Introduction
Stroke is an episode of neurological dysfunction
caused by focal cerebral, spinal or retinal infarction, and has
become the second leading cause of disability and death in adults
worldwide (1,2). Ischemic stroke (IS) is the most common
type of stroke. There are multiple risk factors for IS, which
include age, sex, body mass index (BMI), hyperlipidemia,
hypertension, diabetes, smoking and particulate matter 2.5
pollution (3,4).
Previous studies have examined the roles of
polymorphisms in numerous genes involved in the pathogenesis of IS,
including C-C motif chemokine 11, paraoxonase 1,
angiotensin-converting enzyme (ACE), and methylenetetrahydrofolate
reductase (MTHFR) (5-7).
It has been reported that the ACE I/D polymorphism is significantly
associated with IS in different ethnic groups (8). However, other studies failed to
observe this association (9,10).
Patients with the MTHFR 677TT genotype have vascular occlusion,
infarct and increased levels of blood homocysteine, which form the
basis for the association of this genetic polymorphism with
hypertension (11). Polymorphism of
MTHFR has important roles in hypertension and IS, both of which are
caused by atherosclerotic vascular disease (12). Numerous studies have been performed
to investigate the potential association between polymorphisms of β
fibrinogen (β-Fg) and the risk of IS. However, the results of these
studies were inconsistent, and the sample sizes of individual
studies were inadequate to draw definite conclusions (13-15).
The 4G/5G polymorphism in the promoter of the plasminogen activator
inhibitor-1 gene (PAI-1) is one of the most frequently studied
(16). A single 4G allele is
considered to be a risk factor for coronary artery disease and the
4G/4G genotype is thought to increase the risk of coronary artery
disease (17,18). Several studies addressed the
association between the 4G/5G polymorphism and stroke, but these
results were inconsistent (19,20).
Although PAI-1 may be an important factor in the occurrence of IS,
the association between PAI-1 gene polymorphisms and the risk of IS
has remained to be elucidated. Apolipoprotein E (ApoE)
polymorphism involves a single amino substitution and results in
three major alleles (ε2, ε3 and ε4) with six corresponding
phenotypes (ε2/ε2, ε3/ε3, ε4/ε4, ε2/ε3, ε2/ε4 and ε3/ε4) (21). The relationship between ApoE
genotype and stroke is unclear because of inconsistent study
results. Certain reports indicated a positive association between
ApoE4-containing genotypes and stroke, but others indicated no
relationships between ApoE isoforms and dyslipidemia or stroke
(22,23). The influence of the gene
polymorphisms on clinical laboratory parameters of IS has not been
described in the Chinese Han population, to the best of our
knowledge.
The aim of the present study was to evaluate
potential associations between polymorphisms in six genes, namely
ACE D/I, MTHFR C677T, β-Fg A/G, 455/148T/C, PAI-1, 4G/5G and ApoE
ε2,3,4, and clinical laboratory parameters of IS in the Chinese Han
population. Such data may be used to provide a control range of
laboratory parameters according to genotype for the early
prevention of IS in patients with diabetes and hyperlipidemia.
Patients and methods
Patients
The medical records of all newly diagnosed patients
with proven diabetes and hyperlipidemia who were admitted to
Shanghai Tongji Hospital (Shanghai, China) from October 2016 to
November 2018 were examined. The present study was approved by the
ethics committee of Shanghai Tongji Hospital (Shanghai, China) and
was conducted according to the Declaration of Helsinki. All
participants provided informed consent in written form. For
participants who were unable to communicate, written consent was
obtained from their legal relatives.
Patients who met the following criteria were
included: i) Fasting blood glucose (FBG) ≥7.0 mmol/l or 2-h oral
glucose tolerance test ≥11.1 mmol/l; ii) triglyceride (TG) ≥1.7
mmol/l orlow-density lipoprotein cholesterol (LDL-C) ≥3.37 mmol/l
or total cholesterol (CHOL) ≥5.18 mmol/l; iii) a diagnosis of IS
meeting the standards of Chinese guidelines for the diagnosis and
treatment of acute ischemic stroke in 2014(24), which were issued by Chinese Medical
Association; and iv) Chinese Han population and unrelated to other
participants in the study.
Patients with a diagnosis of any of the following
conditions were excluded: i) Other types of cerebrovascular
disease, including intracranial hemorrhage, subarachnoid
hemorrhage, cerebrovascular malformation or cerebral aneurysm; and
ii) severe systemic diseases, including cancer, severe inflammatory
disease or serious chronic diseases (e.g. hepatic failure or renal
failure).
The control group comprised 336 individuals without
a history of cerebrovascular disease who were physical examined in
the hospital during the same period. Age, sex, use of oral
contraceptives and history of thrombotic events or drug abuse were
recorded. The baseline characteristics of patients and controls are
presented in Table SI.
Serum lipid, glucose and transaminase
measurement
Blood samples (~3 ml fasting blood) were collected
from each study participant and then separated by centrifugation at
840 x g for 10 min in a SPINCHRON™ DLX centrifuge (Beckman
Coulter). Serum levels of FBG, CHOL, TG, LDL-C, high-density
lipoprotein cholesterol (HDL-C), alanine aminotransferase (ALT) and
aspartate aminotransferase (AST) were analyzed by an automatic
biochemical analyzer (DXC800; Beckman Coulter).
DNA extraction and genetic
analysis
Genomic DNA was extracted from peripheral blood
leukocytes using a TIANamp Genomic DNA kit (Tiangen Biotech)
following the protocol of the manufacturer. Polymorphisms were
genotyped using allele-specific PCR and restriction fragment length
polymorphism analysis described in Data S1. PCR primers were synthesized by
Sangon Biotech Co., Ltd.; the sequences are listed in Table SII. Amplified PCR products of
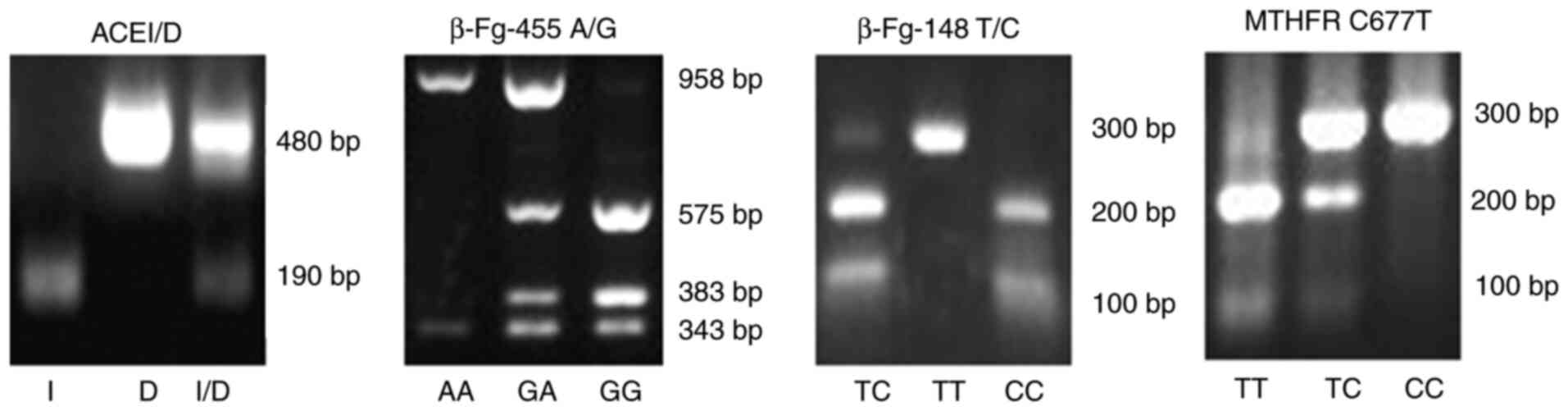
intron 16 of ACE were separated on a 2% agarose gel (Bio-Rad
Laboratories, Inc.) (Fig. 1). The
presence of 191-bp fragments indicated the D allele, 480-bp
fragments represented the I allele and 191- and 480-bp fragments
represented the D/I allele. Amplification products of β-Fg-455 were
digested by HaeIII (FD0154; Thermo Fisher Scientific, Inc.)
and then separated on a 2% agarose gel (Fig. 1). The presence of 343-, 383- and
575-bp fragments represented the GG allele; 343- and 958-bp
fragments represented the AA allele, and 343-, 383-, 575- and
958-bp fragments represented the G/A allele. Amplification products
of β-Fg-148 were digested by HindIII (FD0505; Thermo Fisher
Scientific, Inc.) (Fig. 1). The
presence of 100- and 200-bp fragments represented the CC allele,
the presence of 300-bp fragments represented the TT allele and the
presence of 100-, 200- and 300-bp fragments represented the T/C
allele. Amplification products of MTHFR were digested by
HinfI (FD0804; Thermo Fisher Scientific, Inc.) (Fig. 1). The presence of 100- and 200-bp
fragments represented the TT allele, 300-bp fragments represented
the CC allele and 100-, 200- and 300-bp fragments represented the
T/C allele. Amplification products of PAI-1and ApoE ε2,3,4 gene
alleles were separated on a 2% agarose gel and subjected to
first-generation sequencing by Genewiz, Inc..
Statistical analysis
SPSS statistical software version 21.0 (IBM Corp.)
was used for statistical analysis. Pearson's chi-squared test or
Fisher's exact test were used for statistical comparisons of count
data. Continuous variables are expressed as the mean ± standard
deviation. Receiver operating characteristic (ROC) curve analysis
was used to analyze laboratory parameters related to IS. Logistic
regression was used to analyze the models 1 and 2. P<0.05 was
considered to indicate statistical significance.
Results
Relationships between laboratory
parameters and genotype and allele frequencies
Table I indicates
the associations between laboratory parameters and the ACE I/D,
β-Fg-455 A/G and PAI-1 4G/5G gene polymorphisms. The D allele of
ACE I/D was associated with high levels of TC and LDL-C, as
presented in Table II.
Furthermore, a significant association between the β-Fg-455 A/G
polymorphism and LDL-C levels and an association between PAI-1
4G/5G and TG were observed (Table
I). The frequency of the 4G/5G genotype of PAI-1 4G/5G was
highest in the TG <1.70 mmol/l group and the frequency of the
4G/5G genotype was equal to that of the 4G4G genotype in the TG
≥1.70 mmol/l group. However, there were no significant differences
in genotype frequency distributions of the MTHFR C677T, β-Fg-148T/C
and ApoE ε2-4 polymorphisms for the parameters of FBG, TC, TG and
LDL-C (Table I).
 | Table IAssociation of laboratory parameters
with genotype frequencies of ACE I/D, MTHFR C677T, β-Fg-455A/G,
β-Fg-148T/C, PAI-1 4G/5G and ApoE ε2-4. |
Table I
Association of laboratory parameters
with genotype frequencies of ACE I/D, MTHFR C677T, β-Fg-455A/G,
β-Fg-148T/C, PAI-1 4G/5G and ApoE ε2-4.
| | FBG (mmol/l) | TC (mmol/l) | TG (mmol/l) | LDL-C (mmol/l) |
|---|
| Item | ≥7.00 | <7.00 | ≥5.18 | <5.18 | ≥1.70 | <1.70 | >3.37 | ≤3.37 |
|---|
| ACE I/D | | | | | | | | |
|
II | 23 (51.1) | 142 (48.5) | 14 (32.6) | 151 (51.2) | 21 (42.0) | 144 (50.0) | 15 (31.3) | 150 (51.7) |
|
I/D | 17 (37.8) | 108 (36.8) | 20 (46.5) | 105 (35.6) | 19 (38.0) | 106 (36.8) | 24 (50.0) | 101 (34.8) |
|
DD | 5 (11.1) | 43 (14.7) | 9 (20.9) | 39 (13.2) | 10 (20.0) | 38 (13.2) | 9 (18.8) | 39 (13.4) |
|
χ2/P-value | 0.414/0.813 | 5.448/0.066 | 1.964/0.374 | 6.915/0.032 |
| MTHFR C677T | | | | | | | | |
|
CC | 15 (33.3) | 96 (32.8) | 12 (27.9) | 99 (33.5) | 19 (38.0) | 92 (31.9) | 15 (31.3) | 96 (33.1) |
|
T/C | 25 (55.6) | 144 (49.1) | 21 (48.8) | 148 (50.2) | 21 (42.0) | 148 (51.4) | 23 (47.9) | 146 (50.3) |
|
TT | 5 (11.1) | 53 (18.1) | 10 (23.3) | 48 (16.3) | 10 (20.0) | 48 (16.7) | 10 (20.8) | 48 (16.6) |
|
χ2/P-value | 1.431/0.489 | 1.445/0.485 | 1.503/0.472 | 0.532/0.767 |
| β-Fg-455A/G | | | | | | | | |
|
GG | 22 (48.9) | 187 (63.8) | 22 (51.2) | 187 (63.4) | 30 (60.0) | 179 (62.2) | 25 (52.1) | 184 (63.4) |
|
G/A | 21 (46.7) | 95 (32.4) | 21 (48.8) | 95 (32.2) | 17 (34.0) | 99 (34.4) | 23 (47.9) | 93 (32.1) |
|
AA | 2 (4.4) | 11 (3.8) | 0 (0.0) | 13 (4.4) | 3 (6.0) | 10 (3.5) | 0 (0.0) | 13 (4.5) |
|
χ2/P-value | 3.761/0.152 | 5.828/0.054 | 0.741/0.690 | 6.026/0.049 |
| β-Fg-148T/C | | | | | | | | |
|
CC | 26 (57.8) | 166 (56.7) | 23 (53.5) | 169 (57.3) | 29 (58.0) | 163 (56.6) | 26 (54.2) | 166 (57.2) |
|
T/C | 17 (37.8) | 106 (36.2) | 19 (44.2) | 104 (35.3) | 18 (36.0) | 105 (36.5) | 21 (43.8) | 102 (35.2) |
|
TT | 2 (4.4) | 21 (7.2) | 1 (2.3) | 22 (7.5) | 3 (6.0) | 20 (6.9) | 1 (2.1) | 22 (7.6) |
|
χ2/P-value | 0.461/0.794 | 2.371/0.306 | 0.073/0.964 | 2.734/0.255 |
| PAI-1 4G/5G | | | | | | | | |
|
4G4G | 14 (31.1) | 92 (31.4) | 14 (32.6) | 92 (31.2) | 17 (34.0) | 89 (30.9) | 15 (31.3) | 91 (31.4) |
|
4G/5G | 23 (51.1) | 144 (49.1) | 22 (51.2) | 145 (49.2) | 17 (34.0) | 150 (52.1) | 26 (54.2) | 141 (48.6) |
|
5G5G | 8 (17.8) | 57 (19.5) | 7 (16.2) | 58 (19.7) | 16 (32.0) | 49 (17.0) | 7 (14.5) | 58 (20.0) |
|
χ2/P-value | 0.088/0.957 | 0.276/0.871 | 7.925/0.019 | 0.885/0.642 |
| ApoE ε2-4 | | | | | | | | |
|
E2/2 | 0 (0.0) | 5 (1.7) | 1 (2.3) | 4 (1.4) | 1 (2.0) | 4 (1.4) | 1 (2.1) | 4 (1.4) |
|
E2/3 | 7 (15.6) | 44 (15.0) | 5 (11.6) | 46 (15.6) | 10 (20.0) | 41 (14.2) | 5 (10.4) | 46 (15.9) |
|
E2/4 | 0 (0.0) | 5 (1.7) | 1 (2.3) | 4 (1.4) | 1 (2.0) | 4 (1.4) | 1 (2.1) | 4 (1.4) |
|
E3/3 | 33 (73.3) | 195 (66.6) | 29 (67.5) | 199 (67.5) | 31 (62.0) | 197 (68.4) | 32 (66.7) | 196 (67.6) |
|
E3/4 | 5 (11.1) | 43 (14.7) | 7 (16.3) | 41 (13.9) | 6 (12.0) | 42 (14.6) | 9 (18.7) | 39 (13.4) |
|
E4/4 | 0 (0.0) | 1 (0.3) | 0 (0.0) | 1 (0.3) | 1 (2.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) |
|
χ2/P-value | 1.471/0.965 | 2.631/0.708 | 6.470/0.229 | 3.307/0.622 |
 | Table IIAssociation of laboratory parameters
with allele frequencies of ACE I/D, MTHFR C677T, β-Fg-455A/G,
β-Fg-148T/C, PAI-1 4G/5G and ApoE ε2-4. |
Table II
Association of laboratory parameters
with allele frequencies of ACE I/D, MTHFR C677T, β-Fg-455A/G,
β-Fg-148T/C, PAI-1 4G/5G and ApoE ε2-4.
| | FBG (mmol/l) | TC (mmol/l) | TG (mmol/l) | LDL-C (mmol/l) |
|---|
| Item | ≥7.00 | <7.00 | ≥5.18 | <5.18 | ≥1.70 | <1.70 | >3.37 | ≤3.37 |
|---|
| ACE I/D | | | | | | | | |
|
I | 63 (70.0) | 392 (66.9) | 48 (55.8) | 407 (69.0) | 61 (61.0) | 394 (68.4) | 54 (56.3) | 401 (69.1) |
|
D | 27 (30.0) | 194 (33.1) | 38 (44.2) | 183 (31.0) | 39 (39.0) | 182 (31.6) | 42 (43.7) | 179 (30.9) |
|
χ2/P-value | 0.324/0.559 | 5.916/0.015 | 2.122/0.145 | 6.217/0.013 |
| MTHFR C677T | | | | | | | | |
|
C | 55 (61.1) | 336 (57.3) | 45 (52.3) | 346 (58.6) | 59 (59.0) | 332 (57.6) | 53 (55.2) | 338 (58.3) |
|
T | 35 (38.9) | 250 (42.7) | 41 (47.7) | 244 (41.4) | 41 (41.0) | 244 (42.4) | 43 (44.8) | 242 (41.7) |
|
χ2/P-value | 0.456/0.500 | 1.229/0.268 | 0.065/0.799 | 0.318/0.573 |
| β-Fg-455A/G | | | | | | | | |
|
G | 65 (72.2) | 469 (80.0) | 65 (75.6) | 469 (79.5) | 77 (77.0) | 457 (79.3) | 73 (76.0) | 461 (79.5) |
|
A | 25 (27.8) | 117 (20.0) | 21 (24.4) | 121 (20.5) | 23 (23.0) | 119 (20.7) | 23 (24.0) | 119 (20.5) |
|
χ2/P-value | 2.869/0.090 | 0.692/0.406 | 0.281/0.596 | 0.588/0.443 |
| β-Fg-148T/C | | | | | | | | |
|
C | 69 (76.7) | 438 (74.7) | 65 (75.6) | 442 (74.9) | 76 (76.0) | 431 (74.8) | 73 (76.0) | 434 (74.8) |
|
T | 21 (23.3) | 148 (25.3) | 21 (24.4) | 148 (25.1) | 24 (24.0) | 145 (25.2) | 23 (24.0) | 146 (25.2) |
|
χ2/P-value | 0.154/0.695 | 0.018/0.894 | 0.063/0.802 | 0.065/0.799 |
| PAI-1 4G/5G | | | | | | | | |
|
4G | 51 (56.7) | 328 (56.0) | 50 (58.1) | 329 (55.8) | 51 (51.0) | 328 (56.9) | 56 (58.3) | 323 (55.7) |
|
5G | 39 (43.3) | 258 (44.0) | 36 (41.9) | 261 (44.2) | 49 (49.0) | 248 (43.1) | 40 (41.7) | 257 (44.3) |
|
χ2/P-value | 0.015/0.902 | 0.172/0.678 | 1.222/0.269 | 0.234/0.629 |
| ApoE ε2-4 | | | | | | | | |
|
E2 | 7 (7.8) | 59 (10.1) | 8 (9.3) | 58 (9.8) | 13 (13.0) | 53 (9.2) | 8 (8.3) | 58 (10.0) |
|
E3 | 78 (86.7) | 477 (81.4) | 70 (81.4) | 485 (82.2) | 78 (78.0) | 477 (82.8) | 78 (81.3) | 477 (82.2) |
|
E4 | 5 (5.5) | 50 (8.5) | 8 (9.3) | 47 (8.0) | 9 (9.0) | 46 (8.0) | 10 (10.4) | 45 (7.8) |
|
χ2/P-value | 1.533/0.465 | 0.192/0.908 | 1.607/0.448 | 0.959/0.619 |
Table SIII
indicates the association between the BMI, ACE I/D, β-Fg-455A/G and
PAI-1 4G/5G gene polymorphisms. The frequency of the ACE genotype
DD in obese (BMI≥28.00 kg/m2) and overweight
(28.00>BMI ≥24.00 kg/m2) subjects was higher than
that in normal subjects (24.00>BMI≥18.5 kg/m2). As
presented in Table SIV, the D
allele of the ACE gene was associated with a high BMI. Furthermore,
a significant association between β-Fg-455 gene polymorphisms and
the BMI was observed (Table SIII).
The relationship between the PAI-1 gene and the BMI is presented in
Table SIII. The frequency of the
AA genotype of the β-Fg-455 gene in obese subjects was ~8% higher
than that in normal subjects, while that in overweight subjects was
~6% higher (P<0.05). Furthermore, the frequency of the PAI-1
5G5G genotype in obese and overweight subjects was significantly
higher than that in normal subjects (P<0.05). However, there was
no significant difference in the MTHFR C677T, β-Fg-148T/C and ApoE
ε2-4 genotypes between groups of high/low HDL-C, ALT and AST
(Table SIII).
Predictive value of clinical
laboratory parameters for IS
As presented in Table
III, high levels of FBG, TG and LDL-C were risk factors for IS.
The results suggested that the risk of IS increased by 6.47-, 4.64-
and 7.62-fold along with each 1-mmol/l increment of FBG, TG and
LDL-C, respectively. Age and sex were included for adjusting in
model 2 and these risk values increased to 5.38-, 5.41- and
6.21-fold for FGB, TG and LDL, respectively. The results in
Table SV suggested that a high BMI
is a risk factor for IS. It was indicated that with the increase of
the BMI by 1 kg/m2, the risk increased by 2.26-fold.
When age and sex were included for adjusting in model 2, the risk
increased by 2.35-fold. HDL-C was indicated to be a protective
factor against IS. With the increase of HDL-C by 1 mmol/l, the risk
of disease was reduced to 27% and after adjustment for age and sex,
it was 32%.
 | Table IIIPrediction of ischemic stroke by
laboratory parameters. |
Table III
Prediction of ischemic stroke by
laboratory parameters.
| | Model 1 | Model 2 |
|---|
| | 95% CI | | 95% CI | |
|---|
| Factor | OR | Lower | Upper | P-value | OR | Lower | Upper | P-value |
|---|
| FBG (mmol/l) | 6.47 | 3.95 | 10.61 | <0.001 | 5.38 | 3.25 | 8.91 | <0.001 |
| TC (mmol/l) | 0.34 | 0.10 | 1.13 | 0.077 | 0.44 | 0.15 | 1.28 | 0.130 |
| TG (mmol/l) | 4.64 | 2.09 | 10.32 | <0.001 | 5.41 | 2.28 | 12.81 | <0.001 |
| LDL-C (mmol/l) | 7.62 | 1.60 | 36.40 | 0.011 | 6.21 | 1.59 | 24.16 | 0.008 |
ROC curve analysis of laboratory
parameters for IS
Area under the curve values at the 95% confidence
interval (95% CI) for IS predicted by laboratory parameters were as
follows: FBG, 0.828 (0.784-0.867); TC, 0.595 (0.541-0.648); TG,
0.702 (0.650-0.751); LDL-C, 0.638 (0.584-0.689); and BMI, 0.735
(0.685-0.786) (Tables IV, SVI and Fig.
S1). It was observed that FBG had better specificity (0.833)
and sensitivity (0.694) for predicting IS compared to the other
parameters. The optimal cut-off point of FGB for predicting IS was
5.27 mmol/l. Among the five parameters, LDL-C had the highest
specificity (0.999) with an optimal cut-off point for predicting IS
of 3.36 mmol/l. The sensitivity values of TC, TG, LDL-C and BMI for
predicting IS were poor (0.265, 0.471, 0.284 and 0.591,
respectively), although their specificity values were better.
 | Table IVROC curve analysis of laboratory
parameters for ischemic stroke. |
Table IV
ROC curve analysis of laboratory
parameters for ischemic stroke.
| | 95% CI |
|---|
| Parameter | Cut-off point | Specificity | Sensitivity | AUC | Lower | Upper |
|---|
| FBG (mmol/l) | 5.27 | 0.833 | 0.694 | 0.828 | 0.784 | 0.867 |
| TC (mmol/l) | 5.12 | 0.994 | 0.265 | 0.595 | 0.541 | 0.648 |
| TG (mmol/l) | 1.32 | 0.833 | 0.471 | 0.702 | 0.650 | 0.751 |
| LDL-C (mmol/l) | 3.36 | 0.999 | 0.284 | 0.638 | 0.584 | 0.689 |
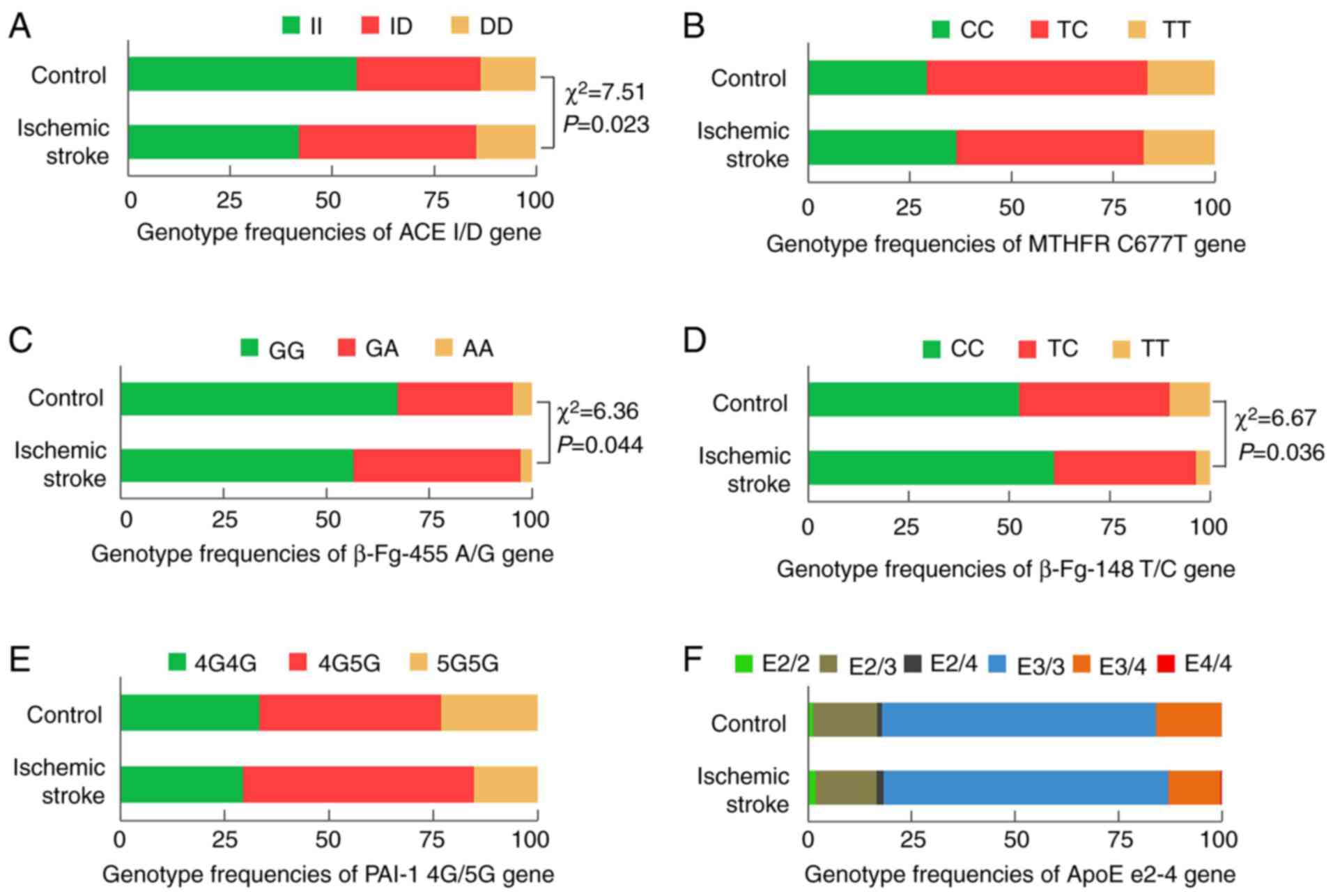
The relationships between IS and allele frequencies
of ACE I/D, MTHFR C677T, β-Fg-455A/G, β-Fg-148T/C, PAI-1 4G/5G and
ApoE ε2-4 genes are presented in Table
V and Fig. 2. Significant
differences in the genotype frequencies of ACE I/D, β-Fg-455 A/G
and β-Fg-148 T/C genes between patients with IS and the control
group were present (Fig. 2).
However, only the allele frequencies of ACE I/D and β-Fg-148 T/C
were significantly different between the two groups (Table V). More specifically, the
frequencies of the D allele of ACE I/D and the C allele of β-Fg-148
T/C were higher in the IS group, whereas the frequencies of the I
allele of ACE I/D and the T allele of β-Fg-148 T/C in the IS group
were lower compared with those in the control group.
 | Table VRelationship between ischemic stroke
and allele frequencies of ACE I/D, MTHFR C677T, β-Fg-455 A/G,
β-Fg-148 T/C, PAI-1 4G/5G and ApoE ε2-4. |
Table V
Relationship between ischemic stroke
and allele frequencies of ACE I/D, MTHFR C677T, β-Fg-455 A/G,
β-Fg-148 T/C, PAI-1 4G/5G and ApoE ε2-4.
| | ACE I/D | MTHFR C677T | β-Fg-455A/G | β-Fg-148T/C | PAI-1 4G/5G | ApoE ε2-4 |
|---|
| Group | I | D | C | T | G | A | C | T | 4G | 5G | E2 | E3 | E4 |
|---|
| IS | 216 (63.5) | 124 (36.5) | 202 (59.4) | 138 (40.6) | 261 (76.8) | 79 (23.2) | 268 (78.8) | 72 (21.2) | 194 (57.1) | 146 (42.9) | 34 (10.0) | 280 (82.4) | 26 (7.6) |
| Control | 239 (71.1) | 97 (28.9) | 189 (56.3) | 147 (43.7) | 273 (79.0) | 63 (21.0) | 239 (71.1) | 97 (28.9) | 185 (55.1) | 151 (44.9) | 32 (9.5) | 275 (81.9) | 29 (8.6) |
| χ2 | 4.438 | 0.693 | 2.049 | 5.334 | 0.274 | 0.246 |
| P-value | 0.035 | 0.405 | 0.152 | 0.021 | 0.601 | 0.884 |
Discussion
Strokes are the second leading cause of death and
the leading cause of permanent disability in adults worldwide
(25), with IS accounting for 85%
of the total number of strokes (26). IS is caused by the occlusion of
major arteries or branches of the brain, which leads to vascular
occlusion and deprivation of oxygen and energy. This is followed by
the formation of reactive oxygen species, then the release of
glutamate, the accumulation of intracellular calcium and the
induction of inflammatory processes (27). Previous studies have reported that
IS is closely related to certain genes, as well as blood glucose
and blood lipid levels (28-31).
However, it is currently not possible to control or treat stroke at
the genetic level. To explore this possibility, the relationships
between IS and genetic and laboratory parameters (FBG, TC, TG,
LDL-C, HDL-C, ALT, AST and BMI) were examined in the present
study.
The results of the present study revealed
correlations between IS and the ACE I/D, β-Fg-455A/G and
β-Fg-148T/C polymorphisms. Frequencies of both the D allele of ACE
I/D and the C allele of β-Fg-148T/C in the IS group were higher
than those in the control group, suggesting that IS is closely
related to the D allele of those two alleles, which is consistent
with the studies performed by Zhao et al (32) and Wu et al (33).
In accordance with the results of a study by Lin
et al (30), the present
results indicated that FBG was an effective parameter for
predicting IS, suggesting that routine monitoring for FBG may
effectively control and prevent the progression of IS. In a study
performed by Anderson et al (34), hyperglycemia was indicated to affect
mitochondrial function in the ischemic penumbra, resulting in
cortical acidosis and cell death. Hyperglycemia also impaired
cerebrovascular reactivity in the microvasculature, which may
disturb reperfusion after recanalization (35). Diabetes or hyperglycemia may alter
blood-brain barrier permeability and induce disruption of the
blood-brain barrier, which may aggravate the formation of brain
edema and lead to hemorrhagic transformation (36). In addition, the results of the
present study suggested that hyperlipidemia is a risk factor for
IS. Lee et al (31) pointed
out that a high TG level is a risk factor for IS and that the risk
is 1.28-fold higher than that of individuals with normal TG levels.
Lee et al (31) also noted
that the presence of LDL-C ≥130 mg/dl may increase the risk of IS.
Furthermore, Pawelczyk et al (37) reported a significant increase in the
plasma concentration of soluble P-selectin (sP-selectin) in
patients with stroke and hyperlipidemia and hyperglycemia compared
with normolipidemic/normoglycemic patients with stroke. On the one
hand, a strong positive correlation was observed between
hyperglycemia and sP-selectin levels, which emphasizes the leading
role of hyperglycemia in atherothrombosis progression. On the other
hand, hyperlipidemia was also associated with an increase in the
plasma sP-selectin level (37).
This glycoprotein has a role in stimulating the release of
procoagulant microparticles, which induces a procoagulant state
(38).
Methods to treat IS mainly comprise initial
treatment with intra-arterial thrombolysis, endovascular mechanical
thrombectomy and antiplatelet treatment (39). However, such treatments rarely
negate the possibility of recurrence of IS. Studying associations
of stroke-related genes with blood glucose and blood lipids may
offer alternative approaches. The present results suggested that
LDL-C and TG were closely related to the ACE I/D, β-Fg-455 A/G and
PAI-1 4G/5G polymorphisms, consistent with the results of Li
(40) and Guney et al
(41). The results of the present
study also indicated that hyperlipidemia was correlated with the D
allele of ACE I/D, which confirmed the results of Suzuki et
al (42) and Lee and Tsai
(43), who demonstrated positive
associations of hyperlipidemia with the DD genotype and D allele
frequency of ACE I/D.
There are certain limitations to the present study.
First, the study enrolled patients with IS at a hospital rather
than patients from a community-based general population.
Furthermore, the study comprised a single population with limited
sample size; thus, the results require to be confirmed in multiple
centers using larger sample sizes and in different ethnic
populations. In addition, the study did not collect data on several
other major risk parameters of IS (such as homocysteine, fibrinogen
or prothrombin), which should also be examined in future research.
Finally, dietary habits exhibited a marked variation among the
participants making it is difficult to assess the impact of dietary
habits on IS in the present study.
In summary, the present study indicated that the D
allele of ACE I/D and the C allele of β-Fg-148 T/C were
significantly associated with IS and that the frequency of the D
allele of ACE I/D was significantly higher in individuals with
hyperlipidemia. High levels of FGB, TG, LDL-C and BMI were risk
factors for IS, with optimal predictive cut-off points of 5.27,
1.32 and 3.36 mmol/l and 23.12 kg/m2, respectively. The
present results suggested that individuals with hyperlipidemia or a
high frequency of the D allele of ACE I/D may be at risk of IS. By
contrast, there were no significant differences in TC levels and
the MTHFR C677T, PAI-1 4G/5G and ApoE ε2-4 gene polymorphisms
between patients with IS and controls. Identifying the
relationships among IS, stroke-related genes and blood lipid and
blood glucose levels may lead to a better understanding of the
pathophysiology of IS in the Chinese Han population and may provide
a reasonable control range of laboratory parameters for the
prevention of IS in patients with diabetes and hyperlipidemia as
early as possible.
Supplementary Material
The experimental details of
allele-specific PCR and restriction fragment length polymorphism
analysis. Polymorphisms were genotyped using allele-specific
PCR and restriction fragment length polymorphism analysis. PCR
primers were synthesized by Sangon Biotech Co., Ltd.; the sequences
are listed in Table SII. PCR
conditions of ACE were as follows: 95˚C for 2 min, followed by 35
cycles at 95˚C for 30 sec, 55˚C for 30 sec, and 72˚C for 30 sec,
final step was performed at 72˚C for 5 min. Amplified PCR products
of intron 16 of ACE were separated on a 2% agarose gel (Bio-Rad
Laboratories, Inc.) (Fig. 1). The
presence of 191-bp fragments indicated the D allele, 480-bp
fragments represented the I allele and 191- and 480-bp fragments
represented the D/I allele. PCR conditions of β-Fg-455 were as
follows: 94˚C for 2 min, followed by 35 cycles at 94˚C for 30 sec,
55˚C for 30 sec, and 72˚C for 80 sec, the final step was performed
at 72˚C for 5 min. Amplification products of β-Fg-455 were digested
by HaeIII (FD0154; Thermo Fisher Scientific, Inc.) and then
separated on a 2% agarose gel (Fig.
1). The presence of 343-, 383- and 575-bp fragments represented
the GG allele; 343- and 958-bp fragments represented the AA allele,
and 343-, 383-, 575- and 958-bp fragments represented the G/A
allele. PCR conditions of β-Fg-148 were as follows: 94˚C for 2 min,
followed by 35 cycles at 94˚C for 30 sec, 58.5˚C for 30 sec, and
72˚C for 30 sec, final step was performed at 72˚C for 5 min.
Amplification products of β-Fg-148 were digested by HindIII
(cat. no. FD0505; Thermo Fisher Scientific, Inc.) (Fig. 1). The presence of 100- and 200-bp
fragments represented the CC allele, the presence of 300-bp
fragments represented the TT allele and the presence of 100-, 200-
and 300-bp fragments represented the T/C allele. PCR conditions of
MTHFR were as follows: 94˚C for 2 min, followed by 35 cycles at
94˚C for 30 sec, 58˚C for 30 sec, and 72˚C for 30 sec, and the
final step was performed at 72˚C for 5 min. Amplification products
of MTHFR were digested by HinfⅠ (FD0804; Thermo Fisher
Scientific, Inc.) (Fig. 1). The
presence of 100- and 200-bp fragments represented the TT allele,
300-bp fragments represented the CC allele and 100-, 200- and
300-bp fragments represented the T/C allele. PCR conditions of
PAI-1 were as follows: 94˚C for 2 min, followed by 35 cycles at
94˚C for 30 sec, 58.5˚C for 45 sec, and 72˚C for 30 sec, and the
final step was performed at 72˚C for 5 min. PCR conditions of ApoE
ε2,3,4 were as follows: 95˚C for 2 min, followed by 35 cycles at
95˚C for 30 sec, 60˚C for 45 sec, and 72˚C for 55 sec, and the
final step was performed at 72˚C for 5 min.
ROC curve analysis of laboratory
parameters to distinguish patients with ischemic stroke from normal
controls. ROC, receiver operating characteristic; FBG, fasting
blood glucose; TC, total cholesterol; TG, triglyceride; LDL-C,
low-density lipoprotein cholesterol; BMI, body mass index.
Characteristics of the two groups at
baseline.
Sequences of PCR primers.
Association of BMI, HDL-C, ALT and AST
with genotype frequencies of ACE I/D, MTHFR C677T, β-Fg-455A/G,
β-Fg-148T/C, PAI-1 4G/5G and ApoE ε2-4 genes.
Association of BMI, HDL-C, ALT and AST
with allele frequencies of ACE I/D, MTHFR C677T, β-Fg-455A/G,
β-Fg-148T/C, PAI-1 4G/5G and ApoE ε2-4 genes.
Prediction of ischemic stroke by the
BMI and the levels of HDL-C, ALT and AST.
ROC curve analysis of BMI for ischemic
stroke.
Acknowledgements
The authors thank Dr Michelle Kahmeyer-Gabbe for
editing the English text of a draft of this manuscript.
Funding
This work was supported by the National Natural Science
Foundation of China (grant nos. 81974314, 81873975, 81802084 and
81902984), the Excellent Academic Leader Training Program of
Shanghai Health System (grant no. 2018BR31), the Medical Guidance
Science and Technology Support Project of Shanghai (grant no.
19411964800) and the Clinical Research and Cultivation Project of
Shanghai Tongji Hospital [grant nos. ITJ(ZD)1803, ITJ(ZD)1905 and
ITJ(QN)1905].
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW and XC collected the data and performed the
statistical analysis and interpretation of the result. ZS analyzed
the patient data, wrote the manuscript and approved the final
version. YY, JW and WQ acquired, analyzed, and interpreted the data
for the work. PN and DL designed the overall study and revised the
manuscript, and also checked and approved the authenticity of the
raw date. All authors read and approved the manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committee of
Shanghai Tongji Hospital (Shanghai, China) and was performed
according to the Declaration of Helsinki. All participants provided
written informed consent. If the subjects were unable to
communicate, written informed consent was obtained from their legal
relatives.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Feigin VL, Lawes CM, Bennett DA,
Barker-Collo SL and Parag V: Worldwide stroke incidence and early
case fatality reported in 56 population-based studies: A systematic
review. Lancet Neurol. 8:355–369. 2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sacco RL, Kasner SE, Broderik JP, Caplan
LR, Connors JJ, Culebras A, Elkind MS, George MG, Hamdan AD,
Higashida RT, et al: An updated definition of stroke for the 21st
century: A statement for healthcare professionals from the American
heart association/American stroke association. Stroke.
44:2064–2089. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liu L, Wang D, Wong KS and Wang Y: Stroke
and stroke care in China: Huge burden, significant workload, and a
national priority. Stroke. 42:3651–3654. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Feigin VL, Roth GA, Naghavi M, Parmar P,
Krishnamurthi R, Chugh S, Mensah GA, Norrving B, Shiue I, Ng M, et
al: Global burden of stroke and risk factors in 188 countries,
during 1990-2013: A systematic analysis for the global burden of
disease study 2013. Lancet Neurol. 15:913–924. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liang C, Ni G, Ma J, Liu H, Mao Z, Sun H
and Zhang X: Impact of tag single nucleotide polymorphisms (SNPs)
in CCL11 gene on risk of subtypes of ischemic stroke in Xinjiang
Han populations. Med Sci Monit. 23:4291–4298. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bersano A, Ballabio E, Bresolin N and
Candelise L: Genetic polymorphisms for the study of multifactorial
stroke. Hum Mutat. 29:776–795. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Huen K, Yousefi P, Street K, Eskenazi B
and Holland N: PON1 as a model for integration of genetic,
epigenetic, and expression data on candidate susceptibility genes.
Environ Epigenet. 1(dvv003)2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang Z, Xu G, Liu D, Fan X, Zhu W and Liu
X: Angiotensin-converting enzyme insertion/deletion polymorphism
contributes to ischemic stroke risk: A meta-analysis of 50
case-control studies. PLoS One. 7(e46495)2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tuncer N, Tuglular S, Kılıç G, Sazci A, Us
O and Kara I: Evaluation of the angiotensin-converting enzyme
insertion/deletion polymorphism and the risk of ischaemic stroke. J
Cli Neurosci. 13:224–227. 2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pera J, Slowik A, Dziedzic T, Wloch D and
Szczudlik A: ACE I/D polymorphism in different etiologies of
ischemic stroke. Acta Neurol Scand. 114:320–322. 2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu J, Sun K, Bai Y, Zhang W, Wang X, Wang
Y, Wang H, Chen J, Song X, Xin Y, et al: Association of three-gene
interaction among MTHFR, ALOX5AP and NOTCH3 with thrombotic stroke:
A multicenter case-control study. Hum Genet. 125:649–656.
2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Arina CA, Amir D, Siregar Y and Sembiring
RJ: The Role of polymorphism gen methylene tetra hydrofolate
reductase (MTHFR) C677T in ischaemic stroke patients with and
without hypertension. Open Access Maced J Med Sci. 7:29–32.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Coen Herak D, Lenicek Krleza J, Radic
Antolic M, Horvat I, Djuranovic V, Zrinski Topic R and Zadro R:
Association of polymorphisms in coagulation factor genes and
enzymes of homocysteine metabolism with arterial ischemic stroke in
children. Clin Appl Thromb Hemost. 23:1042–1051. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kopyta I, Niemiec P, Balcerzyk A,
Emich-Widera E, Pilarska E, Pienczk-Ręcławowicz K, Kaciński M,
Wendorff J, Nowak T, Iwanicki T, et al: Fibrinogen alpha and beta
gene polymorphisms in pediatric stroke-case-control and family
based study. Eur J Paediatr Neurol. 19:176–180. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kumar A, Misra S, Kumar P, Sagar R and
Prasad K: Association between beta-fibrinogen C148T gene
polymorphism and risk of ischemic stroke in a north Indian
population: A case-control study. Pulse (Basel). 4:165–171.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hu X, Zan X, Xie Z, Li Y, Lin S, Li H and
You C: Association between plasminogen activator inhibitor-1
genetic polymorphisms and stroke susceptibility. Mol Neurobiol.
54:328–341. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Eriksson P, Kallin B, Van't Hooft FM,
Båvenholm P and Hamsten A: Allele-specific increase in basal
transcription of the plasminogen-activator inhibitor 1 gene is
associated with myocardial infarction. Proc Natl Acad Sci USA.
92:1851–1855. 1995.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Iacoviello L, Burzotta F, Di Castelnuovo
A, Zito F, Marchioli R and Donati MB: The 4G/5G polymorphism of
PAI-1 promoter gene and the risk of myocardial infarction: A
meta-analysis. Thromb Haemost. 80:1029–1030. 1998.PubMed/NCBI
|
|
19
|
Jood K, Ladenvall P, Tjärnlund-Wolf A,
Ladenvall C, Andersson M, Nilsson S, Blomstrand C and Jern C:
Fibrinolytic gene polymorphism and ischemic stroke. Stroke.
36:2077–2081. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wiklund PG, Nilsson L, Ardnor SN, Eriksson
P, Johansson L, Stegmayr B, Hamsten A, Holmberg D and Asplund K:
Plasminogen activator inhibitor-1 4G/5G polymorphism and risk of
stroke: Replicated findings in two nested case-control studies
based on independent cohorts. Stroke. 36:1661–1665. 2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Mahley RW and Rall SC Jr: Apolipoprotein
E: Far more than a lipid transport protein. Annu Rev Genomics Hum
Genet. 1:507–537. 2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pezzini A, Grassi M, Del Zotto E, Bazzoli
E, Archetti S, Assanelli D, Akkawi NM, Albertini A and Padovani A:
Synergistic effect of apolipoprotein E polymorphisms and cigarette
smoking on risk of ischemic stroke in young adults. Stroke.
35:438–442. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sturgeon JD, Folsom AR, Bray MS,
Boerwinkle E and Ballantyne CM: Atherosclerosis Risk in Communities
Study Investigators. Apolipoprotein E genotype and incident
ischemic stroke: The aherosclerosis rsk in cmmunities sudy. Stroke.
36:2484–2486. 2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chinese Neurology Association. Chinese
guidelines for the diagnosis and treatment of acute ischemic stroke
in 2014. Chin J Neurol. 48:246–257. 2015.
|
|
25
|
Murray CJ and Lopez AD: Measuring the
global burden of disease. N Engl J Med. 369:448–457.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Beal CC: Gender and stroke symptoms: A
review of the current literature. J Neurosci Nurs. 42:80–87.
2010.PubMed/NCBI
|
|
27
|
Fluri F, Schuhmann MK and Kleinschnitz C:
Animal models of ischemic stroke and their application in clinical
research. Drug Des Devel Ther. 9:3445–3454. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li G, Liu Y, Li X, Ning Z, Sun Z, Zhang M,
Lu Y, Wu L and Wang L: Association of PAI-1 4G/5G polymorphism with
ischemic stroke in Cinese patients with type 2 diabetes mellitus.
Genet Test Mol Biomarkers. 22:554–560. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhao LL, Su G, Chen LX, Yan Q, Wang XP,
Yuan W, Wang L and Zhang ZC: Apolipoprotein E polymorphisms are
associated with ischemic stroke susceptibility in a Northwest China
Han population. Biosci Rep. 37(BSR20171088)2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lin CC, Yang CP, Li CI, Liu CS, Chen CC,
Lin WY, Hwang KL, Yang SY and Li TC: Visit-to-visit variability of
fasting plasma glucose as predictor of ischemic stroke: Competing
risk analysis in a national cohort of Taiwan diabetes study. BMC
Med. 12(165)2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lee JS, Chang PY, Zhang Y, Kizer JR, Best
LG and Howard BV: Triglyceride and HDL-C dyslipidemia and risks of
coronary heart disease and ischemic stroke by glycemic
dysregulation status: The strong heart study. Diabetes Care.
40:529–537. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhao J, Qin X, Li S and Zeng Z:
Association between the ACE I/D polymorphism and risk of ischemic
stroke: An updated meta-analysis of 47,026 subjects from 105
case-control studies. J Neuro Sci. 345:37–47. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wu G, Cai H, Cai H, Chen Z, Tan L, Qi H
and Cai Y: Effect of the-148C/T, 448G/A, and -854G/A polymorphisms
of the β-fibrinogen gene on the risk of ischemic stroke in Chinese
population. J Stroke Cerebrovasc Dis. 24:1577–1590. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Anderson RE, Tan WK, Martin HS and Meyer
FB: Effects of glucose and PaO2 modulation on cortical
intracellular acidosis, NADH redox state, and infarction in the
ischemic penumbra. Stroke. 30:160–170. 1999.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kawai N, Keep RF and Betz AL:
Hyperglycemia and the vascular effects of cerebral ischemia.
Stroke. 28:149–154. 1997.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Dietrich WD, Alonso O and Busto R:
Moderate hyperglycemia worsens acute blood-brain barrier injury
after forebrain ischemia in rats. Stroke. 24:111–116.
1993.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Pawelczyk M, Kaczorowska B and Baj Z: The
impact of hyperglycemia and hyperlipidemia on plasma P-selectin and
platelet markers after ischemic stroke. Arch Med Sci. 13:1049–1056.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Nomura S, Inami N, Iwasaka T and Liu Y:
Platelet activation markers, microparticles and soluble adhesion
molecules are elevated in patients with arteriosclerosis
obliterans: Therapeutic effects by cilostazol and potentiation by
dipyridamole. Platelets. 15:167–172. 2004.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Powers WJ, Rabinstein AA, Ackerson T,
Adeoye OM, Bambakidis NC, Becker K, Biller J, Brown M, Demaerschalk
BM, Hoh B, et al: 2018 Guidelines for the early management of
patients with acute ischemic stroke: A guideline for healthcare
professionals from the American heart association/American stroke
association. Stroke. 49:e46–e110. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li YY: Plasminogen activator inhibitor-1
4G/5G gene polymorphism and coronary artery disease in the Chinese
Han population: A meta-analysis. PLoS One. 7(e33511)2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Guney AI, Ergec D, Kirac D, Ozturhan H,
Caner M, Koc G, Kaspar C, Ulucan K and Agirbasli M: Effects of ACE
polymorphisms and other risk factors on the severity of coronary
artery disease. Genet Mol Res. 12:6895–6906. 2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Suzuki T, Yokota H, Yamazaki T, Kitamura
K, Yamaoki K, Nagai R and Yazaki Y: Angiotensin converting enzyme
polymorphism is associated with severity of coronary heart disease
and serum lipids (total cholesterol and triglycerides levels) in
Japanese patients. Coron Artery Dis. 7:371–375. 1996.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lee YJ and Tsai JC: ACE gene
insertion/deletion polymorphism associated with 1998 world health
organization definition of metabolic syndrome in Chinese type 2
diabetic patients. Diabetes Care. 25:1002–1008. 2002.PubMed/NCBI View Article : Google Scholar
|