Introduction
Acute respiratory distress syndrome (ARDS) is a
severe respiratory disease that results in low oxygen levels in the
blood and multiple organ failure (1). Its clinical symptoms include diffuse
alveolar injury, oedema, bleeding, the formation of a transparent
membrane and polymorphonuclear neutrophil infiltration (2,3). The
outcome and risk of ARDS may be influenced by multiple factors and
thus, it is difficult to explain the risk and outcome of ARDS with
only one clinical factor (4).
Furthermore, the characteristics and outcome of ARDS may change
over time (5). This means that
although individuals are exposed to similar environmental factors,
they have different risks of morbidity and survival rates.
Depending on these factors, the mortality rate of patients with
ARDS is 30-40% (6). It has been
speculated that the genome may play an important role in the ARDS
(4).
MicroRNAs (miRNAs/miRs) are 21-24 nt duplex RNAs and
one miRNA may regulate hundreds of targets (7). Most of the human genome was predicted
to be regulated by miRNAs and multiple miRNAs may act cooperatively
to silence the same target gene (8). They have important roles in the
regulation of post-transcriptional gene expression. Mature miRNAs
bind to the 3'-untranslated region of their target gene to control
gene expression, resulting in either reduced protein translation or
degradation of mRNA (9). With the
advances in the current understanding of the genome, miRNAs have
been indicated to have an important role in the regulation of
numerous biological processes (10). Dysregulation of miRNAs has been
identified in various diseases, such as lung cancer (11). In addition, differentially expressed
miRNAs are thought to have distinct regulatory roles in biological
processes (12). miRNAs, which are
non-coding RNA molecules that regulate gene expression at the
post-transcriptional level, have emerged as a novel class of gene
expression modulators and certain miRNAs have important roles in
inflammation and apoptosis, which are common manifestations of ARDS
and diffuse alveolar damage (13).
Progress has been made in the study of lung disease
on a genome-wide scale and numerous miRNAs have been indicated to
be expressed in the lungs (4,14,15).
For instance, Yuan et al (16) inhibited neutrophilic inflammation by
silencing triggering receptor expressed on myeloid cells (TREM-1)
with a nanomicellar approach, suggesting that TREM-1 is a potential
therapeutic target for neutrophilic lung inflammation and ARDS.
However, studies reporting on the discovery of candidate genes are
difficult to replicate due to small sample sizes, population
stratification, variability of the control populations or
heterogeneity of the ARDS phenotype (4). Thus, the progress of research into
biomarkers for ARDS is very slow. However, at the same time, it has
been demonstrated that numerous miRNAs are expressed specifically
in ARDS compared with normal lung tissue in rat model studies
(17,18). For instance, after
trauma/transfusion or the physiological remission of ARDS, miR-223
expression increased in the lungs of methyltetradecanoic
acid-induced mice or patients with ARDS (19). miR-181a and miR-92a are
risk-associated biomarkers for ARDS, whereas miR-424 is a
protective biomarker (20). Liu
et al (21) indicated that
upregulation of miRNA-200c-3p reduced the levels of
angiotensin-converting enzyme 2, which has a crucial role in the
occurrence and development of ARDS.
High-throughput sequencing technology has made it
possible to sequence full human genomes and enhance the
understanding of genetic effects on diseases, treatment outcomes
and public health (22). In the
present study, high-throughput sequencing was used to identify
differentially expressed miRNAs in the serum of patients with ARDS
and reverse transcription-quantitative (RT-q)PCR verification
indicated that miR-584 and miR-146a may have important roles in
ARDS. miR-122 may have a key role in the progression of ARDS from
initiation to death. These genes may be potential targets for the
treatment of ARDS.
Materials and methods
Clinical samples
Serum samples of patients with ARDS (age range,
23-85; average age, 63 years; sex ratio, 8:13) and healthy subjects
(age range, 27-69; average age, 45 years; sex ratio, 9:11) were
collected from December 2013 to February2016, at Xixi Hospital
(Hangzhou, China). Healthy samples underwent routine medical
examinations at Xixi Hospital. All patients with ARDS met the
Berlin diagnostic definition (23):
The time of ARDS was within 1 week of a known clinical insult or
new or worsening respiratory symptoms; chest imaging indicated
bilateral opacities (not fully explained by effusions, lobar/lung
collapse or nodules); respiratory failure was not fully explained
by cardiac failure or fluid overload; and ARDS severity was based
on oxygenation index (arterial oxygen partial pressure/inhaled
oxygen concentration). Detailed information on the patients with
ARDS is presented in Table I. Serum
samples were collected from patients who were grouped according to
the two stages of improvement and death. The criteria for selection
included an age of ≥18 years; absence of cardiac insufficiency and
no pregnancy or lactation. Serum was stored at -80˚C after
collection.
 | Table IInformation of patients with
ARDS. |
Table I
Information of patients with
ARDS.
| Patient ID | Age (years) | Sex | Clinical
diagnosis | Oxygenation index
(mmHg) | Clinical
outcome |
|---|
| 1a | 32 | Male | Severe measles,
severe pneumonia, type I respiratory failure, hepatitis B,
hypernatremia. | 222 | Death |
| 2 | 47 | Male | Measles complicated
with pneumonia, respiratory failure, intestinal obstruction. | 208 | Improvement |
| 3 | 56 | Male | Pulmonary
infection, respiratory failure type I, cholestatic hepatitis,
parapsoriasis, hypoproteinemia. | 161 | Improvement |
| 4 | 58 | Male | PCP, pulmonary
edema, liver dysfunction. | 85 | Death |
| 5 | 66 | Male | Chronic renal
failure, pulmonary infection, liver cancer after interventional
therapy, diabetes mellitus type II, hypertension. | 172 | Improvement |
| 6 | 69 | Male | Posthepatitic
cirrhosis, septic shock, hepatic encephalopathy. | 190 | Death |
| 7a | 72 | Male | Sepsis, septic
shock, severe pulmonary infection, pleural effusion, occupation of
left liver space. | 116 | Improvement |
| 8 | 83 | Male | Cerebral
infarction, pulmonary infection, respiratory failure type I, septic
shock, senile dementia, myocardial infarction, renal failure. | 125 | Death |
| 9 | 23 | Female | Severe pneumonia,
respiratory failure. | 129 |
Transferb |
| 10 | 35 | Female | Pneumonia,
respiratory failure, diabetes mellitus type II. | 180 | Improvement |
| 11 | 56 | Female | H7N9, severe
pneumonia, respiratory failure. | 136 | Death |
| 12a | 63 | Female | H1N1, pneumonia,
post-operative thyroid cancer, diabetes mellitus type II. | 193 | Improvement |
| 13 | 69 | Female | Respiratory failure
type I, severe measles, chronic renal failure. | 153 | Death |
| 14a | 71 | Female | H7N9, respiratory
failure, apoplexy. | 140 | Death |
| 15 | 74 | Female | Right inguinal
hernia, intestinal obstruction, septic shock, cardiac arrest,
aspiration pneumonia. | 56 | Death |
| 16 | 76 | Female | Pulmonary
infection, septic shock, respiratory failure, senile emphysema,
coronary heart disease. | 198 | Improvement |
| 17 | 79 | Female | Renal failure,
heart failure, pulmonary infection, respiratory failure. | 88 | Death |
| 19a | 81 | Female | Community-acquired,
respiratory failure type I, hypertension, post-operative
appendicitis. | 58 | Death |
| 20 | 84 | Female | Hypertension,
respiratory failure, sepsis, septic shock, severe pulmonary
infection. | 76 |
Transferb |
| 21 | 85 | Female | Pulmonary edema,
stroke, liver cirrhosis, respiratory failure. | 103 |
Transferb |
RNA extraction and quality
control
RNA was extracted by TRIzol reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
NanoDrop-2000 (Thermo Fisher Scientific, Inc.) was used to measure
the quantity and quality of RNA. The integrity of RNA was assessed
by standard denaturing agarose gel electrophoresis (1%).
High-throughput sequencing
From the 21 cases from the ARDS group and the 20
cases from the healthy group, five serum samples per group were
selected. Age, sex, oxygenation index and outcome were evenly
distributed among the 5 samples from the ARDS group (Table I). In terms of age, younger (age, 32
years), middle-aged (age, 63 years) and elderly (age, 71, 72 and 81
years) patients were selected. In terms of sex, three females and
two males were selected. For the oxygenation index, patients with
low (index=58), medium (index=116, 140) and high levels (index=193,
222) were selected. For the final outcome, three patients who died
and two patients who finally improved were selected. In terms of
clinical diagnosis, patients with H1N1 (n=1) or H7N9 (n=1)
infection and patients without special infection were selected. In
the healthy group, the age range was 31-69 years and the
male-to-female ratio was 3:2. For each group, these 5 samples were
mixed for high-throughput sequencing to determine differentially
expressed miRNAs. The procedure followed standard Illumina
procedures, including library preparation and sequencing. TruSeq
Small RNA Sample Prep Kits (Illumina, Inc.) were used to prepare
the small RNA sequencing library. After library preparation was
completed, Illumina Hiseq2000/2500 (Illumina, Inc.) was used to
sequence the constructed library with a single-terminal 1x50-bp
reading length.
High-throughput sequencing data
analysis
To obtain the clean sequence, ACGT101-miR v4.2 (LC
Sciences) was used to remove the 3'adaptor and low-quality
sequences. From the remaining sequences, 15 to 27 bp sequences,
which were most likely to contain mature miRNAs, were retained for
further analysis. miRNA sequences were filtered out and clean data
were obtained by comparisons with RFam (http://rfam.janelia.org), Repbase (http://www.girinst.org/repbase) and mRNAbase
(ftp://ftp.ensembl.org/pub/release-90/fasta/homo_sapiens/).
After comparison with miRNAbase (ftp://mirbase.org/pub/mirbase/CURRENT/), the reads
were divided into 4 groups as the follows: ‘Group (gp)1’-reads
mapped to pre-miRNAs in miRbase and the pre-miRNAs further mapped
to the genome and expressed sequence tag; ‘gp2’-the mapped
pre-miRNAs did not map to the genome, but the reads (and the miRNAs
of the pre-miRNAs) mapped to the genome; ‘gp3’-reads mapped to
selected miRNAs/pre-miRNAs in miRbase, but the mapped pre-miRNAs
and the reads did not map to the genome; and ‘gp4’-reads did not
map to selected pre-miRNAs in miRbase, but the reads mapped to the
genome and the extended genome sequences in the genome may form
hairpins.
After screening for valid sequences, the expression
profiles of these sequences were normalized. Valid sequences were
identified in all samples and a reference frame, which consists of
the median of the copy number of the relevant valid sequence in all
samples, was established. Subsequently, the data of all samples and
reference frame data pairs were log2 transformed. The Δlog2 of the
log2-transformed data between each respective sample and the
reference dataset was calculated and sequences with |Δlog2|<2
were selected. Next, a linear regression analysis of the reference
set was performed between samples and subsets to derive the linear
formula y=aix+bi, where ai and
bi are the slope and intercept, respectively. For the
resulting line, x is the log2 of the reference dataset and y is the
expected value log2 of sample i on a corresponding sequence. The
median value of the reference frame, xmid, was then calculated as
xmid=[max(x)-min(x)]/2. The log2 of the relevant sample i was
calculated as yi,
mid=aixmid + bi, such
that yr, mid=xmid and Δyi=yr,
mid-yi, mid, which is the logarithmic correction
factor of sample i. The correction factor algorithm for sample i
fi=2Δyi was then obtained. The number of
copies for each sample was obtained by multiplying the original
copy number with the algorithm correction factor fi.
RT-qPCR
Candidate miRNA selection was based on the following
criterion: Fold-change >2 after data normalization. Patients
were divided into the disease group (n=8) and the death group
(n=10) according to whether the final clinical symptoms were
improved or the patient died, and three patients were transferred
to a different hospital according to their families' wishes, so
they were not included in the disease group or death group. A total
of five randomly selected miRNAs were validated by RT-qPCR. The
stem-loop primers used for RT of miRNAs (http://primer3.ut.ee/) are listed in Table II. Complementary DNA was
synthesized using M-MLV(H-) Reverse Transcriptase obtained from
Vazyme Biotechnology. M-MLV(H-) Reverse Transcriptase (Vazyme
Biotech), 5x RT Buffer (Vazyme Biotech), dNTP Mixture (Vazyme
Biotech), primer (Sangon Biotech), RNase Inhibitor (Vazyme Biotech)
and ddH2O are included in the reverse transcription
reaction system. The temperature protocol for reverse transcription
was as follows: 45 min at 42˚C, 5 min at 85˚C. The reaction was
carried out in a PCR instrument (cat. no. T1000; Bio-Rad
Laboratories, Inc.). qPCR was performed with SYBR Green qPCR
(Toyobo) according to the manufacturer's protocol. The composition
of this reaction mixture included the primers (Sangon Biotech),
SYBR Green qPCR Mix (Toyobo) and ddH2O. The temperature
protocol for reverse transcription was as follows: Initial
denaturation 95˚C for 30 sec, followed by 40 cycles of 95˚C for 10
sec and 60˚C for 30 sec. The reaction was carried out in a qPCR
instrument (7500; Applied Biosystems; Thermo Fisher Scientific,
Inc.). 5S ribosomal RNA was used as an internal control. Relative
mRNA expression of each gene compared to 5S ribosomal RNA was
calculated using the 2-ΔΔCq method (24). The primer information of miRNAs for
qPCR is provided in Table III.
All miRNAs had the same stem-loop sequence. The reverse primer used
for qPCR was designed based on the stem-loop sequence and thus, it
was a universal reverse primer.
 | Table IIStem-loop primers used for
reverse-transcription PCR. |
Table II
Stem-loop primers used for
reverse-transcription PCR.
| Name | Sequence
(5'-3') |
|---|
| miR-584 |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTCAGTC |
| miR-451 |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACTCAG |
| miR-146a |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACCCAT |
| miR-193a |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCATCTC |
| miR-122 |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCAAACAC |
 | Table IIISequences of primers used for
quantitative PCR. |
Table III
Sequences of primers used for
quantitative PCR.
| Name | Forward primer
(5'-3') | Reverse
primera (5'-3') |
|---|
| hsa-miR-584 |
GGTTATGGTTTGCCTGGGA | |
| hsa-miR-451 |
CCCCAAACCGTTACCATTACT | |
| hsa-miR-146a |
GGGGTGAGAACTGAATTCCAT |
CAGTGCGTGTCGTGGAGT |
| hsa-miR-193a |
GGGTCTTTGCGGGCG | |
| hsa-miR-122 |
GGTGGAGTGTGACAATGGTG | |
| 5S rRNA |
GTCTACGGCCATACCACCCTGAA |
AAGCCTACAGCACCCGGTATTCC |
miRNA and mRNA co-expression
network
Based on the results of the sequence analysis, the
following five miRNAs with the largest difference in expression
levels were obtained: miR-584, miR-451, miR-146a, miR-193a and
miR-122. miRDB (http://mirdb.org), miRWalk (http://mirwalk.umm.uni-heidelberg.de)
and TargetScan 7.0 (http://www.targetscan.org/vert_72/) were used for
target gene prediction.
Gene Ontology (GO) enrichment and
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
GO annotations of target genes corresponding to
differentially expressed miRNAs were performed with the WEB-based
GEne SeT AnaLysis Toolkit (http://www.webgestalt.org/option.php). KEGG pathways
were analysed through KEGG Mapper (https://www.kegg.jp/kegg/mapper.html) to identify the
significant pathways of the differentially expressed genes. GO
terms and KEGG pathways with a corrected P-value <0.05, which
was calculated by the hypergeometric test, were considered to be
significantly enriched. Futhermore, -log10(P) was used to determine
the enrichment of each GO term by the differentially expressed
genes and the significance of the pathway associations.
Statistical analysis
All statistical analyses were conducted with SPSS
(20.0 IBM Corp.). Statistical comparisons of the sequencing data
were performed using Student's t-test. ANOVA was used for
comparisons between multiple groups, followed by the
Least-Significant Difference test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Processing of raw miRNA
high-throughput sequencing data
A total of 1.8x107 raw-sequence reads
from the serum of patients with ARDS and 1.9x107
raw-sequence reads from the serum of healthy subjects were
obtained. In the filtering analysis, a total of 679 miRNAs were
identified to be differentially expressed between the ARDS and
healthy samples. Of the 679 miRNAs, 537 miRNAs belonged to the
category gp1, 17 miRNAs to gp2, 24 miRNAs to gp3 and 101 miRNAs to
gp4. In gp1 and gp2, 112 miRNAs were selected according to the
screening conditions fold-change >2 and P<0.05. A total of 55
miRNAs were upregulated, while 57 miRNAs were downregulated. In the
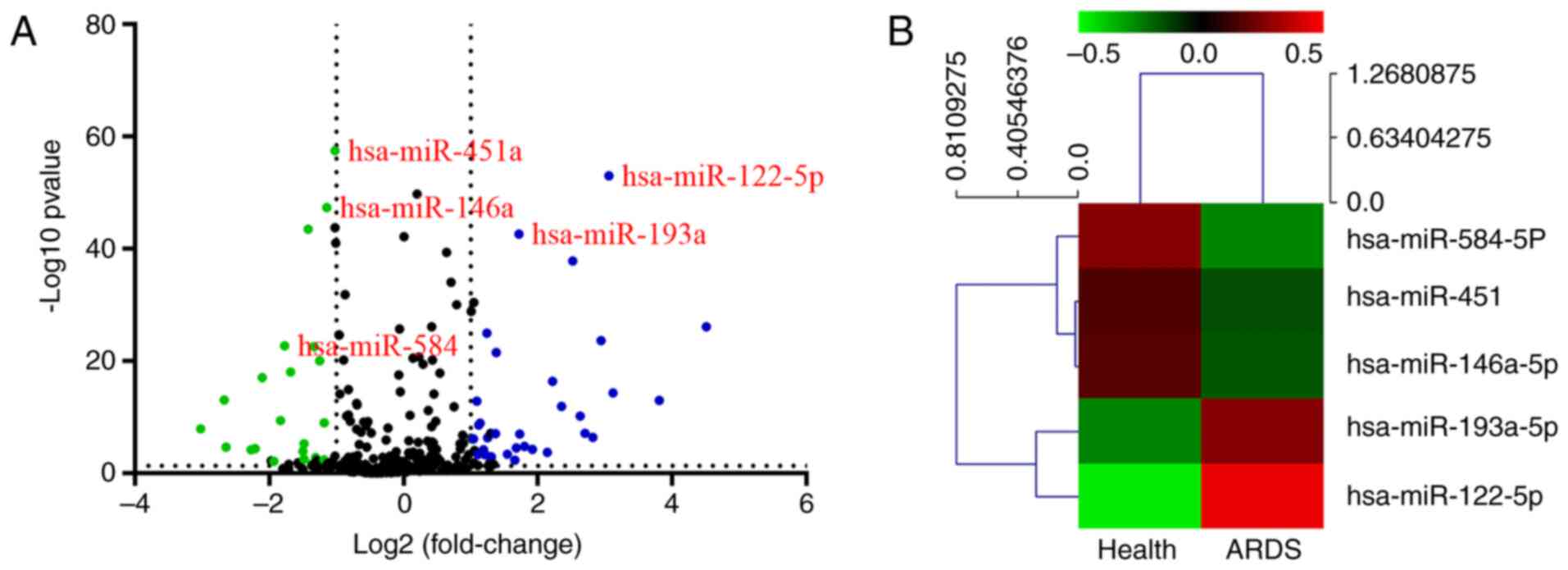
volcano plot analysis of the differentially expressed miRNAs, the
ARDS group was successfully separated from the normal group
(Fig. 1A). The top 5 differentially
expressed miRNAs are presented in a heat map in Fig. 1B. miR-584, miR-451, miR-146a and
miR-193a were the most downregulated miRNAs, while miR-122 was the
most upregulated.
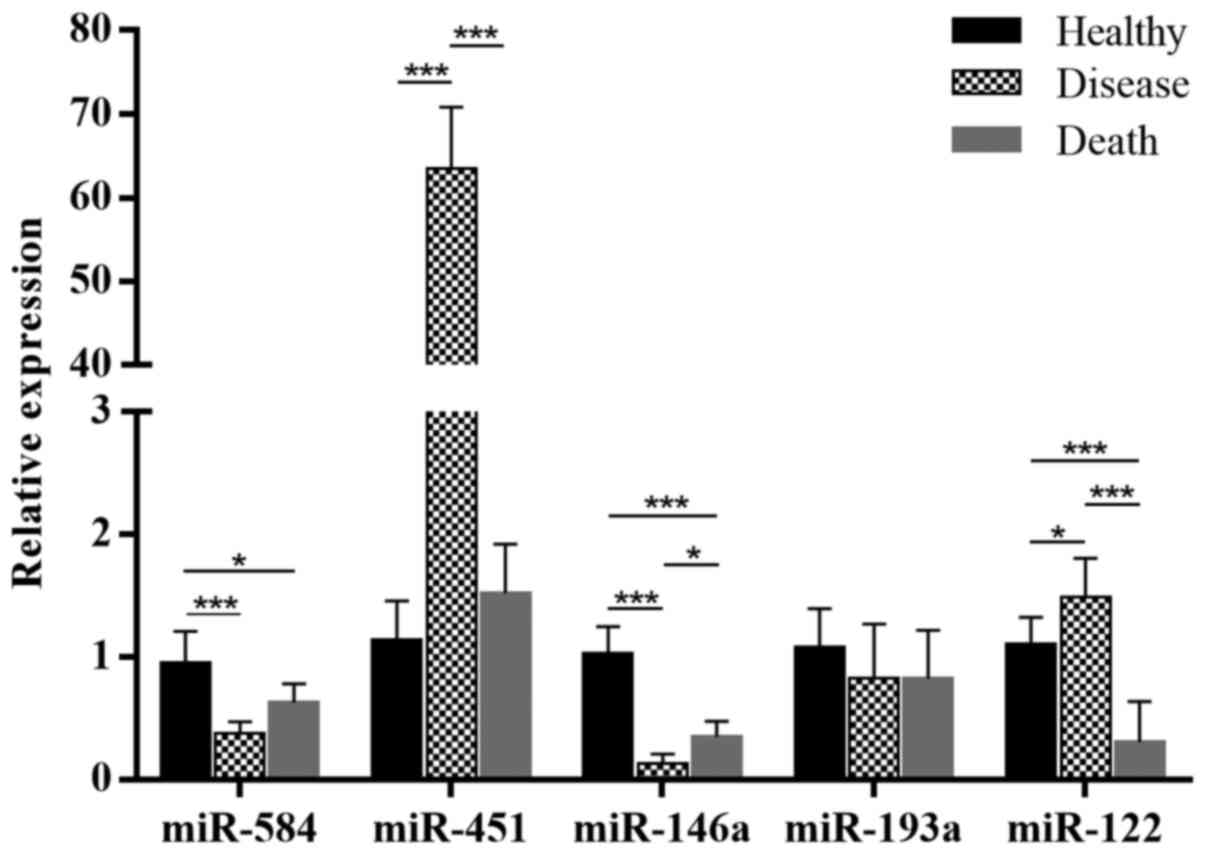
Validation of differentially expressed
miRNAs in ARDS and healthy serum by RT-qPCR
To confirm the previous results and identify the
functions of miRNAs in ARDS, five randomly selected miRNAs
(Table IV), including three
downregulated miRNAs, Homo sapiens (hsa)-miR-584,
hsa-miR-451 and hsa-miR-146a, and two upregulated miRNAs,
hsa-miR-193a and hsa-miR-122, were validated by RT-qPCR (Fig. 2).
 | Table IVDetails of the 5 randomly selected
miRNAs for reverse transcription-quantitative PCR confirmation. |
Table IV
Details of the 5 randomly selected
miRNAs for reverse transcription-quantitative PCR confirmation.
| miRNA name | P-value | Fold-change | Direction of
regulation | Chromosome | Strand | Expression |
|---|
| hsa-miR-584 |
1.93x10-23 | 0.29 | Down | Chr5 | - | Middle |
| hsa-miR-451a |
3.36x10-44 | 0.49 | Down | Chr17 | - | High |
| hsa-miR-146a |
4.86x10-83 | 0.45 | Down | Chr5 | + | High |
| hsa-miR-193a |
2.85x10-110 | 3.29 | Up | Chr17 | + | Middle |
| hsa-miR-122 |
2.94x10-26 | 8.32 | Up | Chr18 | + | High |
The results were similar to those of the
high-throughput sequencing. High-throughput sequencing and RT-qPCR
demonstrated downregulated expression of hsa-miR-584 and
hsa-miR-146a and upregulated expression of hsa-miR-122 in the
disease vs. healthy group (Fig.
2A). However, in the death vs. healthy group, miR-122 was
downregulated (Fig. 2B). In the
comparison of the death vs. healthy group, only the results of
hsa-miR-584 and hsa-miR-146a were similar to those of the
high-throughput sequencing (Fig.
2C). To reduce the individual differences for sequencing, the
samples were pooled in the sequencing analysis, mixing patients
from the disease and death groups; thus, the sequencing results did
not reflect the differences between the disease group and the death
group.
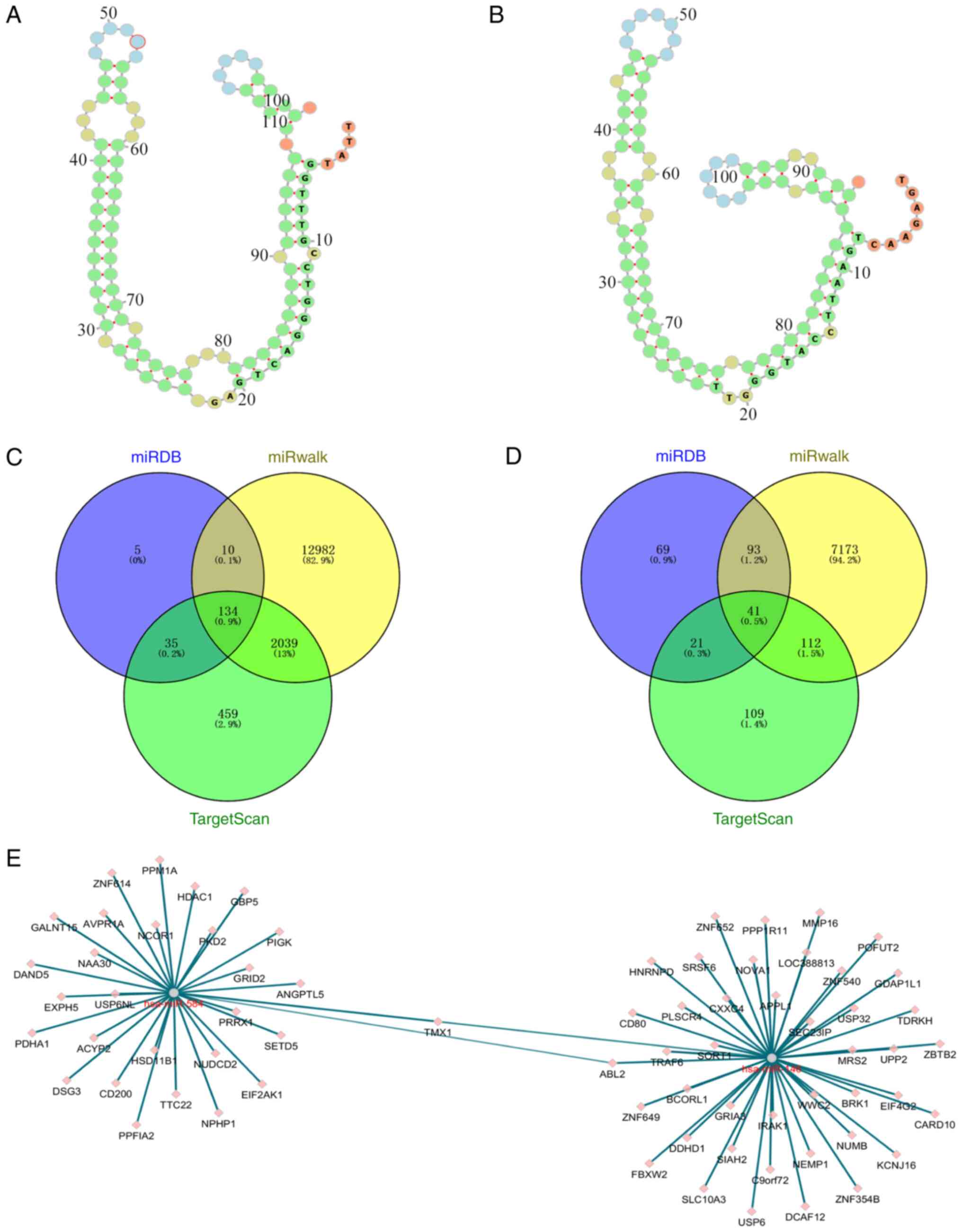
Prediction of potential miRNA
targets
As miRNAs affect gene expression by regulating
mRNAs, the target mRNAs of the miRNAs were predicted in a further
analysis. miR-584 and miR-146a were selected for target gene
prediction due to their significant differences. ViennaRNA Web
Services (http://nibiru.tbi.univie.ac.at/forna/) were used to
plot the secondary structures of the miR-584 and miR-146 precursors
(Fig. 3A and B). MiRDB, miRWalk and TargetScan 7.0 were
used for this prediction and the results were intersected. A total
of 134 target genes were predicted for miR-584 and 41 target genes
for miR-146a (Fig. 3C and D). Analysis of the network map of the
target genes of these two miRNAs identified only two common target
genes (Fig. 3E).
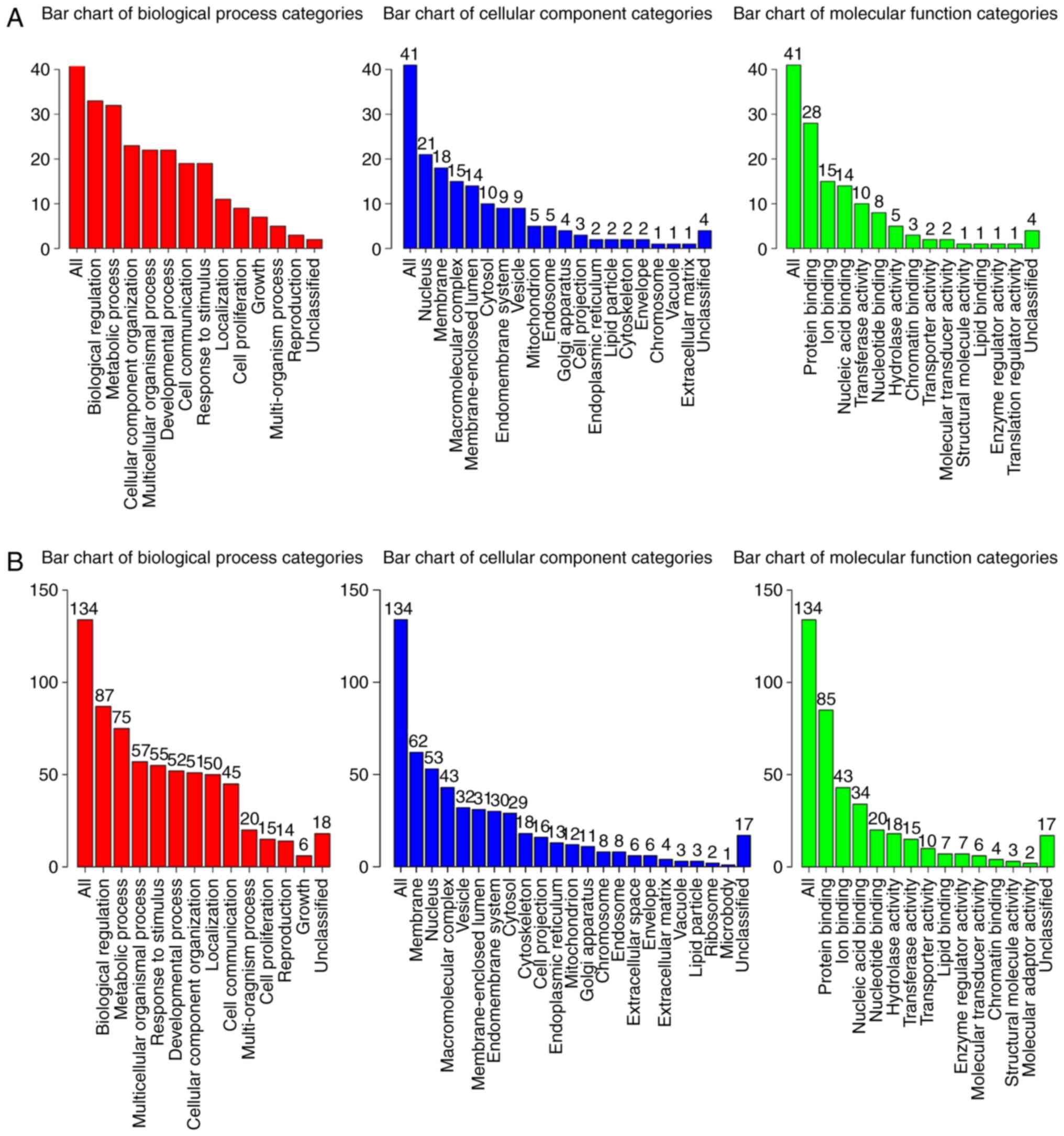
GO analysis
GO enrichment analysis of significantly
differentially expressed mRNAs was performed to determine the
effects of these miRNAs. The WEB-based GEne SeT AnaLysis Toolkit
(webgestalt; http://www.webgestalt.org/option.php) was used to
annotate the potential biological functions and signalling pathways
of the target genes. GO categories of ‘biological process’ (BP),
‘cellular component’ (CC) and ‘molecular function’ (MF) were
analysed to determine gene functions and gene product enrichment.
The results of the GO enrichment analysis are presented in Fig. 4. The analysis revealed that the
majority of the BP terms associated with both miR-146a (Fig. 4A) and miR-584 (Fig. 4B) were involved in the processes of
cell life activity and composition, such as ‘biological
regulation’, ‘metabolic process’, ‘multicellular organismal
process’ and ‘cellular component organization’. In the category CC,
the terms of the target mRNAs of miR-146a and miR-584 included
‘nucleus’, ‘membrane’ and ‘macromolecular complex’. In the MF
category, the target mRNAs were enriched in terms associated with
binding activities, including ‘protein binding’, ‘ion binding’ and
‘nucleic acid binding’.
KEGG analysis
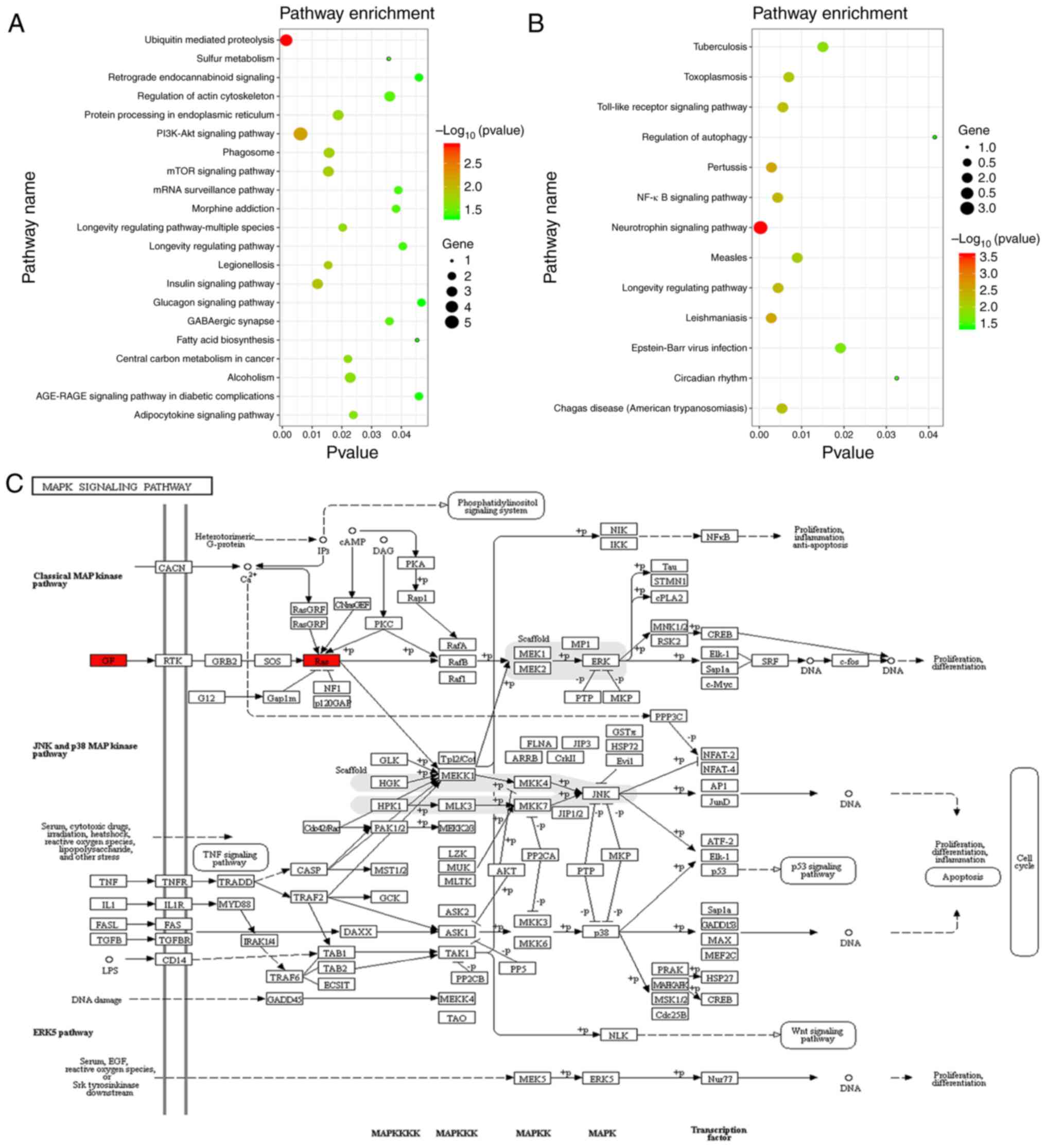
The results of the KEGG pathway enrichment analysis
of target mRNAs are presented in Fig.
5 and pathways with P<0.05 were selected to draw a bubble
diagram. These results demonstrated that the target mRNAs of
hsa-miRNA-584 may be involved in ‘ubiquitin-mediated proteolysis’,
‘sulfur metabolism’, ‘retrograde endocannabinoid signaling’ and
‘regulation of actin cytoskeleton’ (Fig. 5A). In addition, ‘tuberculosis’,
‘toxoplasmosis’, ‘Toll-like receptor (TLR) signaling pathway’ and
‘NF-κB signaling pathway’ were associated with the target mRNAs of
hsa-miR-146a (Fig. 5B). MAPK
cascade activation is the hub of multiple signalling pathways and
NF-κB is one of the downstream effectors of the MAPK signalling
pathway (25). Thus, further
analysis focused on the MAPK signaling pathway. According to the
KEGG prediction, glial cell line-derived neurotrophic factor
(GDNF), which is a target gene of miR-146a, and RAB23, which
belongs to the RAS family and is a target gene of miR-584, are
involved in the MAPK signalling pathway, which is an
inflammation-associated signalling pathway (26) (Fig.
5C).
Discussion
At present, no sensitive or specific biomarker for
the early diagnosis and treatment of ARDS is available. Considering
the high risk of death with ARDS, particularly in children
(27), further investigations into
the molecular mechanisms of ARDS are crucial to improve the
survival rate. ARDS is a multifactorial syndrome with high
morbidity and mortality rates, despite an enhanced understanding of
ARDS pathogenesis, the capacity to predict the development of ARDS
remains limited (28). miRNAs have
emerged as critical molecules in human diseases (13,29-31).
However, a limited number of studies on miRNAs associated with the
pathogenesis and progression of ARDS have been published.
In the present study, using high-throughput
sequencing technology, a preliminary molecular analysis of miRNAs
and mRNAs in ARDS was performed to facilitate further studies on
the pathogenesis of ARDS and to explore whether the pathology of
ARDS may be induced by novel miRNAs. In addition, GO and KEGG
pathway enrichment analyses were performed to identify the
potential functions of differentially expressed mRNAs.
Comparison of the expression profiles of miRNAs in
ARDS and healthy samples provided 679 differentially expressed
miRNAs. The most upregulated or downregulated miRNAs may help
identify molecular markers for the early diagnosis of ARDS. A total
of five miRNAs were randomly selected for RT-qPCR validation and
the results of only 3 miRNAs were consistent with the sequencing
analysis. This may be because a pooled sample strategy was adopted
during sequencing, while in the RT-qPCR analysis, a more detailed
grouping was performed. After excluding the difference between the
disease group and the death group, the sequencing results were
still reliable.
GO enrichment analysis was used to identify the
functions of the miRNAs through the expression patterns of their
target mRNAs. Among the target mRNAs of miR-146a and miR-584, most
of the BP terms were associated with cell activity and composition.
Alveolar macrophages and alveolar epithelial cells are the first
line of host defence and innate immunity (32). In addition to macrophages and
alveolar epithelial cells, leukocytes and endothelial cells are
also essential for the inflammatory response in acute lung injury
(ALI)/ARDS. In addition, regulating the function of macrophages and
monocytes may be a promising therapeutic strategy against ALI/ARDS
(33). This evidence may indicate
that the differentially expressed miRNAs are associated with the
activities and functions of these immune cells.
The enriched CC terms were associated with the
nucleus and membrane, which are key organelles for biological
regulation, protein formation and secretion. As discussed above,
macrophages are important in ARDS, both in terms of
pro-inflammatory and anti-inflammatory activities (34). In addition, NF-κB is a key
transcription factor in the regulation of the innate immune
inflammatory response in activated macrophages (35). Activation of the NF-κB signalling
pathway requires extracellular factors to stimulate receptors on
the cell membrane and free NF-κB is required to enter the nucleus
to initiate transcription (36).
Members of the IL-1 family are key determinants of inflammation.
They mainly participate in the inflammatory response through
membrane processes, including direct plasma membrane translocation,
lysosome exocytosis, exosome formation, membrane vesiculation,
autophagy and pyroptosis (37).
The MF terms were associated with protein binding
and ion binding. After the NF-κB signalling pathway is activated,
NF-κB binds to multiple proteins or factors to mediate its effector
functions (38). In addition,
numerous studies have indicated that zinc modulates the NF-κB
pathway (39). This is consistent
with the results of the GO enrichment analysis in the present
study.
KEGG pathway enrichment analysis revealed that
pathways such as ‘ubiquitin-mediated proteolysis’, ‘sulfur
metabolism’ and ‘regulation of actin cytoskeleton’ were associated
with the target mRNAs of miR-584. NF-κB is a critical transcription
factor for the maximal expression of numerous cytokines that are
involved in the pathogenesis of inflammatory diseases, such as ARDS
and sepsis syndrome. NF-κB activation involves the phosphorylation,
ubiquitination and proteolysis of IκB (40). There is also experimental evidence
that the actin cytoskeleton has an important role in the regulation
of NF-κB activation and inflammatory events in intestinal
epithelial cells (41). Numerous
studies have proven that sulphides directly or indirectly affect
the NF-κB signalling pathway, but they focused on a specific
sulphide, not the entire sulfur metabolism system (42-44).
The enrichment analysis of miR-584 target genes is basically
consistent with previous experiments.
Pathways such as ‘tuberculosis’, ‘toxoplasmosis’,
‘TLR signaling pathway’ and ‘NF-κB signaling pathway’ were
associated with the target mRNAs of miR-146a. The TLR family acts
as a primary sensor of innate immunity and all TLR signalling
pathways culminate with the activation of the transcription factor
NF-κB (45). Numerous studies have
proven that miR-146a is closely related to inflammation (46-48).
There is also considerable evidence that miR-146a is closely linked
to lung diseases, including ARDS (49-51).
The results of the target gene enrichment in the present study are
consistent with previous results.
One of the predicted target genes of miR-146a, GDNF,
is involved in the development of pulmonary innervation (52). Certain studies have proven that the
introduction of the GDNF gene into macrophages may effectively
reduce neuronal inflammation (53).
RAB23 is a predicted target gene of miR-584. Huang et al
(54) detected the expression of
Rab23 protein in the nuclei in lung cancer tissues by
immunohistochemistry. Furthermore, the dysregulation of RAB23 may
affect the NF-κB signalling pathway (55). The predictions of the present study
are consistent with these findings.
A limitation of the present study is the focus on
high-throughput analysis and the related miRNA-mRNA network, GO
analysis and KEGG analysis based on the predicted targeted genes.
Further research on miRNA functions will be performed in future
studies with the aim of obtaining additional evidence to
corroborate the bioinformatics predictions.
In conclusion, the results of the present study
suggested that miRNA-584 and miR-146a may be involved in the
occurrence and development of inflammation in ARDS by affecting
macrophages, and NF-κB may have an important role in this process.
These molecules may be promising therapeutic targets for patients
with ARDS and further studies are required to explore the precise
mechanisms of ARDS.
Overall, it was revealed that miR-584, miR-146a and
miR-122 have a certain association with ARDS and may be potential
therapeutic targets. However, certain limitations of the present
study should also be acknowledged. Of note, the results were only
based on 21 serum samples from patients with ARDS. There may be
certain false-positives and further verification is required.
Future research on these targets is required to validate the
functions of the identified miRNAs in ARDS and provide a more
comprehensive understanding of the underlying mechanisms.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the Department of Health
of Zhejiang Province (grant no. 2019322603) and the Hangzhou
Science and Technology Bureau (grant no. 20191223B75).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
SZ and YH made substantial contributions to the
conception or design of the study and drafting the manuscript or
revising it critically for important intellectual content. HL, JX,
QW, YZ, XZ and YY collected, analysed or interpreted data. XD
edited and critically revised the manuscript. KZ proposed the
experimental idea and participated in the design of the experiment.
XD is the main designer of the experiment specifically performing
the experiments, and data collection. KZ ensured that questions
relating to the accuracy or completeness of any part of the work
are properly investigated and resolved. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Human Ethics
Review Committee at XiXi Hospital of Hangzhou and Zhejiang Sci-Tech
University (Hangzhou, China). All participants provided written
informed consent prior to enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Holland MC, Mackersie RC, Morabito D,
Campbell AR, Kivett VA, Patel R, Erickson VR and Pittet JF: The
development of acute lung injury is associated with worse
neurologic outcome in patients with severe traumatic brain injury.
J Trauma. 55:106–111. 2003.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Katzenstein AL, Bloor CM and Leibow AA:
Diffuse alveolar damage- the role of oxygen, shock, and related
factors. Am J Pathol. 85:209–228. 1976.PubMed/NCBI
|
|
3
|
Ware LB and Matthay MA: The acute
respiratory distress syndrome. N Engl J Med. 342:1334–1349.
2000.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Reilly JP, Christie JD and Meyer NJ: Fifty
years of research in ARDS. Genomic contributions and opportunities.
Am J Respir Crit Care Med. 196:1113–1121. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ferring M and Vincent JL: Is outcome from
ARDS related to the severity of respiratory failure? Eur Respir J.
10:1297–1300. 1997.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rubenfeld GD, Caldwell E, Peabody E,
Weaver J, Martin DP, Neff M, Stern EJ and Hudson LD: Incidence and
outcomes of acute lung injury. N Engl J Med. 8:1685–1693.
2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang Y, Brahmakshatriya V, Zhu H, Lupiani
B, Reddy SM, Yoon BJ, Gunaratne PH, Kim JH, Chen R, Wang J and Zhou
H: Identification of differentially expressed miRNAs in chicken
lung and trachea with avian influenza virus infection by a deep
sequencing approach. BMC Genomics. 10(512)2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lu J and Clark AG: Impact of microRNA
regulation on variation in human gene expression. Genome Res.
22:1243–1254. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lu Y, Okubo T, Rawlins E and Hogan BL:
Epithelial progenitor cells of the embryonic lung and the role of
microRNAs in their proliferation. Proc Am Thorac Soc. 5:300–304.
2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Li J, Aung LH, Long B, Qin D, An S and Li
P: miR-23a binds to p53 and enhances its association with miR-128
promoter. Sci Rep. 5(16422)2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ferruelo A, Penuelas O and Lorente JA:
MicroRNAs as biomarkers of acute lung injury. Ann Transl Med.
6:34–44. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Orwoll BE and Sapru A: Biomarkers in
pediatric ARDS: Future directions. Front Pediatr.
4(55)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Rajasekaran S, Pattarayan D, Rajaguru P,
Sudhakar Gandhi PS and Thimmulappa RK: MicroRNA regulation of acute
lung injury and acute respiratory distress syndrome. J Cell
Physiol. 231:2097–2106. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yuan Z, Syed M, Panchal D, Joo M, Bedi C,
Lim S, Onyuksel H, Rubinstein I, Colonna M and Sadikot RT:
TREM-1-accentuated lung injury via miR-155 is inhibited by LP17
nanomedicine. Am J Physiol Lung Cell Mol Physiol. 310:L426–L438.
2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Huang C, Xiao X, Chintagari NR, Breshears
M, Wang Y and Liu L: MicroRNA and mRNA expression profiling in rat
acute respiratory distress syndrome. BMC Med Genomics.
7(46)2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Guan Y, Jin X, Liu X, Huang Y, Wang M and
Li X: Identification of microRNAs in acute respiratory distress
syndrome based on microRNA expression profile in rats. Mol Med Rep.
16:3357–3362. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Feng Z, Qi S, Zhang Y, Qi Z, Yan L, Zhou
J, He F, Li Q, Yang Y, Chen Q, et al: Ly6G+ neutrophil-derived
miR-223 inhibits the NLRP3 inflammasome in mitochondrial
DAMP-induced acute lung injury. Cell Death Dis.
8(e3170)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhu Z, Liang L, Zhang R, Wei Y, Su L,
Tejera P, Guo Y, Wang Z, Lu Q, Baccarelli AA, et al: Whole blood
microRNA markers are associated with acute respiratory distress
syndrome. Intensive Care Med Exp. 5(38)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu Q, Du J, Yu X, Xu J, Huang F, Li X,
Zhang C, Li X, Chang J, Shang D, et al: miRNA-200c-3p is crucial in
acute respiratory distress syndrome. Cell Discov.
3(17021)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Goldfeder RL, Wall DP, Khoury MJ,
Ioannidis JPA and Ashley EA: Human genome sequencing at the
population scale: A primer on High-throughput DNA sequencing and
analysis. Am J Epidemiol. 186:1000–1009. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
de Luis Cabezón N, Sánchez Castro I,
Bengoetxea Uriarte UX, Rodrigo Casanova MP, García Peña JM and
Aguilera Celorrio L: Acute respiratory distress syndrome: A review
of the Berlin definition. Rev Esp Anestesiol Reanim. 61:319–327.
2014.PubMed/NCBI View Article : Google Scholar : (In Spanish).
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Seger R and Krebs EG: The MAPK signaling
cascade. FASEB J. 9:726–735. 1995.PubMed/NCBI
|
|
26
|
Kaminska B: MAPK signalling pathways as
molecular targets for anti-inflammatory therapy-from molecular
mechanisms to therapeutic benefits. Biochim Biophys Acta.
1754:253–262. 2005.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Monahan LJ: Acute respiratory distress
syndrome. Curr Probl Pediatr Adolesc Health Care. 43:278–284.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Capelozzi VL, Allen TC, Beasley MB, Cagle
PT, Guinee D, Hariri LP, Husain AN, Jain D, Lantuejoul S, Larsen
BT, et al: Molecular and immune biomarkers in acute respiratory
distress syndrome: A perspective from members of the pulmonary
pathology society. Arch Pathol Lab Med. 141:1719–1727.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bekris LM and Leverenz JB: The biomarker
and therapeutic potential of miRNA in Alzheimer's disease.
Neurodegener Dis Manag. 5:61–74. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Qadir MI and Faheem A: miRNA: A diagnostic
and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene
Expr. 27:197–204. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lee H, Abston E, Zhang D, Rai A and Jin Y:
Extracellular vesicle: An emerging mediator of intercellular
crosstalk in lung inflammation and injury. Front Immunol.
9(924)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Huang X, Xiu H, Zhang S and Zhang G: The
Role of Macrophages in the Pathogenesis of ALI/ARDS. Mediators
Inflamm. 2018(1264913)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ye C, Li H, Bao M, Zhuo R, Jiang G and
Wang W: Alveolar macrophage-derived exosomes modulate severity and
outcome of acute lung injury. Aging (Albany NY). 12:6120–6128.
2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ernst O, Vayttaden SJ and Fraser IDC:
Measurement of NF-κB activation in TLR-activated macrophages.
Methods Mol Biol. 1714:67–78. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Napetschnig J and Wu H: Molecular basis of
NF-κB signaling. Annu Rev Biophys. 42:443–468. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sitia R and Rubartelli A: Evolution, role
in inflammation, and redox control of leaderless secretory
proteins. J Biol Chem. 295:7799–7811. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hoesel B and Schmid JA: The complexity of
NF-κB signaling in inflammation and cancer. Mol Cancer.
12(86)2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jarosz M, Olbert M, Wyszogrodzka G,
Młyniec K and Librowski T: Antioxidant and anti-inflammatory
effects of zinc. Zinc-dependent NF-κB signaling.
Inflammopharmacology. 25:11–24. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Blackwell TS and Christman JW: The role of
nuclear factor-kappa B in cytokine gene regulation. Am J Respir
Cell Mol Biol. 17:3–9. 1997.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Németh ZH, Deitch EA, Davidson MT, Szabó
C, Vizi ES and Haskó G: Disruption of the actin cytoskeleton
results in nuclear factor-kappaB activation and inflammatory
mediator production in cultured human intestinal epithelial cells.
J Cell Physiol. 200:71–81. 2004.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chi Q, Wang D, Hu X and Li S and Li S:
Hydrogen sulfide gas exposure induces necroptosis and promotes
inflammation through the MAPK/NF-κB Pathway in Broiler Spleen. Oxid
Med Cell Longev. 2019(8061823)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Benedetti F, Davinelli S, Krishnan S,
Gallo RC, Scapagnini G, Zella D and Curreli S: Sulfur compounds
block MCP-1 production by Mycoplasma fermentans-infected
macrophages through NF-κB inhibition. J Transl Med.
12(145)2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ruff AL and Dillman JF III: Sulfur mustard
induced cytokine production and cell death: Investigating the
potential roles of the p38, p53, and NF-kappaB signaling pathways
with RNA interference. J Biochem Mol Toxicol. 24:155–164.
2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kawai T and Akira S: Signaling to
NF-kappaB by Toll-like receptors. Trends Mol Med. 13:460–469.
2007.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Pfeiffer D, Roßmanith E, Lang I and
Falkenhagen D: miR-146a, miR-146b, and miR-155 increase expression
of IL-6 and IL-8 and support HSP10 in an In vitro sepsis model.
PLoS One. 12(e0179850)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Li Y, Zhu H, Wei X, Li H, Yu Z, Zhang H
and Liu W: LPS induces HUVEC angiogenesis in vitro through
miR-146a-mediated TGF-β1 inhibition. Am J Transl Res. 9:591–600.
2017.PubMed/NCBI
|
|
48
|
Du L, Chen X, Duan Z, Liu C, Zeng R, Chen
Q and Li M: miR-146a negatively regulates dectin-1-induced
inflammatory responses. Oncotarget. 8:37355–37366. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Shi L, Xu Z, Wu G, Chen X, Huang Y, Wang
Y, Jiang W and Ke B: Up-regulation of miR-146a increases the
sensitivity of non-small cell lung cancer to DDP by downregulating
cyclin J. BMC Cancer. 17(138)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yuwen DL, Sheng BB, Liu J, Wenyu W and Shu
YQ: miR-146a-5p level in serum exosomes predicts therapeutic effect
of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol
Sci. 21:2650–2658. 2017.PubMed/NCBI
|
|
51
|
Gan L, Sun T, Li B, Tian J, Zhang J, Chen
X, Zhong J, Yang X and Li Q: Serum miR-146a and miR-150 as
potential new biomarkers for hip Fracture-induced acute lung
injury. Mediators Inflamm. 2018(8101359)2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Freem LJ, Escot S, Tannahill D,
Druckenbrod NR, Thapar N and Burns AJ: The intrinsic innervation of
the lung is derived from neural crest cells as shown by optical
projection tomography in Wnt1-Cre;YFP reporter mice. J Anat.
217:651–664. 2010.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhao Y, Haney MJ, Gupta R, Bohnsack JP, He
Z, Kabanov AV and Batrakova EV: GDNF-transfected macrophages
produce potent neuroprotective effects in Parkinson's disease mouse
model. PLoS One. 9(e106867)2014.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Huang S, Yang L, An Y, Ma X, Zhang C, Xie
G, Chen ZY, Xie J and Zhang H: Expression of hedgehog signaling
molecules in lung cancer. Acta Histochem. 113:564–569.
2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Jiang Y, Han Y, Sun C, Han C, Han N, Zhi W
and Qiao Q: Rab23 is overexpressed in human bladder cancer and
promotes cancer cell proliferation and invasion. Tumour Biol.
37:8131–8138. 2016.PubMed/NCBI View Article : Google Scholar
|