Introduction
Psoriasis is a relatively common chronic
inflammatory skin disease that adversely affects the lives of the
patients (1). Psoriasis affects
2-3% of the population worldwide (2,3).
Previous research indicates that psoriasis is associated with a
number of complications, including psoriatic arthritis,
cardiovascular diseases, Crohn's disease, anxiety and depression,
with the occurrence of these diseases increasing the mortality risk
(4,5). It has been reported that certain
therapeutic strategies may be used to treat psoriasis with good
results, including local therapy, physical therapy, systemic
therapy and new biological agents (such as TNF-α, IL-12/23 and
IL-17 inhibitors), but the recurrence rate of psoriasis remains
high (6-8).
Therefore, identifying a novel therapeutic strategy for psoriasis
would be beneficial. It was previously demonstrated that IL-22
regulates human keratinocyte proliferation and increases IL-22
expression in the serum and skin lesions of patients with psoriasis
(9). Therefore, IL-22 may play a
key role in the development of psoriasis (10). In addition, IL-22-induced skin
inflammation was reported in a mouse model, further suggesting that
IL-22 is associated with psoriasis (10).
Integrated genomic and transcriptome sequencing
results have demonstrated that >90% of DNA sequences are
actively transcribed, 98% of which are transcribed into non-coding
RNA (ncRNA), including microRNAs (miRNAs/miRs) and long ncRNAs
(lncRNAs) (11,12). lncRNAs are transcripts that are
>200 nucleotides in length with limited potential for protein
coding (13). lncRNAs regulate gene
expression at different levels and participate in a variety of
biological processes (14,15). Previous research has indicated that
lncRNA may be abnormally expressed in mammalian and plant cells
(16,17). An increasing number of studies have
demonstrated that lncRNAs may be used as biomarkers for diagnosing
and predicting different types of cancer (18-23).
Additional studies have reported that the abnormal expression of
lncRNAs is closely associated with the occurrence and development
of psoriasis (24,25). Jia et al (26) demonstrated that the lncRNA MEG3
affects the proliferation and apoptosis of psoriatic epidermal
cells via targeting miR-21/caspase-8. Gao et al (27) reported that the lncRNA MIR31HG may
be a potential diagnostic biomarker and therapeutic target for
psoriasis. Qiao et al (28)
reported that the lncRNA msh homeobox 2 pseudogene 1 may
participate in the occurrence of psoriasis. Previous research also
indicated that the stress-induced lncRNA
psoriasis-susceptibility-related RNA gene may play a key role in
psoriasis (29). Tang et al
(30) demonstrated that the lncRNA
homeobox transcript antisense RNA (HOTAIR) acts as an oncogene in
human cancer. Moreover, Yang et al (31) indicated that HOTAIR enhanced liver
cancer cell proliferation via promotion of
epithelial-to-mesenchymal transition. A recent study indicated the
presence of an association between a genomic variant within HOTAIR
and the risk of psoriasis (32).
However, the specific role and related mechanisms of HOTAIR in
psoriasis remain to be further elucidated.
Numerous studies have reported that the expression
of multiple miRNAs, such as miR-125b, miR-155 and miR-26b, is
associated with the progression of psoriasis (33-35).
It has also been demonstrated that miR-126 is highly expressed in
the tissues of patients with psoriasis, but with lower plasma
expression levels. Furthermore, miR-126 expression in the plasma of
patients was found to be inversely correlated with the risk and
severity of psoriasis. In addition, a mutual binding site between
lncRNA HOTAIR and miR-126 has been identified (36,37).
Therefore, it may be hypothesized that the lncRNA HOTAIR is
involved in the occurrence and development of psoriasis through
regulation of miR-126 expression.
The aim of the present study was to explore whether
HOTAIR was involved in psoriasis through modulation of miR-126
expression, so as to provide more strategies and a theoretical
basis for the treatment of psoriasis.
Materials and methods
Cell culture and transfection
Human HaCaT keratinocytes (CLS Cell Lines Service
GmbH) were cultured in DMEM (Hyclone; Cytiva) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin (Beyotime Institute of Biotechnology) at
37˚C in a 5% CO2 incubator. For overexpression of
HOTAIR, full-length HOTAIR was amplified by PCR (primers:
5'-GACTCGCCTGTGCTCTGGAGCT-3' and 5'-TTGAA AATGCATCCAGATTTTT-3') and
then cloned into the multiple cloning site of the pcDNA3.1 vector
(Invitrogen; Thermo Fisher Scientific, Inc.) to construct the
HOTAIR-plasmid. The empty pcDNA3.1 vector (Invitrogen; Thermo
Fisher Scientific, Inc.) was used as the control-plasmid.
Subsequently, HaCaT cells (5x104 cells per well; 6-well
plate) were transfected with 100 ng control-plasmid, 100 ng
HOTAIR-plasmid, 100 nM mimic control (sense: 5'-UUCUCCGAACGUGU
CACGUTT-3'; anti-sense: 5'-ACGUGACACGUUCGGAGA ATT-3'; GenePharma),
100 nM miR-126 mimic (sense: 5'-UCG UACCGUGAGUAAUAAUGCG-3';
antisense: 5'-CAUUA UUACUCACGGUACGAUU-3'; GenePharma), 100 ng
HOTAIR-plasmid + 100 nM mimic control or 100 ng HOTAIR-plasmid +
100 nM miR-126 mimic using Polyplus transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). At 24 h
post-transfection, the cells were used for subsequent
experiments.
Psoriasis model establishment
HaCaT cells (5x104 cells per well) were
seeded into a 6-well plate. After reaching 80-90% confluence, the
medium was replaced with serum-free DMEM at 37˚C for 24 h. HaCaT
cells were then treated with 100 ng/ml IL-22 and serum-starved at
37˚C for an additional 24 h (38).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted using TRIzol™ reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. RNA extraction was considered successful when three bands
were observed in the nucleic acid gel. When RNA extraction was
successful, total RNA was reverse-transcribed into cDNA using a
reverse transcription kit (Vazyme Biotech Co., Ltd.) according to
the manufacturer's instructions. Subsequently, qPCR was performed
using SYBR Green PCR kit (Vazyme Biotech Co., Ltd.). The
thermocycling conditions were as follows: Initial denaturation for
5 min at 95˚C; followed by 38 cycles of denaturation at 94˚C for 1
min, annealing at 60˚C for 1 min and extension at 72˚C for 1 min,
followed by a final extension step at 72˚C for 10 min. mRNA and
miRNA expression levels were quantified using the 2-ΔΔCq
method (39) and normalized to the
internal reference genes GAPDH and U6, respectively. All samples
were assessed in triplicate and all experiments were repeated three
times. The primer sequences used were as follows: GAPDH, forward
5'-ATTCCATGGCA CCGTCAAGGCTGA-3' and reverse 5'-TTCTCCATGGTG
GTGAAGACGCCA-3'; U6, forward 5'-GCTTCGGCA GCACATATACTAAAAT-3' and
reverse 5'-CGCTTCACGAA TTTGCGTGTCAT-3'; HOTAIR, forward
5'-CAGTGGGGA ACTCTGACTCG-3' and reverse 5'-GTGCCTGGTGCTGTC
TTACC-3'; and miR-126 forward 5'-GCTGTCAGT TTGTCAA ATA-3' and
reverse 5'-GTGCAGGGTCCGAGGT-3'.
Western blotting
Total protein was extracted from cells using RIPA
buffer (Beyotime Institute of Biotechnology) and quantified using a
bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific,
Inc.). Equal amounts of proteins (40 µg per lane) were separated
via 12% SDS-PAGE for 40 min and transferred to PVDF membranes (EMD
Millipore). The membranes were blocked for 1.5 h at room
temperature with 5% non-fat milk. Subsequently, the membranes were
incubated at 4˚C overnight with the following primary antibodies:
Anti-cleaved caspase-3 (cat no. ab32042; 1:1,000; Abcam),
anti-pro-caspase-3 (cat no. ab32499; 1:1,000; Abcam) and GAPDH (cat
no. ab9485; 1:1,000; Abcam). Following primary incubation, the
membranes were incubated with an anti-rabbit horseradish peroxidase
conjugated IgG secondary antibody (cat no. 7074; 1:2,000; Cell
Signaling Technology, Inc.) for 2 h. Protein bands were visualized
using the enhanced chemiluminescence method (Cytiva). GAPDH was
used as the loading control.
Flow cytometry
Cell apoptosis was assessed using the Annexin
V/propidium iodide (PI) Apoptosis Detection kit (Beyotime Institute
of Biotechnology). Briefly, cells (5x104 cells per well)
were plated in 6-well plates overnight. On the following day, HaCaT
cells were transfected with plasmid or mimic. Subsequently, cells
were directly collected, centrifuged at 1,000 x g at 4˚C for 5 min,
and resuspended in 100 µl FITC-binding buffer. Subsequently, cells
were incubated with 5 µl ready-to-use Annexin V-FITC (BD
Biosciences) and 5 µl PI in the dark at 4˚C for 30 min. Cell
apoptosis was assessed using a BD FACSCalibur flow cytometer (BD
Biosciences). Data were analyzed using CellQuest™ software, version
5.1 (BD Biosciences).
Dual-luciferase reporter assay
The wild-type (WT) or mutant (MUT) 3' untranslated
region (UTR) of HOTAIR was cloned into the pmiRGLO vector (Promega
Corporation). Recombinant plasmids were acquired using the EndoFree
Plasmid Maxi kit (Vazyme Biotech Co., Ltd.). HaCaT cells
(5x104 cells per well) were co-transfected with 100 nM
miR-126 mimic or 100 nM mimic control and 1 ng MUT-3'UTR-HOTAIR or
1 ng WT-3'UTR-HOTAIR at 37˚C for 48 h. Renilla luciferase
pRL-TK vector (Promega Corporation) was used as the control. At 48
h post-transfection, luciferase activity was measured using a
dual-luciferase reporter assay system (Promega Corporation). Firefly
luciferase activity was normalized to Renilla luciferase
activity.
MTT assay
Cell viability was assessed by performing an MTT
assay. Transfected HaCaT cells were treated with 100 ng/ml IL-22 at
37˚C for 24 h. HaCaT cells were plated into a 96-well plate and
incubated at 37˚C for 24, 48 or 72 h. Subsequently, 20 µl MTT (5
mg/ml; Sigma-Aldrich; Merck KGaA) was added into each well at 37˚C
for 4 h. Absorbance was measured using a multifunctional plate
reader (BD Biosciences) at a wavelength of 570 nm. Data are
presented as the mean ± standard deviations of three separate
experiments.
Caspase-3 activity detection
Caspase-3 activity was detected using a caspase-3
activity detection kit (Beyotime Institute of Biotechnology).
Following transfection, HaCaT cells were treated with 100 ng/ml
IL-22 at 37˚C for 24 h. Subsequently, cells were collected by
centrifugation (600 x g for 5 min at 4˚C) and caspase-3 enzyme
activity in cells was measured at a wavelength of 405 nm using a
multifunctional plate reader (BD Biosciences).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 6; GraphPad Software, Inc.). Comparisons
between two groups were analyzed using the unpaired Student's
t-test. Comparisons among multiple groups were analyzed using
one-way ANOVA followed by Tukey's post hoc test. Data are presented
as the mean ± SD from at least three independent experiments.
P<0.05 was considered to indicate a statistically significant
difference.
Results
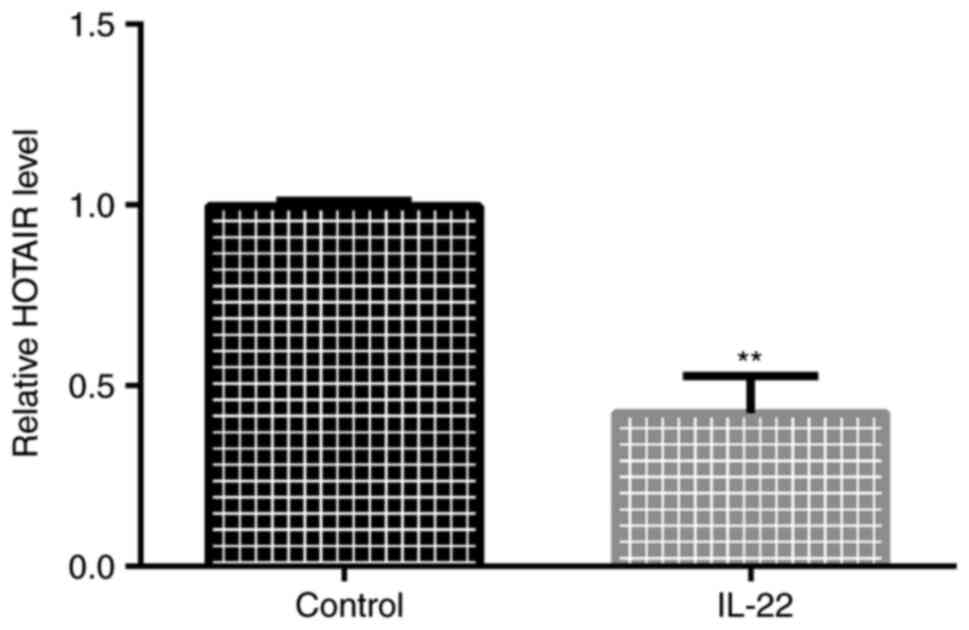
HOTAIR is downregulated in
IL-22-stimulated HaCaT cells
To explore HOTAIR expression in psoriasis, an in
vitro cell model of psoriasis was established. Cells were
starved in serum-free DMEM for 24 h and then stimulated with 100
ng/ml IL-22 in serum-free DMEM for 24 h. The RT-qPCR results
indicated that HOTAIR was downregulated in IL-22-treated HaCaT
cells (Fig. 1).
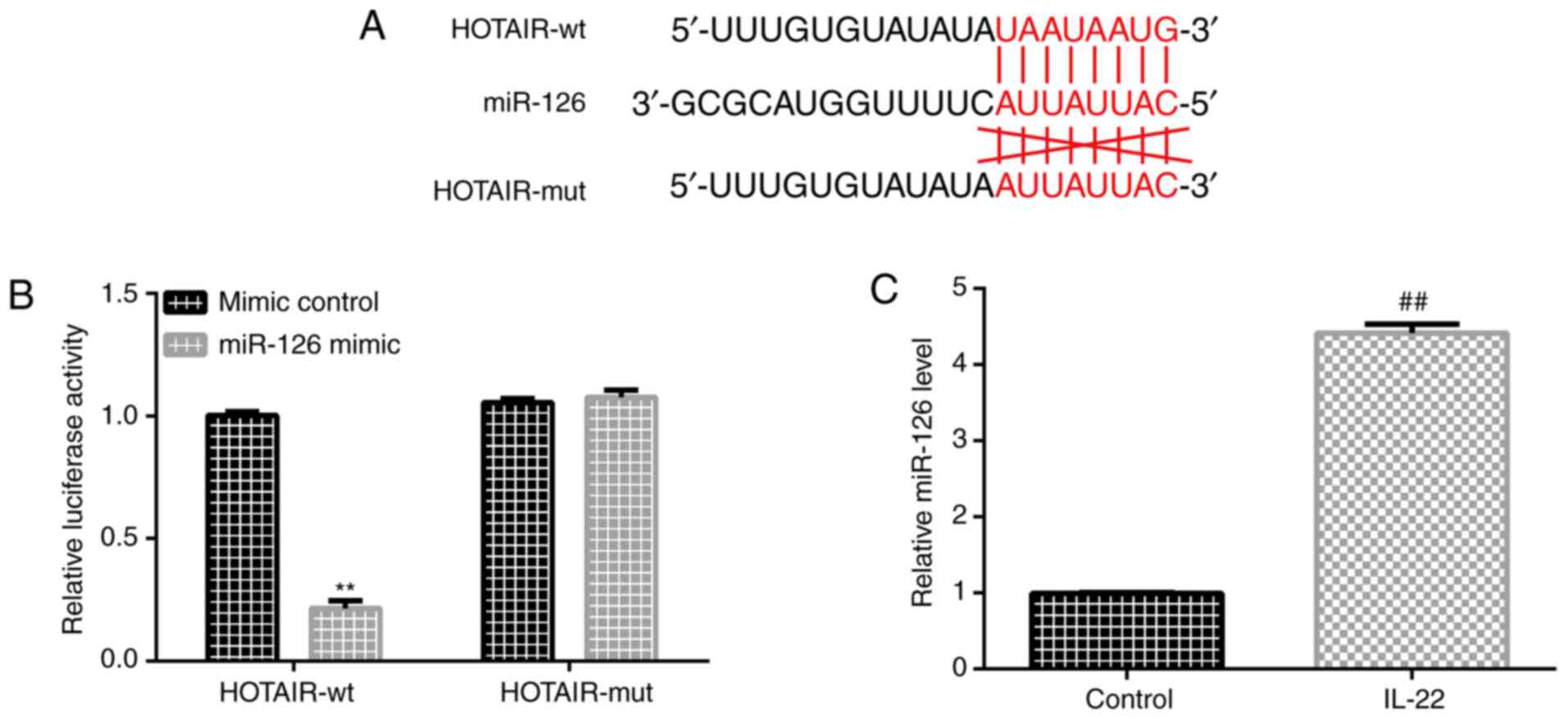
HOTAIR binds to miR-126
Previous studies (36,37)
reported the existence of a mutual binding site between HOTAIR and
miR-126 (Fig. 2A). A luciferase
assay was performed to investigate the interaction between HOTAIR
and miR-126. HaCaT cells were co-transfected with a luciferase
plasmid containing the lncRNA HOTAIR sequence (HOTAIR-WT and
HOTAIR-MUT) and miRNA-126 mimic. The results indicated that miR-126
suppressed the luciferase activity of HOTAIR-WT but did not alter
the luciferase activity of HOTAIR-MUT (Fig. 2B). RT-qPCR analysis was performed to
detect miR-126 expression in HaCaT cells that were treated with
IL-22 and untreated cells. The results indicated that miR-126 was
upregulated in IL-22-stimulated HaCaT cells (Fig. 2C).
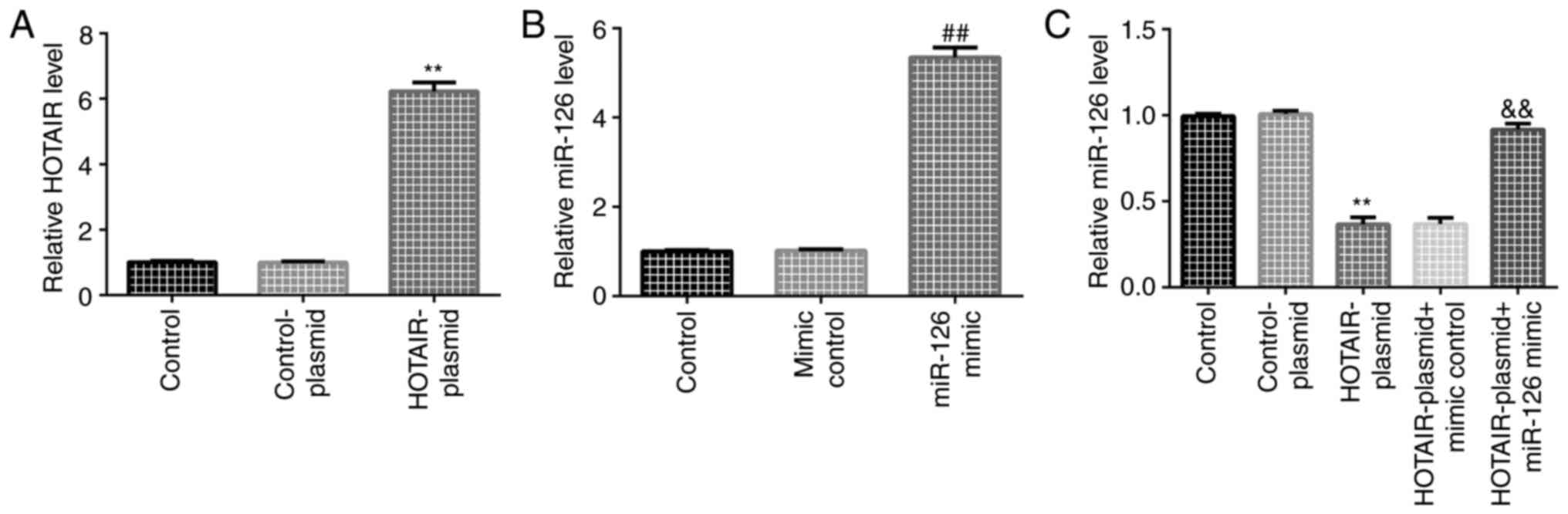
Transfection efficiency of HOTAIR and
miR-126 in HaCaT cells
HaCaT cells were transfected with control-plasmid or
HOTAIR-plasmid. The RT-qPCR results indicated that, compared with
the control-plasmid group, HOTAIR-plasmid increased HOTAIR
expression in HaCaT cells (Fig.
3A). HaCaT cells were transfected with mimic control or miR-126
mimic. The RT-qPCR results indicated that, compared with the mimic
control group, miR-126 mimic increased miR-126 expression in HaCaT
cells (Fig. 3B). HaCaT cells were
also transfected with HOTAIR-plasmid + mimic control or
HOTAIR-plasmid + miR-126 mimic for 24 h. The results suggested that
HOTAIR-plasmid significantly reduced miR-126 expression in HaCaT
cells, which was reversed by miR-126 mimic (Fig. 3C).
HOTAIR suppresses cell proliferation
and induces apoptosis in IL-22-stimulated HaCaT cells by regulating
miR-126 expression
Subsequently, MTT assays and flow cytometry were
performed to detect cell proliferation and apoptosis, respectively.
The MTT assay suggested that, compared with the control group,
HaCaT cell proliferation was increased in the IL-22 group. In
addition, compared with the IL-22 + control-plasmid group,
HOTAIR-plasmid suppressed cell proliferation, which was reversed by
miR-126 mimic (Fig. 4A). The flow
cytometry results indicated that, compared with the control group,
cell apoptosis was decreased in IL-22-induced HaCaT cells (Fig. 4B and C). Furthermore, the caspase-3 activity
assay results indicated that compared with the control group,
caspase-3 activity was significantly decreased in the IL-22 group
(Fig. 4D). The western blotting
results indicated that compared with the control group, cleaved
caspase-3 protein expression and the cleaved caspase-3/caspase-3
ratio was decreased in IL-22-induced HaCaT cells (Fig. 4E and F). Compared with the IL-22 +
control-plasmid group, HOTAIR-plasmid increased cell apoptosis
(Fig. 4B and C), promoted caspase-3 activity (Fig. 4D), increased cleaved caspase-3
protein expression and enhanced the cleaved caspase-3/caspase-3
ratio (Fig. 4E and F), whereas miR-126 mimic reversed these
effects.
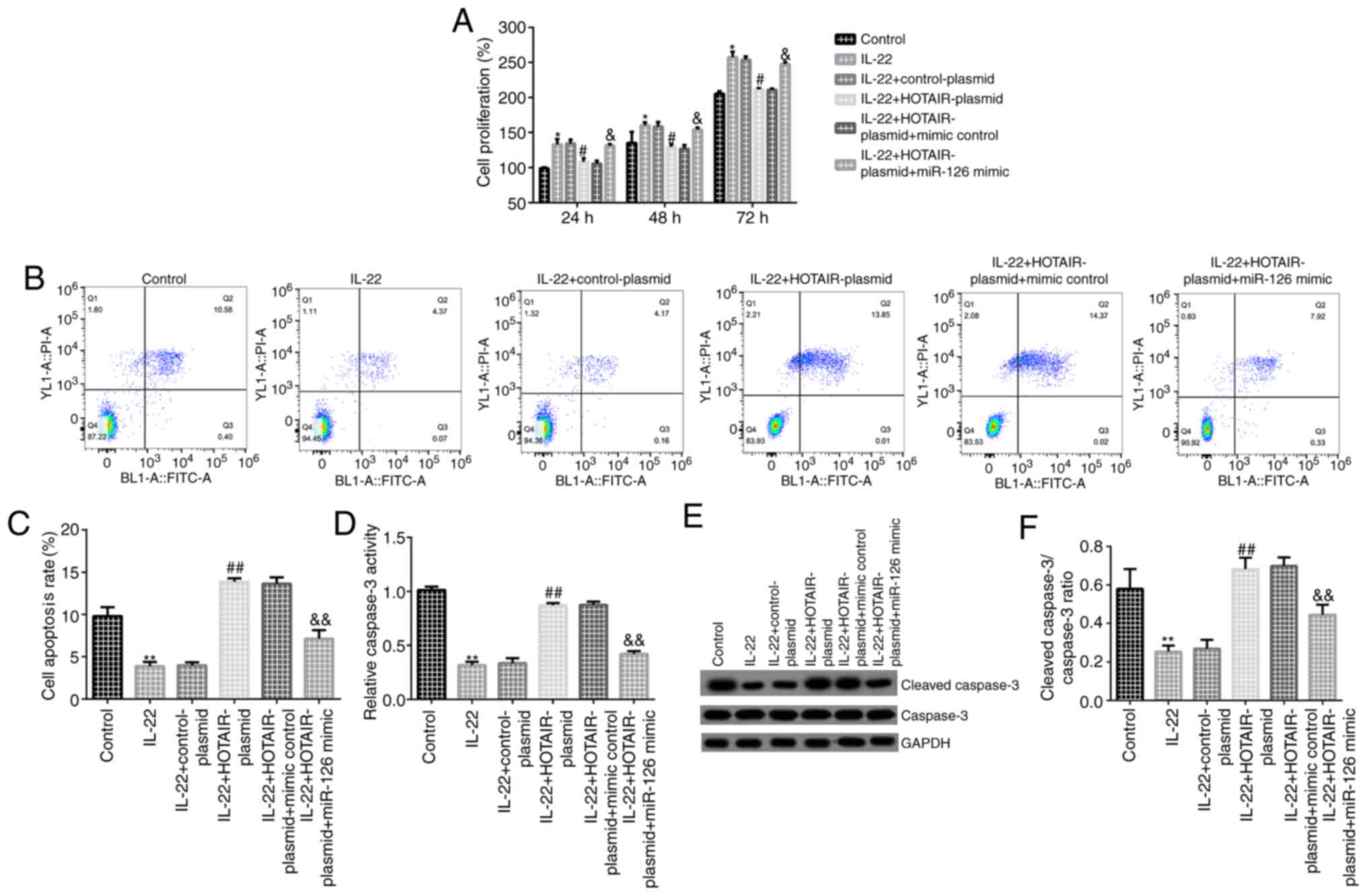 | Figure 4Effects of HOTAIR on IL-22-stimulated
HaCaT cell proliferation and apoptosis. HaCaT cells were
transfected with control-plasmid, HOTAIR-plasmid, HOTAIR-plasmid +
mimic control or HOTAIR-plasmid + miR-126 mimic for 24 h. After 24
h, cells were stimulated with 100 ng/ml IL-22 for a further 24 h.
(A) The MTT assay was conducted to detect cell proliferation at 24,
48 and 72 h. (B) Flow cytometry was performed to detect cell
apoptosis. (C) Cell apoptosis rate. (D) Caspase-3 activity. (E)
Western blotting was performed to measure cleaved caspase-3 and
caspase-3 protein expression levels in different groups. (F)
Cleaved caspase-3/caspase-3 ratio. Data are presented as the mean ±
SD.*P<0.05, **P<0.01 vs. control group;
#P<0.05, ##P<0.01 vs. IL-22 +
control-plasmid group; &P<0.05,
&&P<0.01 vs. IL-22 + HOTAIR-plasmid + mimic
control group.HOTAIR, HOX transcript antisense RNA; miR, microRNA;
IL, interleukin. |
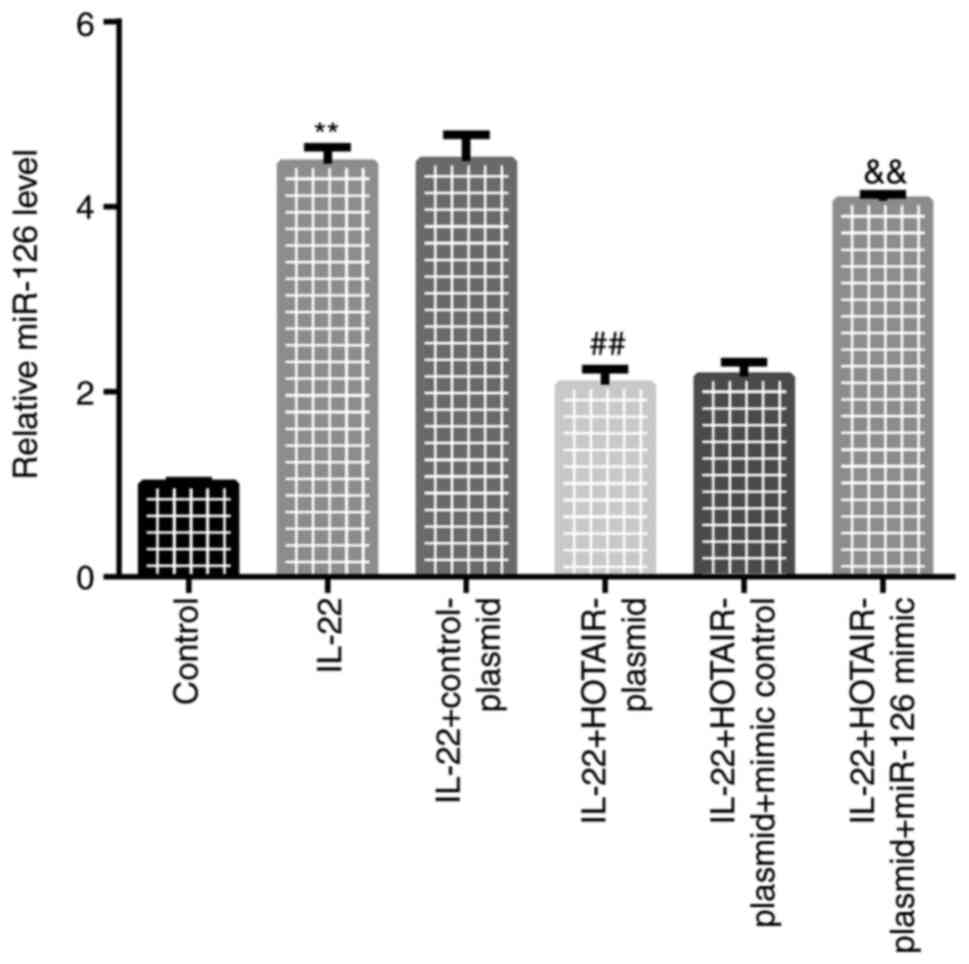
HOTAIR-plasmid reduces miR-126
expression
Finally, RT-qPCR analysis was performed to detect
miR-126 expression. Compared with the control group, miR-126
expression was significantly increased in the IL-22 group. Compared
with the IL-22 + control-plasmid group, HOTAIR-plasmid reduced
miR-126 expression, which was reversed by miR-126 mimic (Fig. 5).
Discussion
lncRNAs have been found to be closely associated
with the occurrence and development of psoriasis (24,25).
The focus of the present study was the lncRNA HOTAIR, which has
been extensively investigated (40). Accumulating evidence suggests that
abnormal expression of HOTAIR is implicated in cancer development,
including lung and gastric cancer, as well as hepatocellular
carcinoma (41-43).
Zhang et al (41) reported
that HOTAIR promoted cell viability, migration and invasion. Gao
et al (43) demonstrated
that reduced HOTAIR expression decreased hepatocellular carcinoma
growth. Liu et al (42)
indicated that HOTAIR overexpression increased cell viability via
miR-331-3p in gastric cancer. Furthermore, a previous study
indicated that HOTAIR participated in psoriasis development
(32). However, the expression and
role of HOTAIR in psoriasis is not completely understood. Psoriasis
is a T-cell-mediated chronic skin condition manifesting clinically
as erythema and scaly skin. The primary pathological process of
psoriasis is excessive keratinocyte proliferation and continuous
skin inflammation (44,45), but the pathogenesis of the condition
has yet to be fully elucidated. Therefore, the aim of the present
study was to investigate the underlying mechanism in order to
identify novel therapeutic targets for psoriasis.
First, the expression of HOTAIR was determined in an
in vitro cell model of psoriasis, which was established by
stimulating HaCaT cells with 100 ng/ml IL-22 for 24 h in serum-free
DMEM. The findings indicated that HOTAIR was downregulated in
IL-22-stimulated HaCaT cells. In addition, Yan et al
(36) demonstrated that HOTAIR had
a binding site with miR-126 in gastric cancer (36). Therefore, a dual-luciferase reporter
assay was performed in the present study to investigate the
interaction between HOTAIR and miR-126. The results also indicated
that there was a mutual binding site between HOTAIR and
miR-126.
miRNAs have been demonstrated to play key roles in
psoriasis (33-35).
Furthermore, miRNAs participate in the development of immune cells,
maintain immune homeostasis and regulate certain immune regulatory
factors (46). miR-126 has been
found to participate in the occurrence and development of multiple
immune diseases, such as rheumatoid arthritis, systemic lupus
erythematosus (SLE), inflammatory bowel disease and psoriasis
(47-50).
Qu et al (47) reported that
miR-126 promoted cell proliferation via the PI3K/AKT signaling
pathway. miR-126 was upregulated in the CD4+ T cells of
patients with SLE and enhanced T-cell autoreactivity to promote SLE
progression. Feng et al (50) demonstrated that miR-126 was
upregulated in patients with psoriasis. Consistent with previous
results, the present study demonstrated that miR-126 was
upregulated in IL-22-stimulated HaCaT cells, and that miR-126
expression was negatively correlated with HOTAIR expression.
Subsequently, the effects of HOTAIR on IL-22-stimulated HaCaT cell
proliferation and apoptosis were investigated. The results
indicated that HOTAIR-plasmid suppressed cell viability and
increased cell apoptosis, and these effects were reversed by
miR-126 mimic. Therefore, the findings of the present study
suggested that HOTAIR/miR-126 may represent promising novel targets
for the treatment of psoriasis.
In conclusion, HOTAIR inhibited IL-22-induced HaCaT
cell proliferation and increased apoptosis by regulating miR-126
expression, indicating that HOTAIR may play a protective role in
psoriasis. However, this was an in vitro basic study of
HOTAIR in psoriasis, and there were certain limitations:
Experiments in vivo were not conducted; the target genes of
miR-126 were not analyzed in depth; in addition, the correlation
between the expression of HOTAIR/miR-126 and clinicopathological
parameters in patients with psoriasis was not analyzed. Despite
these limitations, however, the present study provides new
potential targets for the clinical treatment of psoriasis and a
theoretical basis for the development of psoriasis treatment
strategies. In view of the aforementioned limitations, the role of
HOTAIR/miR-126 in psoriasis will be further investigated in future
studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WZ contributed to study design, data collection,
statistical analysis, data interpretation and manuscript
preparation. BG, SC, JL and YS contributed to data collection and
statistical analysis. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu Q, Wu DH, Han L, Deng JW, Zhou L, He
R, Lu CJ and Mi QS: Roles of microRNAs in psoriasis: Immunological
functions and potential biomarkers. Exp Dermatol. 26:359–367.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Boehncke WH and Schön MP: Psoriasis.
Lancet. 386:983–994. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Parisi R, Symmons DP, Griffiths CE and
Ashcroft DM: Identification and Management of Psoriasis and
Associated ComorbidiTy (IMPACT) project team. Global epidemiology
of psoriasis: A systematic review of incidence and prevalence. J
Invest Dermatol. 133:377–385. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ritchlin CT, Colbert RA and Gladman DD:
Psoriatic arthritis. N Engl J Med. 376:957–970. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liakou AI and Zouboulis CC: Links and
risks associated with psoriasis and metabolic syndrome. Psoriasis
(Auckl). 5:125–128. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Vide J and Magina S: Moderate to severe
psoriasis treatment challenges through the era of biological drugs.
An Bras Dermatol. 92:668–674. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roubille C, Richer V, Starnino T, McCourt
C, McFarlane A, Fleming P, Siu S, Kraft J, Lynde C, Pope J, et al:
The effects of tumour necrosis factor inhibitors, methotrexate,
non-steroidal anti-inflammatory drugs and corticosteroids on
cardiovascular events in rheumatoid arthritis, psoriasis and
psoriatic arthritis: A systematic review and meta-analysis. Ann
Rheum Dis. 74:480–489. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Geller S, Xu H, Lebwohl M, Nardone B,
Lacouture ME and Kheterpal M: Malignancy risk and recurrence with
psoriasis and its treatments: A concise update. Am J Clin Dermatol.
19:363–375. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Boniface K, Bernard FX, Garcia M, Gurney
AL, Lecron JC and Morel F: IL-22 inhibits epidermal differentiation
and induces proinflammatory gene expression and migration of human
keratinocytes. J Immunol. 174:3695–3702. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wolk K, Witte E, Wallace E, Döcke WD, Kunz
S, Asadullah K, Volk HD, Sterry W and Sabat R: IL-22 regulates the
expression of genes responsible for antimicrobial defense, cellular
differentiation, and mobility in keratinocytes: A potential role in
psoriasis. Eur J Immunol. 36:1309–1323. 2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Amaral PP, Dinger ME, Mercer TR and
Mattick JS: The eukaryotic genome as an RNA machine. Science.
319:1787–1789. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181.
2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lekka E and Hall J: Noncoding RNAs in
disease. FEBS Lett. 592:2884–2900. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu Y, Ferguson JF, Xue C, Ballantyne RL,
Silverman IM, Gosai SJ, Serfecz J, Morley MP, Gregory BD, Li M, et
al: Tissue-specific RNA-Seq in human evoked inflammation identifies
blood and adipose LincRNA signatures of cardiometabolic diseases.
Arterioscler Thromb Vasc Biol. 34:902–912. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu YW, Sun M, Xia R, Zhang EB, Liu XH,
Zhang ZH, Xu TP, De W, Liu BR and Wang ZX: LincHOTAIR
epigenetically silences miR34a by binding to PRC2 to promote the
epithelial-to-mesenchymal transition in human gastric cancer. Cell
Death Dis. 6(e1802)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chang S, Chen B, Wang X, Wu K and Sun Y:
Long non-coding RNA XIST regulates PTEN expression by sponging
miR-181a and promotes hepatocellular carcinoma progression. BMC
Cancer. 17(248)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA linc00673
regulated non-small cell lung cancer proliferation, migration,
invasion and epithelial mesenchymal transition by sponging
miR-150-5p. Mol Cancer. 16(118)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zheng J, Huang X, Tan W, Yu D, Du Z, Chang
J, Wei L, Han Y, Wang C, Che X, et al: Pancreatic cancer risk
variant in LINC00673 creates a miR-1231 binding site and interferes
with PTPN11 degradation. Nat Genet. 48:747–757. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Sha M, Lin M, Wang J, Ye J, Xu J, Xu N and
Huang J: Long non-coding RNA MIAT promotes gastric cancer growth
and metastasis through regulation of miR-141/DDX5 pathway. J Exp
Clin Cancer Res. 37(58)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J,
Chen Z and He J: Long non-coding RNA NKILA inhibits migration and
invasion of non-small cell lung cancer via NF-κB/Snail pathway. J
Exp Clin Cancer Res. 36(54)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yan J, Song J, Qiao M, Zhao X, Li R, Jiao
J and Sun Q: Long noncoding RNA expression profile and functional
analysis in psoriasis. Mol Med Rep. 19:3421–3430. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ahn R, Gupta R, Lai K, Chopra N, Arron ST
and Liao W: Network analysis of psoriasis reveals biological
pathways and roles for coding and long non-coding RNAs. BMC
Genomics. 17(841)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jia HY, Zhang K, Lu WJ, Xu GW, Zhang JF
and Tang ZL: lncRNA MEG3 influences the proliferation and apoptosis
of psoriasis epidermal cells by targeting miR-21/caspase-8. BMC Mol
Cell Biol. 20(46)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Gao J, Chen F, Hua M, Guo J, Nong Y, Tang
Q, Zhong F and Qin L: Knockdown of lncRNA MIR31HG inhibits cell
proliferation in human HaCaT keratinocytes. Biol Res.
51(30)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Qiao M, Li R, Zhao X, Yan J and Sun Q:
Up-regulated lncRNA-MSX2P1 promotes the growth of IL-22-stimulated
keratinocytes by inhibiting miR-6731-5p and activating S100A7. Exp
Cell Res. 363:243–254. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Széll M, Danis J, Bata-Csörgő Z and Kemény
L: PRINS, a primate-specific long non-coding RNA, plays a role in
the keratinocyte stress response and psoriasis pathogenesis.
Pflugers Arch. 468:935–943. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tang Q and Hann SS: HOTAIR: An oncogenic
long non-coding RNA in human cancer. Cell Physiol Biochem.
47:893–913. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yang T, He X, Chen A, Tan K and Du X:
lncRNA HOTAIR contributes to the malignancy of hepatocellular
carcinoma by enhancing epithelial-mesenchymal transition via
sponging miR-23b-3p from ZEB1. Gene. 670:114–122. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Rakhshan A, Zarrinpour N, Moradi A, Ahadi
M, Omrani MD, Ghafouri-Fard S and Taheri M: A single nucleotide
polymorphism within HOX Transcript Antisense RNA (HOTAIR) is
associated with risk of psoriasis. Int J Immunogenet. 47:430–434.
2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xu N, Brodin P, Wei T, Meisgen F, Eidsmo
L, Nagy N, Kemeny L, Ståhle M, Sonkoly E and Pivarcsi A: miR-125b,
a microRNA downregulated in psoriasis, modulates keratinocyte
proliferation by targeting FGFR2. J Invest Dermatol. 131:1521–1529.
2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Xu L and Leng H, Shi X, Ji J, Fu J and
Leng H: miR-155 promotes cell proliferation and inhibits apoptosis
by PTEN signaling pathway in the psoriasis. Biomed Pharmacother.
90:524–530. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cheung L, Fisher RM, Kuzmina N, Li D, Li
X, Werngren O, Blomqvist L, Ståhle M and Landén NX: Psoriasis skin
inflammation-induced microRNA-26b targets NCEH1 in underlying
subcutaneous adipose tissue. J Invest Dermatol. 136:640–648.
2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yan J, Dang Y, Liu S, Zhang Y and Zhang G:
lncRNA HOTAIR promotes cisplatin resistance in gastric cancer by
targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol.
37(30)2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jiang B, Tang Y, Wang H, Chen C, Yu W, Sun
H, Duan M, Lin X and Liang P: Down-regulation of long non-coding
RNA HOTAIR promotes angiogenesis via regulating miR-126/SCEL
pathways in burn wound healing. Cell Death Dis.
11(61)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang R, Wang FF, Cao HW and Yang JY:
miR-223 regulates proliferation and apoptosis of IL-22-stimulated
HaCat human keratinocyte cell lines via the PTEN/Akt pathway. Life
Sci. 230:28–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Δ Δ C(T)) Method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang C, Xu L, Deng G, Ding Y, Bi K, Jin
H, Shu J, Yang J, Deng H, Wang Z, et al: Exosomal HOTAIR promotes
proliferation, migration and invasion of lung cancer by sponging
miR-203. Sci China Life Sci. 63:1265–1268. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer.
13(92)2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Gao JZ, Li J, Du JL and Li XL: Long
non-coding RNA HOTAIR is a marker for hepatocellular carcinoma
progression and tumor recurrence. Oncol Lett. 11:1791–1798.
2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lowes MA, Suárez-Fariñas M and Krueger JG:
Immunology of psoriasis. Annu Rev Immunol. 32:227–255.
2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Jadali Z and Eslami MB: T cell immune
responses in psoriasis. Iran J Allergy Asthma Immunol. 13:220–230.
2014.PubMed/NCBI
|
|
46
|
Soltanzadeh-Yamchi M, Shahbazi M, Aslani S
and Mohammadnia-Afrouzi M: MicroRNA signature of regulatory T cells
in health and autoimmunity. Biomed Pharmacother. 100:316–323.
2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Qu Y, Wu J, Deng JX, Zhang YP, Liang WY,
Jiang ZL, Yu QH and Li J: MicroRNA-126 affects rheumatoid arthritis
synovial fibroblast proliferation and apoptosis by targeting PIK3R2
and regulating PI3K-AKT signal pathway. Oncotarget. 7:74217–74226.
2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Liang Y, Zhao S, Liang G, Zhao M and Lu Q:
DNA methylation status of miR-126 and its host gene EGFL7 in
CD4+ T cells from patients with systemic lupus
erythematosus. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 38:793–797.
2013.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
49
|
Thorlacius-Ussing G, Schnack Nielsen B,
Andersen V, Holmstrøm K and Pedersen AE: Expression and
localization of miR-21 and miR-126 in mucosal tissue from patients
with inflammatory bowel disease. Inflamm Bowel Dis. 23:739–752.
2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Feng SK, Wang L, Liu W, Zhong Y and Xu SJ:
miR-126 correlates with increased disease severity and promotes
keratinocytes proliferation and inflammation while suppresses
cells' apoptosis in psoriasis. J Clin Lab Anal.
32(e22588)2018.PubMed/NCBI View Article : Google Scholar
|