Introduction
Glioma has been indicated to be the most common
primary brain tumor, accounting for 75% of all intracranial tumors
in adults from 2010-2014 in USA (1), with a diagnosis of ~350,000 cases per
year. There has been substantial progress in the treatment of
glioma; however, the median survival time of patients with
glioblastoma has been reported to be only 12-15 months worldwide in
2013(2). Therefore, the discovery
of novel molecular targets and therapeutic methods for the
treatment of glioma are required.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
that have been discovered ~20 years ago (3,4).
miRNAs have been associated with a number of important biological
processes, including developmental regulation, organ formation,
cell proliferation and apoptosis (5,6).
Previous studies have indicated that several miRNAs were abnormally
expressed and participated in the occurrence and development of
glioma (7,8). miR-383 has been indicated to be
downregulated in hepatocellular carcinoma and non-small cell lung
cancer, acting either as a tumor suppressor gene (9,10).
Moreover, a previous study has reported that the inhibition of
miR-383 in glioma tissue decreased the proliferation of glioma
cells (11). However, the mechanism
by which miR-383 may affect the proliferation or apoptosis of
glioma cells is still unclear.
Peroxiredoxin 3 (PRDX3) is an
oxidation/reduction-related protein found in the mitochondria,
which has been found to exhibit protective antioxidant effects and
inhibit autophagy (12,13). PRDX3 has been reported to promote
cancer cell survival by aiding cancer cells to escape cell
oxidation, and the protein expression levels of PRDX3 has been
negatively associated with autophagy (12,14).
Previously, PRDX3 has been identified as a target protein of
miR-383, and a negative association between the expression levels
of miR-383 and PRDX3 was found (15). Therefore, based on these studies,
miR-383 may exert antitumor effects via the oxidative stress
pathway to inhibit tumor growth. Mitochondrial reactive oxygen
species (ROS) are considered to be harmful products of oxidative
metabolism, as they cause cell damage and result in diseases,
including diabetes and cardiovascular diseases (16-19).
Previous studies have reported that oxidative stress may disrupt
endoplasmic reticulum function (20), activate endoplasmic reticulum stress
(21,22) and downregulate autophagy (23,24).
Whether an association between miR-383 and autophagy via
PRDX3-mediated oxidative stress exists, remains to be further
elucidated.
The present study examined the effects of
manipulating the expression levels of miR-383 and PRDX3 on
apoptosis, mitochondrial ROS production and autophagy in human
glioma U87 cells. In addition, the present study also explored
whether PRDX3 is a downstream target gene of miR-383, with the aim
to clarify the potential mechanism by which miR-383 may regulate
autophagy in human glioma cells.
Materials and methods
Cell culture
The U87 cell line (accession no. CVCL_0022; American
Type Culture Collection), which is a glioblastoma cell line of
unknown origin, that was authenticated using STR profiling, was
maintained in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
50 U/ml penicillin and 50 mg/ml streptomycin. The cells were
cultured in an atmosphere at 37˚C containing 5% CO2.
Construction of miR-383
mimics/inhibitors and PRDX3 interference/overexpression
vectors
The commercial miR-383 mimics/inhibitors and their
negative controls (NCs) were purchased from Guangzhou RiboBio Co.,
Ltd. (miR-383 mimics, cat. no. miR10000748-1-5; miR-383 inhibitors,
cat. no. miR20000748-1-5; mimics NC, cat. no. miR1N0000001-1-5;
inhibitors NC, cat. no. miR2N0000001-1-5). The sequences are as
follows: miR-383 mimic, 5'-AGAUCAGAAGGUGAUUGUGGCU-3' and miR-383
inhibitor, 5'-AGCCACAAUCACCUUCUGAUCU-3'.
The small interfering RNA Target Finder (https://rnaidesigner.thermofisher.com/rnaiexpress/design.do;
Ambion; Thermo Fisher Scientific, Inc.) was used to select a
segment (5'-AACGAGCTTGACAATCTCTTGAA-3') within the coding region of
PRDX3 (NC_000010.11) for the short hairpin (sh)RNA design. The
corresponding DNA template sequence containing restriction sites
for EcoRI and BamHI was used, as a target sequence.
The template primer sequences were as follows: Sense,
5'-GATCCGGAAGAACGAGCTTGACAATTCAAGAGATTGTCAAGCTCGTTCTTCCTTTTTTACGCGTG-3';
antisense, 5'-AATTCACGCGTAAAAAAGGAAGAACGAGCTTGACAATCTCTTGAA
TTGTCAAGCTCGTTCTTCCG-3'. The shRNA sequence, which was obtained
using PCR, and pSIREN-RetroQ-shN plasmid vector, which was a gift
from Joe Landry (Addgene plasmid no. 73665; http://n2t.net/addgene:73665; RRID:Addgene_73665), was
digested using EcoRI and BamHI enzymes (Thermo Fisher
Scientific, Inc.) and subsequently ligated. The recombinant plasmid
(pSIREN-sh-PRDX3) was transfected into U87 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.).
The pre-PRDX3 plasmid was constructed using the same
method as aforementioned. The corresponding DNA template containing
restriction sites for EcoRI and XhoI was designed as
a target sequence. The template primer sequences were as follows:
Forward primer, 5'-CGGAATTCCGATGGCGGCTGCTGTA-3'; reverse primer,
5'-CCCTCGAGGGTTACCTTCTGAAAGTA-3'. pcDNA3.1-HA plasmid, which was a
gift from Oskar Laur (Addgene plasmid no. 128034; http://n2t.net/addgene:128034; RRID:Addgene_128034),
was digested using EcoRI and XhoI enzymes (Thermo
Fisher Scientific, Inc.) and ligated to the PRDX3 sequence obtained
using PCR from DNA template from U-87 MG cells according to the
following system: 12.5 µl PCR mix (2xTaq Master Mix; Vazyme Biotech
Co., Ltd.), 0.5 µl forward primer, 0.5 µl reverse primer, 1 µl DNA
template and 10.5 µl ddH2O. The thermocycling conditions
are as follows: Initial denaturation at 94˚C for 3 min, followed by
30 cycles of denaturation at 94˚C for 60 sec, annealing at 56˚C for
60 sec, and extension at 72˚C for 60 sec; and final extension at
72˚C for 5 min. The recombinant plasmid (pcDNA3.1-pre-PRDX3) was
transfected into U87 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.).
Cell transfection
U87 cells (1x105 cells/well) were seeded
in 6-well plates at 37˚C overnight and transfected with miR-383
mimics/inhibitors (20 nM) and miR-383 mimics/inhibitors NCs,
pre-/sh-PRDX3 (0.8-1.0 µg), scramble shRNA (sh-NC) or pre-NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). During transfection, the cells were maintained
in Opti-MEM reduced-serum medium (Gibco; Thermo Fisher Scientific,
Inc.) for 6 h at 37˚C, and subsequently the medium was replaced
with DMEM. The cells were analyzed using reverse
transcription-quantitative PCR (RT-qPCR) and western blot analysis
following 48 h of transfection.
Flow cytometry
To detect ROS levels,
2',7'-dichlorodihydrofluorescein diacetate (DCFH-DA; Sigma-Aldrich;
Merck KGaA) was diluted with serum-free DMEM (1:1,000; HyClone;
Cytiva) to a final concentration of 10 µmol/l. U87 cells were
collected by centrifugation at 400 x g, 4˚C for 5 min, suspended in
diluted DCFH-DA (10 µmol/l) at 1x107 cells/ml and
incubated at 37˚C for 20 min. The cell suspension was mixed every
3-5 min to fully integrate the probe with the cells. Following
incubation, the cells were washed three times with serum-free
medium to remove excessive DCFH-DA. The cells (1x106)
were subsequently harvested by centrifugation at 400 x g, 4˚C for 5
min, and ROS levels were detected using flow cytometry (FC500 MCL;
Beckman Coulter, Inc.). The data was analyzed using CXP analysis
version 2.0 (Beckman Coulter, Inc.).
To detect apoptosis, U87 cells (1x106)
were collected and washed with 1 ml pre-cooled PBS. Annexin V-FITC
and propidium iodide (10 µl each; both Beijing Solarbio Science and
Technology Co., Ltd.) were added to the cells and the percentage of
apoptosis in the stained cells was quantified using flow cytometry
(FC500 MCL; Beckman Coulter, Inc.). The data was analyzed using CXP
Analysis version 2.0 (Beckman Coulter, Inc.).
Western blot analysis
U87 cells were lysed in pre-cooled RIPA lysis buffer
(Beyotime Institute of Biotechnology). Protein quantification was
performed using bicinchoninic acid protein concentration
determination assay (Thermo Fisher Scientific, Inc.). Following
incubation in a water bath for 10 min, 25 µg protein/lane were
subjected to SDS-PAGE on 12% gels. The proteins were transferred to
a PVDF membrane (EMD Millipore), blocked in 5% skim milk at 25˚C
for 2 h and incubated with primary antibodies (all 1:1,000) against
PRDX3 (cat. no. ab73349; Abcam), autophagy-related protein 9 (ATG9;
cat. no. ab108338; Abcam), Ras-related protein Rab-1 (RAB1; cat.
no. 13075; Cell Signaling Technology, Inc.), p62 (cat. no. ab91526;
Abcam) and GAPDH (cat. no. 5174; Cell Signaling Technology, Inc.)
overnight at 4˚C. The membranes were subsequently washed 3 times
with PBS containing 5% Tween-20 and incubated with goat anti-rabbit
IgG horseradish peroxidase-conjugated secondary antibody (1:2,000;
cat. no. ab6721; Abcam) at 25˚C for 1 h. The membranes were treated
with ECL reagent (EMD Millipore) to detect the expression of
proteins using an automatic chemiluminescence analyzer (Tanon 5200;
Tanon Science and Technology Co., Ltd.). The band gray values were
read using the Tanon GIS 4.2 software (Tanon Science and Technology
Co., Ltd.).
RT-qPCR
Total RNA was extracted from U87 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse-transcribed into cDNA using the
Advantage® RT-for-PCR Kit (cat. no. 639505; Takara
Biotechnology Co., Ltd.). The reverse transcription condition were
as follows: 42˚C for 15 min and hold at 16˚C. SYBR®
Premix Ex Taq™ II kit (Takara Biotechnology Co., Ltd.) was used for
subsequent qPCR according to the manufacturer's instructions. The
thermocycling conditions were as follows: Initial denaturation at
95˚C for 3 min, followed by 39 cycles of denaturation at 95˚C for 5
sec, annealing at 56˚C for 10 sec and extension at 72˚C for 25 sec,
before final extension at 65˚C for 5 sec and 95˚C for 50 sec. All
primers were purchased from Nanjing Genscript Biological Technology
Co., Ltd., and were as follows: PRDX3 forward,
5'-TTCCAGTCAAGCAAAAT-3' and reverse, 5'-GAAGAAAAGCACCAAAT-3';
miR-383 forward, 5'-GGGAGATCAGAAGGTGA-3' and reverse,
5'-AACTGGTGTCGTGGAGTCGGC-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3'
and reverse, 5'-AACGCTTCACGAATTTGCGT-3' and GAPDH forward,
5'-CCACTCCTCCACCTTTG-3' and reverse, 5'-CACCACCCTGTTGCTGT-3'. The
experiments were performed independently with three biological
replicates. The data was analyzed using the 2-ΔΔCq
method (25), where GAPDH was used
as the reference gene of PRDX3 and U6 was used as the reference
gene of miR-383.
Acridine orange staining
An Acridine Orange detection kit (Beijing Leagene
Biotech Co., Ltd.) was used to detect cell autophagy. A total of
1x106 U87 cells/ml were washed with acridine orange (AO)
stain buffer (1X) and subsequently mixed with AO stain buffer (1X)
at a ratio of 19:1 (v/v). The cells were stained at 25˚C for 15
min, placed on a glass slide and covered with a coverslip.
Subsequently, images were obtained (magnification, x200) using a
fluorescence microscope (excitation filter wavelength, 488 nm;
blocking filter wavelength, 515 nm; IX53; Olympus Corporation).
Interaction between miR-383 and
PRDX3
The 3'-untranslated region (3'-UTR) of the PRDX3
gene and potential targets of miR-383 were predicted using the
TargetScanHuman v7.1 software (http://www.targetscan.org/vert_71/). The QuikChange
Site-Directed Mutagenesis kit (Agilent Technologies, Inc.) was used
to mutate the 3'-UTR of PRDX3 at the putative binding site of
miR-383. Wild-type (WT) and mutant (Mut) PRDX3 3'-UTRs were cloned
into the pmirGLO-vector (Promega Corporation). 293 cells were
co-transfected with pmirGlO-3'-UTR-PRDX3 (50 ng) and miR-383
mimics/inhibitor (50 nM) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) in 24-well plates for
12 h. Following transfection, cell lysates were prepared using
Passive Lysis Buffer (Promega Corporation) and luciferase activity
was measured using a Dual-Luciferase® Reporter assay
system according to manufacturer's protocol (Promega Corporation).
All luciferase activities were normalized to that of Renilla
luciferase activity.
Statistical analysis
SPSS v19.0 (IBM Corp.) was used to analyze all data,
and the data are presented as the mean ± standard deviation from
three experimental repeats. Statistical analysis was performed
using one-way ANOVA followed by Dunnett's or Duncan's test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Efficiency of pre-/sh-PRDX3 vectors
and miR-383 mimics/inhibitor and their effect on the expression of
autophagy-associated proteins
The mRNA and protein expression level of PRDX3 was
increased in the pre-PRDX3 group and decreased in
sh-PRDX3-transfected cells compared with that in the control cells
(P<0.05), indicating that the transfection of pre-/sh-PRDX3
plasmids was successful (Fig. 1A
and C). In addition, the mRNA
expression level of miR-383 was increased following transfection
with miR-383 mimics and decreased in cells transfected with miR-383
inhibitor compared with that in the control cells (P<0.05),
demonstrating that transfection with miR-383 mimics and inhibitor
was also successful (Fig. 1B).
Additionally, neither miR-383 mimics/inhibitors negative controls
or pre-/sh-PRDX3 negative controls exerted an effect on miR-383 or
PRDX3 expression (Fig. S1).
Therefore, the NC groups were not take into consideration in the
follow-up study. Moreover, the autophagy-related proteins, ATG9,
RAB1 and p62, were examined using western blot analysis following
pre-/sh-PRDX3 transfection. ATG9 and RAB1 were downregulated in
cells transfected with pre-PRDX3 (P<0.05 and P<0.01,
respectively), but were upregulated in cells transfected with
sh-PRDX3 (P<0.05 and P<0.01, respectively) compared with that
in the control cells (Fig. 1C). By
contrast, p62 was upregulated by pre-PRDX3 (P<0.01) and
downregulated by sh-PRDX3 (P<0.05) compared with that in the
control group (Fig. 1C).
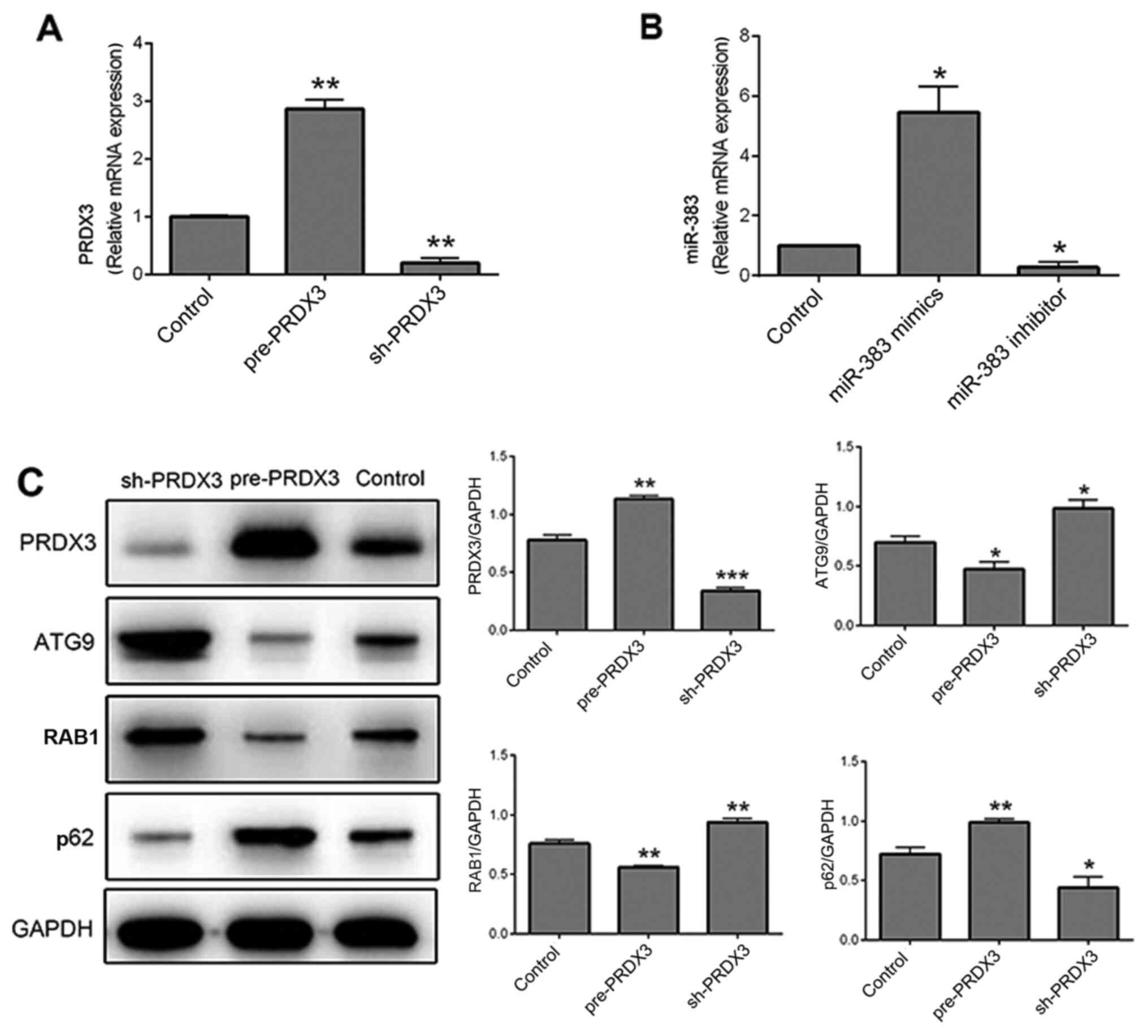 | Figure 1Expression of proteins associated
with autophagy after transfection with pre-/sh-PRDX3 vectors and
miR-383 mimics/inhibitor. (A) mRNA expression level of PRDX3 in
cells transfected with pre-PRDX3, sh-PRDX3 and the control group.
(B) mRNA expression level of miR-383 in cells transfected with
miR-383 mimics, miR-383 inhibitor and the control group. (C)
Protein expression of PRDX3 and the autophagy-related proteins
ATG9, RAB1 and p62 in cells transfected with pre-PRDX3, sh-PRDX3
and the control group. n=3. *P<0.05,
**P<0.01 and ***P<0.001 vs. Control
group. PRDX3, peroxiredoxin 3; miR, microRNA; sh, short hairpin;
ATG9, autophagy-related protein 9; RAB1, Ras-related protein
Rab-1. |
PRDX3 and miR-383 are associated with
ROS and autophagy in human glioma U87 cells
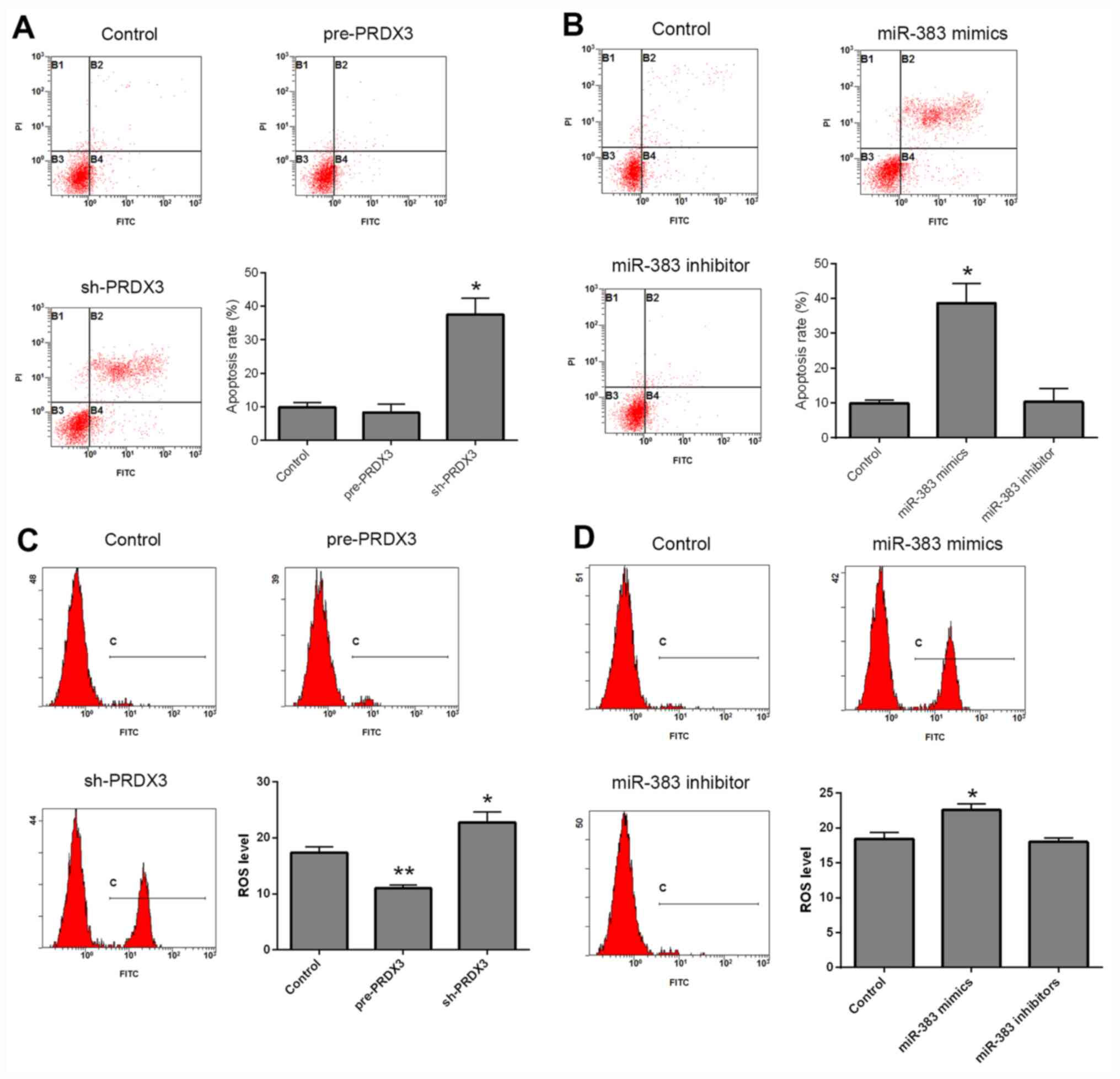
Apoptosis and ROS levels were detected using flow
cytometry, following transfection. Compared with that in the
control group, cells transfected with pre-PRDX3 exhibited no
difference in the rate of apoptosis, whereas sh-PRDX3 significantly
increased apoptosis in transfected cells (Fig. 2A; P<0.05). In addition, the
miR-383 inhibitor did not affect apoptosis, whereas transfection
with miR-383 mimics significantly increased apoptosis compared with
that in the control cells (Fig. 2B;
P<0.05). ROS levels were decreased following transfection with
pre-PRDX3 (P<0.01), but were increased in the
sh-PRDX3-transfected cells (P<0.05) compared with that in the
control group (Fig. 2C). Moreover,
transfection with miR-383 mimics significantly increased ROS levels
(P<0.05), whereas cells transfected with miR-383 inhibitor
exhibited no significant difference compared with that in the
control cells (Fig. 2D). Autophagy
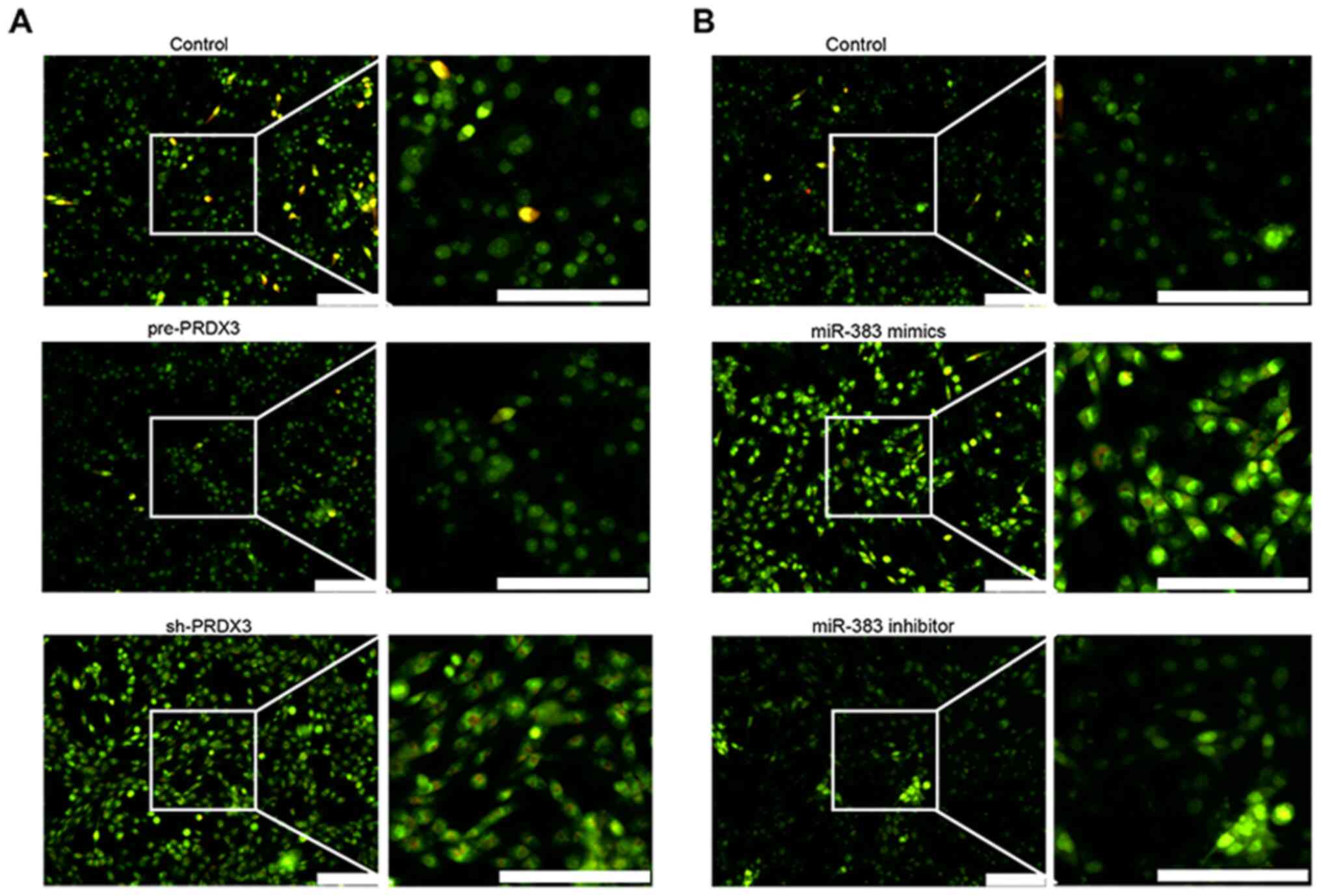
was examined in U87 cells using AO staining (Fig. 3). Cells transfected with sh-PRDX3
presented an increased autophagy compared with that in the control
group and pre-PRDX3-transfected cells, as indicated by the increase
in autolysosome formation (Fig. 3A;
orange red staining). Similarly, autophagy was enhanced in cells
transfected with miR-383 mimics compared with that in the control
group and miR-383 inhibitor-transfected cells (Fig. 3B). In addition, the AO staining can
also be used to evaluate the apoptosis of cells, which shows
yellow-green staining. The increased yellow-green staining in
sh-PRDX3 and miR-383 mimics groups suggested increased levels of
apoptosis, consistent with results that both sh-PRDX3 and miR-383
mimics promoted apoptosis.
Interaction between miR-383 and
PRDX3
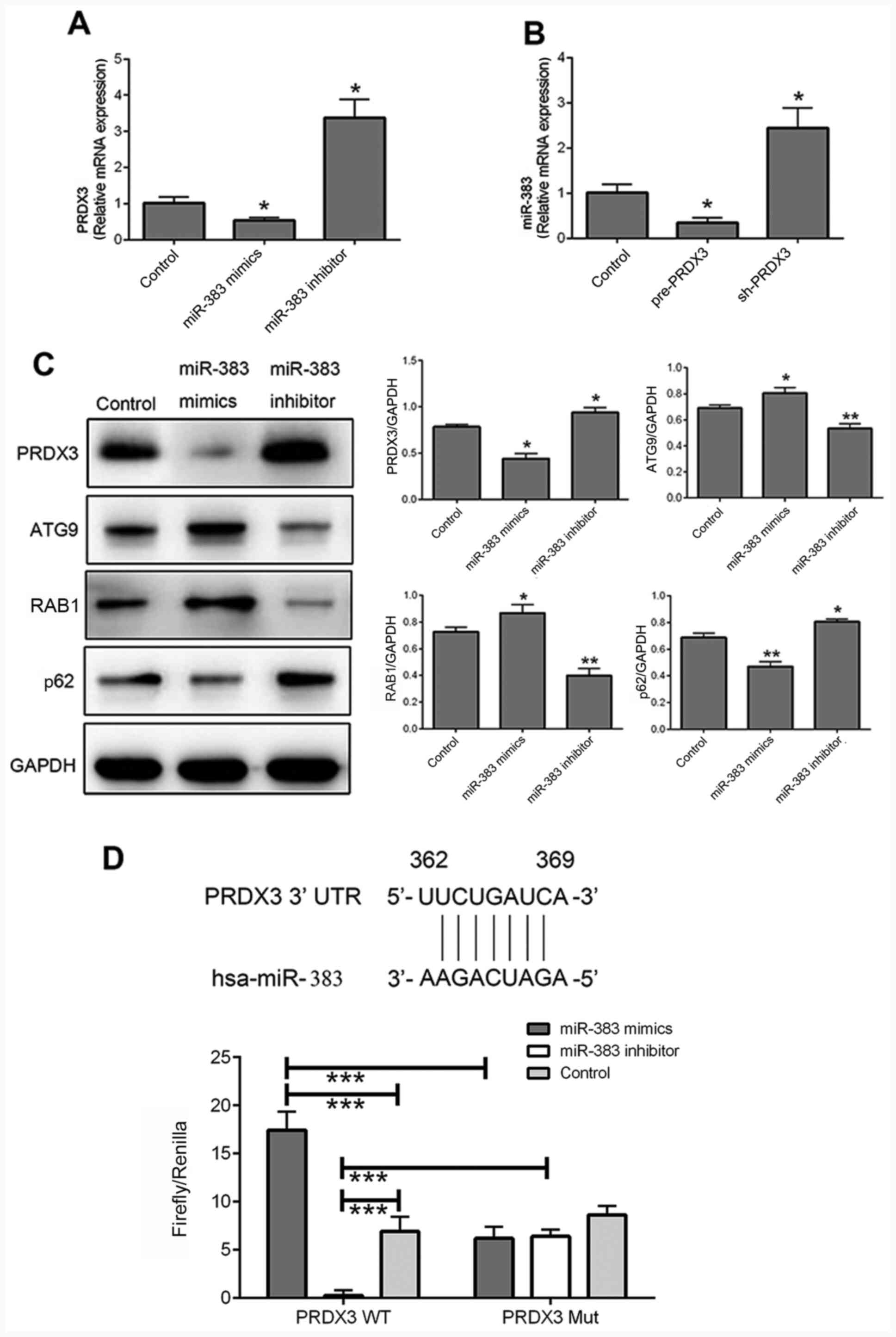
RT-qPCR was used to detect the mRNA expression
levels of PRDX3 and miR-383 following transfection with miR-383
mimics and inhibitors and pre/sh-PRDX3. The mRNA expression levels
of PRDX3 were significantly decreased following transfection with
miR-383 mimics (P<0.05) and significantly increased with miR-383
inhibitor (P<0.05) compared with that in the control cells
(Fig. 4A). In addition, the
expression level of miR-383 was significantly downregulated in
cells transfected with pre-PRDX3 (P<0.05), but was significantly
upregulated following transfection with sh-PRDX3 (P<0.05)
compared with that in the control group (Fig. 4B). The protein expression level of
PRDX3 was similar to the mRNA levels in cells transfected with
miR-383 mimics and inhibitor compared with that in the control
cells (Fig. 4C; both P<0.05).
Moreover, ATG9 and RAB1 were significantly downregulated in cells
transfected with miR-383 inhibitor (both P<0.01), but were
significantly upregulated following transfection with miR-383
mimics (both P<0.05) compared with that in the control group
(Fig. 4C). p62 was also
significantly upregulated following transfection with miR-383
inhibitor (P<0.05), but was significantly downregulated in
miR-383 mimics-transfected cells (P<0.01), compared with that in
the control cells (Fig. 4C). The
effect of miR-383 on the expression level of autophagy-associated
proteins was opposite to that of PRDX3 (Fig. 1C). The potential interaction between
miR-383 and PRDX3 was investigated in U87 cells using a
dual-luciferase assay (Fig. 4D). In
the PRDX3 WT group, luciferase activity was significantly increased
following transfection with miR-383 mimics (P<0.001) and
significantly decreased with miR-383 inhibitor (P<0.001)
compared with that in the control group. Moreover, in cells
transfected with miR-383 mimics, the luciferase activity of the
PRDX3 WT group was significantly increased compared with that in
the PRDX3 Mut group (P<0.001), and in cells transfected with
miR-383 inhibitor, the luciferase activity of the PRDX WT group was
significantly decreased compared with that in the PRDX3 Mut group
(P<0.001).
Discussion
Basu et al (26) previously reported that the
expression level of PRDX3 in cells of patients with prostate cancer
was higher compared with that in normal prostate cells, and
suggested that PRDX3 served a cancer-promoting role. In addition,
elevated expression levels of PRDX3 in the serum may be considered
to be a marker of prostate carcinogenesis (26). In addition, PRDX3 was found to be
overexpressed in glioblastoma and PRDX3 inhibition attenuated
glioma cell growth (9). These
indicated the tumorigenic role of PRDX3. The majority of PRDX3 is
localized in the mitochondria (27); however, PRDX3 expression on the cell
membrane has been shown to be regulated by androgens in LNCaP
cells, which is a prostate cancer cell line (28). PRDX3 is a member of the
peroxiredoxin family, which is responsible for neutralizing ROS
(29), has been reported to be
upregulated in a specific endocrine-regulated tumor (prostate
cancer) (24). In
androgen-resistant LNCaP cells, PRDX3 protein expression levels
have been indicated to be upregulated, thereby preventing
H2O2-induced apoptosis, while PRDX3 knockout
has been demonstrated to restore the sensitivity of
H2O2-induced apoptosis. Therefore, PRDX3 may
serve an important role in inhibiting apoptosis induced by cell
oxidation (24).
Mitochondria have been found to be directly or
indirectly associated with the metabolism of cancer cells (30-32),
which results in an increased amount of ROS and abnormal
mitochondria shape and function (33-35).
Oxidative stress is caused by the imbalance between ROS production
and the antioxidant capacity of the cells (36). Certain cancer treatments, such as
chemotherapy and radiation, have been reported to disrupt
mitochondrial homeostasis and induce the release of cytochrome c,
resulting in the formation and activation of apoptotic bodies
(37,38). This is modulated by the degree of
mitochondrial oxidative stress, and previous reports have suggested
that mitochondrial oxidative stress may serve an important role in
cancer development (39,40).
PRDX3 has been indicated to serve a major role in
controlling mitochondrial ROS production (41). In a previous study (41), PRDX3-knockout mice have been
reported to be lethargic with aging; moreover, compared with WT
mice, PRDX3-knockout mice had increased oxidative damage and a
decreased copy number of mitochondrial DNA in skeletal muscle. In
addition, the apoptosis of brain cells in PRDX3-knockout mice was
increased compared with that in WT mice. These results indicated
that the lack of PRDX3 accelerated the oxidative stress and
mitochondrial damage, resulting in a reduced energy supply and
cellular activity (41). Therefore,
PRDX3 may be associated with the inhibition of the aging process
(41).
In another study, PRDX3 knockdown in hepatoma cells
has been performed to explore whether PRDX3 inhibition may prevent
the oxidation of mitochondrial DNA and the downstream ATP synthesis
and result in the inhibition of tumor cell growth or apoptosis
(42). It was found that PRDX3
mediated mitochondrial oxidative stress to promote tumor growth and
migration (42). In the present
study, cell apoptosis, ROS levels and autophagy were examined
following overexpression and knockdown of PRDX3 (pre-PRDX3 and
sh-PRDX3, respectively). The results indicated that sh-PRDX3
increased apoptosis, ROS levels and autophagy in U87 cells, and it
was hypothesized that sh-PRDX3 may promote U87 cell apoptosis via
ROS-mediated autophagy. These findings are consistent with those of
previous studies aforementioned (42).
miRNAs are non-coding RNAs that regulate gene
expression and have been associated with cancer pathogenesis
(3,4,7,10).
Owing to its abnormal downregulation in certain types of human
cancers, including gastric cancer and non-small cell lung cancer,
miR-383 may be employed for cancer treatment and diagnosis by its
upregulation (43,10). The gene of miR-383 is localized in
the intron of the protein-coding gene SGCZ, which is
dysregulated in various diseases (9,10,43). A
previous study found that miR-383 was downregulated in intestinal
gastric adenocarcinoma and it has been suggested that miR-383 may
be used as a potential tumor marker for the diagnosis of gastric
cancer and as a potential target for gene therapy (43).
The expression level of miR-383 has been reported to
be downregulated in U87 human glioma cells and has been negatively
associated with the pathological grades of gliomas (44). It has been indicated that decreased
miR-383 expression in glioma cells inhibited cell proliferation,
migration and invasion, affected the cell cycle regulation and
induced apoptosis (45).
Overexpression of miR-383 was found to inhibit the growth of U251
and U87 glioma cells, downregulate the protein expression level of
cyclin D1, and induce cell cycle arrest at the
G0/G1 phase (44). This is consistent with previous
studies in different types of tumors, including colorectal cancer
and glioma (11,46,47).
In the present study, cell apoptosis, ROS levels and autophagy were
examined in cells following transfection with miR-383 mimics and
inhibitor. The results indicated that miR-383 mimics increased
apoptosis, ROS levels and autophagy in U87 cells, suggesting that
miR-383 mimics promoted U87 cell apoptosis via ROS-mediated
autophagy. The limitation of this study is that the NC groups were
not taken into consideration in the follow-up study after
transfection efficiency detection.
Li et al (15) reported that PRDX3 was a target
protein of miR-383, and miR-383 has been found to be negatively
associated with PRDX3 expression. In the present study, the
interaction between miR-383 and PRDX3 was also verified. Moreover,
as PRDX3 has been indicated to be a key enzyme in oxidative stress
and autophagy, it may be hypothesized that miR-383 may serve an
antitumor role via regulating the oxidative stress pathway to
induce autophagy.
Supplementary Material
Expression of miR-383 and PRDX3 after
transfection with pre-/sh-PRDX3 vectors, vector NC, miR-383
mimics/inhibitor, miR-NC or inhibitor NC. (A) Expression level of
miR-383; (B) mRNA expression level of PRDX3. n=3;
*P<0.05. PRDX3, peroxiredoxin 3; miR, microRNA; sh,
short hairpin; NC, negative control.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZX, XZ and ML performed the experiments. ZX and JL
analyzed and interpreted the experimental data. ZX and IC designed
the study. ZX was a major contributor to writing the manuscript. IC
revised the manuscript. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lapointe S, Perry A and Butowski NA:
Primary brain tumours in adults. Lancet. 392:432–446.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Piccolo SR and Frey LJ: Clinical and
molecular models of glioblastoma multiforme survival. Int J Data
Min Bioinform. 7:245–265. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fasolo F, Di Gregoli K, Maegdefessel L and
Johnson JL: Non-coding RNAs in cardiovascular cell biology and
atherosclerosis. Cardiovasc Res. 115:1732–1756. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Correia de Sousa M, Gjorgjieva M, Dolicka
D, Sobolewski C and Foti M: Deciphering miRNAs' action through
miRNA editing. Int J Mol Sci. 20(6249)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang F, Liu W, Jin Y, Wang F and Ma J:
Prenatal and neonatal exposure to perfluorooctane sulfonic acid
results in aberrant changes in miRNA expression profile and levels
in developing rat livers. Environ Toxicol. 30:712–723.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Iida A, Shinoe T, Baba Y, Mano H and
Watanabe S: Dicer plays essential roles for retinal development by
regulation of survival and differentiation. Invest Ophthalmol Vis
Sci. 52:3008–3017. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mohammad SM, Emadodin M and Hamed M:
MiR-21: A key player in glioblastoma pathogenesis. J Cell Biochem.
119:1285–1290. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Huang TZ, Wan XC, Alvarez AA, James CD,
Song X, Yang Y, Sastry N, Nakano I, Sulman EP, Hu B and Cheng SY:
MIR93 (microRNA-93) regulates tumorigenicity and therapy response
of glioblastoma by targeting autophagy. Autophagy. 15:1100–1111.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen L, Guan H, Gu C, Cao Y, Shao J and
Wang F: miR-383 inhibits hepatocellular carcinoma cell
proliferation via targeting APRIL. Tumour Biol. 37:2497–2507.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shang Y, Zang A, Li J, Jia Y, Li X, Zhang
L, Huo R, Yang J, Feng J, Ge K, et al: MicroRNA-383 is a tumor
suppressor and potential prognostic biomarker in human non-small
cell lung caner. Biomed Pharmacother. 83:1175–1181. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhao LN, Wang P, Liu YH, Cai H, Ma J, Liu
LB, Xi Z, Li ZQ, Liu XB and Xue YX: MiR-383 inhibits proliferation,
migration and angiogenesis of glioma-exposed endothelial cells in
vitro via VEGF-mediated FAK and Src signaling pathways. Cell
Signal. 30:142–153. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jiang MY, Han ZD, Li W, Yue F, Ye J, Li B,
Cai Z, Lu JM, Dong W, Jiang X, et al: Mitochondrion-associated
protein peroxiredoxin 3 promotes benign prostatic hyperplasia
through autophagy suppression and pyroptosis activation.
Oncotarget. 8:80295–80302. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Magarin M, Pohl T, Lill A, Schulz H,
Blaschke F, Heuser A, Thierfelder L, Donath S and Drenckhahn JD:
Embryonic cardiomyocytes can orchestrate various cell protective
mechanisms to survive mitochondrial stress. J Mol Cell Cardiol.
97:1–14. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Song IS, Jeong YJ, Seo YJ, Byun JM, Kim
YN, Jeong DH, Han J, Kim KT and Jang SW: Peroxiredoxin 3 maintains
the survival of endometrial cancer stem cells by regulating
oxidative stress. Oncotarget. 8:92788–92800. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li KK, Pang JC, Lau KM, Zhou L, Mao Y,
Wang Y, Poon WS and Ng HK: MiR-383 is downregulated in
medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol.
23:413–425. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Srivastava RK, Li C, Ahmad A, Abrams O,
Gorbatyuk MS, Harrod KS, Wek RC, Afaq F and Athar M: ATF4 regulates
arsenic trioxide-mediated NADPH oxidase, ER-mitochondrial crosstalk
and apoptosis. Arch Biochem Biophys. 609:39–50. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen J, Stimpson SE, Fernandez-Bueno GA
and Mathews CE: Mitochondrial reactive oxygen species and type 1
diabetes. Antioxid Redox Signal. 29:1361–1372. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Holzerová E and Prokisch H: Mitochondria:
Much ado about nothing? How dangerous is reactive oxygen species
production? Int J Biochem Cell Biol. 63:16–20. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kornfeld OS, Hwang S, Disatnik MH, Chen
CH, Qvit N and Mochly-Rosen D: Mitochondrial reactive oxygen
species at the heart of the matter: New therapeutic approaches for
cardiovascular diseases. Circ Res. 116:1783–1799. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu KM, Chuang SM, Long CY, Lee YL, Wang
CC, Lu MC, Lin RJ, Lu JH, Jang MY, Wu WJ, et al: Ketamine-induced
ulcerative cystitis and bladder apoptosis involve oxidative stress
mediated by mitochondria and the endoplasmic reticulum. Am J
Physiol Renal Physiol. 309:F318–F331. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Christen V, Camenzind M and Fent K: Silica
nanoparticles induce endoplasmic reticulum stress response,
oxidative stress and activate the mitogen-activated protein kinase
(MAPK) signaling pathway. Toxicol Rep. 1:1143–1151. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cominacini L, Mozzini C, Garbin U, Pasini
A, Stranieri C, Solani E, Vallerio P, Tinelli IA and Fratta Pasini
A: Endoplasmic reticulum stress and Nrf2 signaling in
cardiovascular diseases. Free Radic Biol Med. 88:233–242.
2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Filomeni G, De Zio D and Cecconi F:
Oxidative stress and autophagy: The clash between damage and
metabolic needs. Cell Death Differ. 22:377–388. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Muriach M, Flores-Bellver M, Romero FJ and
Barcia JM: Diabetes and the brain: Oxidative stress, inflammation,
and autophagy. Oxid Med Cell Longev. 2014(102158)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Basu A, Banerjee H, Rojas H, Martinez SR,
Roy S, Jia Z, Lilly MB, De León M and Casiano CA: Differential
expression of peroxiredoxins in prostate cancer: Consistent
upregulation of PRDX3 and PRDX4. Prostate. 71:755–765.
2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Moreira EF, Kantorow M and Rodriguez IR:
Peroxiredoxin 3 (PDRX3) is highly expressed in the primate retina
especially in blue cones. Exp Eye Res. 86:452–455. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Whitaker HC, Patel D, Howat WJ, Warren AY,
Kay JD, Sangan T, Marioni JC, Mitchell J, Aldridge S, Luxton HJ, et
al: Peroxiredoxin-3 is overexpressed in prostate cancer and
promotes cancer cell survival by protecting cells from oxidative
stress. Br J Cancer. 109(983)2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lee YJ: Knockout mouse models for
peroxiredoxins. Antioxidants (Basel). 9(182)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lamb R, Ozsvari B, Lisanti CL, Tanowitz
HB, Howell A, Martinez-Outschoorn UE, Sotgia F and Lisanti MP:
Antibiotics that target mitochondria effectively eradicate cancer
stem cells, across multiple tumor types: Treating cancer like an
infectious disease. Oncotarget. 6:4569–4584. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li JZ, Ke Y, Misra HP, Trush MA, Li YR,
Zhu H and Jia Z: Mechanistic studies of cancer cell mitochondria-
and NQO1-mediated redox activation of beta-lapachone, a potentially
novel anticancer agent. Toxicol Appl Pharmacol. 281:285–293.
2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Vayalil PK, Oh JY, Zhou F, Diers AR, Smith
MR, Golzarian H, Oliver PG, Smith RA, Murphy MP, Velu SE and Landar
A: A novel class of mitochondria-targeted soft electrophiles
modifies mitochondrial proteins and inhibits mitochondrial
metabolism in breast cancer cells through redox mechanisms. PLoS
One. 10(e0120460)2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hrycay EG and Bandiera SM: Involvement of
cytochrome P450 in reactive oxygen species formation and cancer.
Adv Pharmacol. 74:35–84. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Doktorova H, Hrabeta J, Khalil MA and
Eckschlager T: Hypoxia-induced chemoresistance in cancer cells: The
role of not only HIF-1. Biomed Pap Med Fac Univ Palacky Olomouc
Czech Repub. 159:166–177. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Szarek E, Ball ER, Imperiale A, Tsokos M,
Faucz FR, Giubellino A, Moussallieh FM, Namer IJ, Abu-Asab MS,
Pacak K, et al: Carney triad, SDH-deficient tumors, and
Sdhb+/- mice share abnormal mitochondria. Endocr Relat
Cancer. 22:345–352. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Moloney JN and Cotter TG: ROS signalling
in the biology of cancer. Semin Cell Dev Biol. 80:50–64.
2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Pörn-Ares MI, Ares MP and Orrenius S:
Calcium signalling and the regulation of apoptosis. Toxicol In
Vitro. 12:539–543. 1998.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wu CH, Li J, Li L, Sun J, Fabbri M, Wayne
AS, Seeger RC and Jong AY: Extracellular vesicles derived from
natural killer cells use multiple cytotoxic proteins and killing
mechanisms to target cancer cells. J Extracell Vesicles.
8(1588538)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Song IS, Kim HK, Jeong SH, Lee SR, Kim N,
Rhee BD, Ko KS and Han J: Mitochondrial peroxiredoxin III is a
potential target for cancer therapy. Int J Mol Sci. 12:7163–7185.
2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kudryavtseva AV, Krasnov GS, Dmitriev AA,
Alekseev BY, Kardymon OL, Sadritdinova AF, Fedorova MS, Pokrovsky
AV, Melnikova NV, Kaprin AD, et al: Mitochondrial dysfunction and
oxidative stress in aging and cancer. Oncotarget. 7:44879–44905.
2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang YG, Wang L, Kaifu T, Li J, Li X and
Li L: Accelerated decline of physical strength in peroxiredoxin-3
knockout mice. Exp Biol Med (Maywood). 241(1395)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu Z, Hu Y, Liang H, Sun Z, Feng S and
Deng H: Silencing PRDX3 inhibits growth and promotes invasion and
extracellular matrix degradation in hepatocellular carcinoma cells.
J Proteome Res. 15:1506–1514. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Azarbarzin S, Feizi MAH, Safaralizadeh R,
Kazemzadeh M and Fateh A: The value of miR-383, an intronic miRNA,
as a diagnostic and prognostic biomarker in intestinal-type gastric
cancer. Biochem Genet. 55:244–252. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xu Z, Zeng X, Tian D, Xu H, Cai Q, Wang J
and Chen Q: MicroRNA-383 inhibits anchorage-independent growth and
induces cell cycle arrest of glioma cells by targeting CCND1.
Biochem Biophys Res Commun. 453:833–838. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xu D, Ma P, Gao G, Gui Y, Niu X and Jin B:
MicroRNA-383 expression regulates proliferation, migration,
invasion, and apoptosis in human glioma cells. Tumor Biol.
36:7743–7753. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yin M, Wang X, Yao G, Lü M, Liang M, Sun Y
and Sun F: Transactivation of micrornA-320 by microRNA-383
regulates granulosa cell functions by targeting E2F1 and SF-1
proteins. J Biol Chem. 289:18239–18257. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Fateh A, Hosseinpour Feizi MA,
Safaralizadeh R, Somi MH, Ravanbakhsh R, Shokoohi B, Hashemzadeh S
and Azarbarzin S: Prognostic and predictive roles of microRNA-383
in colorectal cancer. Gastroenterol Insights. 7:1–14. 2016.
|