Introduction
Oral squamous cell carcinoma (OSCC) is the most
common type of oral cancer and a major cause of head and neck
squamous cell carcinoma (HNSCC)-related morbidity and mortality
worldwide (1-3).
Although diagnosis and treatment modalities for OSCC, such as
chemoradiotherapy, surgery and photodynamic therapy, have
progressed (4,5), the 5-year survival rate of patients
with OSCC has not substantially improved over recent decades
(2,6,7). In
addition, the mechanisms underlying the onset and progression of
OSCC are not fully understood. Therefore, further studies are
required for the improvement of early diagnosis and targeted
therapy of OSCC.
Previous studies have suggested that microRNAs
(miRNAs; miRs) influence the onset and progression of tumors,
including OSCC, and can often be used as diagnostic and prognostic
biomarkers (8-14).
For example, Wu et al (15)
demonstrated that the miR-375/solute carrier family 7 member 11
axis regulates OSCC cell proliferation and invasion. In addition,
Cao et al (16) suggested
that miR-375 inhibits OSCC cell migration and invasion by targeting
platelet-derived growth factor-A. Among these miRNAs, miR-363-3p
has been identified as a tumor suppressor in several types of
cancer (17-20).
However, whether miR-363-3p also serves a role in OSCC remains
unclear.
Sperm-specific antigen 2 (SSFA2), also referred to
as K-Ras-induced actin-interacting protein, is upregulated in human
colon cancer (21). A previous
study has described the critical roles of SSFA2 as a potential
target in human diseases, particularly in cancer (22). Moreover, P38/MAPK inhibition
downregulates SSFA2 expression and promote the progression of
lymphoma (23).
In the present study, the expression levels and the
function of miR-363-3p were evaluated in OSCC cell lines. The
findings of the present study suggested that miR-363-3p could
inhibit the proliferation, migration and invasion of OSCC cell
lines. The potential downstream target and regulatory mechanism of
miR-363-3p were also examined. miR-363-3p exerted its effect on
OSCC cell lines by negatively regulating SSFA2 expression levels.
Thus, the present study provided insight into the role of
miR-363-3p in OSCC.
Materials and methods
Cell culture
The human SCC-9 (cat. no. CRL-1629), SCC-25 (cat.
no. CRL-1628) and Cal-27 (cat. no. CRL-2095) OSCC cell lines were
purchased from American Type Culture Collection. The immortalized
normal human oral keratinocyte (NHOK) were a gift from Dr Suyin Hu,
Department of Endocrinology, Wuhu Hospital of Traditional Chinese
Medicine's (Wuhu, China) laboratory by dissociated from the tissue
at Passage 4(24). Cells were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C in a humidified atmosphere of 5%
CO2.
Cell transfection
The miR-363-3p mimic, pcDNA3.1-SSFA2 overexpression
vector, as well as and their negative controls were obtained from
Shanghai GenePharma Co., Ltd. SSFA2 overexpression plasmid, empty
plasmid vector, mimic NC or miR-363-3p mimic were transfected into
SCC-9 and SCC-25 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at final concentration
of 10 µM according to the manufacturer's protocols. The miR-363-3p
mimic with the sense strand sequence 5'-AAUUGCACGGUAUCCAUCUGUA-3'
and the mimic NC sense strand sequence 5'-UUGUAC UACACAAAAGUACUG-3'
were used for the transient transfection of glioma cells. Reverse
transcription-quantitative PCR (RT-qPCR) was performed to assess
the transfection efficiency.
RT-qPCR
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from SCC-9
and SCC-25 cells. Equal amounts of RNA were reverse transcribed
into cDNA using the BeyoRT™ II First Strand cDNA Synthesis Kit
(Beyotime Institute of Biotechnology) according to the
manufacturer's protocols. The PCR program was as follows: 42˚C for
15 min, followed by 85˚C for 5 sec. The expression levels of
miR-363-3p and SSFA2 were then quantified by RT-qPCR, using One
Step SYBR PrimeScript RT-PCR kit (cat. no. DRR096A; Takara Bio,
Inc.). The thermocycling conditions consisted of an initial
denaturation at 95˚C for 10 min, followed by 40 cycles at 95˚C for
15 sec and 60˚C for 30 sec. U6 was used as the reference gene for
miR-363-3p and GADPH was used as endogenous control for SSFA2. The
2-ΔΔCq comparative method (25) was used to normalize the gene
expression levels. The specific primers are as follows: miR-363-3p
forward, 5'-GCCGAGAATTGCACGGTATC-3' and reverse,
5'-CTCAACTGGTGTCGTGGA-3'; SSFA2 forward, 5'-TGGCAAGAAAGGCCCCTGTG-3'
and reverse, 5'-GGAGCAGCAGCAGGATCAGG-3'; GAPDH forward,
5'-ACGGATTTGGTCGTATTGGGCG-3' and reverse,
5'-CTCCTGGAAGATGGTGATGG-3' and U6 forward, 5'-CTCGCTTCGGCAGCACA-3'
and reverse, 5'-AACGCTTCACGAATTTGCGT-3'.
Western blotting
After total protein (1 mg) extraction using RIPA
buffer (Thermo Fisher Scientific, Inc.), protein quantification was
determined by the bicinchoninic acid protein assay method. The
equivalent amount of total protein (5 µg) was segregated on 10%
SDS-PAGE and subsequently transferred to PVDF membranes (EMD
Millipore). Thereafter, the membranes were blocked in 5% non-fat
milk at 37˚C 1 h, followed by incubation with primary antibody at
4˚C for overnight and incubation with the secondary horseradish
peroxidase-conjugated antibodies (cat. nos. 7074 and 7076; 1:2,000;
Cell Signaling Technology, Inc.) at room temperature for 1 h. The
bands were visualized using enhanced chemiluminescence reagent
(cat. no. SW2030, Beijing Solarbio Science & Technology Co.,
Ltd.). The signals of target proteins were normalized to that of
β-actin and GAPDH. The relative expression of the proteins was
analyzed using Image J v1.8.0 software (National Institutes of
Health). The primary antibodies were as follows: Cyclin D1
(1:1,000; cat. no. ab238625), PCNA (1:1,000; cat. no. ab18197),
SSFA2 (1:1,000; cat. no. ab195334), Cox-2 (1:1,000; cat. no.
ab15191), MMP-2 (1:1,000; cat. no. ab86607), MMP-9 (1:1,000; cat.
no. ab58803), β-actin (1:1,000; cat. no. ab8226). Antibodies were
purchased from Abcam.
Cell Counting Kit-8 (CCK-8) and
5-ethynyl-2'-deoxyuridine (EdU) assays
Cell proliferation (1x104) was detected
by CCK-8 and EdU assays. Cells (1x104) were cultured in
96-well plates and CCK-8 assay was performed according to the
manufacturer's instructions (Beyotime Institute of Biotechnology).
The optical density values were then measured at 450 nm.
Cells (1x104) were cultured in six-well
plates and cell proliferation was also determined using EdU assays,
which were performed using an EdU Apollo DNA in vitro kit
(Guangzhou RiboBio Co., Ltd.), according to the manufacturer's
protocols. The cells were visualized under a fluorescence
microscope (magnification, x200).
Cell cycle analysis
For cell cycle analysis, OSCC cells were collected
and trypsinized after transfection. The cells were fixed in 75%
ethanol at room temperature for 20 min, treated with propidium
iodide (Sigma-Aldrich; Merck KGaA) at room temperature for 10 min.
The samples were then washed three times with PBS, and cell cycle
distribution was subsequently analyzed by flow cytometry BD
LSRFortessa™ X-20 (BD Biosciences) using the FlowJo V10.0 software
(BD Biosciences).
Cell migration assay
Transfected cells were cultured until 90%
confluence. Before making the wound, the DMEM medium was replaced
with fresh culture medium without FBS. The cells were then
scratched using 10-µl sterile pipette tips and incubated for 48 h
at 37˚C. The images were then captured at the 0 and 48 h timepoints
after wounding under a light microscope at x200 magnification. The
cell migratory ability was quantified by measuring the width of the
advancing margins of cells in three randomly selected microscopic
fields.
Transwell assay
Migration and invasion assays were performed
using Transwell chambers assay as described previously (26). The chambers were coated with
Matrigel (50 µg; BD Biosciences) 37˚C for 4 h. A total of
~1x106 cells were added to the top chamber of 24-well
plates in serum-free medium. The bottom chamber was filled with
complete medium supplemented with 20% FBS. After 24-h incubation,
the invasive or migratory cells were fixed with 4% formaldehyde at
room temperature for 15 min and stained with 0.1% crystal violet at
room temperature for 20 min. Transwell migration assay was
performed using a similar protocol but without Matrigel. The cells
were then counted and imaged from five fields of view per chamber
under a light microscope (magnification, x200). Migration
(Invasion) rate (%)=Migrated (Invaded) cells/total cells x100%.
Luciferase reporter assay
Bioinformatics tool Starbase v2.0 (https://starbase.sysu.edu.cn/) was used to predict
SSFA2 binding-sites for miR-363-3p. The 3'untranslated region (UTR)
of SSFA2 containing the wildtype (WT) or mutant (Mut) binding sites
for miR-363-3p were cloned into pmirGLO reporter vector (Promega
Corporation) to make the SSFA2-WT or SSFA2-Mut plasmids,
respectively. The SCC-9 and SCC-25 cell lines were co-transfected
with 150 ng pmirGLO luciferase reporter vectors (Promega
Corporation) containing SSFA2-wild-type (WT) or SSFA2-mutant (Mut)
(Shanghai GenePharma Co., Ltd.), together with miR-363-3p mimic or
mimic NC at final concentration of 10 µM using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The luciferase activities were detected at 48 h
post-transfection by the Dual-luciferase® reporter assay
system (Promega Corporation)with Renilla luciferase
activities used as internal control.
RNA pull-down assay
Cells were transfected with WT-bio-miR-363-3p,
MUT-bio-miR-363-3p or Bio-NC (GE Healthcare Dharmacon, Inc.)
labeled with 50 nM biotin using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.), which were obtained using biotin
RNA Labeling Mix (Roche Diagnostics). Cells (1x104) were
then pelleted at 1,200 x g for 5 min at room temperature after
transfected for 48 h. The cells were then resuspended in lysis
buffer (20 mM Tris, 5 mM MgCl2, 100 mM KCl, 0.3% NP-40
and 50 U RNase OUT; Invitrogen; Thermo Fisher Scientific, Inc.)
after washing twice with PBS and complete mini-protease inhibitor
cocktail (Roche Applied Science), which was incubated for 5 min on
ice. The cell lysate was isolated and collected by centrifugation
at 12,000 x g for 5 min at room temperature. Moreover, the miRNA
biotin pull-down experiments were conducted according to the
previous reports (27). RT-qPCR was
used to determine the mRNA levels in the precipitates.
Statistical analysis
Data are presented as the mean ± SD. All experiments
were independently conducted three times and statistical analysis
was performed using SPSS 21.0 software (IBM Corp.). Student's
t-test was used to determine the significant differences between
two groups and one-way ANOVA followed by Tukey's post hoc test were
used to analyze differences between > two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-363-3p inhibits the proliferation
of OSCC cells
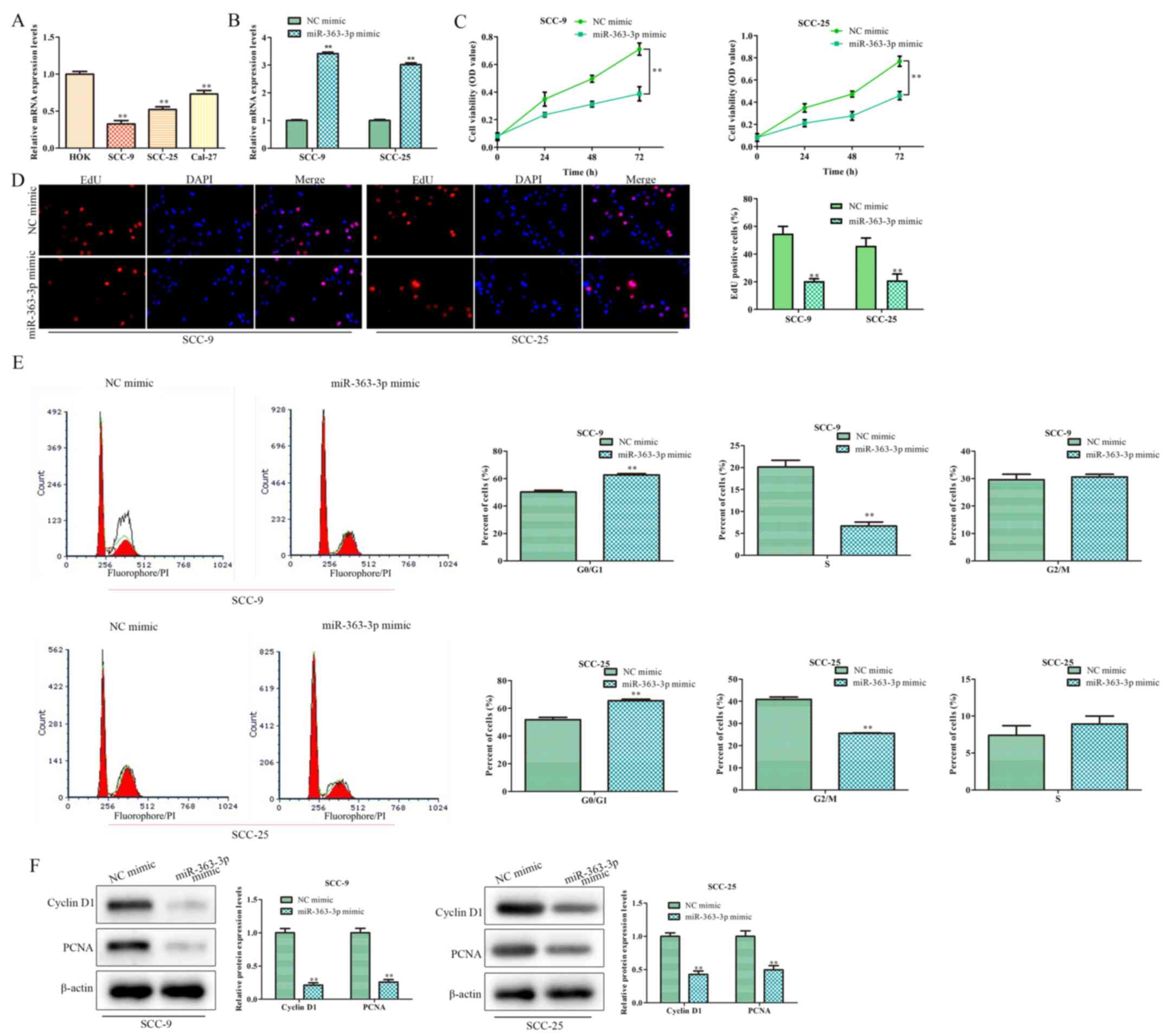
RT-qPCR was performed to evaluate the expression
levels of miR-363-3p in the human SCC-9, SCC-25 and Cal-27 OSCC
cell lines, and in the HOK normal epithelial cell line. The
expression levels of miR-363-3p were significantly lower in OSCC
cell lines, compared with HOK cells (Fig. 1A). As SCC-9 and SCC-25 cells
exhibited the lowest miR-363-3p expression levels, these cell lines
were used in subsequent experiments.
Additional experiments were conducted to determine
the potential effect of miR-363-3p on the proliferation of OSCC
cells. miR-363-3p mimic transfection was used to increase the
expression level of miR-363-3p, and transfection efficiency was
confirmed by RT-qPCR (Fig. 1B).
CCK-8 and EdU assays demonstrated that cell proliferation was
significantly reduced following transfection with the miR-363-3p
mimic, compared with the NC mimic-transfected groups (Fig. 1C and D, respectively). In addition, the number
of cells in G0/G1 phase significantly
increased, whilst the proportion of SCC-9 cells at the S phase was
significantly decreased (Fig. 1E).
No significant differences were observed at the G2 phase
following miR-363-3p overexpression. The proportion of SCC-25 cells
at the G2 phase was significantly decreased but no
significant differences at the S phase when miR-363-3p was
overexpressed (Fig. 1E). Moreover,
the expression levels of proliferation markers cyclin D1 and PCNA
were decreased in the presence of miR-363-3p mimic, compared with
NC mimic (Fig. 1F). Altogether,
these results suggested that miR-363-3p overexpression suppresses
the proliferation of OSCC cells.
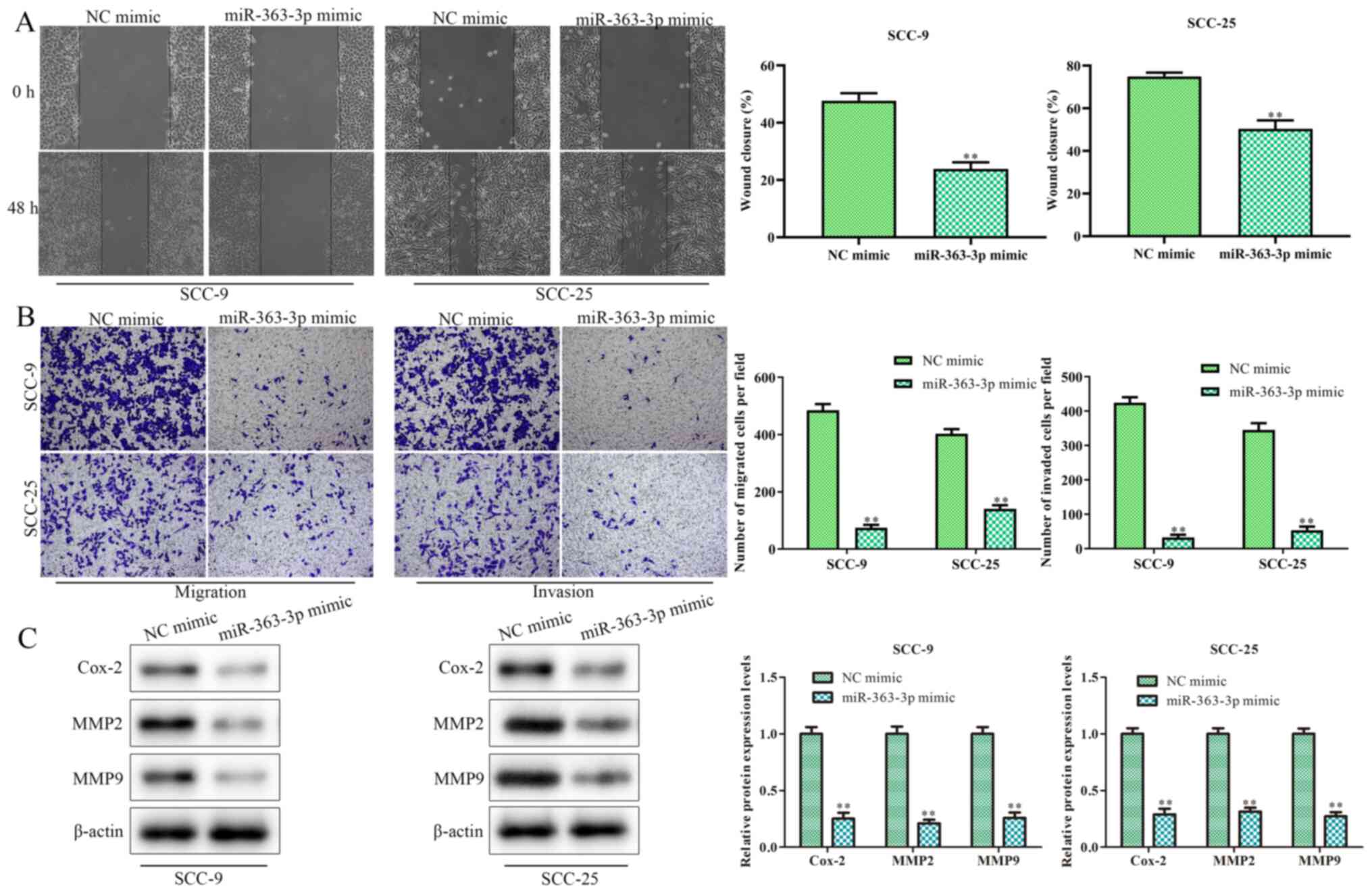
miR-363-3p inhibits OSCC cell
migration and invasion
The role of miR-363-3p in OSCC cell migration and
invasion was assessed using wound healing and Transwell Matrigel
assays, as well as western blotting. miR-363-3p overexpression
significantly decreased cell migration in the OSCC cell lines,
compared with the respective NC mimic-transfected group (Fig. 2A and B). Additionally, miR-363-3p mimic
transfection inhibited the invasive abilities of SCC-9 and SCC-25
cells (Fig. 2B). Furthermore,
western blotting was performed to detect the expression levels of
Cox-2, MMP-2 and MMP-9, proteins implicated in invasion (17). miR-363-3p overexpression resulted in
decreased levels of Cox-2, MMP-2 and MMP-9 in SCC-9 and SCC-25 cell
lines compared with expression levels in the respective control
groups (Fig. 2C). These results
suggested that miR-363-3p may inhibit the migration and invasion
potential of OSCC cells.
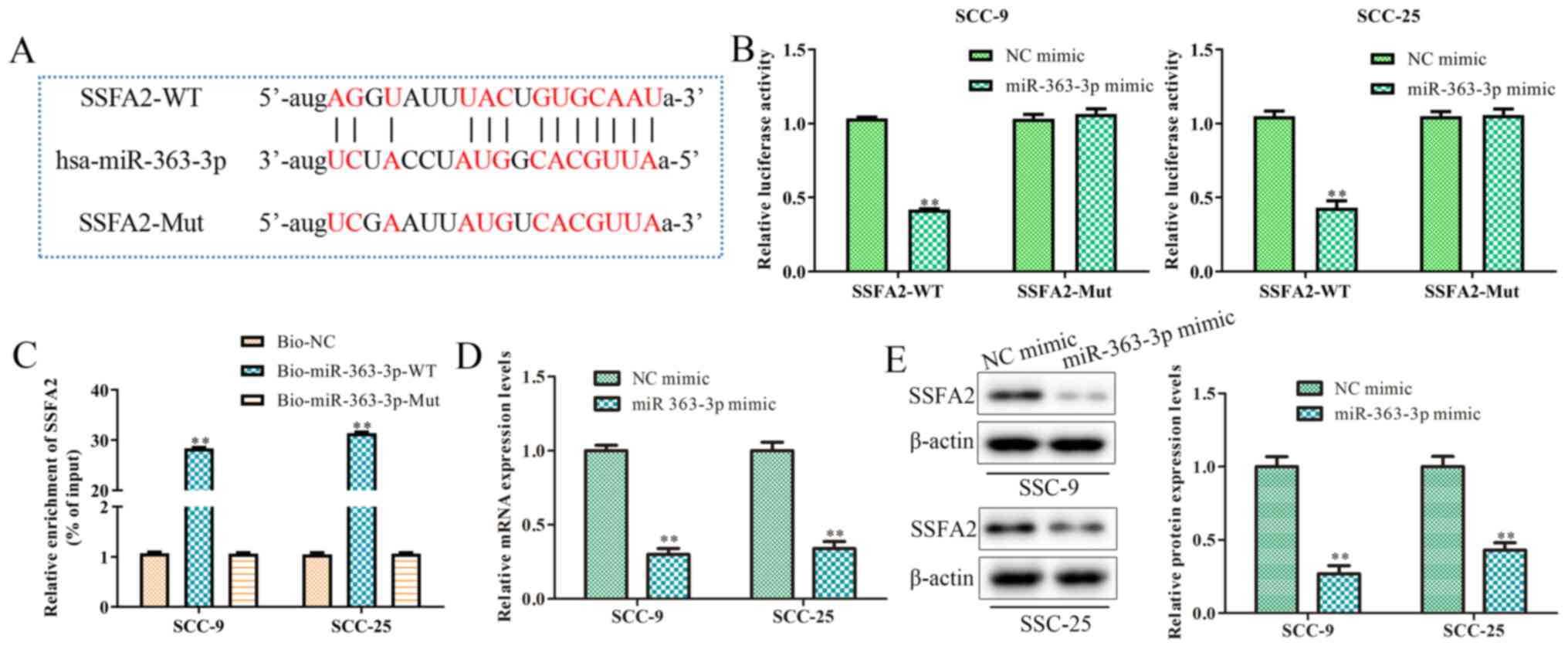
SSFA2 is a target of miR-363-3p
To better understand the mechanism underlying the
effect of miR-363-3p on OSCC cells, Starbase 2.0 was used, where it
was determined that SSFA2 was a potential target of miR-363-3p
(Fig. 3A). To confirm this
potential interaction between SSFA2 and miR-363-3p, a luciferase
reporter assay and an RNA pull-down experiment were performed.
Transfection with the miR-363-3p mimic reduced the activity of
SSFA2-WT by >50%, but not the activity of the SSFA2-Mut reporter
(Fig. 3B). Moreover, the RNA
pull-down assay indicated that SSFA2 was only pulled by
bio-miR-363-3p-WT (Fig. 3C).
Consistent with the aforementioned results, RT-qPCR and western
blotting also suggested that miR-363-3p overexpression
significantly decreased the expression of SSFA2 in SCC-9 and SCC-25
cells, both at the mRNA and the protein levels (Fig. 3D and E, respectively). Collectively, these
findings indicated that SSFA2 is a target of miR-363-3p.
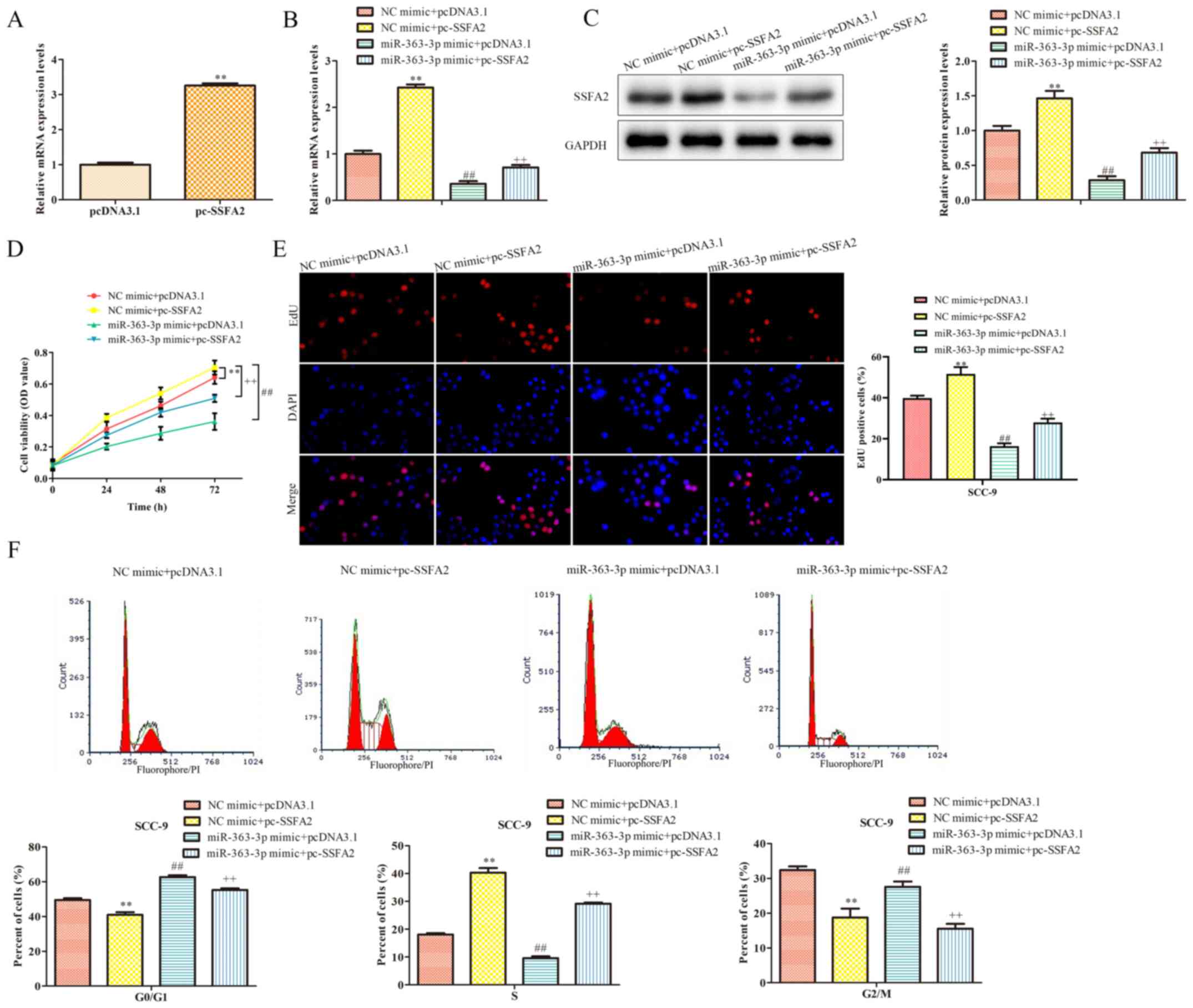
Effect of miR-363-3p on OSCC cell
proliferation by targeting SSFA2
To determine whether the functional role of
miR-363-3p in SSC-9 cells was mediated by SSFA2, co-transfection
experiments were conducted using the miR-363-3p mimic and an SSFA2
overexpression vector; SSFA2 overexpression was confirmed by
RT-qPCR (Fig. 4A). Transfection
with miR-363-3p mimic and empty pcDNA3.1 vector significantly
decreased SSFA2 mRNA and protein expression levels compared with
the control group (Fig. 4B and
C). By contrast, SSFA2
overexpression partially reversed the effect of miR-363-3p on these
expression levels. Moreover, the CCK-8 assay indicated that cell
proliferation was significantly reduced following transfection with
the mimic and empty vector (Fig.
4D); however, proliferation was restored following
co-transfection with pc-SSFA2. Data from the EdU assay confirmed
these results (Fig. 4E). Moreover,
SSFA2 overexpression partially counteracted the inhibitory effects
of miR-363-3p on cell cycle progression (Fig. 4F). Thus, it was demonstrated that
miR-363-3p may inhibit cell proliferation in OSCC cells lines
through SSFA2.
miR-363-3p inhibits the migratory and
invasive abilities of OSCC cells via targeting SSFA2
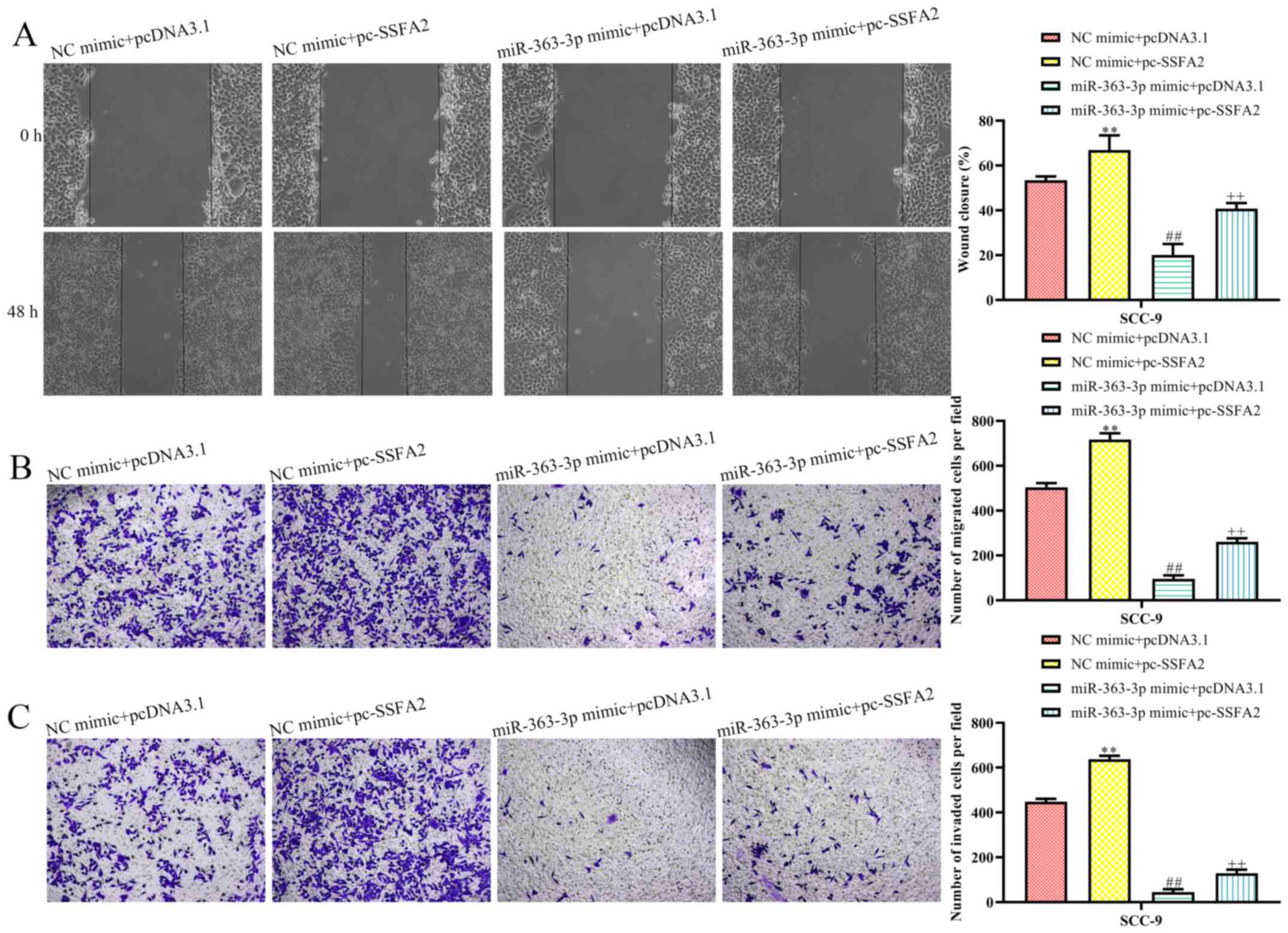
The role of the miR-363-3p/SSFA2 axis in cell
migration and invasion was also examined using co-transfection
experiments. Results from the wound healing assay demonstrated that
the wound closure rate significantly increased following SSFA2
overexpression (Fig. 5A).
Transfection with miR-363-3p mimic and empty pcDNA3.1 vector
significantly inhibited cell migration; however, the overexpression
of SSFA2 reversed the effects of miR-363-3p on cell migration
(Fig. 5A). Similar results were
observed in a Transwell migration assay (Fig. 5B). In addition, cell invasion was
also significantly decreased by miR-363-3p mimic and partially
restored following overexpression of SSFA2 (Fig. 5C). Altogether, these findings
suggested that miR-363-3p may inhibit OSCC cell migration and
invasion by targeting SSFA2.
Discussion
In the present study, miR-363-3p expression was
significantly downregulated in OSCC cells, compared with a normal
epithelial cell line. miR-363-3p overexpression in OSCC cells
significantly inhibited cell proliferation, migration and invasion.
Additionally, SSFA2 was demonstrated to be a direct target of
miR-363-3p, and overexpression of SSFA2 partially reversed the
inhibitory effect of miR-363-3p on OSCC cells. These findings
suggested that miR-363-3p exerted its inhibitory function in OSCC
cells by targeting SSFA2.
Previous studies have described the involvement of
miRNAs in OSCC pathogenesis. For example, miR-101 inhibits OSCC
growth and metastasis by targeting zinc finger E-box binding
homeobox 1. miR-155 regulates proliferation, cell cycle progression
and apoptosis through p27Kip1 in the Tca8113 OSCC cell line
(28). Rastogi et al
(29) demonstrated that knockdown
of miR-377 promoted OSCC growth and migration by targeting histone
deacetylase 9. Downregulated miR-363-3p expression has previously
been reported in colorectal cancer (18), head and neck cancer (27), and laryngeal cancer (28), and may function as a tumor
suppressor in these malignancies (29). However, to the best of our
knowledge, the role of miR-363-3p in OSCC has not yet been studied.
In the present study, the expression levels of miR-363-3p were
assessed in OSCC cell lines, and gain-of-function assays indicated
that overexpression of miR-363-3p reduced OSCC cell proliferation,
migration and invasion, suggesting that miR-363-3p may serve as a
tumor suppressor in OSCC.
Previous studies have demonstrated that miRNAs can
modulate gene expression levels by specifically binding to the 3'
untranslated regions of their target mRNAs, thereby repressing gene
expression at post-transcriptional level (30). Using bioinformatics analysis, it was
demonstrated in the present study that SSFA2 had potential
miR-363-3p target sites. SSFA2 has been described as a potential
target in colon cancer (21). A
recent study has suggested that SSFA2 deletion inhibits cell
proliferation and promotes cell apoptosis in glioma (31). Using co-transfection experiments,
the present study demonstrated that SSFA2 overexpression partly
abrogated the inhibitory effect of miR-363-3p mimic on malignant
OSCC. However, in vivo experiments are required to confirm
the present findings.
In conclusion, miR-363-3p serves an inhibitory role
on the proliferation and invasion of OSCC cells by suppressing
SSFA2 expression. Moreover, the discovery miR-363-3p/SSFA2 axis may
provide a new diagnostic and therapeutic strategy for OSCC.
Acknowledgements
The authors would like to express gratitude to Dr
Suyin Hu, Department of Endocrinology, Wuhu Hospital of Traditional
Chinese Medicine (Wuhu, China), who provided the NHOK cell
line.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PG designed the experiments. LZha, YT, FZ, CW, LZhu
and LX performed the experiments and analyzed the data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shi W, Yang J, Li S, Shan X, Liu X, Hua H,
Zhao C, Feng Z, Cai Z, Zhang L and Zhou D: Potential involvement of
miR-375 in the premalignant progression of oral squamous cell
carcinoma mediated via transcription factor KLF5. Oncotarget.
6:40172–401785. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zini A, Czerninski R and Sgan-Cohen HD:
Oral cancer over four decades: Epidemiology, trends, histology, and
survival by anatomical sites. J Oral Pathol Med. 39:299–305.
2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang Q, Gao P, Wang X and Duan Y: The
early diagnosis and monitoring of squamous cell carcinoma via
saliva metabolomics. Sci Rep. 4(6802)2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Omura K: Current status of oral cancer
treatment strategies: Surgical treatments for oral squamous cell
carcinoma. Int J Clin Oncol. 19:423–430. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
D'Souza G and Robbins HA: Sexual and
relationship health among survivors of oropharyngeal or oral cavity
squamous cell carcinoma. Cancer. 123:1092–1094. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Forastiere A, Koch W, Trotti A and
Sidransky D: Head and neck cancer. N Engl J Med. 345:1890–1900.
2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jadhav KB and Gupta N: Clinicopathological
prognostic implicators of oral squamous cell carcinoma: Need to
understand and revise. N Am J Med Sci. 5:671–679. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhao S, Yan L, Zhao Z and Rong F:
Up-regulation of miR-203 inhibits the growth of cervical cancer
cells by inducing cell cycle arrest and apoptosis. Eur J
Gynaecological Oncol. 40:791–795. 2019.
|
|
9
|
Karamitopoulou E, Haemmig S, Baumgartner
U, Schlup C, Wartenberg M and Vassella E: MicroRNA dysregulation in
the tumor microenvironment influences the phenotype of pancreatic
cancer. Mod Pathol. 30:1116–1125. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Paliouras AR, Monteverde T and Garofalo M:
Oncogene-induced regulation of microRNA expression: Implications
for cancer initiation, progression and therapy. Cancer Lett.
421:152–160. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ganci F, Sacconi A, Manciocco V, Covello
R, Benevolo M, Rollo F, Strano S, Valsoni S, Bicciato S, Spriano G,
et al: Altered peritumoral microRNA expression predicts head and
neck cancer patients with a high risk of recurrence. Mod Pathol.
30:1387–1401. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Katz B, Trope CG, Reich R and Davidson B:
MicroRNAs in ovarian cancer. Hum Pathol. 46:1245–1256.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Szabó Z, Szegedi K, Gombos K, Mahua C,
Flasko T, Harda K and Halmos G: Expression of miRNA-21 and
miRNA-221 in clear cell renal cell carcinoma (ccRCC) and their
possible role in the development of ccRCC. Urol Oncol.
34:533.e21–533.e27. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhao J, Dong X, Liu QC and Lu Q:
Expression of plasma miR-106a in epithelial ovarian cancer and its
diagnostic and prognostic significance. Eur J Gynaecol Oncol.
39:769–772. 2018.
|
|
15
|
Wu Y, Sun X, Song B, Qiu X and Zhao J:
miR-375/SLC7A11 axis regulates oral squamous cell carcinoma
proliferation and invasion. Cancer Med. 6:1686–1697.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cao ZH, Cheng JL, Zhang Y, Bo CX and Li
YL: MicroRNA-375 inhibits oral squamous cell carcinoma cell
migration and invasion by targeting platelet-derived growth
factor-A. Mol Med Rep. 15:922–928. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Dong J, Geng J and Tan W: miR-363-3p
suppresses tumor growth and metastasis of colorectal cancer via
targeting SphK2. Biomed Pharmacother. 105:922–931. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang Y, Chen T, Huang H, Jiang Y, Yang L,
Lin Z, He H, Liu T, Wu B, Chen J, et al: miR-363-3p inhibits tumor
growth by targeting PCNA in lung adenocarcinoma. Oncotarget.
8:20133–20144. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hu F, Min J, Cao X, Liu L, Ge Z, Hu J and
Li X: miR-363-3p inhibits the epithelial-to-mesenchymal transition
and suppresses metastasis in colorectal cancer by targeting Sox4.
Biochem Biophys Res Commun. 474:35–42. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chang J, Gao F, Chu H, Lou L, Wang H and
Chen Y: miR-363-3p inhibits migration, invasion, and
epithelial-mesenchymal transition by targeting NEDD9 and SOX4 in
non-small-cell lung cancer. J Cell Physiol. 235:1808–1820.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Inokuchi J, Komiya M, Baba I, Naito S,
Sasazuki T and Shirasawa S: Deregulated expression of KRAP, a novel
gene encoding actin-interacting protein, in human colon cancer
cells. J Hum Genet. 49:46–52. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kakiuchi S, Daigo Y, Tsunoda T, Yano S,
Sone S and Nakamura Y: Genome-wide analysis of organ-preferential
metastasis of human small cell lung cancer in mice. Mol Canaer Res.
1:485–499. 2003.PubMed/NCBI
|
|
23
|
Lin Z, Crockett DK, Jenson SD, Lim MS and
Elenitoba-Johnson KSJ: Quantitative proteomic and transcriptional
analysis of the response to the p38 mitogen-activated protein
kinase inhibitor SB203580 in transformed follicular lymphoma cells.
Mol Cell Proteomics. 3:820–833. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Min BM, Woo KM, Lee G and Park NH:
Terminal differentiation of normal human oral keratinocytes is
associated with enhanced cellular TGF-beta and phospholipase
C-gamma 1 levels and apoptotic cell death. Exp Cell Res.
249:377–385. 1999.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li L, Zhu X, Shou T, Yang L, Cheng X, Wang
J, Deng L and Zheng Y: MicroRNA-28 promotes cell proliferation and
invasion in gastric cancer via the PTEN/PI3K/AKT signalling
pathway. Mol Med Rep. 17:4003–4010. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lal A, Thomas MP, Altschuler G, Navarro F,
O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, et al:
Capture of microRNA-bound mRNAs identifies the tumor suppressor
miR-34a as a regulator of growth factor signaling. PLoS Genet.
7(e1002363)2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fu S, Chen HH, Cheng P, Zhang CB and Wu Y:
miR-155 regulates oral squamous cell carcinoma Tca8113 cell
proliferation, cycle, and apoptosis via regulating p27Kip1. Eur Rev
Med Pharmacol Sci. 21:937–944. 2017.PubMed/NCBI
|
|
29
|
Rastogi B, Kumar A, Raut SK, Panda NK,
Rattan V, Joshi N and Khullar M: Downregulation of miR-377 promotes
oral squamous cell carcinoma growth and migration by targeting
HDAC9. Cancer Invest. 35:152–162. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lu YC, Cheng AJ, Lee LY, You GR, Li YL,
Chen HY and Chang JT: miR-520b as a novel molecular target for
suppressing stemness phenotype of head-neck cancer by inhibiting
CD44. Sci Rep. 7(2042)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhu A, Li X, Wu H, Miao Z, Yuan F, Zhang
F, Wang B and Zhou Y: Molecular mechanism of SSFA2 deletion
inhibiting cell proliferation and promoting cell apoptosis in
glioma. Pathol Res Pract. 215:600–606. 2019.PubMed/NCBI View Article : Google Scholar
|