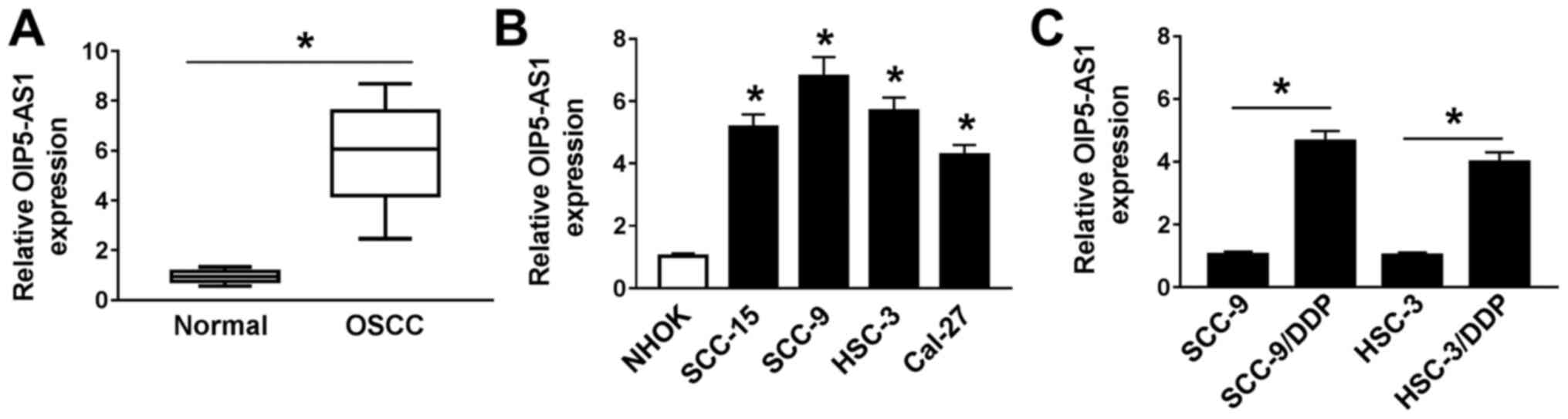
OIP5-AS1 is upregulated in OSCC
tissues and cells, as well as DDP-resistant OSCC cells
To investigate the function of OIP5-AS1 with DDP
resistance in OSCC, its expression was first measured by RT-qPCR
assay. OIP5-AS1 expression was significantly increased in OSCC
tissues in comparison with normal adjacent tissues (n=30; Fig. 1A). Similarly, significantly higher
expression of OIP5-AS1 was observed in OSCC cell lines (SCC-15,
SCC-9, HSC-3 and Cal-27) compared with NHOK cells (Fig. 1B), most notably in SCC-9 and HSC-3
cells. Thus, SCC-9 and HSC-3 cells were selected for the subsequent
analyses.
OIP5-AS1 expression in DDP-resistant OSCC cells was
further examined. The results demonstrated that OIP5-AS1 expression
was significantly upregulated in SCC-9/DDP and HSC-3/DDP cells
compared with their respective parental cells SCC-9 and HSC-3
(Fig. 1C). These data suggested
that dysregulation of OIP5-AS1 maybe associated with DDP resistance
in OSCC cells.
OIP5-AS1 knockdown improves DDP
sensitivity in DDP-resistant OSCC cells
Considering the high expression of OIP5-AS1 in
DDP-resistant OSCC cells, OIP5-AS1 was knocked down in SCC-9/DDP
and HSC-3/DDP cells. The expression of OIP5-AS1 was effectively
downregulated in SCC-9/DDP and HSC-3/DDP cells transfected with
si-OIP5-AS1 compared with cells with si-NC (Fig. 2A). Therefore, this knockdown vector
was used to further evaluate the effect of OIP5-AS1 on DDP
resistance in DDP-resistant OSCC cells. The drug cytotoxicity assay
results suggested that the IC50 value of DDP in
si-OIP5-AS1-transfected DDP-resistant OSCC cells was significantly
decreased compared with the respective si-NC-transfected
DDP-resistant OSCC cells (Fig.
2B-D), indicating that the OIP5-AS1 knockdown could reduce the
resistance of the cells to DDP.
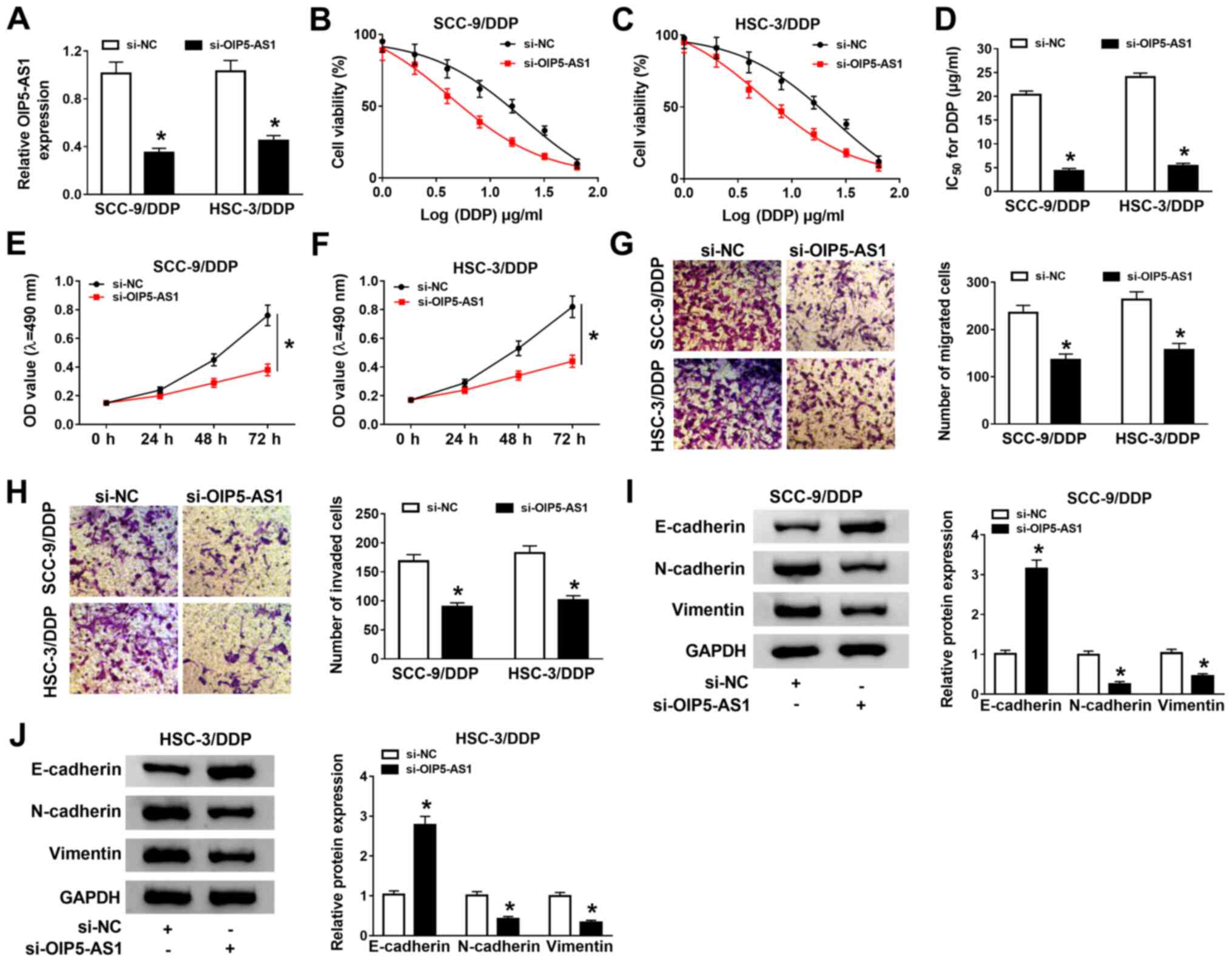 | Figure 2OIP5-AS1 knockdown enhances DDP
sensitivity in DDP-resistant OSCC cells. (A) OIP5-AS1 expression in
SCC-9/DDP and HSC-3/DDP transfected with si-OIP5-AS1 was detected
using RT-qPCR analysis. (B and C) SCC-9/DDP and HSC-3/DDP cells
transfected with transfected were treated with different
concentrations of DDP for 48 h, and then cell viability was
detected by using MTT assay. (D) The IC50 was calculated
using a viability curve. Proliferation rates were analyzed by MTT
assay in si-OIP5-AS1-transfected (E) SCC-9/DDP and (F) HSC-3/DDP
cells. (G) Migration and (H) invasion were analyzed with Transwell
and Matrigel assays (magnification, x100), respectively, in
si-OIP5-AS1-transfected DDP-resistant OSCC cells.
Epithelial-mesenchymal-transition-related protein expression levels
(E-cadherin, N-cadherin and Vimentin) were detected by western blot
analysis in si-OIP5-AS1-transfected (I) SCC-9/DDP and (J) HSC-3/DDP
cells. *P<0.05 vs. si-NC. DDP, cisplatin; NC,
negative control; OD, optical density; OIP5-AS1, opa-interacting
protein 5 antisense RNA 1; OSCC, oral squamous cell carcinoma;
RT-qPCR, reverse transcription-quantitative PCR; siRNA, small
interfering RNA. |
Functional analysis suggested that OIP5-AS1
knockdown significantly repressed proliferation (Fig. 2E and F), migration (Fig. 2G) and invasion (Fig. 2H) in SCC-9/DDP and HSC-3/DDP cells.
Moreover, western blot analysis revealed that OIP5-AS1 knockdown
significantly increased E-cadherin protein expression, but
decreased N-cadherin and Vimentin protein expression levels,
indicating that the knockdown of OIP5-AS1 may suppress
epithelial-mesenchymal transition (EMT) in SCC-9/DDP and HSC-3/DDP
cells (Fig. 2I and J). Collectively, these data demonstrated
that OIP5-AS1 knockdown could decrease DDP resistance, and inhibit
cell growth and metastasis in SCC-9/DDP and HSC-3/DDP cells.
miR-27b-3p directly interacted with
OIP5-AS1
lncRNAs can exert their function by interacting with
miRNAs (27). Hence, the underlying
interacting miRNAs of OIP5-AS1 were predicted using starBase v2.0
software, andmiR-27b-3p was found to possess complementary sites
with OIP5-AS1 (Fig. 3A). The dual
luciferase reporter assay was used to further verify this predicted
outcome. It was demonstrated that miR-27b-3p overexpression
significantly decreased the luciferase activity of OIP5-AS1 WT
reporter plasmid, but had no notable effect on the luciferase
activity of OIP5-AS1 MUT reporter plasmid in SCC-9/DDP and
HSC-3/DDP cells (Fig. 3B and
C).
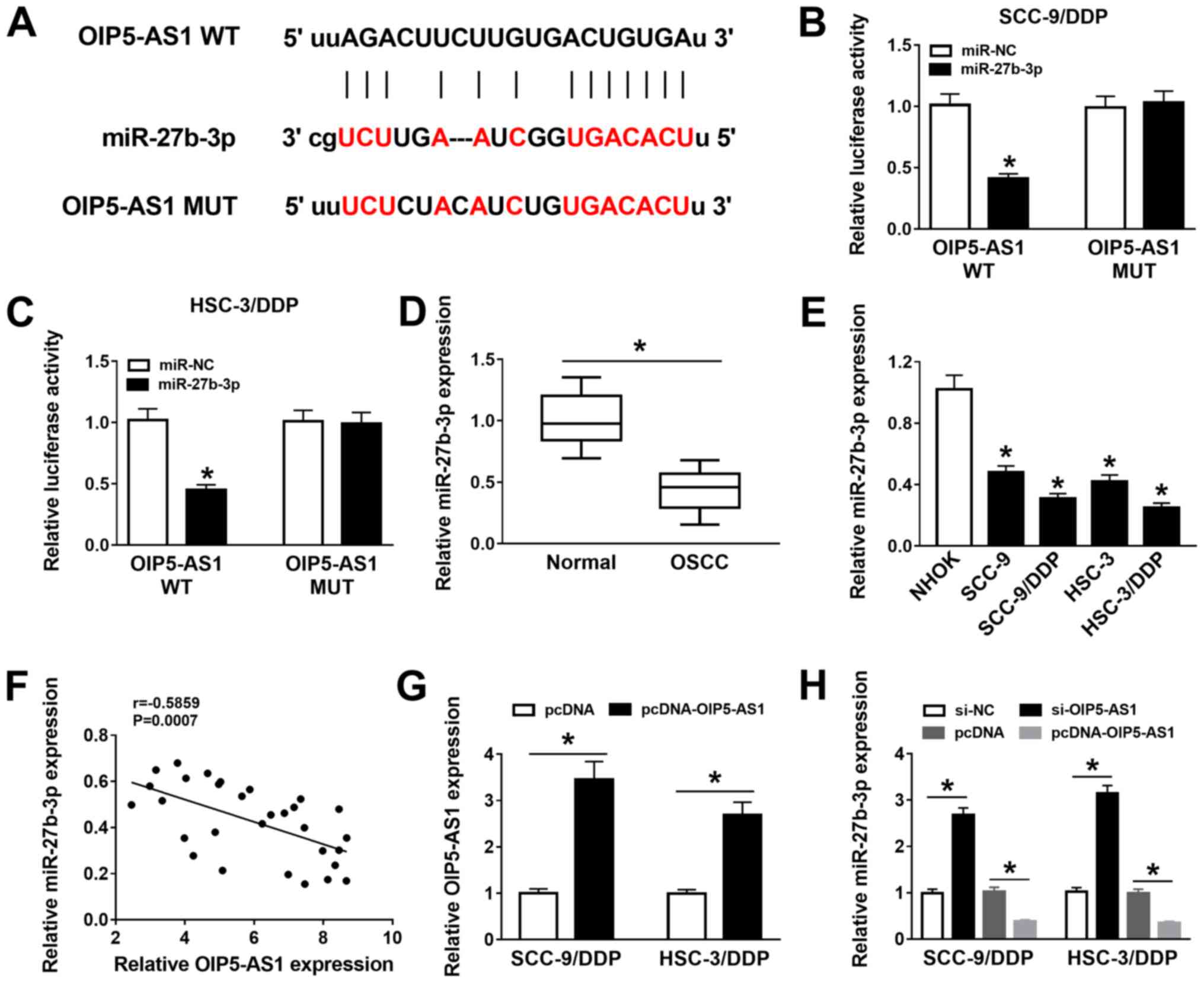 | Figure 3miR-27b-3p is a target of
OIP5-AS1.(A) Binding sites between OIP5-AS1 and miR-27b-3p were
predicted using starBase 2.0 software. Effects of miR-27b-3p
overexpression on luciferase activity of OIP5-AS1 WT and OIP5-AS1
MUT reporters were measured by dual-luciferase reporter assay in
(B) SCC-9/DDP and (C) HSC-3/DDP. (D) miR-27b-3p expression was
detected using RT-qPCR in 30 pairs of OSCC tumor tissues and normal
adjacent tissues. (E) miR-27b-3p expression in NHOK, SCC-9, HSC-3,
SCC-9/DDP and HSC-3/DDP cells was assessed by RT-qPCR. (F)
Correlation between OIP5-AS1 and miR-27b-3p expression levels in
OSCC tissues was analyzed using Pearson correlation analysis. (G)
OIP5-AS1 expression was measured in SCC-9/DDP and HSC-3/DDP cells
transfected with pcDNA and pcDNA-OIP5-AS1. (H) RT-qPCR was
performed to assess the expression levels of miR-27b-3p in
SCC-9/DDP and HSC-3/DDP cells transfected with si-NC, si-OIP5-AS1,
pcDNA orpcDNA-OIP5-AS1. *P<0.05 vs. si-NC or pcDNA.
DDP, cisplatin; siRNA, small interfering RNA; miR, microRNA; MUT,
mutant; NC, negative control; OIP5-AS1, opa-interacting protein 5
antisense RNA 1; OSCC, oral squamous cell carcinoma; RT-qPCR,
reverse transcription-quantitative PCR; WT, wild-type. |
miR-27b-3p was demonstrated to be expressed at
significantly lower levels in OSCC tumors and cell lines compared
with the respective control groups (Fig. 3D and E). In addition, the expression of
miR-27b-3p was moderately negatively correlated with OIP5-AS1
expression in OSCC tumors (Fig. 3F)
(28). The transfection efficiency
of pcDNA-OIP5-AS1 overexpression vector in SCC-9/DDP and HSC-3/DDP
cells was detected (Fig. 3G).
RT-qPCR results showed that miR-27b-3p expression was increased in
si-OIP5-AS1-transfected DDP-resistant OSCC cells but was decreased
in pcDNA-OIP5-AS1-transfected cells (Fig. 3H). Thus, it was indicated that
OIP5-AS1 interacted with miR-27b-3p to hinder its expression.
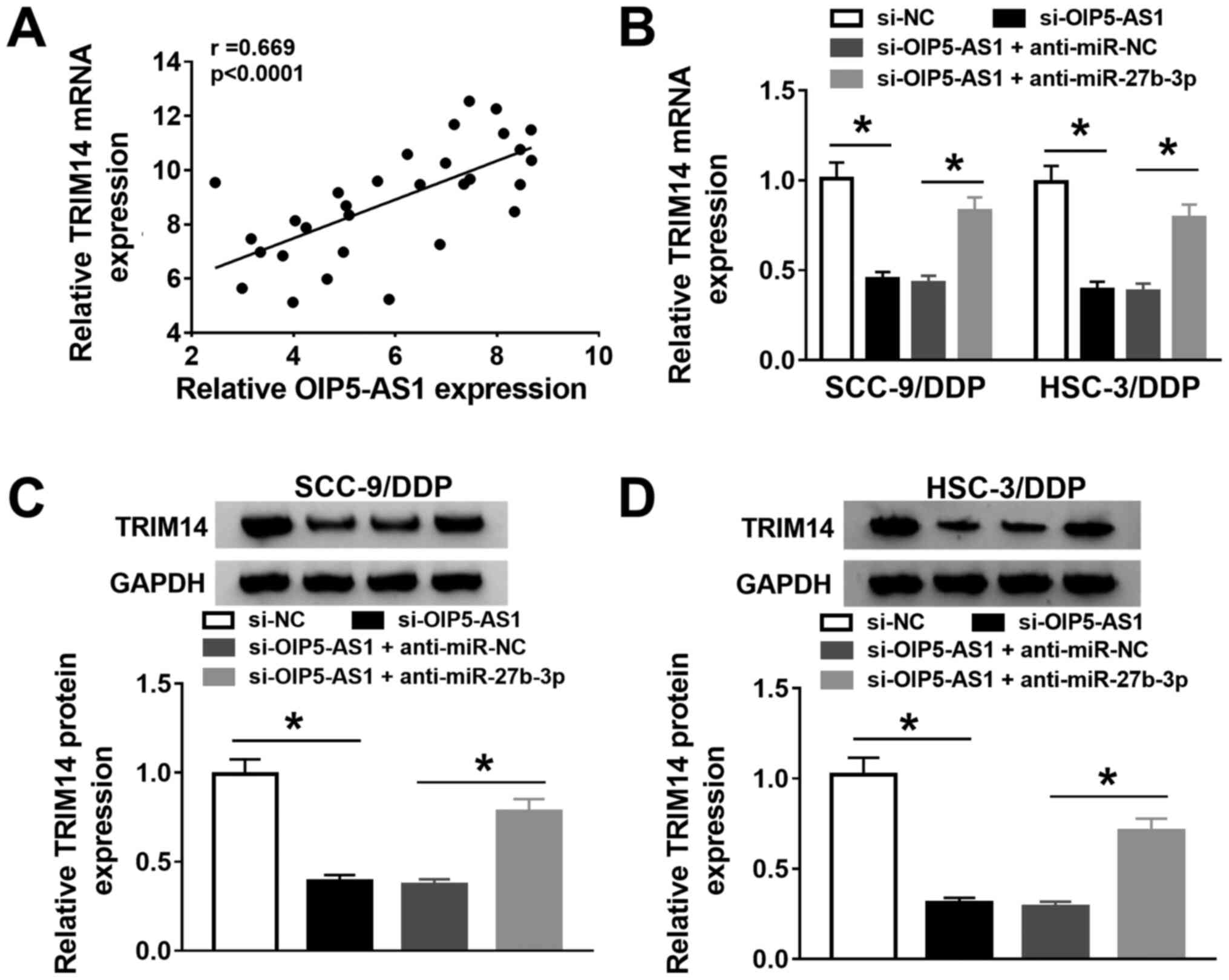
OIP5-AS1 knockdown increases DDP
sensitivity in DDP-resistant OSCC cells by negatively regulating
miR-27b-3p
As an interaction between OIP5-AS1 and miR-27b-3p in
DDP-resistant OSCC cells was indicated, it was further investigated
whether the effect of OIP5-AS1 on DDP resistance was associated
withmiR-27b-3p. Knockdown of OIP5-AS1 could upregulatemiR-27b-3p
expression, which was subsequently downregulated after
co-transfection with anti-miR-27b-3p (Fig. 4A). Furthermore, the results of
IC50 determination suggested that the silencing of
miR-27b-3p partly abolished the inhibitory effect of OIP5-AS1
knockdown on DDP resistance in SCC-9/DDP and HSC-3/DDP cells
(Fig. 4B-D).
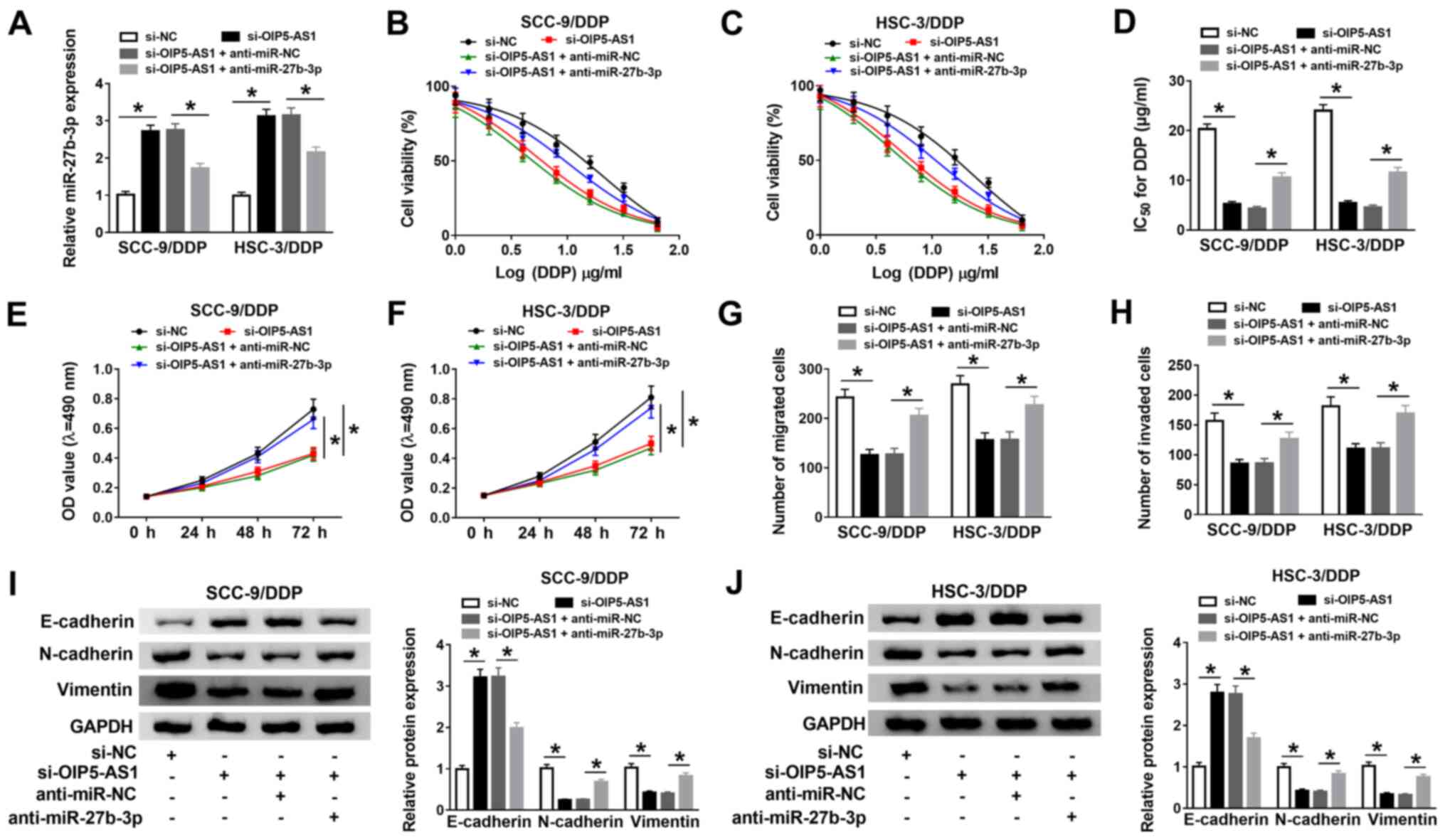 | Figure 4OIP5-AS1 knockdown improves DDP
sensitivity in DDP-resistant oral squamous cell carcinoma cells by
negatively regulating miR-27b-3p. (A) miR-27b-3p expression was
detected using reverse transcription-quantitative PCR in SCC-9/DDP
and HSC-3/DDP cells transfected with si-NC, si-OIP5-AS1,
si-OIP5-AS1 + anti-miR-NC orsi-OIP5-AS1 + anti-miR-27b-3p. (B and
C) Transfected SCC-9/DDP and HSC-3/DDP cells were treated with
various doses of DDP for 48 h. And then, MTT assay was performed to
measure cell viability in treated cells. (D) The viability curve
was applied to calculate the IC50. Proliferation in
transfected (E) SCC-9/DDP and (F) HSC-3/DDP cells was assessed
using a MTT assay. (G) Migration and (H) invasion in transfected
SCC-9/DDP and HSC-3/DDP cells were measured by Transwell and
Matrigel assays, respectively. Protein expression levels of
E-cadherin, N-cadherin and Vimentin in transfected (I) SCC-9/DDP
and (J) HSC-3/DDP cells were detected by western blot analysis.
*P<0.05 vs. si-NC or si-OIP5-AS1 + anti-miR-NC. DDP,
cisplatin; miR, microRNA; NC, negative control; OD, optical
density; OIP5-AS1, opa-interacting protein 5 antisense RNA 1;
siRNA, small interfering RNA. |
Functionally, the knockdown of OIP5-AS1 inhibited
proliferation (Fig. 4E and F), migration (Fig. 4G), and invasion (Fig. 4H) in SCC-9/DDP and HSC-3/DDP cells,
while miR-27b-3p silencing significantly reversed the suppressive
effect of si-OIP5-AS1 on these biological processes. Meanwhile,
cell images of the migration and invasion assays were presented in
Fig. S2A and B. Western blotting results demonstrated
that silencing of miR-27b-3p reversed the
si-OIP5-AS1-inducedenhancement in E-cadherin protein expression, as
well as the reduction in N-cadherin and Vimentin protein expression
levels in SCC-9/DDP and HSC-3/DDP cells (Fig. 4I and J). Taken together, these results suggested
that silencing of miR-27b-3p partly reversed the promotion effect
of OIP5-AS1 knockdown on DDP sensitivity in DDP-resistant OSCC
cells.
TRIM14 is a target of miR-27b-3p
It has been widely reported that miRNA can perform
its function by specifically binding to the 3'-UTR of the
downstream gene (29). Using the
web-based tool starBase, the 3'-UTR of TRIM14 was found to have
complementary target sites to miR-27b-3p (Fig. 5A). To verify this prediction, a
dual-luciferase reporter assay was conducted in SCC-9/DDP and
HSC-3/DDP cells. The results demonstrated that the luciferase
activity was significantly decreased in cells co-transfected with
TRIM14 3'-UTR-WT and miR-27b-3p, whereas there was little effect in
cells co-transfected withTRIM14 3'-UTR-MUT and miR-27b-3p (Fig. 5B and C).
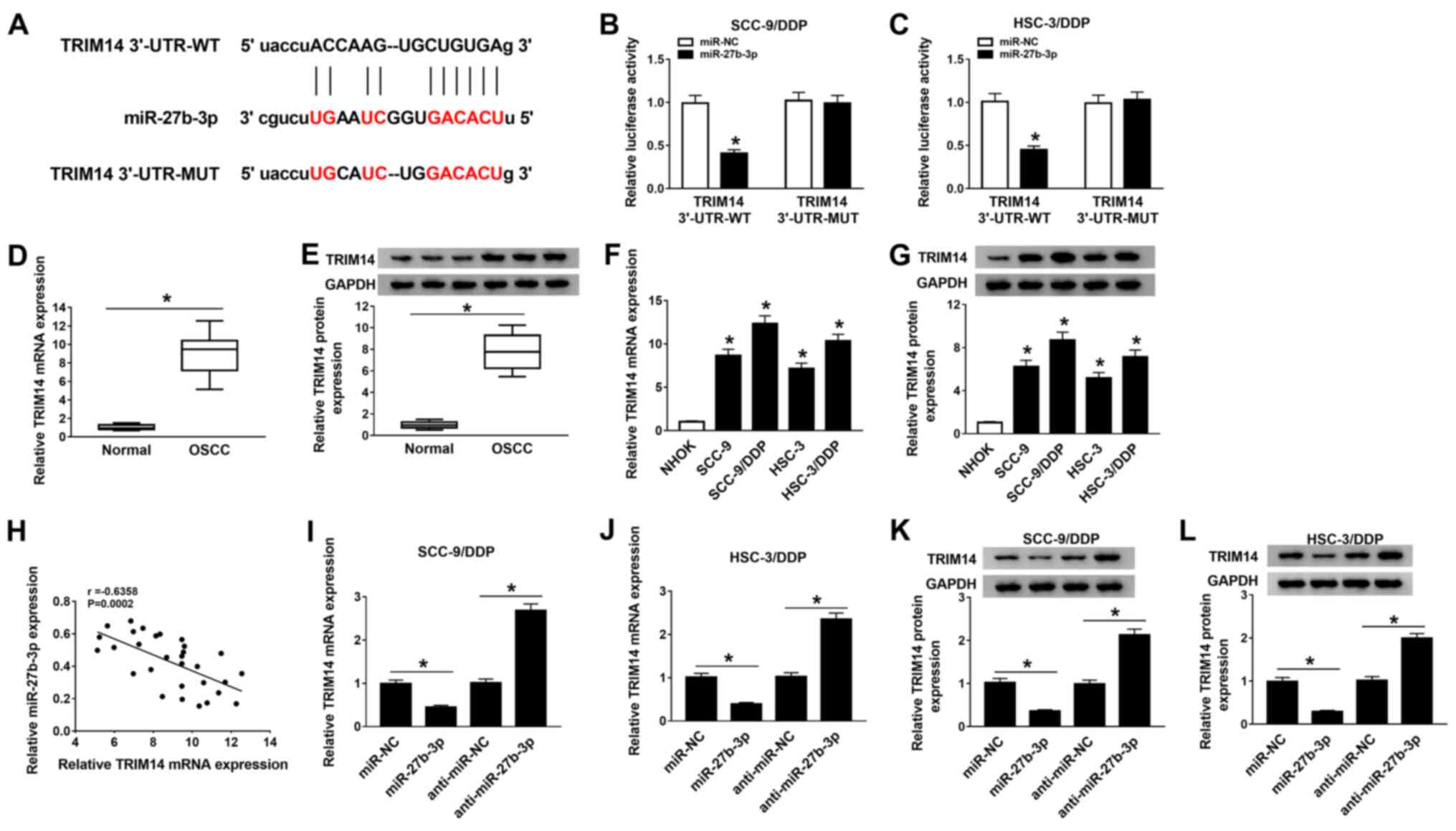 | Figure 5TRIM14 is a direct target of
miR-27b-3p. (A) Putative binding sites between miR-27b-3p and
TRIM14 3'-UTR were predicted using starBase 2.0 software. Relative
luciferase activity was determined using dual-luciferase reporter
assays in (B) SCC-9/DDP and (C) HSC-3/DDP cells co-transfected with
reporter plasmid (TRIM14 3'-UTR-WT or TRIM14 3'-UTR-MUT) and
miR-27b-3p or miR-NC. (D) mRNA and (E) protein expression levels of
TRIM14 in 30 pairs of OSCC tumor tissues and normal adjacent
tissues were measured using RT-qPCR and western blot analysis,
respectively; representative western blotting images of three
normal and three tumoral tissues are presented. (F) RT-qPCR and (G)
western blotting were conducted to evaluate the mRNA and protein
expression levels of TRIM14 in in NHOK, SCC-9, HSC-3, SCC-9/DDP and
HSC-3/DDP cells. (H) Pearson correlation analysis was performed to
determine the correlation between miR-27b-3p and TRIM14 expression
levels in OSCC tissues. TRIM14 mRNA expression was assessed by
RT-qPCR in transfected (I) SCC-9/DDP and (J) HSC-3/DDP cells
transfected with miR-NC, miR-27b-3p, anti-miR-NC oranti-miR-27b-3p.
TRIM14 protein expression was examined using western blotting in
transfected (K) SCC-9/DDP and (L) HSC-3/DDP cells.
*P<0.05 vs. miR-NC or anti-miR-NC. DDP, cisplatin;
miR, microRNA; MUT, mutant; NC, negative control; NHOK, normal
human oral keratinocyte; OIP5-AS1, opa-interacting protein 5
antisense RNA 1; OSCC, oral squamous cell carcinoma; RT-qPCR,
reverse transcription-quantitative PCR; TRIM14, tripartite
motif-containing 14; UTR, untranslated region; WT, wild-type. |
It was demonstrated that the mRNA and protein
expression levels of TRIM14 were significantly upregulated in OSCC
tumor tissues compared with normal adjacent tissues (Fig. 5D and E, respectively). Moreover, TRIM14 was
expressed at a higher level in SCC-9/DDP and HSC-3/DDP cells
compared with SCC-9 and HSC-3 cells (Fig. 5F and G). It was found that TRIM14 expression was
moderately negatively correlated with the expression of miR-27b-3p
in OSCC tumor tissues (Fig.
5H).
The transfection efficiency of miR-27b-3p
overexpression or knockdown was examined and presented in Fig. S1A. RT-qPCR and western blotting
results indicated that at both the mRNA (Fig. 5I and J) and protein (Fig. 5K and L) levels TRIM14 expression was
significantly decreasedaftermiR-27b-3p overexpression, whereas
expression levels were increased by anti-miR-27b-3p transfection in
SCC-9/DDP and HSC-3/DDP cells. Therefore, it was suggested that
miR-27b-3p could interact with TRIM14 to inhibit its
expression.
TRIM14 overexpression partially
reverses the effects of miR-27b-3p on DDP sensitivity in
DDP-resistant OSCC cells
As miR-27b-3p was shown to negatively regulate
TRIM14 expression, whether the effect of miR-27b-3p on DDP
sensitivity was mediated by regulating TRIM14 expression was
investigated. The overexpression of miR-27b-3p decreasedTRIM14
expression, which was significantly reversed by co-transfection
with TRIM14 overexpression vectors in SCC-9/DDP and HSC-3/DDP cells
(Fig. 6A-C). The overexpression
efficiency of pcDNA-TRIM14 was examined and presented in Fig. S1B and SC.
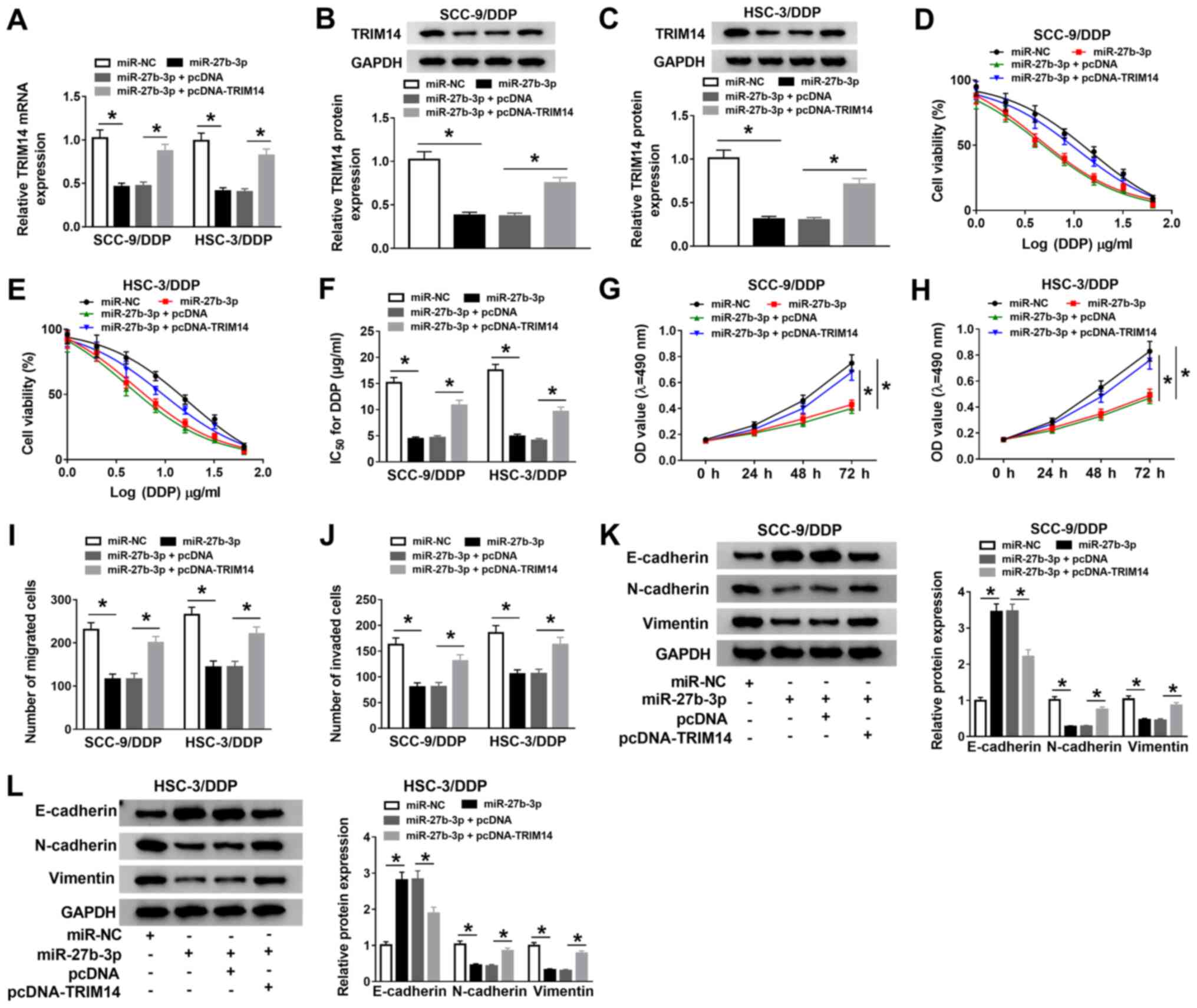 | Figure 6TRIM14 overexpression partly reverses
the effects of miR-27b-3p on DDP sensitivity in DDP-resistant oral
squamous cell carcinoma cells. (A) Reverse
transcription-quantitative PCR was conducted to detect the mRNA
expression levels of TRIM14 in SCC-9/DDP and HSC-3/DDP cells
transfected with miR-NC, miR-27b-3p, miR-27b-3p + pcDNA
ormiR-27b-3p + pcDNA-TRIM14. Western blotting was performed to
assess the protein expression levelsofTRIM14 in transfected (B)
SCC-9/DDP and (C) HSC-3/DDP cells. (D and E) Transfected SCC-9/DDP
and HSC-3/DDP cells were treated with various concentrations of DDP
for 48 h. MTT assay was subsequently carried out to assess cell
viability in treated cells. (F) The viability curve was used to
calculate the IC50. MTT assay was performed to test
proliferation rates in transfected (G) SCC-9/DDP and (H) HSC-3/DDP
cells. Transwell and Matrigel assays were used to measure (I)
migration and (J) invasion, respectively, in transfected SCC-9/DDP
and HSC-3/DDP cells. Western blotting was conducted to assess the
protein expression levels of E-cadherin, N-cadherin and Vimentin in
transfected (K) SCC-9/DDP and (L) HSC-3/DDP cells.
*P<0.05. DDP, cisplatin; miR, microRNA; NC, negative
control; OD, optical density; OIP5-AS1, opa-interacting protein 5
antisense RNA 1; TRIM14, tripartite motif-containing 14. |
The IC50 value of DDP suggested that the
overexpression of TRIM14 effectively abolished the suppressive
effect of miR-27b-3p mimic on DDP resistance in SCC-9/DDP and
HSC-3/DDP cells (Fig. 6D-F).
Moreover, transfection of miR-27b-3p inhibited the proliferation
(Fig. 6G and H), migration (Figs. 6I and S2C) and invasion (Figs. 6J and S2D) in SCC-9/DDP and HSC-3/DDP cells, and
the overexpression of TRIM14 significantly reversed these effects.
It was demonstrated that increased E-cadherin protein expression,
as well as decreased N-cadherin and Vimentin protein expression
levels induced by miR-27b-3p mimic were reversed by
pcDNA-TRIM14co-transfection in SCC-9/DDP and HSC-3/DDP cells
(Fig. 6K and L). Thus, these results suggested that
miR-27b-3p may facilitate DDP sensitivity in DDP-resistant OSCC
cells by regulating TRIM14.
OIP5-AS1 enhances TRIM14 expression by
sponging miR-27b-3p in DDP-resistant OSCC cells
Based on the aforementioned findings, it was
hypothesized that OIP5-AS1 may affect the expression of TRIM14 by
modulating miR-27b-3p in DDP-resistant OSCC cells. A moderate
positive correlation was identified between OIP5-AS1 and TRIM14
expression levels (Fig. 7A).
RT-qPCR results demonstrated that knockdown of OIP5-AS1
decreasedTRIM14 expression, and introduction of anti-miR-27b-3p
effectively reversed this trend in SCC-9/DDP and HSC-3/DDP cells
(Fig. 7B). Similar to the RT-qPCR
results, the protein expression levels of TRIM14 were significantly
suppressed in si-OIP5-AS1-transfected SCC-9/DDP and HSC-3/DDP
cells, whereas silencing of miR-27b-3p mitigated this inhibitory
effect of OIP5-AS1 knockdown (Fig.
7C and D). Taken together,
these results suggested that OIP5-AS1 may serve as a molecular
sponge of miR-27b-3p to upregulate TRIM14 expression in
DDP-resistant OSCC cells.
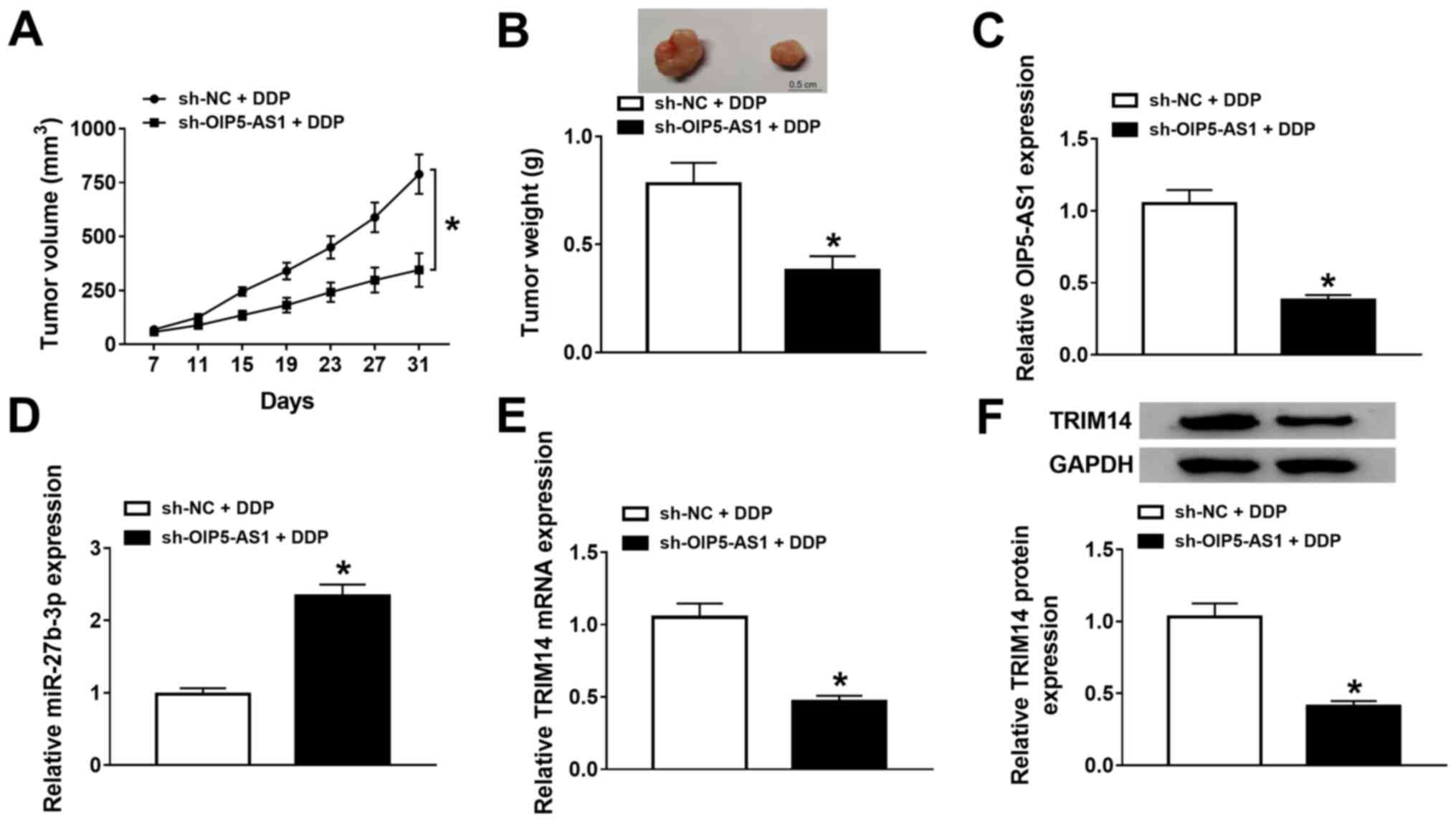
OIP5-AS1 knockdown increases DDP
sensitivity of OSCC in vivo
To further evaluate the functional effects of
OIP5-AS1 on DDP resistance in vivo, a mouse xenograft model
of OSCC was established. The knockdown efficiency of sh-OIP5-AS1 in
SCC-9 cells was measured and presented in Fig. S1D. The results demonstrated that
the tumor size and weight were significantly lower in the
sh-OIP5-AS1 group treated DDP compared with the sh-NC group treated
DDP, indicating that OIP5-AS1 knockdown hindered tumor growth in
OSCC in vivo (Fig. 8A and
B). Furthermore, RT-qPCR and
western blotting results revealed that OIP5-AS1 mRNA (Fig. 8C) and TRIM14 mRNA and protein
(Fig. 8E and F, respectively)expression levels were
decreased, whereasmiR-27b-3p expression was enhanced (Fig. 8D) in tumor tissues from mice
injected with sh-OIP5-AS1-transfected SCC-9 cells compared with
mice injected with sh-NC-transfected SCC-9 cells. Collectively,
these results indicated that knockdown of OIP5-AS1 repressed tumor
growth and enhanced DDP sensitivity partly by regulating the
miR-27b-3p/TRIM14 axis in OSCC in vivo.
Discussion
DDP-based chemotherapy is effective in the clinical
treatment of most cancer types, but the development of drug
resistance leads to poor clinical effectiveness (30). Previous studies have reported that
lncRNAs are essential regulators in development and drug resistance
of various cancer types (31,32).
For example, it was previously reported that the high expression of
OIP5-AS1 was associated with undifferentiated oral tumors and
indicated a poor prognosis (33).
Furthermore, it has been shown that OIP5-AS1 knockdown can reduce
the resistance of osteosarcoma cells to DDP (10). However, the mechanism of OIP5-AS1 in
DDP resistance is yet to be fully elucidated in OSCC.
In the present study, OIP5-AS1 was demonstrated to
be highly expressed in DDP-resistant OSCC cells compared with
parental cells, suggesting that OIP5-AS1 may exert an oncogenic
role in the DDP chemoresistance of OSCC cells. Subsequently, the
biological function of OIP5-AS1 on DDP resistance in OSCC cells was
further evaluated. The results showed that OIP5-AS1 knockdown
increased DDP sensitivity and repressed proliferation, migration,
invasion and EMT in DDP-resistant OSCC cells in vitro.
Moreover, the current study demonstrated that knockdown of OIP5-AS1
hindered OSCC cell growth, and improved DDP sensitivity in
vivo. Therefore, it was suggested that OIP5-AS1 knockdown may
contribute to DDP sensitivity in vitro and in
vivo.
Previous studies have revealed that lncRNAs can
exert their roles by interacting with miRNA (34,35).
In the present study, miR-27b-3p was demonstrated to be a target A
of OIP5-AS1. It has been shown that miR-27b-3p could exert an
inductive effect on drug sensitivity in breast cancer (17). The present results indicated that
miR-27b-3p expression was downregulated and negatively correlated
with the expression of OIP5-AS1 in OSCC tissues and DDP-resistant
OSCC cells. Functionally, silencing miR-27b-3p
reversedOIP5-AS1-knockdown-induced enhancement of DDP sensitivity,
demonstrating that the knockdown of OIP5-AS1 increased DDP
sensitivity partly by interacting with miR-27b-3p in DDP-resistant
OSCC cells.
lncRNAs may act as sponges to reduce mRNA expression
(27,36). In the present research, TRIM14 was
identified as the target of miR-27b-3p using bioinformatics
analysis and dual-luciferase reporter assays. According to previous
literature, TRIM14 can affect the activity of Wnt/β-catenin to
promote the drug resistance of tumors (23). Moreover, the Wnt/β-catenin signaling
pathway contributes to oxaliplatin (OXA) resistance of liver cancer
by enhancing the expression of multidrug resistance mutation 1
(MDR1) (37), and the Wnt/β-catenin
signaling pathway increases DDP resistance of lung adenocarcinoma
by increasing the expression of ATP-binding cassette (ABC)
transporter (38). Therefore, we
hypothesize that TRIM14 could induce ABC transporter and MDR1
expression by activating the Wnt/β-catenin signaling pathway, thus
enhancing OSCC resistance to DDP.
Previous studies have reported that TRIM14 could
increase progression in OSCC and enhance DDP resistance in oral
tongue squamous cell cancer (21,22).
The present study results demonstrated that TRIM14 was upregulated,
and inversely correlated with miR-27b-3p level in OSCC. Functional
analysis demonstrated that TRIM14 overexpression reserved the
promotion role of miR-27b-3p on DDP sensitivity. The inductive
effect ofTRIM14 on DDP resistance was also verified in tongue
squamous cell carcinoma (39).
Additionally, to further assess whether OIP5-AS1 could act as a
miR-27b-3p sponge to impact TRIM14 expression, rescue assays were
conducted. In the present study, the results demonstrated that the
downregulation of miR-27b-3p could partly reversed the suppressive
action of OIP5-AS1 knockdown on TRIM14 expression in DDP-resistant
OSCC cells, verifying the regulatory role of
OIP5-AS1/miR-27b-3p/TRIM14 in OSCC. The current study is limited by
the small sample size and the verification of other associated
signaling pathways (23).
Therefore, in future studies, the sample size should be expanded,
and whether the regulatory role of OIP5-AS1/miR-27b-3p/TRIM14 axis
on DDP resistance was mediated by the Wnt/β-catenin signaling
pathway should be explored further.
In conclusion, the present results suggested that
OIP5-AS1 may act as a sponge of miR-27b-3p to upregulate TRIM14
expression, thus promoting DDP resistance of OSCC cells. Therefore,
targeting OIP5-AS1 maybe a potential therapeutic target for OSCC
treatment.
Supplementary Material
Transfection efficiency of miR-27b-3p
mimic, miR-27b-3p inhibitor, and pcDNA-TRIM14in DDP-resistant OSCC
cells or sh-OIP5-AS1 in OSCC cells was shown. (A) miR-27b-3p
expression levels were detected in SCC-9/DDP and HSC-3/DDP cells
transfected with miR-NC, miR-27b-3p, anti-miR-NC oranti-miR-27b-3p.
(B and C) TRIM14 expression was examined in SCC-9/DDP and HSC-3/DDP
cells transfected with pcDNA and pcDNA-TRIM14 using reverse
transcription-quantitative PCR or western blot analysis. (D)
OIP5-AS1 expression was detected in SCC-9 cells transfected with
sh-NC and sh-OIP5-AS1. *P<0.05 vs. miR-NC,
anti-miR-NC, pcDNA, sh-NC. miR, microRNA; TRIM14, tripartite
motif-containing 14; shRNA, short hairpin RNA; DDP, cisplatin; NC,
negative control.
Cell images of the migration and
invasion assay results. (A) Migration and (B) invasion were
measured by Transwell and Matrigel assays, respectively, in
SCC-9/DDP and HSC-3/DDP cells transfected with si-NC, si-OIP5-AS1,
si-OIP5-AS1 + anti-miR-NC or si-OIP5-AS1 + anti-miR-27b-3p. (C)
Migration and (D) invasion were measured by Transwell and Matrigel
assays (magnification, x100), respectively, in SCC-9/DDP and
HSC-3/DDP cells transfected with miR-NC, miR-27b-3p, miR-27b-3p +
pcDNA or miR-27b-3p + pcDNA-TRIM14. DDP, cisplatin; miR, microRNA;
NC, negative control; OIP5-AS1, opa-interacting protein 5 antisense
RNA 1; TRIM14, tripartite motif-containing 14; siRNA, small
interfering RNA.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZX conceived and designed the study. JL, QJ and DL
performed the experiments, and performed data mining, acquisition
and analysis. ZX wrote and approved the final manuscript. All
authors read and approved the final manuscript.
Ethical approval and consent to
participate
The study was performed with the approval of the
Ethical Committee of The First Hospital of Qiqihar (Qiqihar,
China). Written informed consents were obtained from every
participant. The animal experiments were performed as per the
protocol approved by the Institutional Committee for Animal
Research of The First Hospital of Qiqihar.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jin LJ, Lamster IB, Greenspan JS, Pitts
NB, Scully C and Warnakulasuriya S: Global burden of oral diseases:
Emerging concepts, management and interplay with systemic health.
Oral Dis. 22:609–619. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Montero PH and Patel SG: Cancer of the
oral cavity. Surg Oncol Clin N Am. 24:491–508. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Troiano G, Mastrangelo F, Caponio VCA,
Laino L, Cirillo N and Lo Muzio L: Predictive prognostic value of
tissue-based MicroRNA expression in oral squamous cell carcinoma: A
systematic review and meta-analysis. J Dent Res. 97:759–766.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang D and Lippard SJ: Cellular processing
of platinum anticancer drugs. Nat Rev Drug Discov. 4:307–320.
2005.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y,
Ren X, Wu T, Tao X, Chen X, et al: LncRNA-p23154 promotes the
invasion-metastasis potential of oral squamous cell carcinoma by
regulating Glut1-mediated glycolysis. Cancer Lett. 434:172–183.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang C, Bao C, Zhang X, Lin X, Pan D and
Chen Y: Knockdown of lncRNA LEF1-AS1 inhibited the progression of
oral squamous cell carcinoma (OSCC) via Hippo signaling pathway.
Cancer Biol Ther. 20:1213–1222. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li M, Ning J, Li Z, Fei Q, Zhao C, Ge Y
and Wang L: Long noncoding RNA OIP5-AS1 promotes the progression of
oral squamous cell carcinoma via regulating miR-338-3p/NRP1 axis.
Biomed Pharmacother. 118(109259)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Song L, Zhou Z, Gan Y, Li P, Xu Y, Zhang
Z, Luo F, Xu J, Zhou Q and Dai F: Long noncoding RNA OIP5-AS1
causes cisplatin resistance in osteosarcoma through inducing the
LPAATbeta/PI3K/AKT/mTOR signaling pathway by sponging the
miR-340-5p. J Cell Biochem. 120:9656–9666. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nagai H, Hasegawa S, Uchida F, Terabe T,
Ishibashi Kanno N, Kato K, Yamagata K, Sakai S, Kawashiri S, Sato
H, et al: MicroRNA-205-5p suppresses the invasiveness of oral
squamous cell carcinoma by inhibiting TIMP2 expression. Int J
Oncol. 52:841–850. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Feng X, Luo Q, Wang H, Zhang H and Chen F:
MicroRNA-22 suppresses cell proliferation, migration and invasion
in oral squamous cell carcinoma by targeting NLRP3. J Cell Physiol.
233:6705–6713. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Peng M and Pang C: MicroRNA-140-5p
inhibits the tumorigenesis of oral squamous cell carcinoma by
targeting p21-activated kinase 4. Cell Biol Int: Aug 8, 2019 (Epub
ahead of print). doi: 10.1002/cbin.11213.2019.
|
|
15
|
Fukumoto I, Koshizuka K, Hanazawa T,
Kikkawa N, Matsushita R, Kurozumi A, Kato M, Okato A, Okamoto Y and
Seki N: The tumor-suppressive microRNA-23b/27b cluster regulates
the MET oncogene in oral squamous cell carcinoma. Int J Oncol.
49:1119–1129. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu B, Chen W, Cao G, Dong Z, Xu J, Luo T
and Zhang S: MicroRNA-27b inhibits cell proliferation in oral
squamous cell carcinoma by targeting FZD7 and Wnt signaling
pathway. Arch Oral Biol. 83:92–96. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhu J, Zou Z, Nie P, Kou X, Wu B, Wang S,
Song Z and He J: Downregulation of microRNA-27b-3p enhances
tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1
expression. Cell Death Dis. 7(e2454)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang G, Tian X, Li Y, Wang Z, Li X and
Zhu C: miR-27b and miR-34a enhance docetaxel sensitivity of
prostate cancer cells through inhibiting epithelial-to-mesenchymal
transition by targeting ZEB1. Biomed Pharmacother. 97:736–744.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu B, Cao G, Dong Z and Guo T: Effect of
microRNA-27b on cisplatin chemotherapy sensitivity of oral squamous
cell carcinoma via FZD7 signaling pathway. Oncol Lett. 18:667–673.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Nenasheva VV, Nikolaev AI, Martynenko AV,
Kaplanskaya IB, Bodemer W, Hunsmann G and Tarantul VZ: Differential
gene expression in HIV/SIV-associated and spontaneous lymphomas.
Int J Med Sci. 2:122–128. 2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang T, Ren Y, Liu R, Ma J, Shi Y, Zhang L
and Bu R: miR-195-5p suppresses the proliferation, migration, and
invasion of oral squamous cell carcinoma by targeting TRIM14.
Biomed Res Int. 2017(7378148)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang X, Guo H, Yao B and Helms J: miR-15b
inhibits cancer-initiating cell phenotypes and chemoresistance of
cisplatin by targeting TRIM14 in oral tongue squamous cell cancer.
Oncol Rep. 37:2720–2726. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tan Z, Song L, Wu W, Zhou Y, Zhu J, Wu G,
Cao L, Song J, Li J and Zhang W: TRIM14 promotes chemoresistance in
gliomas by activating Wnt/β-catenin signaling via stabilizing Dvl2.
Oncogene. 37:5403–5415. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Song L, Duan P, Gan Y, Li P, Zhao C, Xu J,
Zhang Z and Zhou Q: Silencing LPAATβ inhibits tumor growth of
cisplatin-resistant human osteosarcoma in vivo and in
vitro. Int J Oncol. 50:535–544. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu W, Chang J, Du X and Hou J: Long
non-coding RNA PCAT-1 contributes to tumorigenesis by regulating
FSCN1 via miR-145-5p in prostate cancer. Biomed Pharmacother.
95:1112–1118. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li S, Chen X, Liu X, Yu Y, Pan H, Haak R,
Schmidt J, Ziebolz D and Schmalz G: Complex integrated analysis of
lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol.
73:1–9. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Schober P, Boer C and Schwarte LA:
Correlation coefficients: Appropriate use and interpretation.
Anesth Analg. 126:1763–1768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: MicroRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Stordal B, Pavlakis N and Davey R: A
systematic review of platinum and taxane resistance from bench to
clinic: An inverse relationship. Cancer Treat Rev. 33:688–703.
2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fang Z, Chen W, Yuan Z, Liu X and Jiang H:
LncRNA-MALAT1 contributes to the cisplatin-resistance of lung
cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed
Pharmacother. 101:536–542. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Arunkumar G, Anand S, Raksha P,
Dhamodharan S, Prasanna Srinivasa Rao H, Subbiah S, Murugan AK and
Munirajan AK: LncRNA OIP5-AS1 is overexpressed in undifferentiated
oral tumors and integrated analysis identifies as a downstream
effector of stemness-associated transcription factors. Sci Rep.
8(7018)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun CC, Zhang L, Li G, Li SJ, Chen ZL, Fu
YF, Gong FY, Bai T, Zhang DY, Wu QM and Li DJ: The lncRNA PDIA3P
interacts with miR-185-5p to modulate oral squamous cell carcinoma
progression by targeting cyclin D2. Mol Ther Nucleic Acids.
9:100–110. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chang SM and Hu WW: Long non-coding RNA
MALAT1 promotes oral squamous cell carcinoma development via
microRNA-125b/STAT3 axis. J Cell Physiol. 233:3384–3396.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou RS, Zhang EX, Sun QF, Ye ZJ, Liu JW,
Zhou DH and Tang Y: Integrated analysis of lncRNA-miRNA-mRNA ceRNA
network in squamous cell carcinoma of tongue. BMC Cancer.
19(779)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cao F and Yin LX: miR-122 enhances
sensitivity of hepatocellular carcinoma to oxaliplatin via
inhibiting MDR1 by targeting Wnt/β-catenin pathway. Exp Mol Pathol.
106:34–43. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang Q, Geng F, Zhou H, Chen Y, Du J,
Zhang X, Song D and Zhao H: MDIG promotes cisplatin resistance of
lung adenocarcinoma by regulating ABC transporter expression via
activation of the WNT/β-catenin signaling pathway. Oncol Lett.
18:4294–4307. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Qiao CY, Qiao TY, Jin H, Liu LL, Zheng MD
and Wang ZL: LncRNA KCNQ1OT1 contributes to the cisplatin
resistance of tongue cancer through the KCNQ1OT1/miR-124-3p/TRIM14
axis. Eur Rev Med Pharmacol Sci. 241:200–212. 2020.PubMed/NCBI View Article : Google Scholar
|