Introduction
Gestational diabetes mellitus (GDM) is defined as
any degree of abnormal glucose tolerance that occurs during
pregnancy (1,2). Glucose intolerance is diagnosed during
pregnancy and GDM is one of the most common complications of
pregnancy, affecting 3-8% of pregnancies (3). The mechanisms of GDM remain to be
fully elucidated. Binding insulin resistance and impaired insulin
secretion lead to an underlying pathophysiology that may be
influenced by interactions between genetic and environmental
factors (4,5). The prevalence of GDM has increased in
recent decades due to the increased mean age of pregnant females
and increased susceptibility to obesity (6). Human and animal studies suggested that
type 2 diabetes mellitus (T2D) and GDM may have certain
pathological changes in common, including insulin resistance,
cellular dysfunction and insulin deficiency (7). Studies such as genome-wide association
studies (GWASs) and single nucleotide polymorphism (SNP) strategies
have identified numerous susceptibility genes associated with
increased T2D sensitivity, including protein tyrosine phosphatase
receptor type D (PTPRD) (8,9).
PTPRD is a member of the PTP family (10). PTPs are considered to regulate
signaling molecules in a variety of cellular processes, including
cell growth, differentiation, mitotic cycle and oncogenic
transformation (11). The
extracellular domain contains the meprin-A antigen-PTP domain
(12). Transformation of the PTPRD
gene may lead to the replacement of the highly conserved residues
in the second tyrosine phosphatase catalytic domain Thr1365-to-Met,
which is highly associated with the pathogenesis of cancer
(13,14). Wang et al (15) determined that the mutant tyrosine
phosphatase is a tumor suppressor gene that regulates cell pathways
and may potentially be utilized for therapeutic intervention in
colorectal cancer. In addition, Chen et al (16) reported that silencing of PTPRD was
caused by DNA methylation in a mouse model of T2D and in patients,
and was correlated with DNA (cytosine-5)-methyltransferase 1
expression. The difference of postprandial blood glucose in the
PTPRD rs17584499 CT+TT genotype was significantly lower than that
of the rs17584499 CC genotype (17).
MicroRNAs (miRNAs/miRs) are small non-coding RNA
molecules consisting of 19-25 nucleotides that have been identified
to have an important role in various human diseases (18). Studies have suggested that miRNA
expression was tissue-specific and it has been reported that
certain miRNAs were specifically expressed in the placenta
(placental-specific miRNA). miRNAs negatively regulated the
expression of target genes at the post-transcriptional level by
binding to the 3'-UTRs of the target information RNA (19,20).
Increasing evidence proved that SNPs located at miRNA binding sites
may lead to decreased or increased target mRNA translation. This
dysfunction was associated with cancer susceptibility (21,22).
For instance, the CC genotype of cyclin-dependent kinase inhibitor
2B rs1063192 in the miR-323b-5p binding site may increase the
susceptibility of pregnant Chinese Han females to GDM (23).
The present study focused on the SNPs in the 3'UTR
of PTPRD by using the miRNASNP-v3 bioinformatics software
(http://bioinfo.life.hust.edu.cn/miRNASNP/#!/) with the
loss-or-gain strategy (24). A
total of 17 potential SNPs were obtained as candidate SNPs, which
may be associated with certain miRNAs and with the pathogenesis of
GDM. Based on this, the association between the allele distribution
and the susceptibility of GDM was further investigated in a
case-control study.
Materials and methods
Study subjects
The study plan was approved by the Ethics Committee
of The Red Cross Hospital of Qinghai Province (Xining, China) and
written informed consent was obtained from all participants prior
to data collection. The subjects provided written informed consent
prior to specimen collection. The study was approved by the ethics
committee of Qinghai Red-Cross Hospital (Xining, China). All
methods were in accordance with the approved guidelines. Subjects
who had been diagnosed with diabetes, had taken drugs that affect
glucose metabolism or had other pregnancy complications were
excluded from the study. GDM was diagnosed according to the
standards of the American Diabetes Association (25). Subjects with negative results of the
50-g glucose stimulation test or a normal glucose tolerance test
were used as controls. Demographic data were collected for all
individuals, including age, body height and weight, and resting
blood pressure at 24-28 weeks of gestation during the first
pregnancy. The pre-pregnancy body mass index was calculated as
[weight (kg)/height (m2)]. Routine laboratory
evaluations were performed, including analysis of glycosylated
hemoglobin (HbA1C), triglycerides, total cholesterol, low-density
lipoprotein cholesterol and high-density lipoprotein (HDL)
cholesterol. Blood glucose and serum insulin concentrations were
measured at 0, 60 and 120 min after the 100-g oral glucose
tolerance test (OGTT). A total of 1,100 subjects were recruited,
including 500 patients with GDM and 600 controls (Table I).
 | Table IFrequency distributions of selected
variables in patients with GDM and healthy controls. |
Table I
Frequency distributions of selected
variables in patients with GDM and healthy controls.
| Variables | GDM (n=500) | Control
(n=600) | P-value |
|---|
| Age (years) | 32±4 | 32±3 | 0.320000 |
| Pregestational BMI
(kg/m2) | 21.55±1.84 | 21.61±1.92 | 0.410000 |
| Fasting plasma
glucosea (mmol/l) | 5.02±0.78 | 4.67±0.12 | 0.000280 |
| OGTT (mmol/l) | | | |
|
0 h | 5.04±0.89 | 4.50±0.11 | 0.000350 |
|
1 h | 10.66±1.25 | 8.01±0.84 | 0.000410 |
|
2 h | 8.99±2.11 | 7.02±0.34 | 0.000521 |
| Physical activity
(number) | | | 0.000010 |
|
0-150 min
per week | 341 | 201 | |
|
≥150 min per
week | 159 | 399 | |
| Glycosylated
hemoglobin (%) | 5.8±0.22 | 4.7±0.12 | 0.000320 |
| Cholesterol
(mmol/l) | 6.09±0.45 | 6.08±0.33 | 0.440000 |
| Triglyceride
(mmol/l) | 2.55±0.98 | 2.45±0.63 | 0.420000 |
| HDL cholesterol
(mmol/l) | 1.97±0.11 | 2.01±0.21 | 0.220000 |
| LDL cholesterol
(mmol/l) | 3.01±0.24 | 3.07±0.28 | 0.290000 |
SNP selection
First, SNPs from the 3'-UTR of the PTPRD gene were
obtained from the National Center for Bioinformatics (NCBI) SNP
database (dbSNP; http://www.ncbi.nlm.nih.gov/snp/) and ENSEMBL v58
(https://www.ensembl.org/index.html).
From the 1000 genome browser (http://www.1000genomes.org/), SNPs in the Chinese
population were identified. Subsequently, the miRanda (http://www.microrna.org), TargetScan (http://www.targetscan.org/), PolymiRTS (http://compbio.uthsc.edu/miRSNP/) and miRDB
software package (http://mirdb.org/) were used to study
the miRNA targets of the SNPs of the binding sites. A
bioinformatics analysis (http://bioinfo.life.hust.edu.cn/miRNASNP2/index.php)
was used to determine the function of the miRNA targets of the SNPs
of the 3'-UTR of PTPRD. If the SNP was in a high linkage
disequilibrium (r2>0.8), only one SNP was genotyped.
For the gain-loss strategy analysis, the miRNA wild-type (wild)
sequence and SNP allele sequence was obtained (24). Subsequently, two target prediction
tools, TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org), were respectively used to
estimate the target site. A total of four results were recorded,
namely wild TargetScan (WT), wild miRanda (WM), SNP TargetScan (ST)
and SNP miRanda (SM). If the target gene of a miRNA was present in
both the WT and WM, but not in the ST and SM, the miRNA was
considered to have a loss of this target gene. Conversely, if a
target was present in both ST and SM, but not WT and WM, the miRNA
was considered to have gained one target gene (26).
Genotyping
Genomic DNA was extracted from leukocyte
microspheres of human plasma samples by traditional protease K
(Beyotime Institute of Biotechnology) digestion and then extracted
by phenol-chloroform and ethanol precipitation. The TaqMan SNP
genotyping test was used for genotyping. PCR was performed in a
total volume of 5 µl containing TaqMan General Master Mix (Beyotime
Institute of Biotechnology), 80X SNP genotyping Mix (Beyotime
Institute of Biotechnology), DNase-free Water and 10 ng genomic
DNA. The PCR conditions were 2 min at 50˚C, 10 min at 95˚C and 40
cycles at 95˚C for 15 sec and 60˚C for 1 min in an ABI 7900HT
Real-Time PCR system (Thermo Fisher Scientific, Inc.). The PCR
products were visualized by 1% agarose gel electrophoresis using
ethidium bromide (MilliporeSigma).
Cell lines and culture
The 293T cell line was purchased from the cell bank
of the Chinese Academy of Sciences. Cells were cultured in RMI-1640
(Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.) in a humidified atmosphere with 5%
CO2 at 37˚C. miR-450a mimics
(5'-TTTTTGCGATGTGTTCCTAATG-3') and miR mimics control
(5'-ACGUGACACGUUCGGAGAATT-3') were obtained from GenScript.
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection in accordance with the
manufacturer's protocol.
Reverse transcription-quantitative
(RT-q)PCR assay
Total RNA was obtained from placental tissues
harvested after birth by using TRIzol® reagent as
described by the manufacturer (Invitrogen; Thermo Fisher
Scientific, Inc.). For mRNA detection, total RNAs (500 ng) were
reverse transcribed using an RT kit (cat. no. D350A; Takara Bio,
Inc.). GAPDH was used as an internal control. qPCR was performed
using ABI Prism 7900HT (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol of
Mir-XTM miRNA qRT-PCR SYBR® kit (cat. no. 638314; Takara
Bio, Inc.). The amplification conditions were 95˚C for 10 min,
followed by 40 cycles of 95˚C for 30 sec, 55˚C for 40 sec and 72˚C
for 30 sec, and finally 4˚C for 30 min for cooling. The primers
used were as follows: PTPRD forward, 5'-CTCCAAGGTTTACACGAACACC-3';
and reverse, 5'-AGTCCGTAAGGGTTGTATTCTGA-3'; GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAAT-3'; and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'. The results were assessed using the
2-∆∆Cq method (27,28).
Construction of luciferase-based
reporter plasmids
The fragments containing the 3'-UTR with the G or C
alleles of SNP rs56407701 were amplified by GenScript. The PCR
product was cloned into the pMIR-REPORT luciferase system (Ambion;
Thermo Fisher Scientific, Inc.). The amplified fragment was
verified by DNA sequencing by GenScript. The pRL-TK vector
containing Renilla luciferase was used as a normalization
control.
Dual-luciferase reporter assay
The 3'-UTR sequence of PTPRD predicted to interact
with miR-450a or the mutant sequence of the predicted target
(synthesized by GenScript) were inserted into the pMIR-REPORT
vector at the restriction enzyme cutting site of HindIII and
SacI (provided by GenScript). 293T cells (1x105)
cultured on 24-well plates were co-transfected with pMIR reporter
vectors containing wild-type or mutant PTPRD 3'-UTR fragments and
miR-450a or control, and pRL-TK containing Renilla
luciferase was used for normalization.
Statistical analysis
Data were examined regarding whether they followed a
normal distribution in order to select an appropriate parametric or
non-parametric test. The chi-square test was applied to compare
differences in categorical variables. The skewness coefficient and
kurtosis coefficient were determined by 0 and the
Kolmogorov-Smirnov test was performed by P>0.05. For continuous
variables with a normal distribution, the results were expressed as
the mean ± SD or the mean ± SEM. For continuous variables with a
normal distribution, an unpaired Student's t-test was used to
compare between two groups. For comparison of multiple groups,
ANOVA was applied. In the case of significant results obtained by
ANOVA, Bonferroni's post hoc test was employed as a
multiple-comparisons test with 0.05 as the significance level. The
Hardy-Weinberg equilibrium for each SNP genotype was determined by
the chi-square test. The relationship between SNPs and the
susceptibility to GDM was calculated by multivariate logistic
regression analysis with the odds ratio (OR) and 95% CI. All
statistical tests were bilateral and P<0.05 was considered to
indicate statistical significance.
Results
Patient characteristics
The clinical characteristics of patients with GDM
and the control group are presented in Table I. No difference was obtained in age
and gender distribution. After fasting and in the OGTT, blood
glucose, HbA1C and triglycerides of patients with GDM were higher
than those in the control group (P<0.001). The HDL cholesterol
level in the GDM group was significantly lower than that in the
control group (P<0.05). Sufficient physical activity during
pregnancy was associated with a significantly reduced incidence of
GDM.
Although the original rationale of the present study
was focused on genetics, according to a recent report the food
intake was also associated with GDM (29). Differences in food intake between
pregnant females with and without GDM, including cereals,
vegetables, fruit, dairy products and sweet beverages, were also
analyzed. Furthermore, ORs and respective 95% CIs of developing GDM
between the two categories of food consumption were calculated,
i.e. ‘once a day or more’ vs. ‘less than once a day’. Regarding
daily consumption, females with GDM exhibited a significantly more
frequent daily intake of dairy products and sweet beverages
(Table SI).
miR SNPs in the 3'-UTR of the PTPRD
gene and genotype and allele analysis
As presented in Table
II, 17 SNPs were predicted that may be bound by candidate
miRNAs. Further genotyping was performed to identify the 17 SNP
alleles.
 | Table IISNPs located in the 3'-UTR of the
protein tyrosine phosphatase receptor type D gene and the predicted
miRNAs. |
Table II
SNPs located in the 3'-UTR of the
protein tyrosine phosphatase receptor type D gene and the predicted
miRNAs.
| SNP | Chromosome | 3'-UTR
position | Associated
miRNA | Allele |
|---|
| rs62536166 | 9 | 3007-3030 |
miR-148a/148b/152/153 | A/C |
| rs77547574 | 9 | 2348-2371 | miR-515/1283 | C/A |
| rs73428138 | 9 | 761-782 | miR-548/1323 | C/T |
| rs117224071 | 9 | 1419-1441 | miR-802 | A/G |
| rs77185985 | 9 | 2682-2703 | miR-759/2673 | T/G |
| rs114870484 | 9 | 2290-2312 | miR-942/3182 | C/G |
| rs73428138 | 9 | 760-782 |
miR-548e/f/o/t/3609 | C/T |
| rs10976945 | 9 | 1020-1037 |
miR-4311/3688/31 | A/C |
| rs1064270 | 9 | 444-464 | miR-3679/4313 | C/T |
| rs56407701 | 9 | 3073-3095 | miR-450a | G/C |
| rs116361362 | 9 | 435-454 | miR-409 | A/G |
| rs75115513 | 9 | 821-842 | miR-1321/548o | A/T |
| rs117795823 | 9 | 1676-1695 | miR-670 | C/T |
| rs74775961 | 9 | 2596-2617 | miR-1298 | A/G |
| rs28554480 | 9 | 1157-1173 | miR-3201 | A/G |
| rs11542527 | 9 | 1576-1597 | miR-3606 | A/G |
| rs79554842 | 9 | 819-842 | miR-3613 | A/C |
First, the influence of the 17 candidate recognition
SNPs on GDM susceptibility in cases vs. control subjects was
explored. The results suggested that GDM was only associated with
SNP rs56407701. As presented in Tables III and SII, chi-square statistical analysis
indicated that the rs56407701 genotype exhibited a Hardy-Weinberg
equilibrium pattern in healthy controls (P=0.22). Logistic
regression analysis revealed that the susceptibility of the GC and
CC genotypes to GDM was significantly higher than that of the GG
genotype (OR=1.09, 95% CI, 1.02-1.72; OR=3.93, 95% CI, 1.11-3.94,
respectively). All ORs were adjusted for fasting plasma glucose,
OGTT, physical activity and glycosylated hemoglobin.
 | Table IIIGenotype frequencies of the protein
tyrosine phosphatase receptor type D rs56407701 polymorphism among
GDM cases and controls. |
Table III
Genotype frequencies of the protein
tyrosine phosphatase receptor type D rs56407701 polymorphism among
GDM cases and controls.
| Genotype | GDM (n=500) | Controls
(n=600) | OR (95%
CI)a |
P-valuea |
|---|
| GG | 411 (82.2) | 522 (87.0) | 1.00 | |
| GC | 54 (10.8) | 63 (10.5) | 1.09
(1.02-1.72) | 0.040 |
| CC | 35 (7.0) | 15 (2.5) | 3.93
(1.11-3.94) | 0.002 |
| C carrier | 89 (17.8) | 78 (13.0) | 1.58
(1.12-1.63) | 0.027 |
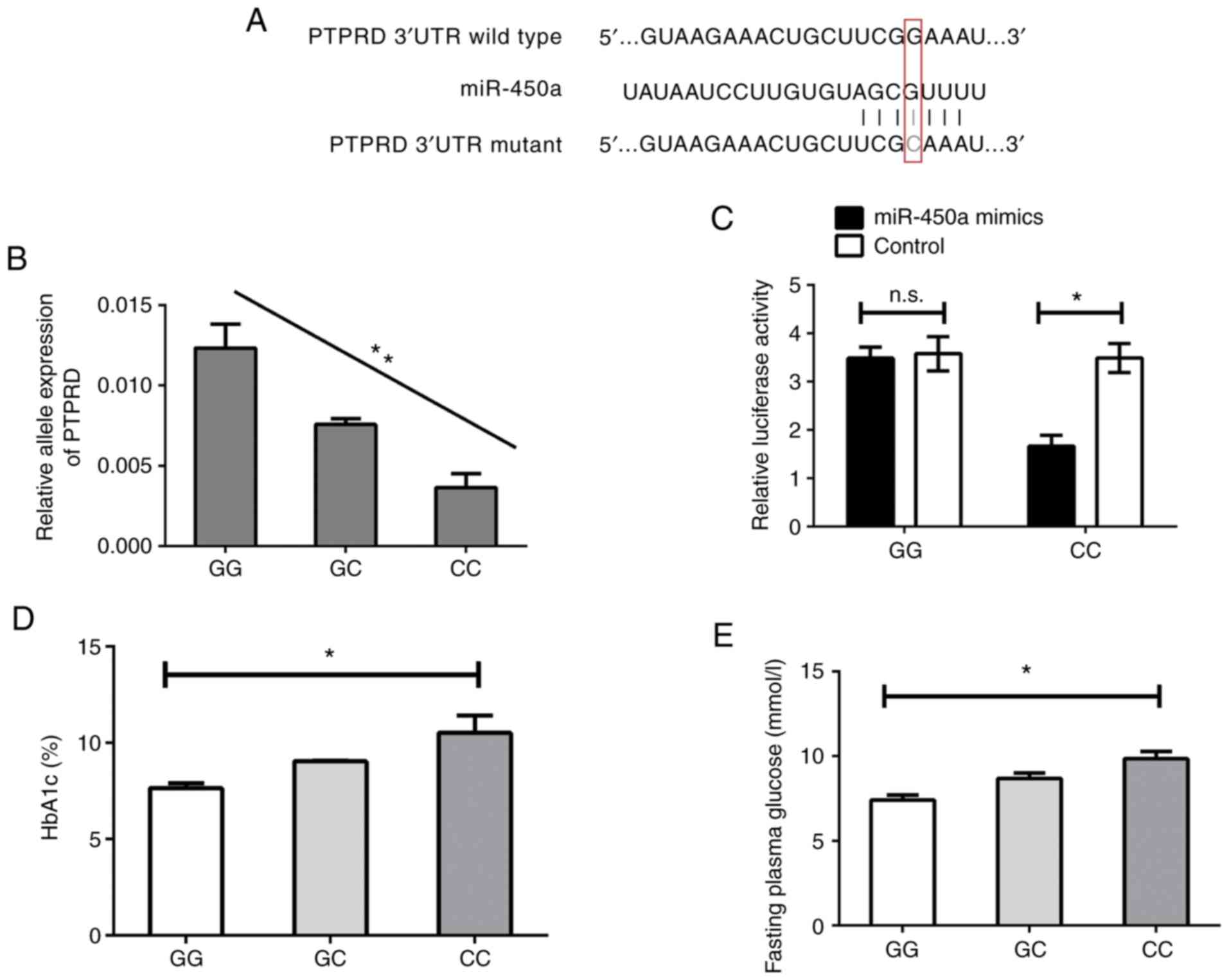
miR-450a binds to the 3'UTR of PTPRD
with the C allele
The potential binding site of miR-450a in the 3'UTR
of PTPRD predicted by the bioinformatics analysis is presented in
Fig. 1A. According to this, in
patients with the CC genotype, the binding ability was
comparatively greater than in the other subjects with more binding
sites. It was hypothesized that the expression of PTPRD may be
regulated by miR-450a, which may be impacted by the SNP rs56407701.
To confirm this, the expression of PTPRD mRNA expression in the
placenta samples of the GDM patients mentioned above was further
investigated. As presented in Fig.
1B, patients with GDM of the CC genotype had suppressed
expression of PTPRD mRNA compared with those of the GC and GG
genotype. Next, vectors including the allele-specific binding
sequences were constructed and then co-transfected with miR-450a or
the respective controls into the 293T cell line. As presented in
Fig. 1C, the luciferase reporter
assay confirmed that miR-450a was able to bind with the 3'UTR of
PTPRD with the CC genotype of the rs56407701 SNP resulting in the
suppression of luciferase activity, which indicated that PTPRD
expression was suppressed. However, the binding ability was
abolished with the GG genotype of the rs56407701 SNP
(wild-type).
Since it is widely known that HbA1c is closely
associated with GDM (30),
information regarding the plasma levels of HbA1c as well as the
fasting plasma glucose levels of the patients was collected. These
values were compared between patients with different genotypes of
the rs56407701 SNP of PTPRD. As presented in Fig. 1D and E, GDM patients with the C allele exhibited
higher HbA1c and fasting plasma glucose levels compared with both
GG and GC groups.
Discussion
GDM is a complex disease caused by the combination
of genetic factors and environmental exposure (31). miR-binding SNPs may serve as novel
targets or destroy existing recognition sites, promoting disease
susceptibility and important disease characteristics, particularly
in cancer and human GDM (32). In
the present study, the relationship between miR SNPs in the 3'-UTR
of PTPRD and GDM susceptibility was investigated. It was observed
that PTPRD rs56407701 was significantly associated with increased
susceptibility and the SNP was located at the binding site of
miR-450, interfering with the inhibition of PTPRD expression by
miR-450, which had an important role in the occurrence and
development of GDM.
To date, no direct evidence has been provided to
support the role of miR-450a in GDM. Abnormal miRNA expression is
associated with a variety of diseases. Research has identified the
potential function of miR-450a in human disease. Upregulated
miR-450a eliminated methylglyoxal-induced insulin resistance via
targeting cyclic AMP response element binding protein and may
therefore be used as a potential target to improve insulin
resistance and treat patients with diabetes-associated diseases
(33). Furthermore, after glutamine
discontinuation, miR-450a overexpression decreased the
mitochondrial membrane potential but increased glucose uptake and
cell viability, which are characteristic of less invasive cancer
cells. In summary, by regulating glutamine decomposition-associated
targets, miR-450a may reduce the production of lipids, amino acids
and nucleic acids and finally influence in the development of
diabetes (34).
PTPs are signaling molecules that regulate a variety
of cellular processes, including differentiation, cell
proliferation, mitotic cycle and oncogenic transformation (35). PTPRD, which is associated with T2D
and involved in the insulin signaling pathway, was first reported
in a GWAS a Chinese Han population (36). Tsai et al (36) indicated that the PTPRD gene was
related to T2D susceptibility in a Chinese Han population.
Subsequently, Below et al (37) performed another GWAS with 837 T2D
cases and 436 normoglycemic controls, followed by a meta-analysis,
revealing such an association with another SNP, rs649891, in PTPRD
in Mexican-Americans (17,36). In a replication study, the PTPRD
genetic variant was suggested to be associated with progression to
diabetes in Han Chinese, most likely through increased insulin
resistance. Recently, Chen et al reported that the levels of
PTPRD were significantly decreased in patients with T2D and that
this protein is involved in the insulin signaling pathway. Chen
et al (16) revealed that
the rs10511544, rs10756026 and rs10809070 SNPs in PTPRD may
contribute to a decreased susceptibility to GDM in Han Chinese
subjects. The SNP rs17584499 located in PTPRD was reported to
interact with the therapeutic efficacy of pioglitazone (38). In the present study, the role of the
PTPRD rs56407701 polymorphism in human GDM was first investigated
and the inhibition of PTPRD in human GDM by miR-binding of an SNP
was first reported. PTPRD mRNA levels were also suppressed in
patients with GDM. These results were similar to those of patients
with T2D (16). Furthermore, PTPRD
was reported to be involved in the insulin signaling pathway:
STAT3, a well-known oncoprotein, was inactivated by PTPRD
activation and STAT3 was overexpressed while PTPRD was inhibited
(39).
The abnormal distribution of PTPRD polymorphisms in
GDM in the present study suggested a strong relationship with the
occurrence of GDM. GDM patients with The C allele had a higher risk
of developing GDM in the presence of hyperglycemic factors than
those with the G allele. In addition, patients who had taken in too
much protein (above the recommended daily intake) and
high-carbohydrate food during pregnancy have an elevated risk of
developing GDM (40).
Analysis of the allele distribution of rs56407701 in
the population of the present study suggested a higher frequency of
homozygote CC compared with the frequency reported in the 1000
Genomes Project (https://www.internationalgenome.org/). The frequency
for homozygote CC controls is 2.5%, according to the 1000 Genomes
Project. This may be due to limitations regarding the sample size
of the present study. The background of the internal association
between these biomarkers (PTPRD gene and miR-450a) and GDM require
further study.
In conclusion, the present study provided the first
evidence that the SNP rs56407701 in the 3'-UTR of PTPRD was
associated with increased susceptibility to GDM. The function the
SNP was regulated by miR-450a, which caused suppression of PTPRD
expression in patients with the GC and CC genotype. The present
study provided evidence that SNPs in this miRNA-binding site may be
a novel source of susceptibility loci for human GDM.
Supplementary Material
Influence of eating habits on the risk
of GDM in pregnant females.
Allele distribution of additional
candidate SNPs located in the 3'-untranslated region of protein
tyrosine phosphatase receptor type D.
Acknowledgements
Not applicable.
Funding
Funding: No funding received.
Availability of data and materials
The datasets generated and analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
YK conducted the experiment, YKand HMH: manuscript
writing, literature search and data analysis; HPL and WPS: data
analysis and statistical analysis. YK: research design. All authors
read and approved the final manuscript
Ethics approval and consent to
participate
The subjects provided written informed consent prior
to specimen collection. The study was approved by the Ethics
Committee of Qinghai Red-Cross Hospital (Xining, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reulen RC, Bright CJ, Winter DL, Fidler
MM, Wong K, Guha J, Kelly JS, Frobisher C, Edgar AB, Skinner R, et
al: Pregnancy and labor complications in female survivors of
childhood cancer: The British childhood cancer survivor study. J
Natl Cancer Inst. 109(djx056)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Garnaes KK, Mørkved S, Salvesen Ø and
Moholdt T: Exercise training and weight gain in obese pregnant
women: A randomized controlled trial (ETIP Trial). PLoS Med.
13(e1002079)2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bao W, Tobias DK, Bowers K, Chavarro J,
Vaag A, Grunnet LG, Strøm M, Mills J, Liu A, Kiely M and Zhang C:
Physical activity and sedentary behaviors associated with risk of
progression from gestational diabetes mellitus to type 2 diabetes
mellitus: A prospective cohort study. JAMA Intern Med.
174:1047–1055. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhang C, Bao W, Rong Y, Yang H, Bowers K,
Yeung E and Kiely M: Genetic variants and the risk of gestational
diabetes mellitus: A systematic review. Hum Reprod Update.
19:376–390. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Luoto R, Kinnunen TI, Aittasalo M, Kolu P,
Raitanen J, Ojala K, Mansikkamäki K, Lamberg S, Vasankari T,
Komulainen T and Tulokas S: Primary prevention of gestational
diabetes mellitus and large-for-gestational-age newborns by
lifestyle counseling: A cluster-randomized controlled trial. PLoS
Med. 8(e1001036)2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Buchanan TA and Xiang AH: Gestational
diabetes mellitus. J Clin Invest. 115:485–491. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Chen L, Magliano DJ and Zimmet PZ: The
worldwide epidemiology of type 2 diabetes mellitus-present and
future perspectives. Nat Rev Endocrinol. 8:228–236. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Retnakaran R, Ye C, Hanley AJ, Connelly
PW, Sermer M, Zinman B and Hamilton JK: Effect of maternal weight,
adipokines, glucose intolerance and lipids on infant birth weight
among women without gestational diabetes mellitus. CMAJ.
184:1353–1360. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hou Z, Li M and Cao Y: TCF7L2, CAPN10
polymorphisms are associated with gestational diabetes mellitus
(GDM) risks: A meta-analysis. Gynecol Endocrinol. 33:399–404.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Besco JA, Frostholm A, Popesco MC, Burghes
AH and Rotter A: Genomic organization and alternative splicing of
the human and mouse RPTPrho genes. BMC Genomics.
2(1)2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang P, Becka S, Craig SE, Lodowski DT,
Brady-Kalnay SM and Wang Z: Cancer-derived mutations in the
fibronectin III repeats of PTPRT/PTPrho inhibit cell-cell
aggregation. Cell Commun Adhes. 16:146–153. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
McAndrew PE, Frostholm A, White RA, Rotter
A and Burghes AH: Identification and characterization of RPTP rho,
a novel RPTP mu/kappa-like receptor protein tyrosine phosphatase
whose expression is restricted to the central nervous system. Brain
Res Mol Brain Res. 56:9–21. 1998.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lee JW, Jeong EG and Lee SH, Nam SW, Kim
SH, Lee JY, Yoo NJ and Lee SH: Mutational analysis of PTPRT
phosphatase domains in common human cancers. APMIS. 115:47–51.
2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yu J, Becka S, Zhang P, Zhang X,
Brady-Kalnay SM and Wang Z: Tumor-derived extracellular mutations
of PTPRT/PTPrho are defective in cell adhesion. Mol Cancer Res.
6:1106–1113. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang D, Cheng Z, Zhao M, Jiao C, Meng Q,
Pan H, Xie Y, Li L, Zhu Y, Wang W, et al: PTPN9 induces cell
apoptosis by mitigating the activation of Stat3 and acts as a tumor
suppressor in colorectal cancer. Cancer Manag Res. 11:1309–1319.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen YT, Lin WD, Liao WL, Lin YJ, Chang JG
and Tsai FJ: PTPRD silencing by DNA hypermethylation decreases
insulin receptor signaling and leads to type 2 diabetes.
Oncotarget. 6:12997–13005. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yu H, Wang L, Lv L, Ma C, Du B, Lu T, Jin
C, Yan H, Yang Y, Li W, et al: Genome-wide association study
suggested the PTPRD polymorphisms were associated with weight gain
effects of atypical antipsychotic medications. Schizophr Bull.
42:814–823. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Plasterk RH: Micro RNAs in animal
development. Cell. 124:877–881. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hyun S, Lee JH, Jin H, Nam J, Namkoong B,
Lee G, Chung J and Kim VN: Conserved MicroRNA miR-8/miR-200 and its
target USH/FOG2 control growth by regulating PI3K. Cell.
139:1096–1108. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang Y, Zhou L, Chen J, Li J, He L, Wu P,
Wang M, Tong N, Zhang Z and Fang Y: Association of the 3'UTR FOXO3a
polymorphism rs4946936 with an increased risk of childhood acute
lymphoblastic leukemia in a Chinese population. Cell Physiol
Biochem. 34:325–332. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang X, Li W, Ma L, Gao J, Liu J, Ping F
and Nie M: Association study of the miRNA-binding site
polymorphisms of CDKN2A/B genes with gestational diabetes mellitus
susceptibility. Acta Diabetol. 52:951–958. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liu CJ, Fu X, Xia M, Zhang Q, Gu Z and Guo
AY: miRNASNP-v3: A comprehensive database for SNPs and
disease-related variations in miRNAs and miRNA targets. Nucleic
Acids Res: Sep 29, 2020 (Online ahead of print).
|
|
25
|
Berger H, Crane J, Farine D, Armson A, De
La Ronde S, Keenan-Lindsay L, Leduc L, Reid G and Van Aerde J:
Maternal-Fetal Medicine Committee; Executive and Coundil fo the
Society of Obstetricians and Gynaecologists of Canada. Screening
for gestational diabetes mellitus. J Obstet Gynaecol Can.
24:894–912. 2002.PubMed/NCBI View Article : Google Scholar : (In English,
French).
|
|
26
|
Gong J, Tong Y, Zhang HM, Wang K, Hu T,
Shan G, Sun J and Guo AY: Genome-wide identification of SNPs in
microRNA genes and the SNP effects on microRNA target binding and
biogenesis. Hum Mutat. 33:254–263. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang Y, Wang L, Yu X and Duan J:
Overexpression of miR-450 affects the biological behavior of HepG2
cells by targeting DNMT3a. Onco Targets Ther. 12:5069–5076.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li M, Li F, Lin Q, Shi J, Luo J, Long Q,
Yang Q, Ouyang Y, Liu H, Bell RC and Guo J: Cultural adaptation,
validation, and primary application of a questionnaire to assess
intentions to eat low-glycemic index foods among rural Chinese
women. Int J Environ Res Public Health. 17(7577)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fatima SS, Chaudhry B, Khan TA and Farooq
S: KCNQ1 rs2237895 polymorphism is associated with gestational
diabetes in Pakistani women. Pak J Med Sci. 32:1380–1385.
2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Poulsen P, Levin K, Petersen I,
Christensen K, Beck-Nielsen H and Vaag A: Heritability of insulin
secretion, peripheral and hepatic insulin action, and intracellular
glucose partitioning in young and old Danish twins. Diabetes.
54:275–283. 2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liu Y, Chai Y, Zhang J and Tang J: A
function variant at miR-501 alters susceptibility to hepatocellular
carcinoma in a Chinese Han population. Cell Physiol Biochem.
38:2500–2508. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wei C, Meng L and Zhang Y: miR-450a-5p
eliminates MGO-Induced insulin resistance via targeting CREB. Int J
Stem Cells. 13:46–54. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Muys BR, Sousa JF, Plaça JR, de Araújo LF,
Sarshad AA, Anastasakis DG, Wang X, Li XL, de Molfetta GA, Ramão A,
et al: miR-450a acts as a tumor suppressor in ovarian cancer by
regulating energy metabolism. Cancer Res. 79:3294–3305.
2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Luo L, Xie Y, Wang A, Liu X, Xiao F, Zhong
X and Zhong C: Desipramine ameliorates Cr(VI)-induced
hepatocellular apoptosis via the inhibition of ceramide channel
formation and mitochondrial PTP opening. Cell Physiol Biochem.
34:2128–2136. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu
CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM, et al: A
genome-wide association study identifies susceptibility variants
for type 2 diabetes in Han Chinese. PLoS Genet.
6(e1000847)2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Below JE, Gamazon ER, Morrison JV,
Konkashbaev A, Pluzhnikov A, McKeigue PM, Parra EJ, Elbein SC,
Hallman DM, Nicolae DL, et al: Genome-wide association and
meta-analysis in populations from Starr County, Texas, and Mexico
City identify type 2 diabetes susceptibility loci and enrichment
for expression quantitative trait loci in top signals.
Diabetologia. 54:2047–2055. 2011.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Pei Q, Huang Q, Yang GP, Zhao YC, Yin JY,
Song M, Zheng Y, Mo ZH, Zhou HH and Liu ZQ: PPAR-γ2 and PTPRD gene
polymorphisms influence type 2 diabetes patients' response to
pioglitazone in China. Acta Pharmacol Sin. 34:255–261.
2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gong K, Qu B, Wang C, Zhou J, Liao D,
Zheng W and Pan X: Peroxisome proliferator-activated receptor α
facilitates osteogenic differentiation in MC3T3-E1 cells via the
sirtuin 1-dependent signaling pathway. Mol Cells. 40:393–400.
2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Atakora L, Poston L, Hayes L, Flynn AC and
White SL: Influence of GDM diagnosis and treatment on weight gain,
dietary intake and physical activity in pregnant women with
obesity: Secondary analysis of the UPBEAT study. Nutrients.
12(359)2020.PubMed/NCBI View Article : Google Scholar
|