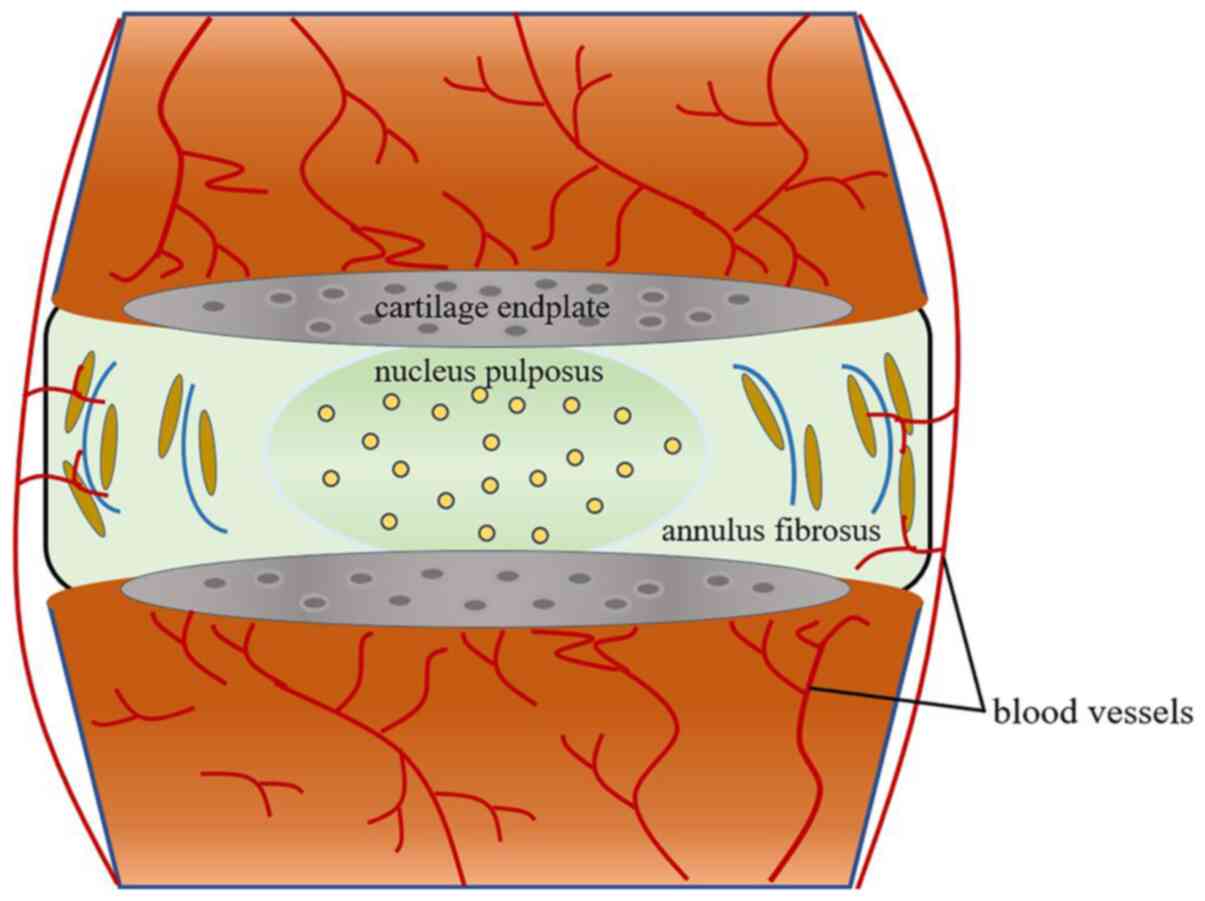
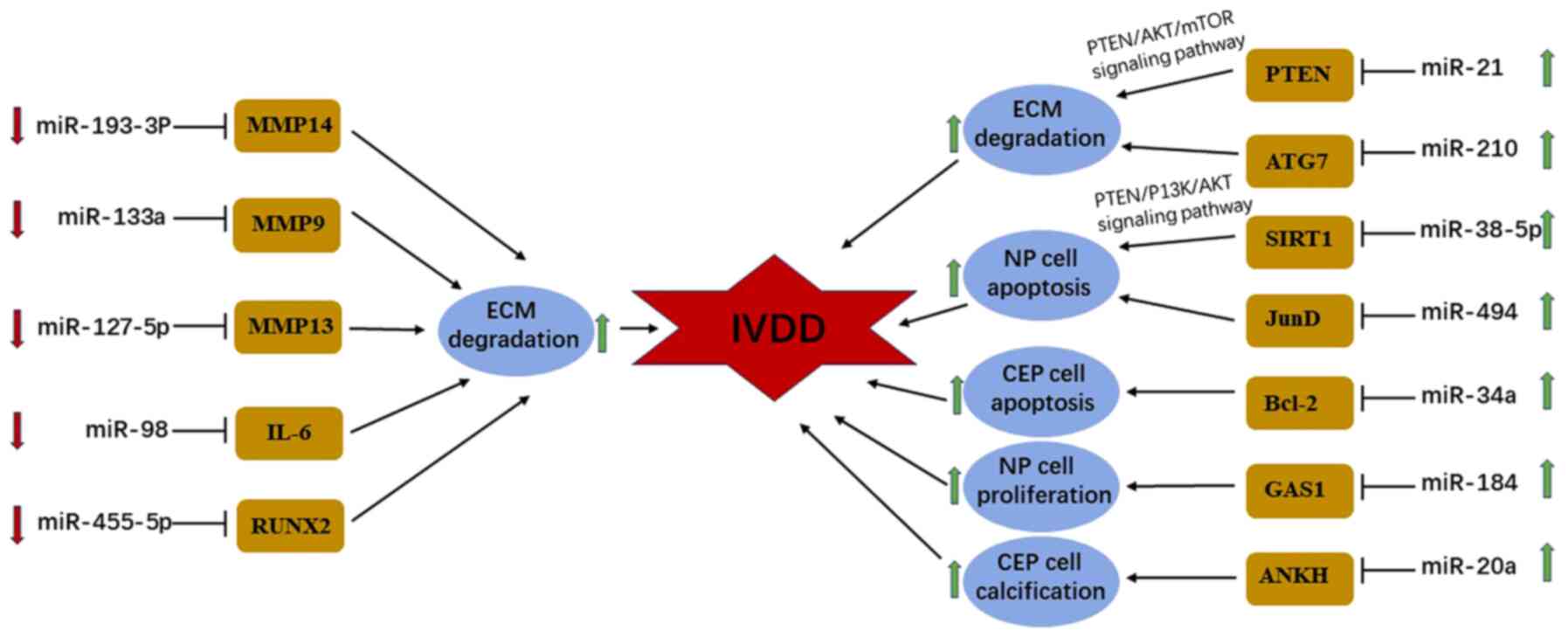
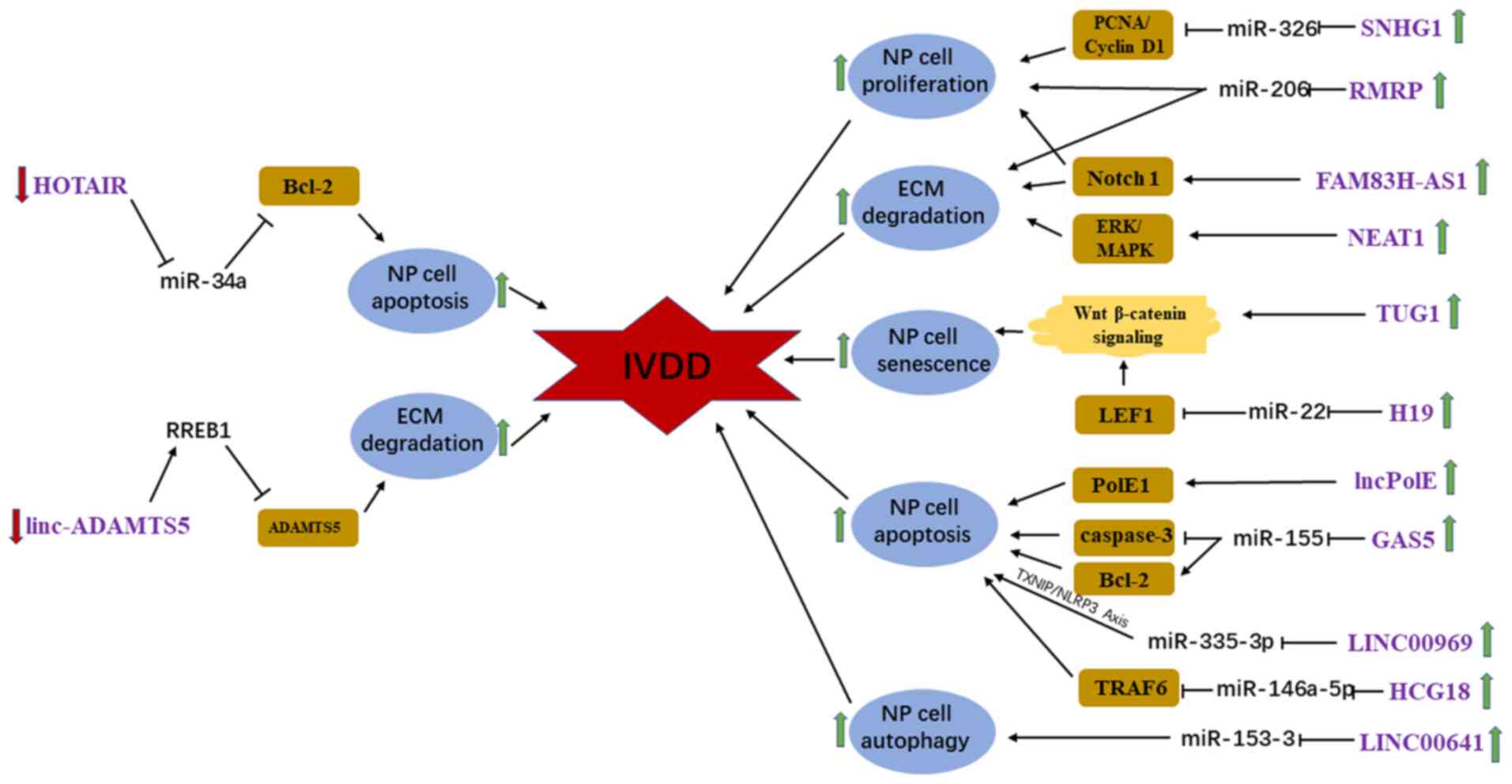
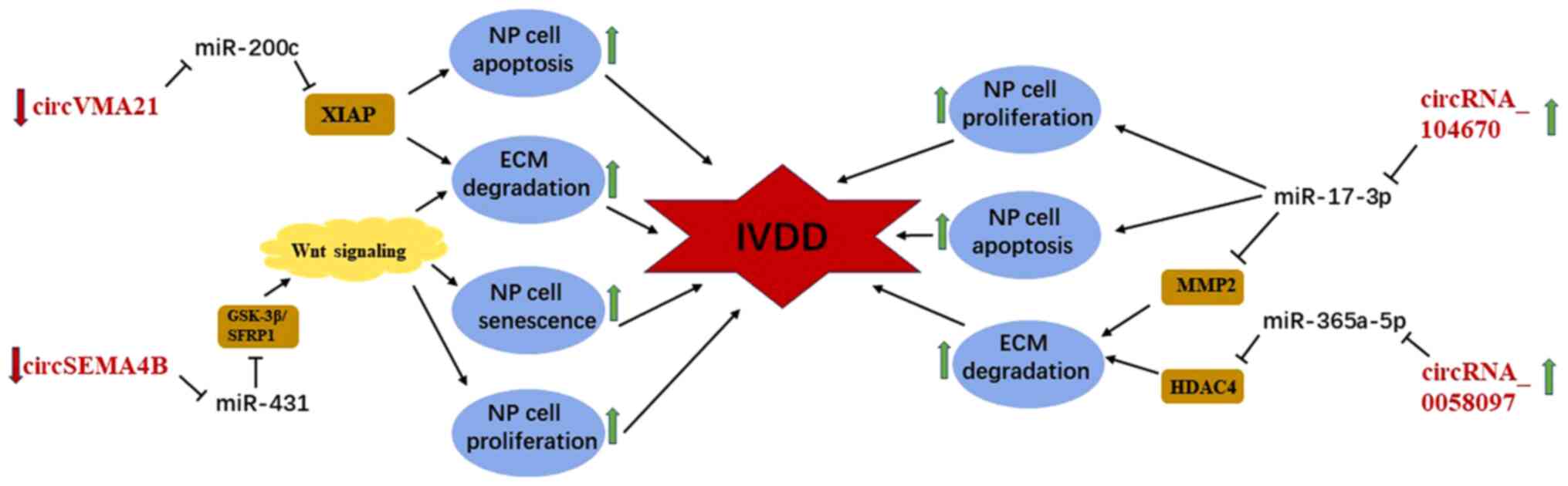
|
1
|
Maher C, Underwood M and Buchbinder R:
Non-specific low back pain. Lancet. 389:736–747. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Dudli S, Fields AJ, Samartzis D, Karppinen
J and Lotz JC: Pathobiology of modic changes. Eur Spine J.
25:3723–3734. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vos T, Flaxman AD, Naghavi M, Lozano R,
Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V,
et al: Years lived with disability (YLDs) for 1160 sequelae of 289
diseases and injuries 1990-2010: A systematic analysis for the
global burden of disease study 2010. Lancet. 380:2163–2196.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hartvigsen J, Hancock MJ, Kongsted A, Louw
Q, Ferreira ML, Genevay S, Hoy D, Karppinen J, Pransky G, Sieper J,
et al: What low back pain is and why we need to pay attention.
Lancet. 391:2356–2367. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yelin E, Weinstein S and King T: The
burden of musculoskeletal diseases in the United States. Semin
Arthritis Rheum. 46:259–260. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Foster NE, Anema JR, Cherkin D, Chou R,
Cohen SP, Gross DP, Ferreira PH, Fritz JM, Koes BW, Peul W, et al:
Prevention and treatment of low back pain: Evidence, challenges,
and promising directions. Lancet. 391:2368–2383. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration, and what causes it? Spine (Phila
Pa 1976). 31:2151–2161. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vo NV, Hartman RA, Patil PR, Risbud MV,
Kletsas D, Iatridis JC, Hoyland JA, Le Maitre CL, Sowa GA and Kang
JD: Molecular mechanisms of biological aging in intervertebral
discs. J Orthop Res. 34:1289–1306. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bian Q, Jain A, Xu X, Kebaish K, Crane JL,
Zhang Z, Wan M, Ma L, Riley LH, Sponseller PD, et al: Excessive
activation of TGFβ by spinal instability causes vertebral endplate
sclerosis. Sci Rep. 6(27093)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Huang YC, Urban JP and Luk KD:
Intervertebral disc regeneration: Do nutrients lead the way? Nat
Rev Rheumatol. 10:561–566. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liao Z, Wu X, Song Y, Luo R, Yin H, Zhan
S, Li S, Wang K, Zhang Y and Yang C: Angiopoietin-like protein 8
expression and association with extracellular matrix metabolism and
inflammation during intervertebral disc degeneration. J Cell Mol
Med. 23:5737–5750. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu X, Zhuang J, Wang D, Lv L, Zhu F, Yao
A and Xu T: Glycyrrhizin suppresses inflammation and cell apoptosis
by inhibition of HMGB1 via p38/p-JUK signaling pathway in
attenuating intervertebral disc degeneration. Am J Transl Res.
11:5105–5113. 2019.PubMed/NCBI
|
|
14
|
Grant MP, Epure LM, Bokhari R, Roughley P,
Antoniou J and Mwale F: Human cartilaginous endplate degeneration
is induced by calcium and the extracellular calcium-sensing
receptor in the intervertebral disc. Eur Cell Mater. 32:137–151.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yao Y, Deng Q, Song W, Zhang H, Li Y, Yang
Y, Fan X, Liu M, Shang J, Sun C, et al: MIF plays a key role in
regulating tissue-specific chondro-osteogenic differentiation fate
of human cartilage endplate stem cells under hypoxia. Stem Cell
Reports. 7:249–262. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tang Z, Hu B, Zang F, Wang J, Zhang X and
Chen H: Nrf2 drives oxidative stress-induced autophagy in nucleus
pulposus cells via a Keap1/Nrf2/p62 feedback loop to protect
intervertebral disc from degeneration. Cell Death Dis.
10(510)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang G, Huang K, Dong Y, Chen S, Zhang J,
Wang J, Xie Z, Lin X, Fang X and Fan S: Lycorine suppresses
endplate-chondrocyte degeneration and prevents intervertebral disc
degeneration by inhibiting NF-κB signalling pathway. Cell Physiol
Biochem. 45:1252–1269. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Matsui M and Corey DR: Non-coding RNAs as
drug targets. Nat Rev Drug Discov. 16:167–179. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ning B, Yu D and Yu AM: Advances and
challenges in studying noncoding RNA regulation of drug metabolism
and development of RNA therapeutics. Biochem Pharmacol.
169(113638)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang WT, Han C, Sun YM, Chen TQ and Chen
YQ: Noncoding RNAs in cancer therapy resistance and targeted drug
development. J Hematol Oncol. 12(55)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346.
2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sato-Kuwabara Y, Melo SA, Soares FA and
Calin GA: The fusion of two worlds: Non-coding RNAs and
extracellular vesicles-diagnostic and therapeutic implications
(Review). Int J Oncol. 46:17–27. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhou X, Chen L, Grad S, Alini M, Pan H,
Yang D, Zhen W, Li Z, Huang S and Peng S: The roles and
perspectives of microRNAs as biomarkers for intervertebral disc
degeneration. J Tissue Eng Regen Med. 11:3481–3487. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou L, Dong S, Deng Y, Yang P, Zheng Y,
Yao L, Zhang M, Yang S, Wu Y, Zhai Z, et al: GOLGA7 rs11337, a
polymorphism at the MicroRNA binding site, is associated with
glioma prognosis. Mol Ther Nucleic Acids. 18:56–65. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye
X, Zhang H, Yang P, Cui X, Ren Y, et al: A noncoding regulatory
RNAs network driven by Circ-CDYL acts specifically in the early
stages hepatocellular carcinoma. Hepatology. 71:130–147.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Shang Q, Yang Z, Jia R and Ge S: The novel
roles of circRNAs in human cancer. Mol Cancer. 18(6)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wu K, Liao X, Gong Y, He J, Zhou JK, Tan
S, Pu W, Huang C, Wei YQ and Peng Y: Circular RNA F-circSR derived
from SLC34A2-ROS1 fusion gene promotes cell migration in non-small
cell lung cancer. Mol Cancer. 18(98)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y,
Huang K, Wang G, Wang J, Ma J, et al: Circular RNA circTADA2A
promotes osteosarcoma progression and metastasis by sponging
miR-203a-3p and regulating CREB3 expression. Mol Cancer.
18(73)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lange S, Banerjee I, Carrion K, Serrano R,
Habich L, Kameny R, Lengenfelder L, Dalton N, Meili R, Börgeson E,
et al: miR-486 is modulated by stretch and increases ventricular
growth. JCI Insight. 4(e125507)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Calderon-Dominguez M, Belmonte T,
Quezada-Feijoo M, Ramos-Sánchez M, Fernández-Armenta J,
Pérez-Navarro A, Cesar S, Peña-Peña L, Vea À, Llorente-Cortés V, et
al: Emerging role of microRNAs in dilated cardiomyopathy: Evidence
regarding etiology. Transl Res. 215:86–101. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Cai B, Zhang Y, Zhao Y, Wang J, Li T,
Zhang Y, Jiang Y, Jin X, Xue G, Li P, et al: Long noncoding
RNA-DACH1 (Dachshund Homolog 1) regulates cardiac function by
inhibiting SERCA2a (Sarcoplasmic Reticulum Calcium ATPase 2a).
Hypertension. 74:833–842. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
super-enhancer-regulated circRNA Nfix induces cardiac regeneration
after myocardial infarction in adult mice. Circulation.
139:2857–2876. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hasvik E, Schjølberg T, Jacobsen DP,
Haugen AJ, Grøvle L, Schistad EI and Gjerstad J: Up-regulation of
circulating microRNA-17 is associated with lumbar radicular pain
following disc herniation. Arthritis Res Ther.
21(186)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang C, Zhang ZZ, Yang W, Ouyang ZH, Xue
JB, Li XL, Zhang J, Chen WK, Yan YG and Wang WJ: MiR-210
facilitates ECM degradation by suppressing autophagy via silencing
of ATG7 in human degenerated NP cells. Biomed Pharmacother.
93:470–479. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Shao T, Hu Y, Tang W, Shen H, Yu Z and Gu
J: The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge
to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated
mechanism. Gene. 715(144029)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tan H, Zhao L, Song R, Liu Y and Wang L:
The long noncoding RNA SNHG1 promotes nucleus pulposus cell
proliferation through regulating miR-326 and CCND1. Am J Physiol
Cell Physiol. 315:C21–C27. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Song J, Wang HL, Song KH, Ding ZW, Wang
HL, Ma XS, Lu FZ, Xia XL, Wang YW, Fei-Zou and Jiang JY:
CircularRNA_104670 plays a critical role in intervertebral disc
degeneration by functioning as a ceRNA. Exp Mol Med.
50(94)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang B, Wang D, Yan T and Yuan H:
miR-138-5p promotes TNF-α-induced apoptosis in human intervertebral
disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt
signaling. Exp Cell Res. 345:199–205. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang C, Wang WJ, Yan YG, Xiang YX, Zhang
J, Tang ZH and Jiang ZS: MicroRNAs: New players in intervertebral
disc degeneration. Clin Chim Acta. 450:333–341. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Mo YY: MicroRNA regulatory networks and
human disease. Cell Mol Life Sci. 69:3529–3531. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ivey KN and Srivastava D: microRNAs as
developmental regulators. Cold Spring Harb Perspect Biol.
7(a008144)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Liu B, Li J and Cairns MJ: Identifying
miRNAs, targets and functions. Brief Bioinform. 15:1–19.
2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8(1015)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Robinson EK, Covarrubias S and Carpenter
S: The how and why of lncRNA function: An innate immune
perspective. Biochim Biophys Acta Gene Regul Mech.
1863(194419)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ji E, Kim C, Kim W and Lee EK: Role of
long non-coding RNAs in metabolic control. Biochim Biophys Acta
Gene Regul Mech. 1863(194348)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan
YG, Zhang J and Wang WJ: lncRNAs: Novel players in intervertebral
disc degeneration and osteoarthritis. Cell Prolif.
50(e12313)2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46.
2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Santer L, Bär C and Thum T: Circular RNAs:
A novel class of functional RNA molecules with a therapeutic
perspective. Mol Ther. 27:1350–1363. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Suzuki H, Zuo Y, Wang J, Zhang MQ,
Malhotra A and Mayeda A: Characterization of RNase R-digested
cellular RNA source that consists of lariat and circular RNAs from
pre-mRNA splicing. Nucleic Acids Res. 34(e63)2006.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461.
2014.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691.
2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Li W, Wang P, Zhang Z, Wang W, Liu Y and
Qi Q: miR-184 regulates proliferation in nucleus pulposus cells by
targeting GAS1. World Neurosurg. 97:710–715.e1. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Che YJ, Guo JB, Liang T, Chen X, Zhang W,
Yang HL and Luo ZP: Assessment of changes in the micro-nano
environment of intervertebral disc degeneration based on Pfirrmann
grade. Spine J. 19:1242–1253. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Yang SD, Yang DL, Sun YP, Wang BL, Ma L,
Feng SQ and Ding WY: 17β-estradiol protects against apoptosis
induced by interleukin-1β in rat nucleus pulposus cells by
down-regulating MMP-3 and MMP-13. Apoptosis. 20:348–357.
2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wang T, Li P, Ma X, Tian P, Han C, Zang J,
Kong J and Yan H: MicroRNA-494 inhibition protects nucleus pulposus
cells from TNF-α-induced apoptosis by targeting JunD. Biochimie.
115:1–7. 2015.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Wang WJ, Yang W, Ouyang ZH, Xue JB, Li XL,
Zhang J, He WS, Chen WK, Yan YG and Wang C: MiR-21 promotes ECM
degradation through inhibiting autophagy via the PTEN/akt/mTOR
signaling pathway in human degenerated NP cells. Biomed
Pharmacother. 99:725–734. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50.
2014.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ji ML, Zhang XJ, Shi PL, Lu J, Wang SZ,
Chang Q, Chen H and Wang C: Downregulation of microRNA-193a-3p is
involved in invertebral disc degeneration by targeting MMP14. J Mol
Med (Berl). 94:457–468. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Xu YQ, Zhang ZH, Zheng YF and Feng SQ:
Dysregulated miR-133a mediates loss of type II collagen by directly
targeting matrix metalloproteinase 9 (MMP9) in human intervertebral
disc degeneration. Spine (Phila Pa 1976). 41:E717–E724.
2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Hua WB, Wu XH, Zhang YK, Song Y, Tu J,
Kang L, Zhao KC, Li S, Wang K, Liu W, et al: Dysregulated
miR-127-5p contributes to type II collagen degradation by targeting
matrix metalloproteinase-13 in human intervertebral disc
degeneration. Biochimie. 139:74–80. 2017.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ji ML, Lu J, Shi PL, Zhang XJ, Wang SZ,
Chang Q, Chen H and Wang C: Dysregulated miR-98 contributes to
extracellular matrix degradation by targeting IL-6/STAT3 signaling
pathway in human intervertebral disc degeneration. J Bone Miner
Res. 31:900–909. 2016.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Chen H, Wang J, Hu B, Wu X, Chen Y, Li R
and Yuan W: miR-34a promotes Fas-mediated cartilage endplate
chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem.
406:21–30. 2015.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Liu MH, Sun C, Yao Y, Fan X, Liu H, Cui
YH, Bian XW, Huang B and Zhou Y: Matrix stiffness promotes
cartilage endplate chondrocyte calcification in disc degeneration
via miR-20a targeting ANKH expression. Sci Rep.
6(25401)2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Xiao L, Xu S, Xu Y, Liu C, Yang B, Wang J
and Xu H: TGF-β/SMAD signaling inhibits intermittent cyclic
mechanical tension-induced degeneration of endplate chondrocytes by
regulating the miR-455-5p/RUNX2 axis. J Cell Biochem.
119:10415–10425. 2018.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Li Z, Li X, Chen C, Li S, Shen J, Tse G,
Chan MTV and Wu WKK: Long non-coding RNAs in nucleus pulposus cell
function and intervertebral disc degeneration. Cell Prolif.
51(e12483)2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Yu Y, Zhang X, Li Z, Kong L and Huang Y:
LncRNA HOTAIR suppresses TNF-α induced apoptosis of nucleus
pulposus cells by regulating miR-34a/Bcl-2 axis. Biomed
Pharmacother. 107:729–737. 2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Li X, Lou Z, Liu J, Li H, Lei Y, Zhao X
and Zhang F: Upregulation of the long noncoding RNA lncPolE
contributes to intervertebral disc degeneration by negatively
regulating DNA polymerase epsilon. Am J Transl Res. 11:2843–2854.
2019.PubMed/NCBI
|
|
78
|
Wang Y, Song Q, Huang X, Chen Z, Zhang F,
Wang K, Huang G and Shen H: Long noncoding RNA GAS5 promotes
apoptosis in primary nucleus pulposus cells derived from the human
intervertebral disc via Bcl-2 downregulation and caspase3
upregulation. Mol Med Rep. 19:2164–2172. 2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Yu L, Hao Y, Xu C, Zhu G and Cai Y:
LINC00969 promotes the degeneration of intervertebral disk by
sponging miR-335-3p and regulating NLRP3 inflammasome activation.
IUBMB life. 71:611–618. 2019.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X
and He Y: Long non-coding HCG18 promotes intervertebral disc
degeneration by sponging miR-146a-5p and regulating TRAF6
expression. Sci Rep. 7(13234)2017.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Wang X, Peng L, Gong X, Zhang X, Sun R and
Du J: lncRNA-RMRP promotes nucleus pulposus cell proliferation
through regulating miR-206 expression. J Cell Mol Med.
22:5468–5476. 2018.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Wei R, Chen Y, Zhao Z, Gu Q and Wu J:
LncRNA FAM83H-AS1 induces nucleus pulposus cell growth via
targeting the Notch signaling pathway. J Cell Physiol.
234:22163–22171. 2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Ruan Z, Ma H, Li J, Liu H, Jia H and Li F:
The long non-coding RNA NEAT1 contributes to extracellular matrix
degradation in degenerative human nucleus pulposus cells. Exp Biol
Med (Maywood). 243:595–600. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Wang K, Song Y, Liu W, Wu X, Zhang Y, Li
S, Kang L, Tu J, Zhao K, Hua W and Yang C: The noncoding RNA
linc-ADAMTS5 cooperates with RREB1 to protect from intervertebral
disc degeneration through inhibiting ADAMTS5 expression. Clin Sci
(Lond). 131:965–979. 2017.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Chen J, Jia YS, Liu GZ, Sun Q, Zhang F, Ma
S and Wang YJ: Role of LncRNA TUG1 in intervertebral disc
degeneration and nucleus pulposus cells via regulating
Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun.
491:668–674. 2017.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wang X, Zou M, Li J, Wang B, Zhang Q, Liu
F and Lü G: lncRNA H19 targets miR-22 to modulate H2
O2-induced deregulation in nucleus pulposus cell
senescence, proliferation, and ECM synthesis through Wnt signaling.
J Cell Biochem. 119:4990–5002. 2018.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Wang XB, Wang H, Long HQ, Li DY and Zheng
X: LINC00641 regulates autophagy and intervertebral disc
degeneration by acting as a competitive endogenous RNA of
miR-153-3p under nutrition deprivation stress. J Cell Physiol.
234:7115–7127. 2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Sampara P, Banala RR, Vemuri SK, Av GR and
Gpv S: Understanding the molecular biology of intervertebral disc
degeneration and potential gene therapy strategies for
regeneration: A review. Gene Ther. 25:67–82. 2018.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Wang X, Wang B, Zou M, Li J, Lü G, Zhang
Q, Liu F and Lu C: CircSEMA4B targets miR-431 modulating
IL-1β-induced degradative changes in nucleus pulposus cells in
intervertebral disc degeneration via Wnt pathway. Biochim Biophys
Acta Mol Basis Dis. 1864:3754–3768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Xiao L, Ding B, Xu S, Gao J, Yang B, Wang
J and Xu H: circRNA_0058097 promotes tension-induced degeneration
of endplate chondrocytes by regulating HDAC4 expression through
sponge adsorption of miR-365a-5p. J Cell Biochem. 121:418–429.
2019.PubMed/NCBI View Article : Google Scholar
|