Introduction
Type 2 difabetes (T2D) is considered a major
contributor to cardiovascular disease (CVD), which is the most
common cause of T2D-associated death worldwide (1). Diabetic cardiomyopathy, a specific
form of CVD, is facilitated by resistance to insulin metabolism in
cardiac tissues, compensatory hyperinsulinemia and the progression
of hyperglycemia (2). Considering
the increasing population of diagnosed, undiagnosed and
pre-diagnosed patients with diabetes (3), there is an imperative demand to
determine an effective therapy for diabetic cardiomyopathy. Damaged
electron transport and hyperglycemia partly result in the increased
production of reactive oxygen species (ROS), leading to
mitochondrial dysfunction and DNA damage, and consequently causing
the development of CVD (4).
Imbalance between ROS production and elimination exacerbates
oxidative stress (OS) and inflammation (5). OS is hypothesized to be a major
damaging factor for insulin resistance, impairment of insulin
secretion from β-cells in pancreatic islets and T2D pathogenesis
(including subsequent complications) by inducing several causative
factors, including the release of proinflammatory cytokines and the
generation of ROS and reactive nitrogen species (6). Additionally, inflammation has been
regarded as the critical regulator of atrial fibrillation in
obesity and diabetes (4),
indicating the role of the inflammatory response in diabetic
cardiomyopathy. However, the mechanism underlying the regulation of
OS and inflammation in cardiomyocytes remain undetermined.
For years, microRNAs (miRs), which regulate gene
expression transcriptionally or post-transcriptionally, have been
considered to be novel antioxidants due to their potent regulatory
effects on OS (7). For instance,
miR-223-3p protects against hypoxia-induced cardiomyocyte apoptosis
and OS by targeting Kruppel-like factor 15(8). Previous data has demonstrated that
miR-145 serves an important role in the regulation of
cardiomyocytes. Geniposide protects H9c2 cells against
lipopolysaccharide (LPS)-induced apoptosis and inflammation by
regulating miR-145 expression and the MEK/ERK signaling pathway
(9). In human aortic endothelial
cells, TNF-α treatment markedly induced the release of inflammatory
cytokines, including monocyte chemoattractant protein-1 (MCP-1),
IL-6 and IL-8, whilst decreasing the expression of miR-145(10). Moreover, He et al (11) demonstrated that LPS treatment
markedly elevated TNF-α levels, which strongly inhibited miR-145
expression under hyperglycemic conditions. Yuan et al
(12) reported that the
introduction of miR-145-5p is effective in suppressing the
production of inflammatory factors, including IL-1β, TNF-α and IL-6
in hypoxia-induced cardiomyocytes. Additionally, an increase in
miR-145 ameliorated OS in retinal endothelial cells under
hyperglycemic conditions by suppressing ROS generation and
malondialdehyde (MDA) activity, and increasing superoxide dismutase
(SOD) activity (13). All these
results indicated the pivotal role of miR-145 in suppressing
inflammation and OS. However, the mechanism of miR-145 on high
glucose (HG)-induced OS and the inflammatory response in
cardiomyocytes remains to be elucidated.
ADP ribosylation factor 6 (ARF6) is a member of the
Ras superfamily of GTP-bound proteins that serves a specific role
in the membrane translocation of protein kinase C βI in glomerular
mesangial cells under high-glucose conditions (14). The function of ARF6 is determined by
the duration of the GTP-bound active state, which is activated by
GTPase-activating protein and GTP-GDP exchange factor (15). In HeLa and H9c2 cells, ARF6 is
involved in clathrin-independent endocytosis of the human
ether-a-go-go-related gene (16).
Notably, miR-145 has been reported to improve macrophage-mediated
inflammation by targeting ARF6(17). The current study used the online
database StarBase and dual luciferase reporter assays to evaluate
the association between miR-145 and ARF6.
The present study aimed to investigate the mechanism
via which miR-145 regulates the inflammatory response and OS injury
in cardiomyocytes exposed to HG. The results of the current study
demonstrated that the overexpression of miR-145 mitigated the
inflammatory response and OS injury induced by HG treatment in
cardiomyocytes. Additionally, the results confirmed that miR-145
exerts its protective role in cardiomyocytes by negatively
targeting ARF6. These findings indicated the potential role of
miR-145 in the treatment of diabetic cardiomyopathy.
Materials and methods
Cell culture
H9c2 cardiomyocytes were obtained from the American
Type Culture Collection and cultured in complete DMEM
(MilliporeSigma) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin.
The cells were placed in an incubator at 5% CO2 and
37˚C.
Cell transfection
H9c2 cells in the log phase were treated with a high
concentration of glucose (33 mmol/l; HG group) or co-treated with
mannitol (24.5 mmol/l) and glucose (5.5 mmol/l) as the control
(hypertonic group) for 2 h at 37˚C. H9c2 cells in the normal
glucose group were treated with a normal concentration of glucose
(5.5 mmol/l; NG group) for 2 h at 37˚C. Following this, cells were
transfected with pcDNA3.1-ARF6 (2 µg), short hairpin (sh)-ARF6 (2
µg), miR-145 mimics (100 nM) or the corresponding negative controls
(NCs; pcDNA3.1, sh-NC or mimic NC) using the
Lipofectamine® 2000 kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. All transfected materials
were obtained from Guangzhou RiboBio Co., Ltd. The sequence of
miR-145 mimic was 5'-GUCCAGUUUUCCCAGGAAUCCCU-3' and that of NC
mimic was 5'-UUCUCCGAACGUGUCACGUUU-3'. The cells were accordingly
grouped into the NG, hypertonic, HG, HG + mimic NC, HG + miR-145
mimic, HG + pcDNA3.1-ARF6, HG + sh-NC, HG + sh-ARF6, HG + control
and HG + miR-145 mimic + pcDNA3.1-ARF6 groups. Cells in the control
group were transfected with pcDNA3.1 and sh-NC (1:1; total mass, 2
µg). H9c2 cells in the blank group did not receive any treatment.
The transfected cells were incubated in serum-free DMEM and
subsequently incubated with 5% CO2 at 37˚C for 48 h
before later use.
Reverse transcription-quantitative PCR
(RT-qPCR)
Following transfection, H9c2 cells were lysed in 1
ml TRIzol® (Thermo Fisher Scientific, Inc.). RNA was
extracted in accordance with the instructions of the TRIzol
reagent® and reverse transcribed into cDNA using
PrimeScript RT reagent kit (Takara Bio, Inc.). The reverse
transcription was performed as follows: 50˚C for 15 min, 85˚C for 5
sec and preservation at 4˚C. The PCR reaction conditions and
reaction system were performed using SYBR® Premix Ex
Taq™ II (Takara Bio, Inc.) kit, according to the manufacturer's
protocol. An ABI 7500 instrument (AB-4351107; Applied Biosystems;
Thermo Fisher Scientific, Inc.) was used for RT-qPCR. The
thermocycling conditions were as follows: Pre-denaturation at 95˚C
for 10 min, followed by 40 cycles of denaturation for 10 sec at
95˚C, annealing at 60˚C for 20 sec and extension at 72˚C for 34
sec. Following this, the expression levels of miR-145 and ARF6 were
assessed. All primers for RT-qPCR (Table I) were synthesized by Genewiz, Inc.
Relative mRNA and miR expression levels were normalized to GAPDH
and U6 levels, respectively. All numerical data were analyzed using
the 2-ΔΔCq method (18).
ΔΔCt=[Ct(target gene)-Ct(internal gene)] experimental
group-[Ct(target gene)-Ct(internal gene)] control group.
 | Table IPrimer sequences of the genes. |
Table I
Primer sequences of the genes.
| Primer | Forward sequence
(5'-3') | Reverse sequence
(5'-3') |
|---|
| miR-145 |
ATCGTCCAGTTTTCCCAGG |
CGCCTCCACACACTCACC |
| ARF6 |
AGCCGATTCACCATCTTCTATAAC |
CAGCATCCTAAACCGCATACC |
| GAPDH |
ACCACAGTCCATGCCATCAC |
TCCACCACCCTGTTGCTGTA |
| U6 |
TCGCTTCGGCAGCACATATAC |
GCGTGTCATCCTTGCGCAG |
Western blotting
H9c2 cells were washed with precooled PBS three
times at 48 h post-transfection. Cell lysis buffer [50 mM Tris (pH
7.4), 150 mM NaCl and 1% NP-40, 0.1% SDS; 100 µl/50 ml] was added
for protein extraction. The lysed cells were placed on ice for 30
min prior to centrifugation at 13,040 x g and 4˚C for 10 min. The
supernatant was placed in centrifugation tubes (0.5 ml) and
maintained at -20˚C or quantified using a BCA kit (Beyotime
Institute of Biotechnology). Following quantification, the proteins
(30 µg) were subjected to denaturation with 6x SDS loading buffer
at 100˚C and separation by 10% SDS-PAGE electrophoresis. The
proteins were then transferred onto PVDF membranes with 4˚C
precooled transfer buffer for 1.5 h and blocked with 5% skim milk
powder dissolved in TBST for 1 h at room temperature. The membranes
were incubated with TBST containing primary antibodies against ARF6
(cat. no. ab77581; 1:1,000; Abcam) and β-actin (cat. no. 4970S;
1:1,000; Cell Signaling Technology, Inc.) at 4˚C overnight.
Following this, membranes were washed three times with TBST (10
min/wash). Goat anti-rabbit IgG antibodies (cat. no. CW0103S,
1:5,000; Beijing ComWin Biotech Co., Ltd.) were added and incubated
at room temperature for 2 h. After washing with TBST, the membranes
were developed using ECL reagent (Thermo Fisher Scientific, Inc.)
and the protein expression levels were detected. To quantify the
protein expression, the X-ray films were scanned and analyzed with
ImageJ v1.48 software (National Institutes of Health). The
experiment was performed in triplicate.
ELISA
Concentrations of IL-6, TNF-α and MCP-1 in the
supernatant of H9c2 cells were assessed using Rat IL-6 ELISA Kit
(cat. no. ab234570), Rat TNF-α ELISA Kit (cat. no. ab236712) and
Rat MCP-1 ELISA Kit (cat. no. ab219045; all from Abcam), according
to the manufacturer's protocol.
Lactate dehydrogenase (LDH)
An LDH kit (Beyotime Institute of Biotechnology) was
used to determine the release of LDH in H9c2 cells. The supernatant
of H9c2 cells (50 µl) was collected from each well and incubated
for 30 min with reduced nicotinamide adenine dinucleotide and
pyruvic acid at 37˚C. Following ~15 min, 0.4 mol/l NaOH was added
to stop the reaction. The absorbance values of the samples were
measured at 440 nm using a microplate reader to calculate LDH
activity. The absorbance of uncultured cells was detected as the
background absorbance value. The maximum LDH content was obtained
after the cells were treated with 1% Triton X-100 (MilliporeSigma)
for 60 min at room temperature.
Detection of ROS
The production of ROS was detected with a
fluorescence probe redox-sensitive-fluoroprobe;
2',7'-dichlorofluorescein-diacetate (DCFDA; cat. no. 287810;
MilliporeSigma) using a ROS detection kit (cat. no. 88-5930-74;
Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. DCFDA was diluted with DMEM (1:1,000)
to a final concentration of 10 µM. After the medium was removed,
diluted DCFDA was added until H9c2 cells were fully covered. The
cells were then cultured in an incubator with 0.5% CO2
at 37˚C in the dark for 1 h and washed with PBS three times to
remove excess DCFDA. Fluorescence intensity was monitored in real
time with a laser confocal microscope (magnification, x400; FV1000;
Olympus Corporation) at an excitation wavelength of 480 nm and
emission wavelength of 530 nm. The fluorescence intensity was
measured as follows: λex=480/λem=530 nm.
Detection of MDA
An MDA kit (cat. no. S0131M; Beyotime Institute of
Biotechnology) was used for the detection of MDA content in H9c2
cells according to the manufacturer's protocol. The absorbance of
the samples was detected at 532 nm using a microplate and the
content of MDA in the samples was calculated according to the
standard curve.
Antioxidant enzyme activities
The activities of SOD, catalase (CAT) and
glutathione peroxidase (GPx) in H9c2 cells were detected using
commercial detection kits (cat. nos. 706002, 707002 and 703102,
respectively; Cayman Chemical Company), according to the
manufacturer's protocol. A BCA kit (Beyotime Institute of
Biotechnology) was used to determine the concentration of protein
in cells. The enzyme activity was expressed based on the content of
the original enzymes.
Flow cytometry (FCM)
Following transfection, H9c2 cells were washed twice
with PBS and digested with pancreatin. Following this, the cells
were subjected to centrifugation at 1,200 x g for 5 min at room
temperature and the supernatant was removed. Subsequently, the
cells were resuspended in PBS and centrifuged at 1,200 x g for 5
min at room temperature. After the supernatant was removed, the
cells (105-106/ml) were resuspended in 490 µl
of precooled 1x binding buffer. Annexin V-FITC (5 µl) and propidium
iodide (5 µl) (cat. no. ab14085; Abcam) were added to the cell
suspension and mixed. Following 10 min of incubation on ice, cell
apoptosis was detected by BD FACSVerse™ Flow Cytometer (BD
Biosciences), and the data were analyzed using FlowJo vX10 software
(FlowJo LLC).
Dual luciferase reporter assay
StarBase (http://starbase.sysu.edu.cn/) predicted the potential
binding sites between miR-145 and ARF6. According to the
prediction, the wild-type (WT) and mutant (MT) sequences of the
binding sites between miR-145 and ARF6 were designed separately by
Genewiz, Inc. The WT or MT sequence was inserted into a luciferase
reporter vector (pGL3-Basic; Promega Corporation) to generate the
vectors WT-ARF6 and MT-ARF6. Following this, WT-ARF6 and MT-ARF6
were co-transfected with miR-145 mimics into 293T cells (American
Type Culture Collection) using Lipofectamine® 2000 kit
(Thermo Fisher Scientific, Inc.). At ~48 h post-transfection,
Dual-Luciferase® Reporter Assay System (Promega
Corporation) was used to detect the activities of firefly and
Renilla luciferase. Renilla luciferase activity was
used as the internal control and the ratio between the activities
of firefly luciferase and Renilla luciferase was calculated
as the relative activity.
Statistical analysis
Statistical analyses were performed using SPSS
(version no. 18.0; SPSS, Inc.) and GraphPad Prism (version no. 6.0;
GraphPad Software, Inc.) software. Each experiment was performed in
triplicate. Numerical data are presented as the mean ± standard
deviation. Comparisons between two groups were assessed using
unpaired Student's t-test, while comparisons among multiple groups
conducted using one-way ANOVA with Bonferroni post-hoc analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
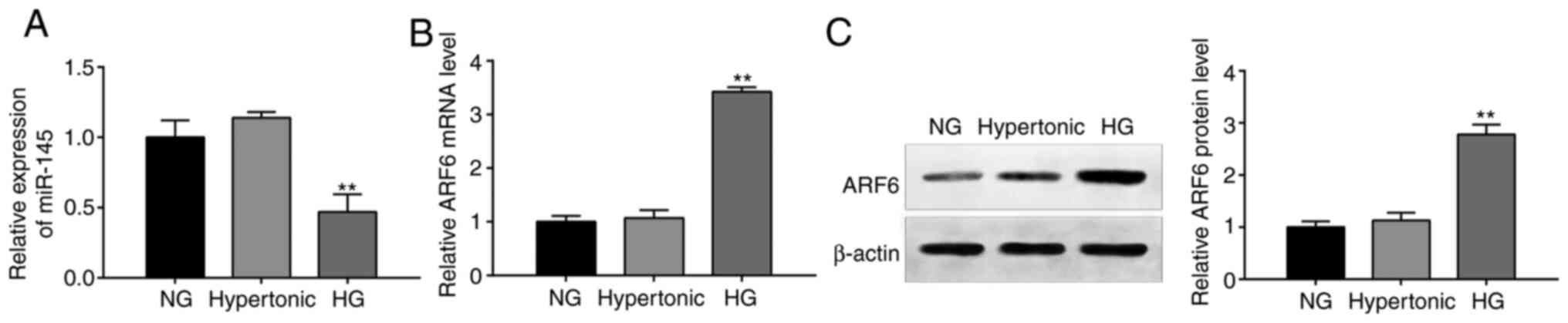
Dysregulation of miR-145 and ARF6 in
HG-treated H9c2 cells
RT-qPCR and western blotting revealed that miR-145
expression (Fig. 1A) and the mRNA
(Fig. 1B) and protein (Fig. 1C) expression of ARF6 in the
hypertonic group differed slightly compared with the NG group;
however, this difference was not significant (all, P>0.05). In
the HG group, miR-145 expression was significantly decreased, while
the protein and mRNA expression of ARF6 was significantly increased
compared with the NG group (all, P<0.01). The results indicated
that miR-145 and ARF6 served crucial roles in HG-treated
cardiomyocytes.
Protective effect of miR-145
overexpression against the inflammatory response and OS injury in
HG-treated H9c2 cells
H9c2 cells were transfected with miR-145 mimics or
mimic NCs to analyze the effect of miR-145 on HG-treated H9c2
cells. The transfection efficiency of the miR-145 mimics group was
detected by RT-qPCR. The results demonstrated that miR-145
expression in the miR-145 mimic group was significantly higher
compared with the blank group (Fig.
2A; P<0.001), indicating the successful transfection of
miR-145 mimics.
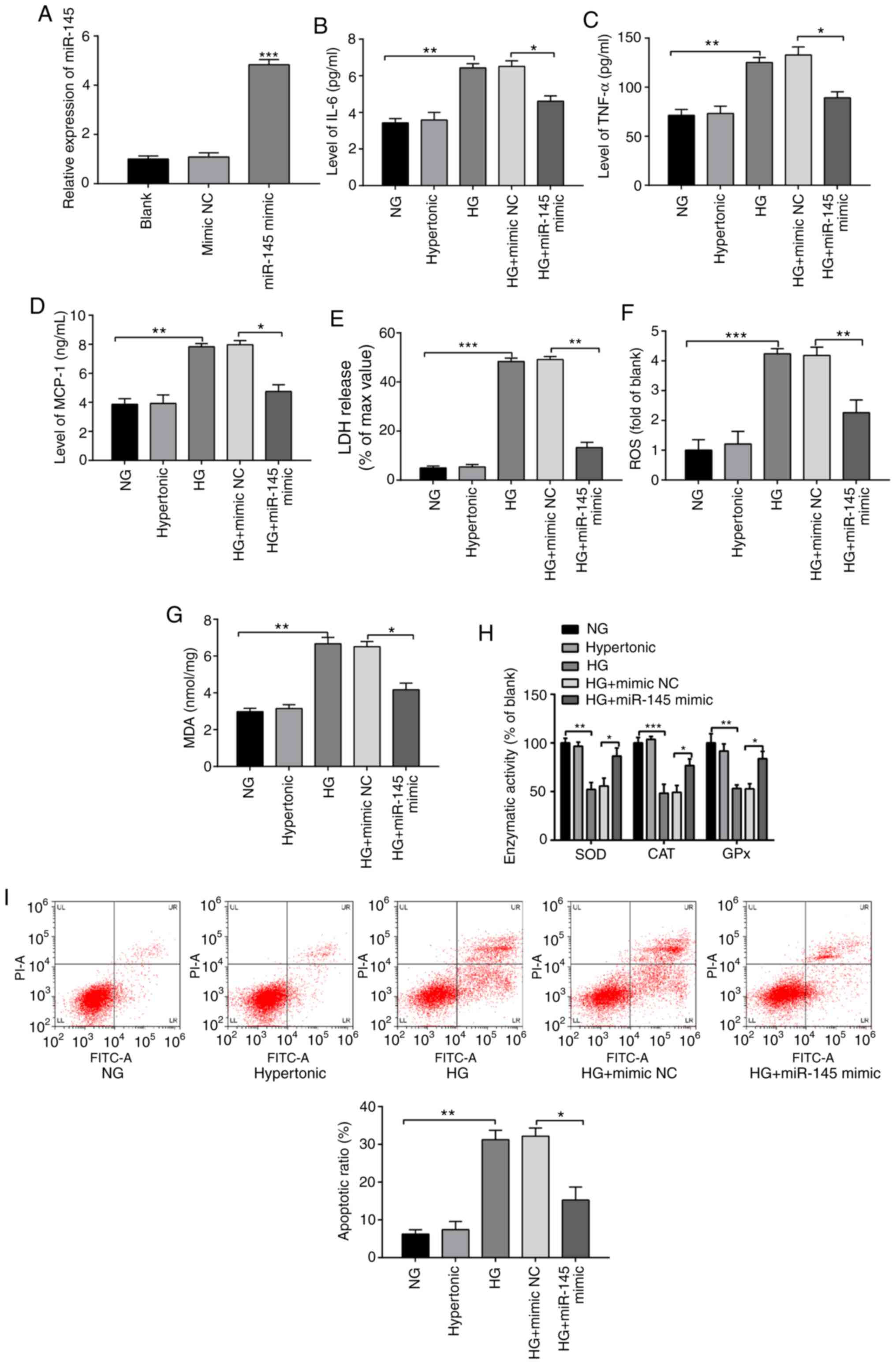 | Figure 2miR-145 ameliorates the inflammatory
response and oxidative stress injury in HG-treated H9c2 cells.
Following treatment with 5.5 or 33 mmol/l glucose for 2 h, H9c2
cells were transfected with miR-145 mimics or mimic NCs. (A)
miR-145 expression was detected by reverse
transcription-quantitative PCR. The expression of (B) IL-6, (C)
TNF-α and (D) MCP-1 was determined via ELISA. (E) LDH content was
detected using an LDH assay. (F) ROS content was examined using an
ROS detection kit. (G) ELISA was performed to determine MDA content
using a lipid peroxidation (MDA) assay kit. (H) The activities of
antioxidant enzymes were evaluated by ELISA. (I) Flow cytometry
detected the apoptotic rate of H9c2 cells. *P<0.05,
**P<0.01 and ***P<0.001 vs. indicated
groups. miR, microRNA; HG, high glucose; NC, negative controls;
MCP-1, monocyte chemoattractant protein 1; LDH, lactate
dehydrogenase; ROS, reactive oxygen species; MDA, malondialdehyde;
NG, normal glucose; SOD, superoxide dismutase; CAT, catalase; GPx,
glutathione peroxidase. |
Compared with the NG group, the levels of IL-6,
TNF-α and MCP-1 in the HG group were significantly increased
(Fig. 2B-D; P<0.01), while the
levels of these inflammatory factors were significantly decreased
in the HG + miR-145 mimic group compared with the HG + mimic NC
group (P<0.05). The results demonstrated that miR-145
overexpression reduced levels of inflammatory factors in H9c2 cells
induced following HG treatment.
The HG group exhibited significantly increased
levels of LDH, ROS (Fig. 2E and
F; P<0.001) and MDA (Fig. 2G; P<0.01), as well as
significantly decreased activities of SOD (P<0.01), CAT
(P<0.001) and GPx (P<0.01) (Fig.
2H), compared with the NG group. Transfection with miR-145
mimics significantly reduced the levels of LDH (P<0.01), MDA
(P<0.05) and ROS (P<0.01) release, and increased the
activities of SOD, CAT and GPx (P<0.05) in HG-treated H9c2 cells
compared with the HG + mimic NC group. The results indicated that
miR-145 overexpression alleviated OS in HG-induced H9c2 cells.
The results of FCM demonstrated that the apoptotic
rate of the hypertonic group was not significantly different
compared with the NG group (Fig.
2I), while the apoptotic rate of the HG group was significantly
increased compared with the NG group (P<0.01). The apoptotic
rate of the HG + miR-145 mimic group was significantly lower
compared with the HG + mimic NC group (P<0.05). Overall, HG
treatment induced OS and an inflammatory response in H9c2 cells,
while overexpression of miR-145 attenuated these effects in
HG-induced H9c2 cells.
Inhibitory effects of ARF6 silencing
on the inflammatory response and OS injury in HG-treated H9c2
cells
H9c2 cells were transfected with pcDNA3.1-ARF6,
sh-ARF6 or their NCs (pcDNA3.1 or sh-NC, respectively) to
investigate the effect of ARF6 on HG-treated H9c2 cells. Following
transfection, the mRNA (Fig. 3A)
and protein expression (Fig. 3B) of
ARF6 was significantly increased in the pcDNA3.1-ARF6 group
(P<0.001) and significantly decreased in the sh-ARF6 group
compared with the blank group (P<0.01), indicating the effective
transfection of the ARF6 reagents.
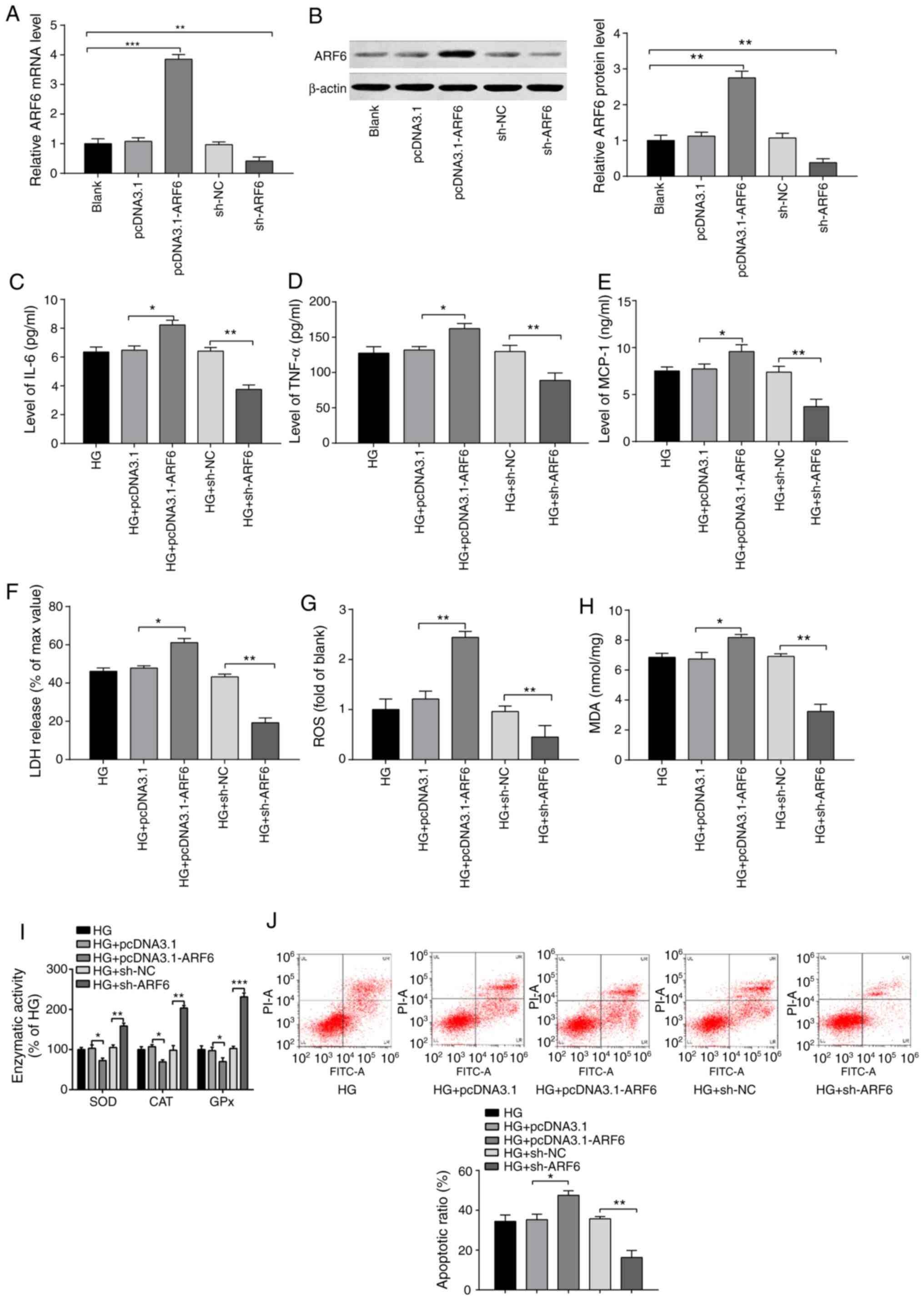 | Figure 3Inhibitory effect of ARF6 knockdown
on the inflammatory response and oxidative stress injury in
HG-treated H9c2 cells. Following 2 h of 33 mmol/l glucose
treatment, H9c2 cells were transfected with pcDNA3.1-ARF6, sh-ARF6
or corresponding negative controls, and the transfection efficiency
was examined by (A) reverse transcription-quantitative PCR and (B)
western blotting. Levels of (C) IL-6, (D) TNF-α and (E) MCP-1 were
assessed by ELISA. (F) LDH content was determined via LDH assays.
(G) ROS content was measured by ROS kits. (H) MDA content and (I)
activities of antioxidant enzymes were evaluated by ELISA. (J)
Detection of H9c2 cell apoptotic rate by flow cytometry.
*P<0.05, **P<0.01 and
***P<0.001 vs. indicated groups. ARF6, ADP
ribosylation factor 6; HG, high glucose; sh, short hairpin; MCP-1,
monocyte chemoattractant protein 1; LDH, lactate dehydrogenase;
ROS, reactive oxygen species; MDA, malondialdehyde; NC, negative
control. |
The expression of IL-6, TNF-α and MCP-1 were
significantly increased in the HG + pcDNA3.1-ARF6 group compared
with the HG + pcDNA3.1 group (Fig.
3C-E; P<0.05). The expression of these proinflammatory
cytokines was also significantly decreased in the HG + sh-ARF6
group compared with the HG + sh-NC group (all, P<0.01). No
significant difference was identified between the HG + pcDNA3.1
group/HG + sh-NC group and the HG group. The results indicated that
ARF6 overexpression increased levels of inflammatory factors and
conversely, the inhibition of ARF6 decreased these levels.
Compared with those in the HG + pcDNA3.1 group, the
levels of LDH (P<0.05), ROS (P<0.01) release and MDA
(P<0.05) were significantly increased (Fig. 3F-H), while the activities of SOD,
CAT and GPx were significantly decreased (Fig. 3I; P<0.05) in the HG +
pcDNA3.1-ARF6 group. The levels of LDH, ROS and MDA were decreased
(Fig. 3F-H; all P<0.01), while
the activities of SOD (P<0.01), CAT (P<0.01) and GPx
(P<0.001) were increased (Fig.
3I) in the HG + sh-ARF6 group compared with the HG + sh-NC
group. The results confirmed that ARF6 overexpression facilitated
OS in H9c2 cells induced by HG treatment.
Moreover, the apoptotic rate of H9c2 cells was
increased in the HG + pcDNA3.1-ARF6 group (Fig. 3J; P<0.05 vs. the HG + pcDNA3.1
group) and decreased in the HG + sh-ARF6 group (P<0.01 vs. the
HG + sh-NC group). The results indicated that ARF6 overexpression
enhanced the inflammatory response and OS injury in HG-treated H9c2
cells, while ARF6 inhibition reversed this effect.
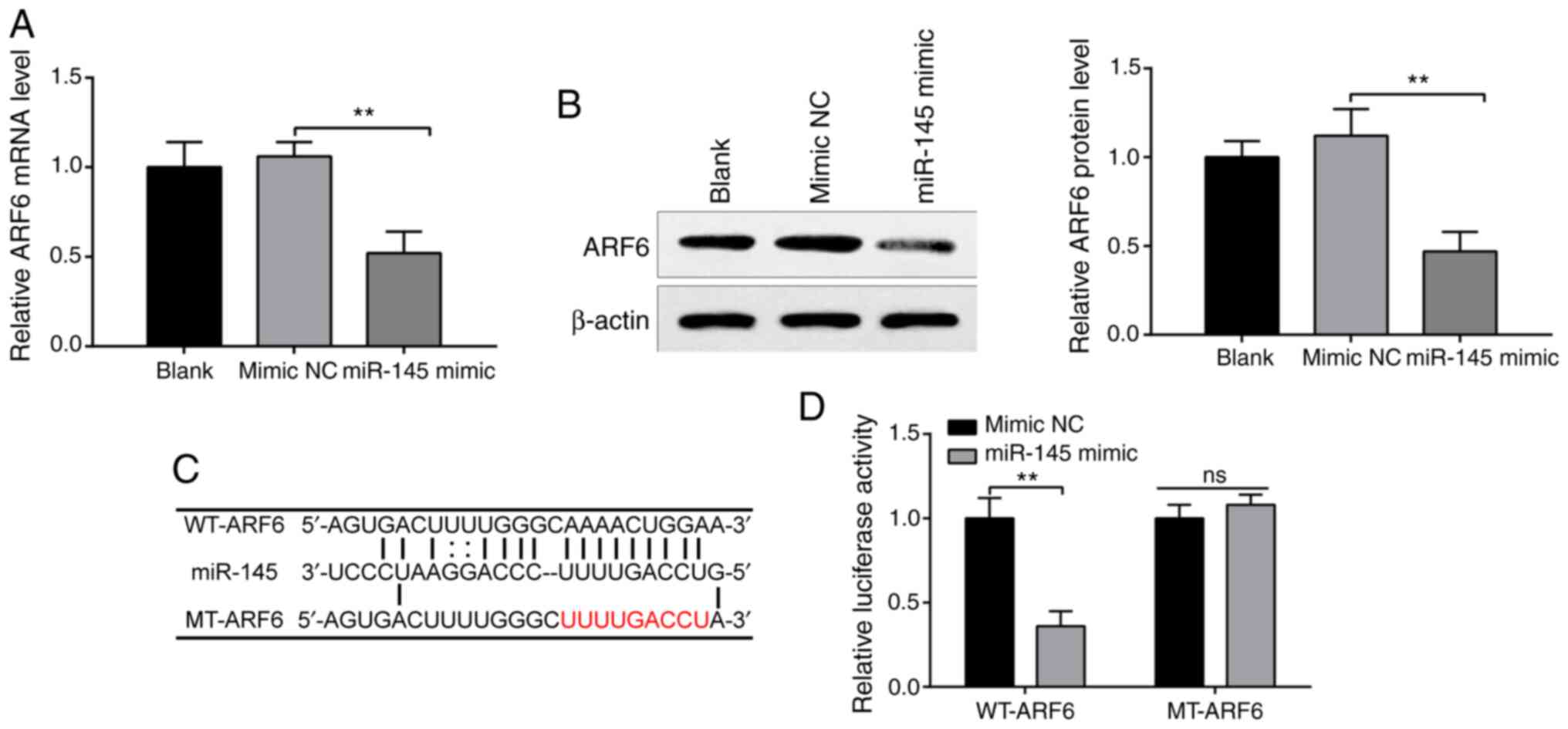
miR-145 negatively targets ARF6
StarBase predicted the potential binding sites
between miR-145 and ARF6. H9c2 cells were transfected with miR-145
mimics or mimic NCs, and the expression of ARF6 was measured to
investigate the interaction between miR-145 and ARF6. RT-qPCR
(Fig. 4A) and western blotting
(Fig. 4B) confirmed that
transfection with miR-145 mimics significantly decreased the
expression of ARF6 compared with the mimic NC group (both
P<0.01), while ARF6 expression in the mimic NC group was not
significantly different compared with the blank group, indicating
that miR-145 negatively regulated ARF6.
Accordingly, it was hypothesized that miR-145
inhibited ARF6 expression by binding to the 3' untranslated region
(3'UTR) of ARF6. The predicted binding sites between miR-145 and
ARF6 and the designed WT and MT sites are presented in Fig. 4C. To test this hypothesis, WT-ARF6
and MT-ARF6 were established. According to the results detected by
luciferase reporter assays, co-transfection with WT-ARF6 and
miR-145 mimics significantly reduced luciferase activity in 293T
cells compared with the mimic NS group (Fig. 4D; P<0.01), while co-transfection
with MT-ARF6 and miR-145 mimic had no significant effect on
luciferase activity. These results indicated that miR-145 acted as
a sponge of ARF6 and bound to the 3'UTR of ARF6.
miR-145 attenuates the HG-induced
inflammatory response and OS injury in cardiomyocytes by regulating
ARF6
To confirm the effect of the miR-145-ARF6
interaction on the inflammatory response and OS injury in
HG-induced cardiomyocytes, H9c2 cells were transfected with miR-145
mimics, miR-145 mimics + pcDNA3.1-ARF6 or their NCs. RT-qPCR and
western blotting was performed to determine the expression levels
of miR-145 and ARF6, the results of which exhibited a significant
increase in miR-145 expression (Fig.
5A; P<0.001) and a significant decrease in ARF6 expression
(Fig. 5B and C; P<0.01) in the HG + miR-145 mimic
group compared with the HG + control group. No significant
difference was observed in miR-145 expression between the HG +
miR-145 mimic + pcDNA3.1-ARF6 group and the HG + miR-145 mimic
group. However, upregulated ARF6 expression was revealed in the HG
+ miR-145 mimic + pcDNA3.1-ARF6 group compared with the HG +
miR-145 mimic group at the mRNA and protein expression levels
(both, P<0.01). The results confirmed successful transfections.
Furthermore, the results demonstrated that the expression levels of
the inflammatory factors IL-6, TNF-α and MCP-1 in the HG + miR-145
mimic group were significantly decreased (Fig. 5D-F; P<0.05 vs. the HG + control
group) and co-transfection with miR-145 mimics and pcDNA3.1-ARF6
significantly upregulated the expression levels of these
inflammatory factors (P<0.05 vs. HG + miR-145 mimic group). The
levels of these inflammatory factors were not significantly
different between the HG + control and HG groups. The results
suggested that ARF6 overexpression inhibited the effect of miR-145
on reducing the secretion of inflammatory factors in H9c2
cells.
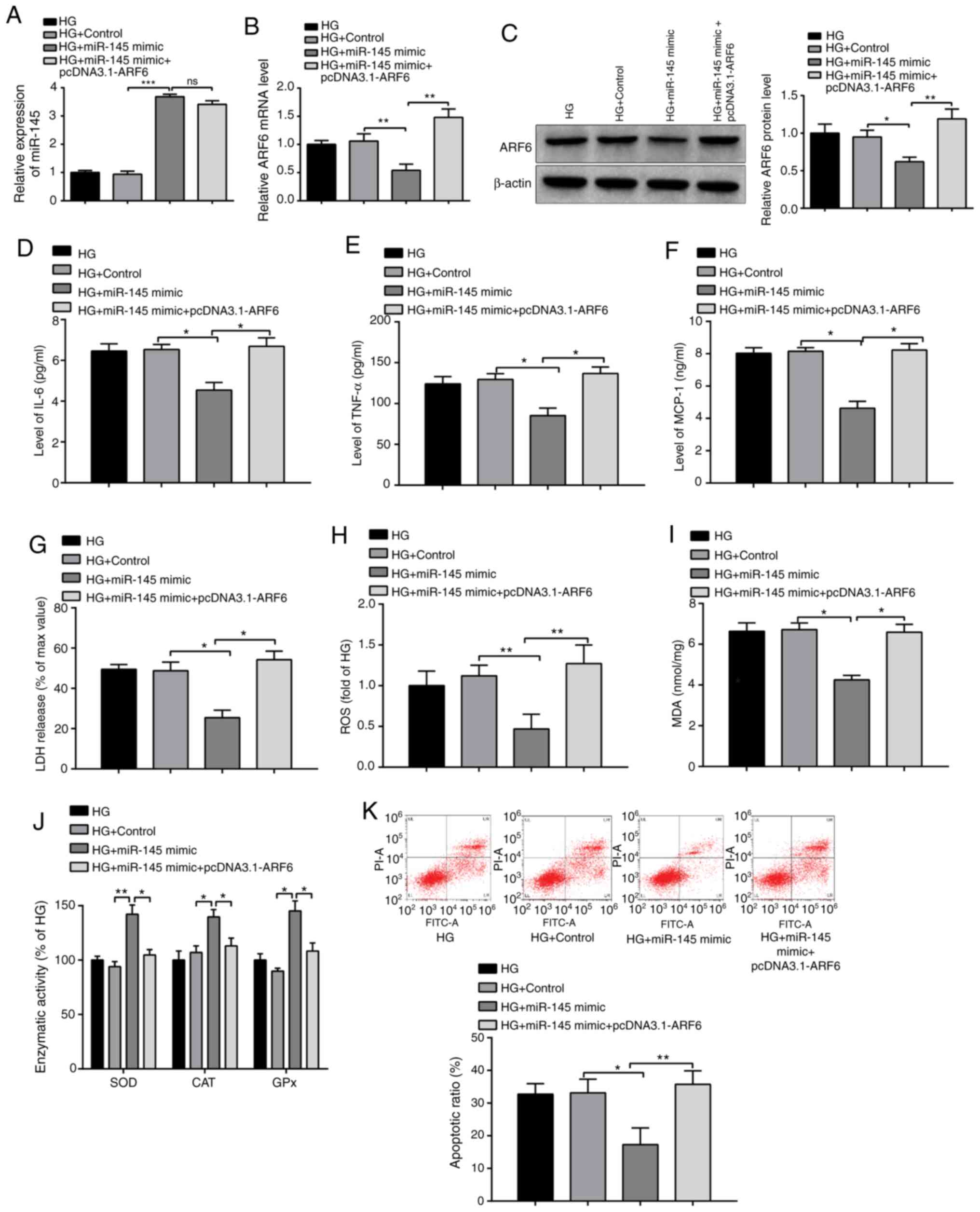 | Figure 5miR-145 protects H9c2 cells from the
HG-induced inflammatory response and oxidative stress injury.
Following 2 h of 33 mmol/l glucose treatment, H9c2 cells were
transfected with miR-145 mimics, pcDNA3.1-ARF6, miR-145 +
pcDNA3.1-ARF6 or corresponding negative controls, and the
expression levels of (A) miR-145 and ARF6 were determined by (B)
reverse transcription-quantitative PCR and (C) western blotting.
Levels of (D) IL-6, (E) TNF-α and (F) MCP-1 were detected by ELISA.
(G) LDH content was detected by a LDH kit. (H) ROS content was
determined using an ROS kit. (I) MDA content and (J) activities of
antioxidant enzymes were evaluated by ELISA. (K) Detection of H9c2
cell apoptotic rate by flow cytometry. *P<0.05,
**P<0.01 and ***P<0.001 vs. indicated
groups. miR, microRNA; HG, high glucose; MCP-1, monocyte
chemoattractant protein 1; LDH, lactate dehydrogenase; ROS,
reactive oxygen species; MDA, malondialdehyde; ns, not
significant. |
Moreover, the levels of LDH (P<0.05), ROS
(P<0.01) and MDA (P<0.05) were significantly downregulated
(Fig. 5G-I) and the activities of
SOD (P<0.01), CAT (P<0.05) and GPx (P<0.05) were
significantly upregulated (Fig. 5J)
in the HG + miR-145 mimic group compared with the HG + control
group. Additionally, the levels of LDH (P<0.05), ROS (P<0.01)
and MDA (P<0.05) in the HG + miR-145 mimic + pcDNA3.1-ARF6 group
were increased and the activities of the antioxidant enzymes were
decreased (P<0.05) compared with the HG + miR-145 mimic group.
The expression levels of these factors were not significantly
different between the HG + control and HG groups. These results
confirmed that the overexpression of ARF6 reversed the protective
effect of miR-145 mimics on HG-induced OS in H9c2 cells.
The apoptotic rate of H9c2 cells was significantly
decreased in the HG + miR-145 mimic group (Fig. 5K; P<0.05 vs. the HG + control
group) and significantly elevated in the HG + miR-145 mimic +
pcDNA3.1-ARF6 group (P<0.01 vs. the HG + miR-145 mimic group).
In summary, ARF6 overexpression exacerbated the inflammatory
response and OS injury in HG-induced H9c2 cells, which was
suppressed by miR-145 overexpression, supporting the hypothesis
that miR-145 alleviated the H9c2 cell inflammatory response and OS
injury by regulating ARF6.
Discussion
miRs, small noncoding RNA molecules whose altered
expression has been reported in cardiomyocytes of experimental
diabetes models, could be considered as potential treatment targets
for diabetic cardiomyopathy in future studies (19). Numerous miRs, such as miR-320,
miR-214 and miR-203, have been demonstrated to be associated with
diabetes-induced cardiomyopathy and may serve as targets of
diabetic cardiomyopathy treatment (20-22).
The present study observed downregulated miR-145 and upregulated
ARF6 expression in HG-induced cardiomyocytes. Overexpression of
miR-145 efficiently protected cardiomyocytes against an HG-induced
inflammatory response and OS injury by inhibiting ARF6, indicating
a novel therapeutic target against diabetic cardiomyopathy.
Previous studies have extensively documented the
antioxidant and anti-inflammatory roles of miR-145. Introduction of
miR-145 overexpression in LPS-induced human fibroblast-like
synoviocytes inhibited the release of proinflammatory cytokines
(23). Injection of miR-145 mimics
markedly alleviated mechanical allodynia and thermal hyperalgesia
in vivo by suppressing the inflammatory response (24). Upregulation of miR-145 inhibited
apoptosis, OS and inflammation in vascular smooth muscle cells, the
dominant subunits of the arterial wall, which provided a potential
direction for the treatment of abdominal aortic aneurysms (25). The current study demonstrated that
miR-145 expression was significantly decreased in response to HG
treatment in cardiomyocytes. Overexpression of miR-145 attenuated
OS and the inflammatory response following HG treatment, as
evidenced by significantly decreasing expression levels of IL-6,
TNF-α, MCP-1, ROS and MDA, as well as significantly increasing
activities of SOD, CAT and GPx.
A previous study reported that the inhibition of
ARF6 prevented the proinflammatory effects of chloride
intracellular channel 4 in human pulmonary artery endothelial cells
(26). To date, the role of ARF6 in
OS remains undetermined. A previous study indicated that blockade
of ARF6 and its effector, c-Myc-binding protein-associated
protein-1, increased the generation of ROS in the breast cancer
cell line MDA-MB-231(27). In
contrast, a second study demonstrated that ARF6 inhibition induced
by secinH3 in insulin-secreting cells (INS-1 832/13 cells)
significantly reduced the glucose-induced production of ROS
(28). In the current study, ARF6
expression was significantly increased in response to HG treatment,
indicating the involvement of ARF6 in HG-treated cardiomyocytes.
Following ARF6 silencing, the inflammatory response and OS injury
in cardiomyocytes were alleviated.
ROS activate poly (ADP-ribose) polymerase (PARP)-1
in the nucleus, resulting in the depletion of cellular nicotinamide
adenine dinucleotide (NAD+) and an increase in
mitochondrial permeability, mitochondrial dysfunction and cell
death (29). PARPs catalyze
mono-ADP-ribose or poly-ADP-ribose (PAR) attachment to target
proteins via NAD+ as a donor (30). Covalently PARylating target proteins
and proteins noncovalently binding with PAR regulate various
biological processes, including inflammation (31). The activated GTP-bound form of ARF6
can be ADP-ribosylated on an arginine residue and the ADP
ribosylation of ARF6 is an important component of a novel
regulatory pathway (32).
Therefore, the current study hypothesized that PARP-1 activated by
ROS catalyzed the ADP ribosylation of ARF6 GTPase and, therefore,
promoted inflammation. Moreover, ARF6 GTPase is required for the
polarization of Rabphilin-3A, Ras-related protein Rab-21 and type I
phosphatidylinositol 4-phosphate 5-kinase isoform γ (PIP5K1C90),
and polarization serves an important role in neutrophil adhesion to
endothelia during inflammation (33).
The current study hypothesized that miR-145 rescued
the inflammatory response and OS injury in HG-treated
cardiomyocytes by binding to the 3'UTR of ARF6. The hypothesis was
tested using the online database StarBase and dual luciferase
reporter assays. miR-145 overexpression reduced the luciferase
activity of 293T cells transfected with WT-ARF6; however, this
reduction was not observed in cells transfected with MT-ARF6.
Therefore, the results confirmed that miR-145 negatively targeted
ARF6. Additionally, targeting between miR-145 and ARF6 has been
reported in breast cancer (34) and
upper tract urothelial carcinoma (35). In subsequent experiments, miR-145
and ARF6 were simultaneously overexpressed to determine the effect
of the miR-145-ARF6 interaction on the cardiomyocyte inflammatory
response and OS injury in cells treated with HG. The experiments
demonstrated that the protective effect of miR-145 overexpression
on the HG-induced inflammatory response and OS injury in
cardiomyocytes was reversed by ARF6 overexpression, indicating that
miR-145 modulated the inflammatory response and OS injury in
HG-treated cardiomyocytes by regulating ARF6 expression.
The present study determined that the expression of
miR-145 and ARF6 were dysregulated in HG-treated cardiomyocytes.
Overexpression of miR-145 or ARF6 silencing in cardiomyocytes
attenuated the inflammatory response and OS injury in HG-treated
cardiomyocytes. Additionally, the current study demonstrated that
miR-145 negatively targeted ARF6, thus protecting cardiomyocytes
against the inflammatory response and OS injury induced by HG
treatment. The results demonstrated that miR-145 may be used as a
novel therapeutic target for diabetic cardiomyopathy. Furthermore,
a limitation of the present study was the limited experimental
data; therefore, future studies should confirm the current results
in animal models.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WZ conceived the study and designed the experiments.
TL, JW and YL performed the experiments. WZ, YZ and QZ analyzed the
data. QZ, WZ and YL provided critical materials. WZ and TL wrote
the manuscript. WZ supervised the study. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Newman JD, Vani AK, Aleman JO, Weintraub
HS, Berger JS and Schwartzbard AZ: The changing landscape of
diabetes therapy for cardiovascular risk reduction: JACC
state-of-the-art review. J Am Coll Cardiol. 72:1856–1869.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jia G, Whaley-Connell A and Sowers JR:
Diabetic cardiomyopathy: A hyperglycaemia- and
insulin-resistance-induced heart disease. Diabetologia. 61:21–28.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bugger H and Abel ED: Molecular mechanisms
of diabetic cardiomyopathy. Diabetologia. 57:660–671.
2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Karam BS, Chavez-Moreno A, Koh W, Akar JG
and Akar FG: Oxidative stress and inflammation as central mediators
of atrial fibrillation in obesity and diabetes. Cardiovasc
Diabetol. 16(120)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Newsholme P, Cruzat VF, Keane KN, Carlessi
R and de Bittencourt PI Jr: Molecular mechanisms of ROS production
and oxidative stress in diabetes. Biochem J. 473:4527–4550.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Qadir MMF, Klein D, Alvarez-Cubela S,
Dominguez-Bendala J and Pastori RL: The role of MicroRNAs in
diabetes-related oxidative stress. Int J Mol Sci.
20(5423)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kyrychenko S, Kyrychenko V, Badr MA, Ikeda
Y, Sadoshima J and Shirokova N: Pivotal role of miR-448 in the
development of ROS-induced cardiomyopathy. Cardiovasc Res.
108:324–334. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tang Q, Li MY, Su YF, Fu J, Zou ZY, Wang Y
and Li SN: Absence of miR-223-3p ameliorates hypoxia-induced injury
through repressing cardiomyocyte apoptosis and oxidative stress by
targeting KLF15. Eur J Pharmacol. 841:67–74. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Su Q, Yao J and Sheng C: Geniposide
attenuates LPS-induced injury via up-regulation of miR-145 in H9c2
cells. Inflammation. 41:1229–1237. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li L, Liu M, He L, Wang S and Cui S:
Baicalin relieves TNF-α-evoked injury in human aortic endothelial
cells by up-regulation of miR-145. Phytother Res. 34:836–845.
2020.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
He M, Wu N, Leong MC, Zhang W, Ye Z, Li R,
Huang J, Zhang Z, Li L, Yao X, et al: miR-145 improves metabolic
inflammatory disease through multiple pathways. J Mol Cell Biol.
12:152–162. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yuan M, Zhang L, You F, Zhou J, Ma Y, Yang
F and Tao L: MiR-145-5p regulates hypoxia-induced inflammatory
response and apoptosis in cardiomyocytes by targeting CD40. Mol
Cell Biochem. 431:123–131. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hui Y and Yin Y: MicroRNA-145 attenuates
high glucose-induced oxidative stress and inflammation in retinal
endothelial cells through regulating TLR4/NF-κB signaling. Life
Sci. 207:212–218. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Padival AK, Hawkins KS and Huang C: High
glucose-induced membrane translocation of PKC betaI is associated
with Arf6 in glomerular mesangial cells. Mol Cell Biochem.
258:129–135. 2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fang Z, Miao Y, Ding X, Deng H, Liu S,
Wang F, Zhou R, Watson C, Fu C, Hu Q, et al: Proteomic
identification and functional characterization of a novel ARF6
GTPase-activating protein, ACAP4. Mol Cell Proteomics. 5:1437–1449.
2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Karnik R, Ludlow MJ, Abuarab N, Smith AJ,
Hardy ME, Elliott DJ and Sivaprasadarao A: Endocytosis of HERG is
clathrin-independent and involves arf6. PLoS One.
8(e85630)2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li R, Shen Q, Wu N, He M, Liu N, Huang J,
Lu B, Yao Q, Yang Y and Hu R: MiR-145 improves macrophage-mediated
inflammation through targeting Arf6. Endocrine. 60:73–82.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gilca GE, Stefanescu G, Badulescu O,
Tanase DM, Bararu I and Ciocoiu M: Diabetic cardiomyopathy: Current
approach and potential diagnostic and therapeutic targets. J
Diabetes Res. 2017(1310265)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li H, Fan J, Zhao Y, Zhang X, Dai B, Zhan
J, Yin Z, Nie X, Fu XD, Chen C and Wang DW: Nuclear miR-320
mediates diabetes-induced cardiac dysfunction by activating
transcription of fatty acid metabolic genes to cause lipotoxicity
in the heart. Circ Res. 125:1106–1120. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang Y, Zhao RZ, Chen PK, Xu GX, Liu ZJ,
Long XP, Qiu ZM and Shi B: Impact and related mechanism on the
improvement of hyperglycemia-induced pyroptosis in H9c2 cells by
mircoRNA-214. Zhonghua Xin Xue Guan Bing Za Zhi. 47:820–828.
2019.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
22
|
Yang X, Li X, Lin Q and Xu Q:
Up-regulation of microRNA-203 inhibits myocardial fibrosis and
oxidative stress in mice with diabetic cardiomyopathy through the
inhibition of PI3K/Akt signaling pathway via PIK3CA. Gene.
715(143995)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhong F, Xu J, Yang X, Zhang Q, Gao Z,
Deng Y, Zhang L and Yu C: miR-145 eliminates
lipopolysaccharides-induced inflammatory injury in human
fibroblast-like synoviocyte MH7A cells. J Cell Biochem.
119:10059–10066. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shi J, Jiang K and Li Z: MiR-145
ameliorates neuropathic pain via inhibiting inflammatory responses
and mTOR signaling pathway by targeting Akt3 in a rat model.
Neurosci Res. 134:10–17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lin H, You B, Lin X, Wang X, Zhou D, Chen
Z, Chen Y and Wang R: Silencing of long non-coding RNA Sox2ot
inhibits oxidative stress and inflammation of vascular smooth
muscle cells in abdominal aortic aneurysm via microRNA-145-mediated
Egr1 inhibition. Aging (Albany NY). 12:12684–12702. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Abdul-Salam VB, Russomanno G, Chien-Nien
C, Mahomed AS, Yates LA, Wilkins MR, Zhao L, Gierula M, Dubois O,
Schaeper U, et al: CLIC4/Arf6 pathway. Circ Res. 124:52–65.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Onodera Y, Nam JM, Horikawa M, Shirato H
and Sabe H: Arf6-driven cell invasion is intrinsically linked to
TRAK1-mediated mitochondrial anterograde trafficking to avoid
oxidative catastrophe. Nat Commun. 9(2682)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jayaram B and Kowluru A: Phagocytic NADPH
oxidase links ARNO-Arf6 signaling pathway in glucose-stimulated
insulin secretion from the pancreatic β-cell. Cell Physiol Biochem.
30:1351–1362. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li C, Zhang J, Xue M, Li X, Han F, Liu X,
Xu L, Lu Y, Cheng Y, Li T, et al: SGLT2 inhibition with
empagliflozin attenuates myocardial oxidative stress and fibrosis
in diabetic mice heart. Cardiovasc Diabetol. 18(15)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ke Y, Wang C, Zhang J, Zhong X, Wang R,
Zeng X and Ba X: The role of PARPs in inflammation-and
metabolic-related diseases: Molecular mechanisms and beyond. Cells.
8(1047)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fehr AR, Singh SA, Kerr CM, Mukai S,
Higashi H and Aikawa M: The impact of PARPs and ADP-ribosylation on
inflammation and host-pathogen interactions. Genes Dev. 34:341–359.
2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Dani N, Barbosa AJ, Del Rio A and Di
Girolamo M: ADP-ribosylated proteins as old and new drug targets
for anticancer therapy: The example of ARF6. Curr Pharm Des.
19:624–633. 2013.PubMed/NCBI
|
|
33
|
Ren C, Yuan Q, Jian X, Randazzo PA, Tang W
and Wu D: Small GTPase ARF6 is a coincidence-detection code for
RPH3A polarization in neutrophil polarization. J Immunol.
204:1012–1021. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
13:330–338. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hsu WC, Li WM, Lee YC, Huang AM, Chang LL,
Lin HH, Wu WJ, Li CC, Liang PI and Ke HL: MicroRNA-145 suppresses
cell migration and invasion in upper tract urothelial carcinoma by
targeting ARF6. FASEB J. 34:5975–5992. 2020.PubMed/NCBI View Article : Google Scholar
|