Non-specific low back pain (NLBP) is a prevalent and
debilitating musculoskeletal disorder that affects people globally
(1). NLBP causes a substantial
burden on medical resources and the economy (2-4).
It is estimated that the medical costs of NLBP in the USA alone are
~$253 billion per year (5). Efforts
have been made by clinicians and researchers to investigate the
pathogenesis of NLBP and develop effective treatment strategies.
Increasing evidence suggests that one of the major causes of NLBP
is intervertebral disc degeneration (IVDD) (6), which can be caused by inflammatory
factors (7), genetic factors
(8), aging (9), intervertebral instability (10) and metabolic disorders (11). Currently, the underlying molecular
mechanisms between these factors and IVDD have not yet been
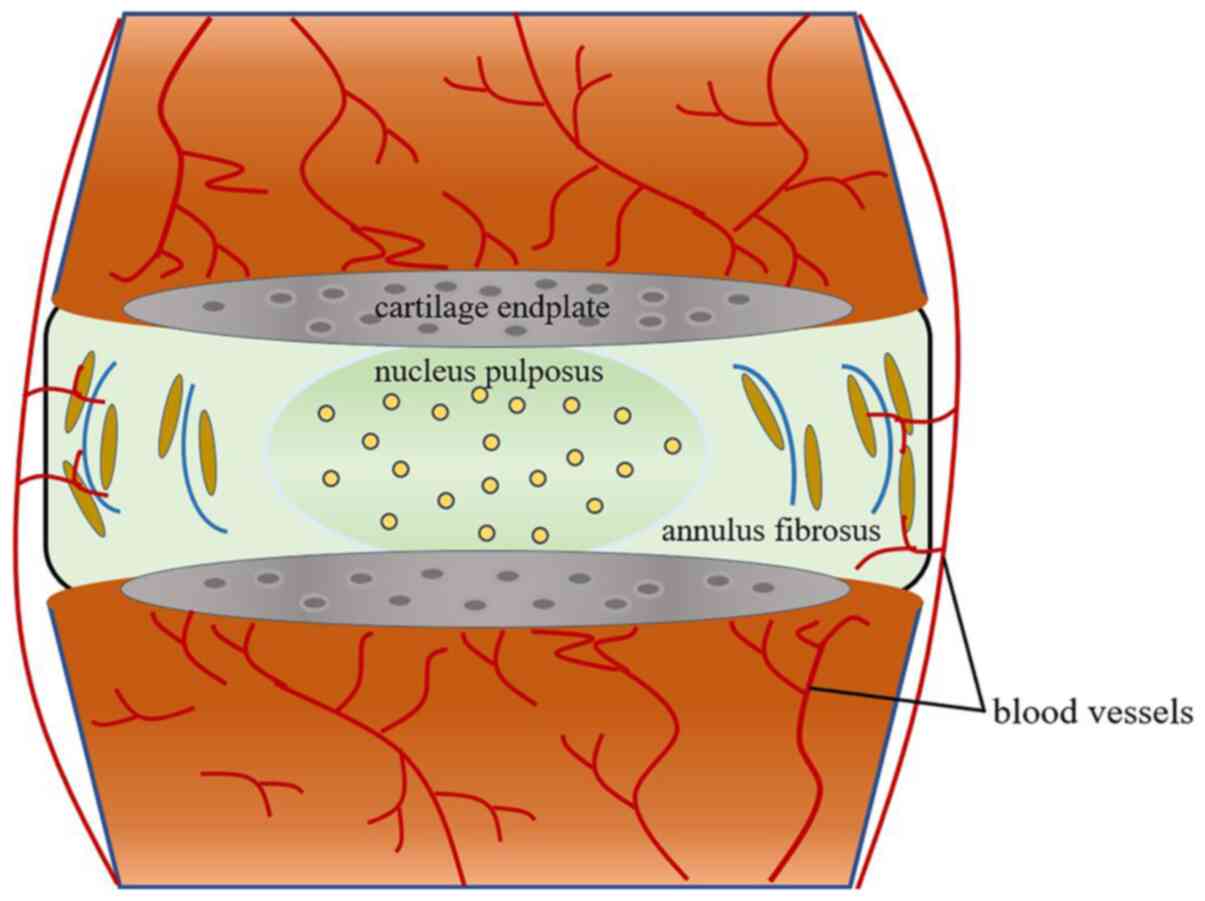
investigated. It is well known that the IVDD is composed of the
inner nucleus pulposus (NP); a proteoglycan-rich gelatinous
substance, the outer annulus fibrosus, as well as the upper and
lower cartilage endplates (CEP) (12) (Fig.
1). NP cells play an important role in secreting extracellular
matrix (ECM) components, such as type II collagen and aggrecan, in
addition to retaining water (13).
CEP, a crucial nutrition and metabolic exchange channel, maintains
the balance between catabolism and anabolism within IVDD (14,15).
Therefore, dysfunction of NP cells and CEP cells, including
apoptosis, senescence and abnormal cell proliferation, may cause an
imbalance between catabolism and anabolism, which is known to be
involved in the pathology of IVDD (16,17).
Non-coding RNAs form a large segment of RNA
molecules that are transcribed from DNA, but lack the potential to
be translated into proteins or peptides (18). Non-coding RNAs include short hairpin
RNA, small interfering RNA, antisense RNA, microRNA (miRNA), long
non-coding RNA (lncRNA), circular RNA (circRNA) and extracellular
RNAs (18-25).
Increasing evidence suggests that miRNAs, lncRNAs and circRNAs have
a vital regulatory function in the pathological process of several
diseases, such as cancer (26-31),
cardiac disease (32-35)
and IVDD (36-41).
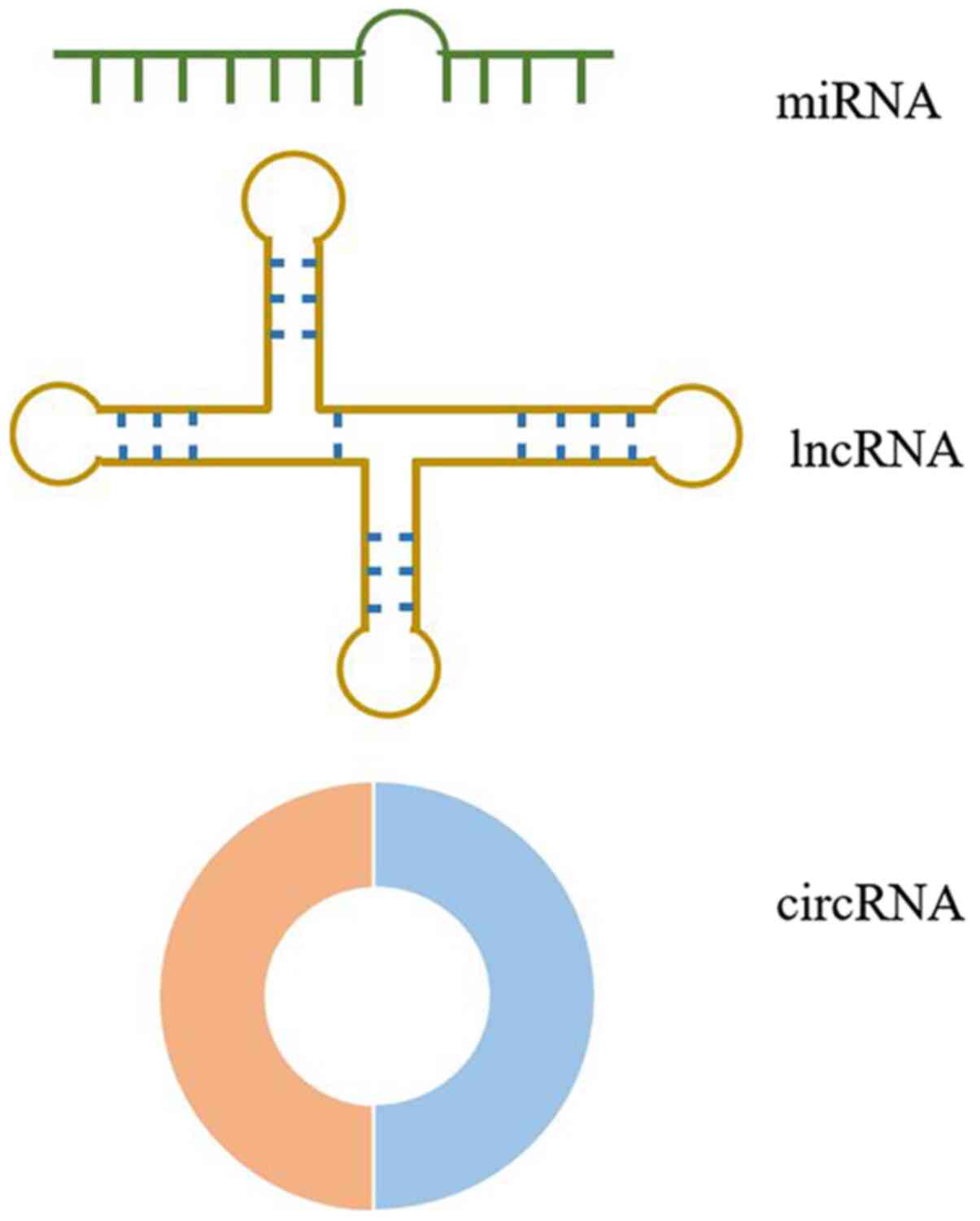
The structures of miRNAs, lncRNAs and circRNAs are presented in
Fig. 2 miRNAs, a class of small
non-coding RNAs that are 19-25 nucleotides in length, suppress gene
expression by directly binding to the 3'-untranslated regions
(UTR), 5'-UTR and coding sequence regions of their target mRNAs,
leading to translational repression and/or cleavage (42). This direct binding to 3'-UTR is the
primary method by which miRNAs regulate target genes (43-49).
Conversely, lncRNAs are the largest non-coding RNAs (>200
nucleotides in length) without an open reading frame (50). lncRNAs exert physiological functions
by modulating gene expression at multiple levels, including DNA
methylation, recruitment of transcriptional factors, miRNA sponges
and protein-protein interactions (50-55).
Unlike the linear structures of miRNAs and lncRNAs, circRNAs are
characterized by covalently closed single-stranded loop structures
without free 3' and 5' ends (56).
This structure hinders the digestion by ribonucleases R and
exonucleases (56-58).
circRNAs are produced by a precursor mRNA back-splicing mechanism
(59). Furthermore, they are widely
expressed in eukaryotes with cell type- and tissue-specific
patterns, acting as competing endogenous RNAs (ceRNAs) and
transcriptional regulators (29,60,61).
The present review article provides an overview of the role of
miRNAs, lncRNAs and circRNAs in the pathological process of IVDD
based on recent studies, in an attempt to clarify the diagnosis and
treatment of IVDD.
Evidence indicates that abnormal proliferation of NP
cells and formation of cell clusters are implicated in IVDD
pathogenesis (45). Li et al
(62) demonstrated that the
expression of miR-184 was positively associated with Pfirrmann
scores (63) and upregulated in
degenerative NP samples compared with that in normal NP samples.
Furthermore, luciferase assays from the same study indicated that
growth arrest specific gene 1 (GAS1) is a target of miR-184, and
degenerative NP tissues present low expression of GAS1 compared
with normal NP samples (59).
Functionally, overexpression of miR-184 can promote abnormal
proliferation and cluster formation of NP cells by inducing AKT
phosphorylation, which plays an important role in the development
of IVDD (62). However, unequivocal
evidence demonstrates that apoptosis exists in diverse biological
processes, including IVDD (64).
The expression levels of miR-138-5p and miR-494 in degenerated NP
tissues compared with normal tissue controls, and their effects on
apoptosis were investigated by Wang et al and Wang et
al (43,65). The aim of their research was to
identify the role of miRNAs in the pathogenesis of IVDD. A total of
two signaling pathways (miR-138-5p/SIRT1/PTEN/PI3K/Akt and
miR-494/JunD) were discovered through gain- and loss-of-function
studies. The results demonstrated that miR-138-5p (43) and miR-494(65) promote tumor necrosis factor-α
(TNF-α)-induced apoptosis of NP cells in IVDD by targeting silent
mating type information regulation 2 homolog-1 and the
transcription factor jun-D via the PTEN/PI3K/AKT signaling pathway
and cytochrome c apoptotic signaling, respectively.
Recent studies have reported an imbalance between
anabolism and catabolism of ECM in the development of IVDD,
predominantly due to excessive ECM degradation. Wang et al
and Wang et al (37,66) investigated whether miR-210 and
miR-21 facilitate the degradation of ECM components, such as type
II collagen and aggrecan within NP tissues. The results indicated
that the expression levels of miR-210 and miR-21 are significantly
upregulated in degenerated NP specimens compared with healthy
controls. Furthermore, miR-210 and miR-21 expression exhibited a
positive association with the grade of IVDD disease, using miRNA
microarray and reverse transcription-quantitative (RT-q)PCR
validation assays. Knockdown and overexpression of miR-210/miR-21
were followed by observation of downstream target genes and
ECM-related gene expression compared with the control group. The
aforementioned gain- and loss-of-function studies demonstrated that
miR-210 and miR-21 promote ECM degradation by suppressing
autophagy, targeting both the autophagy-related protein 7 and the
PTEN/AKT/mTOR signaling pathway in human NP cells. Conversely,
several miRNAs are downregulated in degenerative NP tissues,
indicating that miRNAs may exert a protective effect on normal NP
tissues against degeneration (45).
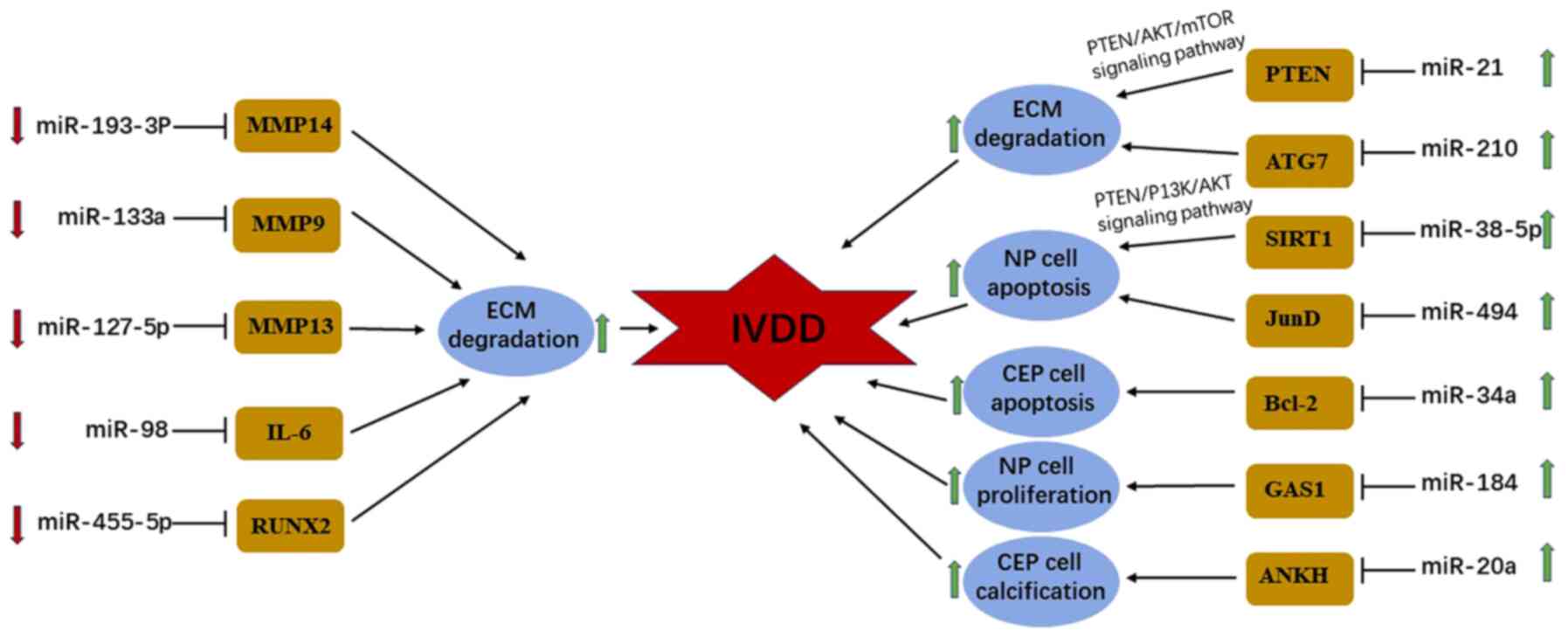
Studies have indicated that 51 miRNAs are differentially expressed
in degenerated intervertebral discs compared with normal
intervertebral discs (67). Of
these, downregulation of miR-127-5p, miR-193a-3p, miR-133a and
miR-98 induce loss of ECM components by targeting matrix
metalloproteinase (MMP)-13, MMP-14, MMP-9 and interleukin-6,
respectively (68-71).
Other miRNAs that have not been studied further may be found to
have no differential expression using (RT-q)PCR.
The dysregulation of cell proliferation, matrix
hardness and ECM degradation of CEP are also involved in the
progression of IVDD (72). Chen
et al (72) performed a
RT-qPCR analysis, which verified that the expression of miR-34a is
markedly elevated in the CEP samples obtained from patients with
IVDD compared with samples of healthy donors. Functionally,
apoptosis and proliferation of CEP cells are facilitated by
upregulating miR-34a through targeting Bcl-2. Liu et al and
Xiao et al (73,74) investigated the underlying molecular
mechanisms of miR-20a and miR-455-5p in the pathogenesis of IVDD.
The results demonstrated that matrix stiffness and ECM loss of CEP
are positively associated with the degree of IVDD. Overexpression
of miR-20a, which is upregulated in degenerative CEP tissues,
accelerates the development of IVDD and facilitates calcification
in CEP cells resulting in matrix stiffness by suppressing the
expression of ankylosis protein homolog. Similarly, enforced
expression of miR-455-5p, which is downregulated in degenerative
CEP samples, promotes the progression of IVDD and increases ECM
loss by targeting Runt-related transcription factor 2(73). Based on these findings, it is
speculated that miRNA may serve as a potential novel therapeutic
target for IVDD (Fig. 3).
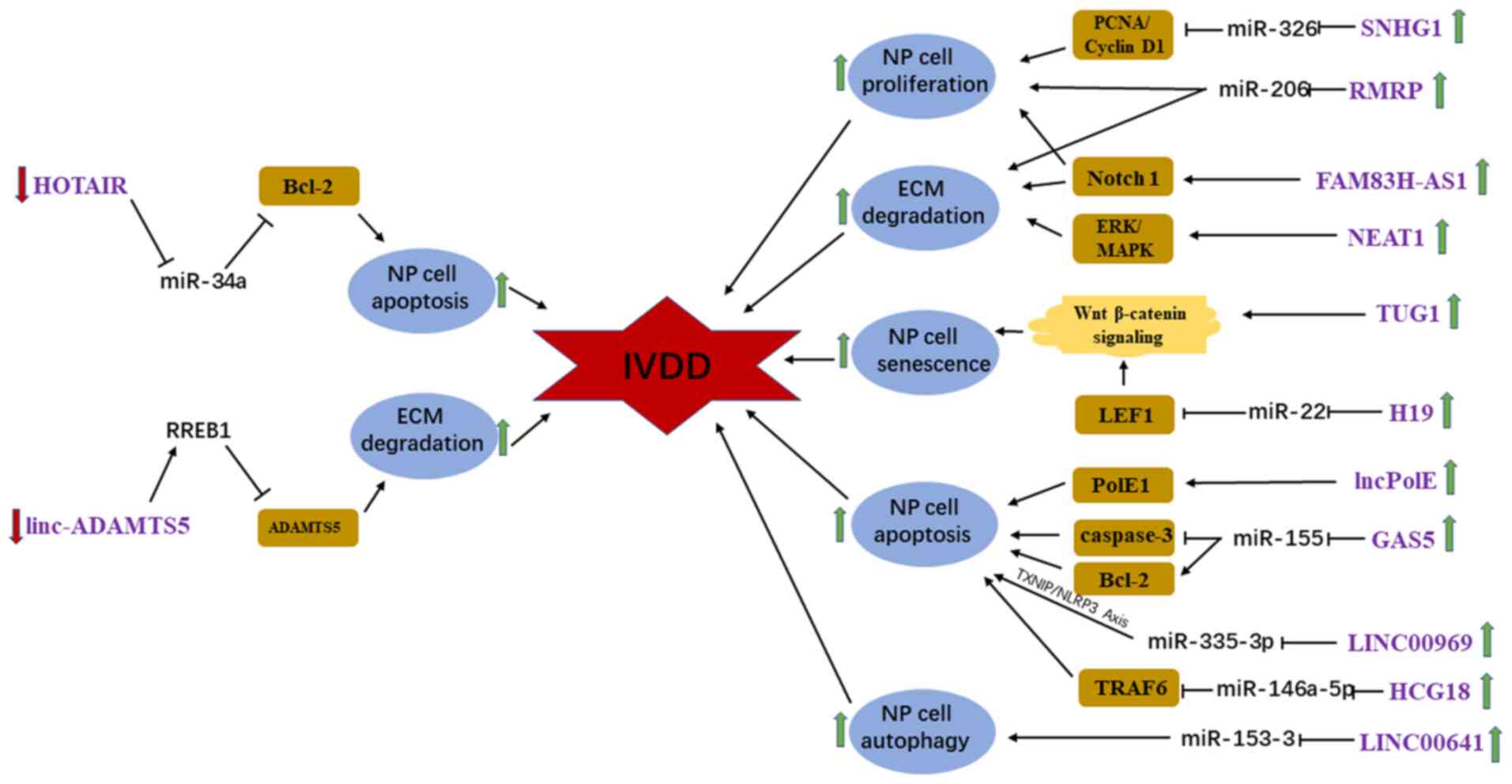
Evidence indicates that lncRNAs are involved in the
pathological process of IVDD and play a key role in relevant
signaling axes (75). Previous
studies have demonstrated that the ectopic expression of homeobox
transcript antisense intergenic RNA (HOTAIR), lncPolE, growth
arrest specific 5, long intergenic non-protein coding RNA 969 and
HLA complex group 18 (HCG18) contributes to initiation of IVDD by
inducing apoptosis of NP cells through diverse signaling pathways
(76-80).
Of these lncRNAs, HOTAIR is downregulated in degenerative NP
samples and inhibits TNF-α-induced apoptosis of NP cells by
regulating Bcl-2 through sponging miR-34a (76). However, the other aforementioned
lncRNAs are markedly upregulated in degenerative NP tissues and
promote apoptosis of NP cells by targeting DNA polymerase Ε
catalytic subunit A, miR-155, miR-335-3p and miR-146a-5p,
respectively (77-80).
HCG18 increases the rate of apoptosis of NP cells and inhibits the
proliferation of NP cells through the miR-146a-5p/TNF
receptor-associated factor 6/NFκB axis (80). Tan et al, Wang et al
and Wei et al (39,81,82)
first demonstrated that ectopic expression of small nucleolar RNA
host gene 1, RNA component of mitochondrial RNA processing
endoribonuclease (RMRP) and IQ motif and ankyrin repeat containing
(FAM83H-AS1), which are substantially upregulated in IVDD samples
compared with control samples, promote the progression of IVDD by
enhancing NP cell proliferation. Mechanistically, they suppress the
expression of miR-326, miR-206 and Notch1 to promote NP cell
proliferation. Furthermore, RMRP and FAM83H-AS1 also demonstrate
the ability to modulate the expression of ECM components, including
type II collagen and aggrecan (39,81,82).
Ruan et al and Wang et al (83,84)
confirmed that nuclear paraspeckle assembly transcript 1 (NEAT1)
and long intergenic non-protein coding RNA (linc)-A disintegrin and
MMP with thrombospondin motifs (ADAMTS)5 play crucial roles in the
progression of IVDD by regulating the balance between synthesis and
degradation of the ECM. However, the expression levels of NEAT1 and
linc-ADAMTS5 are different in NP tissues isolated from patients
with IVDD. In IVDD, NEAT1 and linc-ADAMTS5 are notably upregulated
and downregulated, respectively. Functionally, NEAT1 promotes ECM
degradation by upregulating MMP-13 and ADAMTS4 (genes encoding
ECM-associated enzymes), and downregulating collagen II and
aggrecan through the ERK/mitogen-activated protein kinase signaling
pathway. Linc-ADAMTS5 interacts with Ras-responsive element-binding
protein 1 to suppress the degradation of ECM and inhibit the
expression of ADAMTS5 (83,84). Notably, it was identified that two
different lncRNAs, taurine upregulated gene 1 (TUG1) and H19
imprint maternally expressed transcript (H19), modulate NP cell
senescence, apoptosis and ECM synthesis through the Wnt/β-catenin
signaling pathway (85,86). Functionally, TUG1 and H19, which are
both upregulated in degenerative NP tissues, promote NP cell
senescence, apoptosis and ECM degradation by targeting
Wnt/β-catenin and miR-22, respectively (85,86). A
recent study by Wang et al (87) focused on the role of autophagy in
the pathogenesis of IVDD and demonstrated that the long intergenic
non-protein coding RNA 641, which is markedly upregulated in NP
samples obtained from patients with IVDD compared with controls,
regulate the development of IVDD by inducing autophagic cell death
through targeting miR-153-3p and autophagy-related gene 5. In
addition, some treatments can be used to target lncRNAs in IVDD,
such as silencing of lncRNAs, locked nucleic acid GapmeRs, small
molecule inhibitors, antisense nucleotides and zinc-finger
nucleases (88). lncRNAs may
represent potential effective novel targets for the treatment of
IVDD (Fig. 4).
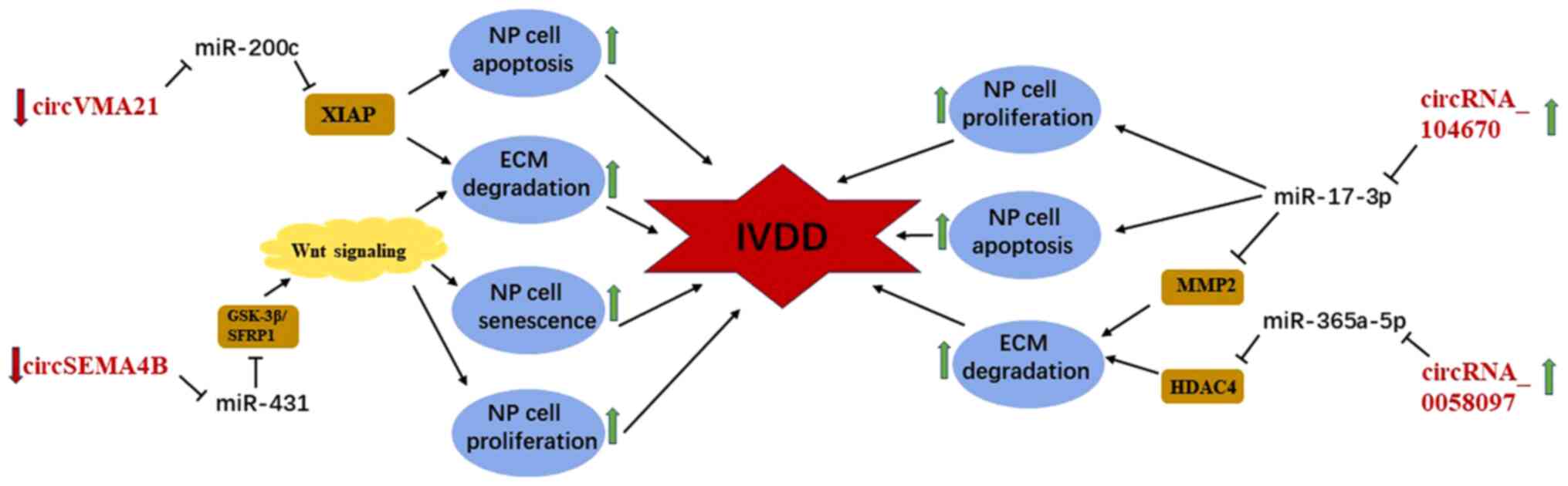
circRNAs are involved in the regulation of manifold
diseases as a novel subtype of non-coding RNAs. Cheng et al
(40) were the first to demonstrate
that circVMA21 derived from vacuolar ATPase assembly factor gene is
markedly decreased in the degenerative NP specimens compared with
the normal NP tissues based on RT-qPCR analyses. Functionally,
circVMA21 is able to protect against IVDD by suppressing
inflammatory cytokine-induced NP cell apoptosis, downregulating the
expression of catabolic enzymes (MMP-3, MMP-13, ADAMTS4 and
ADAMTS5) and promoting synthesis of ECM. Mechanistically, circVMA21
is expected to function as ceRNAs to modulate the pathological
process of IVDD through sponging miR-200c and targeting X-linked
inhibitor-of-apoptosis protein. Recently, Wang et al
(89) analyzed the expression
profiling of human lumbar disc circRNAs based on an online database
and reported that circSEMA4B is substantially downregulated in
degenerative lumbar disc tissues. Functionally, circSEMA4B can
inhibit the development of IVDD by enhancing NP cell proliferation
and alleviating cell senescence and ECM degradation.
Mechanistically, circSEMA4B is a potential therapeutic target for
IVDD as it represses miR-431 via the Wnt/β-catenin signaling
pathway (89). However,
circRNA_104670 and circRNA_0058097 are upregulated in degenerative
NP tissues and tension-induced degenerative endplate chondrocytes,
and it has been reported that they promote the progression of IVDD
by acting as ceRNAs (41,90). Furthermore, Song et al
(41) confirmed via the
dual-luciferase and EGFP/RFP reporter assays that circRNA_104670
directly binds to miR-17-3p, while MMP-2 is the direct target of
miR-17-3p. Knockdown and overexpression of circRNA 104670 was
followed by the observation of proliferation and apoptosis of NP
cells and the expression of miR-17-3p and ECM-related gene compared
with the control group. Functionally, through gain- and
loss-of-function studies, circRNA_104670 was demonstrated to
inhibit proliferation of NP cells and expression of collagen II,
and promote apoptosis and the expression of MMP-2 by targeting
miR-17-3p and MMP-2. Xiao et al (90) reported that circRNA_0058097 may
promote morphological changes of endplate chondrocytes and enhance
ECM degradation and degeneration of IVDs by upregulating the
expression of histone deacetylase 4 through sponging miR-365a-5p.
Thus, circRNA_0058097 promotes the pathological process of IVDD by
regulating tension-induced degeneration of endplate chondrocytes.
CircRNAs modulate the development of IVDD by functioning as ceRNAs
(90) and may serve as a potential
novel therapeutic target of IVDD, similar to miRNAs and lncRNAs
(Fig. 5).
As one of the most prevalent diseases among the
elderly population, NLBP has caused tremendous pressure on medical
resources and the economy. Several studies have demonstrated that
IVDD is responsible for the pathogenesis of NLBP; however, its
underlying molecular and cellular mechanisms remain unclear.
Recently, the role of non-coding RNAs in several diseases emerged,
including IVDD.
In the present review, the role of miRNAs, lncRNAs
and circRNAs in the progression of IVDD is summarized. Furthermore,
it presents a summary of how to modulate the proliferation,
senescence, apoptosis and ECM degradation of NP and CEP by
regulating downstream target genes (Tables
I-III). The data presented in the current review provide novel
insights into the etiology of IVDD and identifies non-coding RNAs
as a potential novel target for the treatment of IVDD. However,
there is still a lack of relevant studies on miRNAs and circRNAs as
therapeutic targets for IVDD. With the development of nanoparticle
technology and an in-depth understanding of the pathogenesis of
IVDD, research on non-coding RNAs, particularly miRNAs, lncRNAs and
circRNAs as therapeutic targets for the treatment of IVDD have
potential to become a novel research focus.
Not applicable.
Funding: The present review was supported by The National
Natural Science Foundation of China (NSFC; grant no. 31971275).
Not applicable.
HW and LW designed the present review. JJ, YS and GX
performed the literature review and drafted the initial manuscript.
HW and LW critically revised the manuscript for important
intellectual content. All authors have read and approved the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Maher C, Underwood M and Buchbinder R:
Non-specific low back pain. Lancet. 389:736–747. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Dudli S, Fields AJ, Samartzis D, Karppinen
J and Lotz JC: Pathobiology of modic changes. Eur Spine J.
25:3723–3734. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vos T, Flaxman AD, Naghavi M, Lozano R,
Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V,
et al: Years lived with disability (YLDs) for 1160 sequelae of 289
diseases and injuries 1990-2010: A systematic analysis for the
global burden of disease study 2010. Lancet. 380:2163–2196.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hartvigsen J, Hancock MJ, Kongsted A, Louw
Q, Ferreira ML, Genevay S, Hoy D, Karppinen J, Pransky G, Sieper J,
et al: What low back pain is and why we need to pay attention.
Lancet. 391:2356–2367. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yelin E, Weinstein S and King T: The
burden of musculoskeletal diseases in the United States. Semin
Arthritis Rheum. 46:259–260. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Foster NE, Anema JR, Cherkin D, Chou R,
Cohen SP, Gross DP, Ferreira PH, Fritz JM, Koes BW, Peul W, et al:
Prevention and treatment of low back pain: Evidence, challenges,
and promising directions. Lancet. 391:2368–2383. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration, and what causes it? Spine (Phila
Pa 1976). 31:2151–2161. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vo NV, Hartman RA, Patil PR, Risbud MV,
Kletsas D, Iatridis JC, Hoyland JA, Le Maitre CL, Sowa GA and Kang
JD: Molecular mechanisms of biological aging in intervertebral
discs. J Orthop Res. 34:1289–1306. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bian Q, Jain A, Xu X, Kebaish K, Crane JL,
Zhang Z, Wan M, Ma L, Riley LH, Sponseller PD, et al: Excessive
activation of TGFβ by spinal instability causes vertebral endplate
sclerosis. Sci Rep. 6(27093)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Huang YC, Urban JP and Luk KD:
Intervertebral disc regeneration: Do nutrients lead the way? Nat
Rev Rheumatol. 10:561–566. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liao Z, Wu X, Song Y, Luo R, Yin H, Zhan
S, Li S, Wang K, Zhang Y and Yang C: Angiopoietin-like protein 8
expression and association with extracellular matrix metabolism and
inflammation during intervertebral disc degeneration. J Cell Mol
Med. 23:5737–5750. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu X, Zhuang J, Wang D, Lv L, Zhu F, Yao
A and Xu T: Glycyrrhizin suppresses inflammation and cell apoptosis
by inhibition of HMGB1 via p38/p-JUK signaling pathway in
attenuating intervertebral disc degeneration. Am J Transl Res.
11:5105–5113. 2019.PubMed/NCBI
|
|
14
|
Grant MP, Epure LM, Bokhari R, Roughley P,
Antoniou J and Mwale F: Human cartilaginous endplate degeneration
is induced by calcium and the extracellular calcium-sensing
receptor in the intervertebral disc. Eur Cell Mater. 32:137–151.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yao Y, Deng Q, Song W, Zhang H, Li Y, Yang
Y, Fan X, Liu M, Shang J, Sun C, et al: MIF plays a key role in
regulating tissue-specific chondro-osteogenic differentiation fate
of human cartilage endplate stem cells under hypoxia. Stem Cell
Reports. 7:249–262. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tang Z, Hu B, Zang F, Wang J, Zhang X and
Chen H: Nrf2 drives oxidative stress-induced autophagy in nucleus
pulposus cells via a Keap1/Nrf2/p62 feedback loop to protect
intervertebral disc from degeneration. Cell Death Dis.
10(510)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang G, Huang K, Dong Y, Chen S, Zhang J,
Wang J, Xie Z, Lin X, Fang X and Fan S: Lycorine suppresses
endplate-chondrocyte degeneration and prevents intervertebral disc
degeneration by inhibiting NF-κB signalling pathway. Cell Physiol
Biochem. 45:1252–1269. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Matsui M and Corey DR: Non-coding RNAs as
drug targets. Nat Rev Drug Discov. 16:167–179. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ning B, Yu D and Yu AM: Advances and
challenges in studying noncoding RNA regulation of drug metabolism
and development of RNA therapeutics. Biochem Pharmacol.
169(113638)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang WT, Han C, Sun YM, Chen TQ and Chen
YQ: Noncoding RNAs in cancer therapy resistance and targeted drug
development. J Hematol Oncol. 12(55)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346.
2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sato-Kuwabara Y, Melo SA, Soares FA and
Calin GA: The fusion of two worlds: Non-coding RNAs and
extracellular vesicles-diagnostic and therapeutic implications
(Review). Int J Oncol. 46:17–27. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhou X, Chen L, Grad S, Alini M, Pan H,
Yang D, Zhen W, Li Z, Huang S and Peng S: The roles and
perspectives of microRNAs as biomarkers for intervertebral disc
degeneration. J Tissue Eng Regen Med. 11:3481–3487. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou L, Dong S, Deng Y, Yang P, Zheng Y,
Yao L, Zhang M, Yang S, Wu Y, Zhai Z, et al: GOLGA7 rs11337, a
polymorphism at the MicroRNA binding site, is associated with
glioma prognosis. Mol Ther Nucleic Acids. 18:56–65. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye
X, Zhang H, Yang P, Cui X, Ren Y, et al: A noncoding regulatory
RNAs network driven by Circ-CDYL acts specifically in the early
stages hepatocellular carcinoma. Hepatology. 71:130–147.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Shang Q, Yang Z, Jia R and Ge S: The novel
roles of circRNAs in human cancer. Mol Cancer. 18(6)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wu K, Liao X, Gong Y, He J, Zhou JK, Tan
S, Pu W, Huang C, Wei YQ and Peng Y: Circular RNA F-circSR derived
from SLC34A2-ROS1 fusion gene promotes cell migration in non-small
cell lung cancer. Mol Cancer. 18(98)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y,
Huang K, Wang G, Wang J, Ma J, et al: Circular RNA circTADA2A
promotes osteosarcoma progression and metastasis by sponging
miR-203a-3p and regulating CREB3 expression. Mol Cancer.
18(73)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lange S, Banerjee I, Carrion K, Serrano R,
Habich L, Kameny R, Lengenfelder L, Dalton N, Meili R, Börgeson E,
et al: miR-486 is modulated by stretch and increases ventricular
growth. JCI Insight. 4(e125507)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Calderon-Dominguez M, Belmonte T,
Quezada-Feijoo M, Ramos-Sánchez M, Fernández-Armenta J,
Pérez-Navarro A, Cesar S, Peña-Peña L, Vea À, Llorente-Cortés V, et
al: Emerging role of microRNAs in dilated cardiomyopathy: Evidence
regarding etiology. Transl Res. 215:86–101. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Cai B, Zhang Y, Zhao Y, Wang J, Li T,
Zhang Y, Jiang Y, Jin X, Xue G, Li P, et al: Long noncoding
RNA-DACH1 (Dachshund Homolog 1) regulates cardiac function by
inhibiting SERCA2a (Sarcoplasmic Reticulum Calcium ATPase 2a).
Hypertension. 74:833–842. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
super-enhancer-regulated circRNA Nfix induces cardiac regeneration
after myocardial infarction in adult mice. Circulation.
139:2857–2876. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hasvik E, Schjølberg T, Jacobsen DP,
Haugen AJ, Grøvle L, Schistad EI and Gjerstad J: Up-regulation of
circulating microRNA-17 is associated with lumbar radicular pain
following disc herniation. Arthritis Res Ther.
21(186)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang C, Zhang ZZ, Yang W, Ouyang ZH, Xue
JB, Li XL, Zhang J, Chen WK, Yan YG and Wang WJ: MiR-210
facilitates ECM degradation by suppressing autophagy via silencing
of ATG7 in human degenerated NP cells. Biomed Pharmacother.
93:470–479. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Shao T, Hu Y, Tang W, Shen H, Yu Z and Gu
J: The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge
to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated
mechanism. Gene. 715(144029)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tan H, Zhao L, Song R, Liu Y and Wang L:
The long noncoding RNA SNHG1 promotes nucleus pulposus cell
proliferation through regulating miR-326 and CCND1. Am J Physiol
Cell Physiol. 315:C21–C27. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Song J, Wang HL, Song KH, Ding ZW, Wang
HL, Ma XS, Lu FZ, Xia XL, Wang YW, Fei-Zou and Jiang JY:
CircularRNA_104670 plays a critical role in intervertebral disc
degeneration by functioning as a ceRNA. Exp Mol Med.
50(94)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang B, Wang D, Yan T and Yuan H:
miR-138-5p promotes TNF-α-induced apoptosis in human intervertebral
disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt
signaling. Exp Cell Res. 345:199–205. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang C, Wang WJ, Yan YG, Xiang YX, Zhang
J, Tang ZH and Jiang ZS: MicroRNAs: New players in intervertebral
disc degeneration. Clin Chim Acta. 450:333–341. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Mo YY: MicroRNA regulatory networks and
human disease. Cell Mol Life Sci. 69:3529–3531. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ivey KN and Srivastava D: microRNAs as
developmental regulators. Cold Spring Harb Perspect Biol.
7(a008144)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Liu B, Li J and Cairns MJ: Identifying
miRNAs, targets and functions. Brief Bioinform. 15:1–19.
2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8(1015)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Robinson EK, Covarrubias S and Carpenter
S: The how and why of lncRNA function: An innate immune
perspective. Biochim Biophys Acta Gene Regul Mech.
1863(194419)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ji E, Kim C, Kim W and Lee EK: Role of
long non-coding RNAs in metabolic control. Biochim Biophys Acta
Gene Regul Mech. 1863(194348)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan
YG, Zhang J and Wang WJ: lncRNAs: Novel players in intervertebral
disc degeneration and osteoarthritis. Cell Prolif.
50(e12313)2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46.
2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Santer L, Bär C and Thum T: Circular RNAs:
A novel class of functional RNA molecules with a therapeutic
perspective. Mol Ther. 27:1350–1363. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Suzuki H, Zuo Y, Wang J, Zhang MQ,
Malhotra A and Mayeda A: Characterization of RNase R-digested
cellular RNA source that consists of lariat and circular RNAs from
pre-mRNA splicing. Nucleic Acids Res. 34(e63)2006.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461.
2014.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691.
2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Li W, Wang P, Zhang Z, Wang W, Liu Y and
Qi Q: miR-184 regulates proliferation in nucleus pulposus cells by
targeting GAS1. World Neurosurg. 97:710–715.e1. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Che YJ, Guo JB, Liang T, Chen X, Zhang W,
Yang HL and Luo ZP: Assessment of changes in the micro-nano
environment of intervertebral disc degeneration based on Pfirrmann
grade. Spine J. 19:1242–1253. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Yang SD, Yang DL, Sun YP, Wang BL, Ma L,
Feng SQ and Ding WY: 17β-estradiol protects against apoptosis
induced by interleukin-1β in rat nucleus pulposus cells by
down-regulating MMP-3 and MMP-13. Apoptosis. 20:348–357.
2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wang T, Li P, Ma X, Tian P, Han C, Zang J,
Kong J and Yan H: MicroRNA-494 inhibition protects nucleus pulposus
cells from TNF-α-induced apoptosis by targeting JunD. Biochimie.
115:1–7. 2015.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Wang WJ, Yang W, Ouyang ZH, Xue JB, Li XL,
Zhang J, He WS, Chen WK, Yan YG and Wang C: MiR-21 promotes ECM
degradation through inhibiting autophagy via the PTEN/akt/mTOR
signaling pathway in human degenerated NP cells. Biomed
Pharmacother. 99:725–734. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50.
2014.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ji ML, Zhang XJ, Shi PL, Lu J, Wang SZ,
Chang Q, Chen H and Wang C: Downregulation of microRNA-193a-3p is
involved in invertebral disc degeneration by targeting MMP14. J Mol
Med (Berl). 94:457–468. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Xu YQ, Zhang ZH, Zheng YF and Feng SQ:
Dysregulated miR-133a mediates loss of type II collagen by directly
targeting matrix metalloproteinase 9 (MMP9) in human intervertebral
disc degeneration. Spine (Phila Pa 1976). 41:E717–E724.
2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Hua WB, Wu XH, Zhang YK, Song Y, Tu J,
Kang L, Zhao KC, Li S, Wang K, Liu W, et al: Dysregulated
miR-127-5p contributes to type II collagen degradation by targeting
matrix metalloproteinase-13 in human intervertebral disc
degeneration. Biochimie. 139:74–80. 2017.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ji ML, Lu J, Shi PL, Zhang XJ, Wang SZ,
Chang Q, Chen H and Wang C: Dysregulated miR-98 contributes to
extracellular matrix degradation by targeting IL-6/STAT3 signaling
pathway in human intervertebral disc degeneration. J Bone Miner
Res. 31:900–909. 2016.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Chen H, Wang J, Hu B, Wu X, Chen Y, Li R
and Yuan W: miR-34a promotes Fas-mediated cartilage endplate
chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem.
406:21–30. 2015.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Liu MH, Sun C, Yao Y, Fan X, Liu H, Cui
YH, Bian XW, Huang B and Zhou Y: Matrix stiffness promotes
cartilage endplate chondrocyte calcification in disc degeneration
via miR-20a targeting ANKH expression. Sci Rep.
6(25401)2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Xiao L, Xu S, Xu Y, Liu C, Yang B, Wang J
and Xu H: TGF-β/SMAD signaling inhibits intermittent cyclic
mechanical tension-induced degeneration of endplate chondrocytes by
regulating the miR-455-5p/RUNX2 axis. J Cell Biochem.
119:10415–10425. 2018.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Li Z, Li X, Chen C, Li S, Shen J, Tse G,
Chan MTV and Wu WKK: Long non-coding RNAs in nucleus pulposus cell
function and intervertebral disc degeneration. Cell Prolif.
51(e12483)2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Yu Y, Zhang X, Li Z, Kong L and Huang Y:
LncRNA HOTAIR suppresses TNF-α induced apoptosis of nucleus
pulposus cells by regulating miR-34a/Bcl-2 axis. Biomed
Pharmacother. 107:729–737. 2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Li X, Lou Z, Liu J, Li H, Lei Y, Zhao X
and Zhang F: Upregulation of the long noncoding RNA lncPolE
contributes to intervertebral disc degeneration by negatively
regulating DNA polymerase epsilon. Am J Transl Res. 11:2843–2854.
2019.PubMed/NCBI
|
|
78
|
Wang Y, Song Q, Huang X, Chen Z, Zhang F,
Wang K, Huang G and Shen H: Long noncoding RNA GAS5 promotes
apoptosis in primary nucleus pulposus cells derived from the human
intervertebral disc via Bcl-2 downregulation and caspase3
upregulation. Mol Med Rep. 19:2164–2172. 2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Yu L, Hao Y, Xu C, Zhu G and Cai Y:
LINC00969 promotes the degeneration of intervertebral disk by
sponging miR-335-3p and regulating NLRP3 inflammasome activation.
IUBMB life. 71:611–618. 2019.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X
and He Y: Long non-coding HCG18 promotes intervertebral disc
degeneration by sponging miR-146a-5p and regulating TRAF6
expression. Sci Rep. 7(13234)2017.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Wang X, Peng L, Gong X, Zhang X, Sun R and
Du J: lncRNA-RMRP promotes nucleus pulposus cell proliferation
through regulating miR-206 expression. J Cell Mol Med.
22:5468–5476. 2018.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Wei R, Chen Y, Zhao Z, Gu Q and Wu J:
LncRNA FAM83H-AS1 induces nucleus pulposus cell growth via
targeting the Notch signaling pathway. J Cell Physiol.
234:22163–22171. 2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Ruan Z, Ma H, Li J, Liu H, Jia H and Li F:
The long non-coding RNA NEAT1 contributes to extracellular matrix
degradation in degenerative human nucleus pulposus cells. Exp Biol
Med (Maywood). 243:595–600. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Wang K, Song Y, Liu W, Wu X, Zhang Y, Li
S, Kang L, Tu J, Zhao K, Hua W and Yang C: The noncoding RNA
linc-ADAMTS5 cooperates with RREB1 to protect from intervertebral
disc degeneration through inhibiting ADAMTS5 expression. Clin Sci
(Lond). 131:965–979. 2017.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Chen J, Jia YS, Liu GZ, Sun Q, Zhang F, Ma
S and Wang YJ: Role of LncRNA TUG1 in intervertebral disc
degeneration and nucleus pulposus cells via regulating
Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun.
491:668–674. 2017.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wang X, Zou M, Li J, Wang B, Zhang Q, Liu
F and Lü G: lncRNA H19 targets miR-22 to modulate H2
O2-induced deregulation in nucleus pulposus cell
senescence, proliferation, and ECM synthesis through Wnt signaling.
J Cell Biochem. 119:4990–5002. 2018.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Wang XB, Wang H, Long HQ, Li DY and Zheng
X: LINC00641 regulates autophagy and intervertebral disc
degeneration by acting as a competitive endogenous RNA of
miR-153-3p under nutrition deprivation stress. J Cell Physiol.
234:7115–7127. 2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Sampara P, Banala RR, Vemuri SK, Av GR and
Gpv S: Understanding the molecular biology of intervertebral disc
degeneration and potential gene therapy strategies for
regeneration: A review. Gene Ther. 25:67–82. 2018.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Wang X, Wang B, Zou M, Li J, Lü G, Zhang
Q, Liu F and Lu C: CircSEMA4B targets miR-431 modulating
IL-1β-induced degradative changes in nucleus pulposus cells in
intervertebral disc degeneration via Wnt pathway. Biochim Biophys
Acta Mol Basis Dis. 1864:3754–3768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Xiao L, Ding B, Xu S, Gao J, Yang B, Wang
J and Xu H: circRNA_0058097 promotes tension-induced degeneration
of endplate chondrocytes by regulating HDAC4 expression through
sponge adsorption of miR-365a-5p. J Cell Biochem. 121:418–429.
2019.PubMed/NCBI View Article : Google Scholar
|