Introduction
The maxillofacial region is important in maintaining
normal feeding, chewing, breathing and protecting the connected
brain, cervical vertebra and other important anatomical structures.
As the most prominent part of the human body, the maxillofacial
region is susceptible to fractures and corresponding soft tissue
injuries (1,2). Maxillofacial fractures primarily
include maxillary and mandible fractures. Patients with maxillary
fractures experience damage to the maxillary sinus, which can
affect the adjacent bone and tissue. If left untreated, fractures
can impact facial appearance and occlusion (3,4).
Maxillofacial fracture repair is a complex physiological process
that is influenced by individual and environmental factors, as well
as cytokines, somatic cells and endocrine factors (5,6).
Osteoblast activity and differentiation also play an important role
in the process of fracture healing (7).
Long non-coding RNA (lncRNA) is a class of
transcripts >200 nt in length that regulate gene expression at
epigenetic, transcriptional and post-transcriptional levels
(8,9). Previous research has found abnormal
lncRNAs to be involved in the regulation of osteoblasts activity.
For example, maternally expressed gene 3 promotes osteoblast
proliferation and differentiation during fracture healing (10); lncRNA AK016739 promotes osteogenic
differentiation in skeletal disorders (11); and lncRNA Crnde mediates bone
formation in animal models (12).
The lncRNA taurine upregulated gene 1 (TUG1) was first identified
in mouse retinal cells treated with taurine (13). Research on TUG1 focuses on its role
in the development of malignant tumors (14,15).
In the current study, the aim was to investigate the effect of TUG1
on maxillofacial fracture development.
Materials and methods
Patients
Between January 2018 and December 2019, a total of
50 patients (range, 18-60 years old) diagnosed with maxillary
fracture at The First Affiliated Hospital Xinjiang Medical
University (Urumqi, China) were registered to participate in this
study. There were 27 male and 23 female patients (age range, 18-60
years old; median, 43.25±5.32). The type and severity of maxillary
fracture were classified according to the LeFort classification
(16): type I (n=15), type II
(n=25) and type III (n=10). During the same period, 50 healthy
volunteers in the physical examination center were enrolled as
controls. There were 27 male and 23 female volunteers (age range,
23-58 years old; median, 40.32±6.78). Written consent was obtained
from all participants prior to enrollment, and the present study
was approved by the Ethics Committee of The First Affiliated
Hospital Xinjiang Medical University. Serum samples (5 ml) from
participants were collected and centrifuged at 12,000 x g for 3 min
at 37˚C, and the supernatant was immediately frozen in liquid
nitrogen for further analysis.
Cell culture
Mouse osteoblast cell line MC3T3-E1 (obtained from
the Cell Bank of the Chinese Academy) was cultured in Dulbecco's
modified Eagle's medium (DMEM) (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% Fetal Bovine Serum (FBS) (Invitrogen; Thermo
Fisher Scientific, Inc.) and 1% penicillin/streptomycin under an
atmosphere of humidified air and 5% CO2 at 37˚C.
Cell transfection
Small interfering RNA (siRNA) targeting TUG1
(si-TUG1; 20 nM), siRNA-negative control (si-NC; 20 nM), microRNA
(miR)-214 mimic (30 nM), miR-NC (final concentration, 30 nM),
miR-214 inhibitor (15 nM), NC inhibitor (15 nM), bone morphogenetic
protein 2 (BMP2) mimics (30 nM) and NC mimic (30 nM) were
synthesized by Shanghai GenePharma Co., Ltd. Transfection was
performed using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37˚C for 48 h, after which the cells
were harvested for reverse transcription-quantitative (RT-q)PCR
analysis or western blotting. The sequences were shown as follows:
si-TUG1 5'-GCUUGGCUUCUAUUCUGAAUCCUUU-3'; scrambled si-NC
5'-TTCTCCGAACGTGTCACGTTT-3'; miR-214 mimic
5'-ACAGCAGGCACAGACAGGCAGU-3'; scrambled miR-NC
5'-TTCTCCGAACGTGTCACGTTT-3'; miR-214 inhibitor
5'-ACGTGACACGTTCGGAGAATT-3'; scrambled NC inhibitor
5'-CAGTACTTTTGTGTAGTACAA-3'; BMP2 mimic 5'-ACCCGCTGTCTTCTAGCGT-3';
and scrambled NC mimic 5'-ACGUGACACGUUCGGAGAATT-3'.
Cell viability
MC3T3-E1 cells (1x105 cells/well) with
different transfections were seeded in 96-well plates and cultured
for at 37˚C for 0, 24, 48, 72 and 96 h. After cells were treated
with 500 µg/ml MTT solution for 3 h at 37˚C, 200 µl
dimethylsulfoxide was added to dissolve precipitates. The optical
density of each well was measured at 540 nm under a microplate
spectrophotometer.
Cell migration
A total of 1x105 MC3T3-E1 cells were
plated into the upper chambers containing 200 µl serum-free DMEM.
DMEM containing 20% FBS was added to the lower chamber. Following
incubation for 24 h at 37˚C, the migrated cells were fixed with
100% methanol for 30 min and stained with 0.1% crystal violet for
30 min (both at room temperature). Finally, images were captured
using an inverted microscope at x200 magnification (Olympus
Corporation), and the mean number of migrated or invaded cells in
five random microscopic fields per Transwell chamber was
counted.
Cell apoptosis assay
After transfection, 1x105 harvested cells
were washed two times with PBS and then were resuspended using 100
µl binding buffer. Transfected cells were double stained with
propidium iodide and Annexin V-FITC (20 mg/ml; BD Pharmingen; BD
Biosciences) at room temperature for 10 min in line with the
manufacturer's instructions according to the manufacturer's
protocol. Apoptosis was evaluated using a flow cytometer BD
FACSCalibur™ (BD Biosciences) and BD FACS™ software
(v1.0.0.650; BD Biosciences).
Reverse transcription-quantitative
(RT-q)PCR analysis
Total RNAs in serums and MC3T3-E1 cells were
isolated using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
cDNA synthesis was performed using the TaqMan MicroRNA Reverse
Transcription Kit (Applied Biosystems; Thermo fisher Scientific,
Inc.). qPCR was performed using the Mx3000p qPCR system (Agilent
Technologies). GAPDH was an internal parameter of TUG1 and BMP2,
while U6 was an internal parameter of miR-214. The following
thermocycling conditions were used: Initial denaturation at 95˚C
for 10 min; followed by 40 cycles of denaturation at 95˚C for 15
sec, and annealing at 60˚C for 1 min; and a final extension of 10
min at 72˚C. The sequences of primers the are as follows: TUG1
forward, 5'-TAGCAGTTCCCCAATCCTTG-3' and reverse,
5'-CACAAATTCCCATCATTCCC-3'; miR-214 forward,
5'-AGCATAATACAGCAGGCACAGAC-3' and reverse,
5'-AAAGGTTGTTCTCCACTCTCTCAC-3'; BMP2 forward,
5'-GCAAAGAAAAGGAACGGACATT-3'; and reverse,
5'-AAAGGTTGTTCTCCACTCTCTCAC-3'; GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3';
and reverse, 5'-ACGCTTCACGAATTTGCGTGTC-3'. The relative expression
was analysis using 2-ΔΔCq (17).
Dual-Luciferase reporter assay
The TUG1 fragments and BMP2 3'-UTR containing
wild-type (WT) or mutant (MUT) miR-214 binding sites were obtained
from Shanghai GenePharma Co., Ltd. Cells (1x106 per
well) were co-transfected with 100 ng TUG1-WT, TUG1-MUT, BMP2-WT or
BMP-2 MUT, and 100 nM miR-214 mimic or miR-NC using
Lipofectamine® 2000. Following incubation for 48 h,
luciferase activities were assessed using a Dual-Luciferase
reporter assay kit (Promega Corporation), according to the
manufacturer's protocol. Firefly luciferase activity was normalized
to that of Renilla luciferase.
Western blotting analysis
Proteins were extracted from MC3T3-E1 cells using
RIPA buffer (Sigma-Aldrich; Merck KGaA) containing a mixture of
protease inhibitors. Total protein was quantified using a BCA
protein assay kit (Pierce; Thermo Fisher Scientific, Inc.).
Proteins (50 µg per lane) were separated using 10% SDS-PAGE and
electrotransferred to nitrocellulose membranes (EMD Millipore). The
membranes were blocked with 5% non-fat milk at room temperature for
1 h. Subsequently, the membranes were incubated with the following
primary antibodies at 4˚C overnight: Anti-runt-related
transcription factor 2 (RUNX2; 1:1,000; cat. no. ab192256),
anti-osteocalcin (OCN; 1:1,000; cat. no. ab133612) and anti-GAPDH
(1:1,000; cat. no. ab9485) all purchased from Abcam. The membranes
were then incubated with anti-rabbit horseradish
peroxidase-conjugated secondary antibody (1:20,000; cat. no.
ab6721; Abcam). Finally, the protein bands were visualized with an
ECL kit (Cytiva) and analyzed using ImageJ v1.8.0 software
(National Institutes of Health).
Bioinformatics analysis
The binding sites between miR-214 and TUG1, as well
as miR-214 and BMP2 were predicted by StarBase v2.0 (http://www.microrna.gr/lncBase) and TargetScan
software v7.1 (http://www.targetscan.org/vert_71/), respectively.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Differences between the two groups were analyzed using
an unpaired Student's t-test. Multigroup comparisons were performed
using a one-way ANOVA with a Tukey's post hoc test. All statistical
analyses were performed using GraphPad Prism 5.0 (GraphPad
Software, Inc.) and SPSS 14.0 (SPSS, Inc.). A statistically
significant difference was defined as P<0.05.
Results
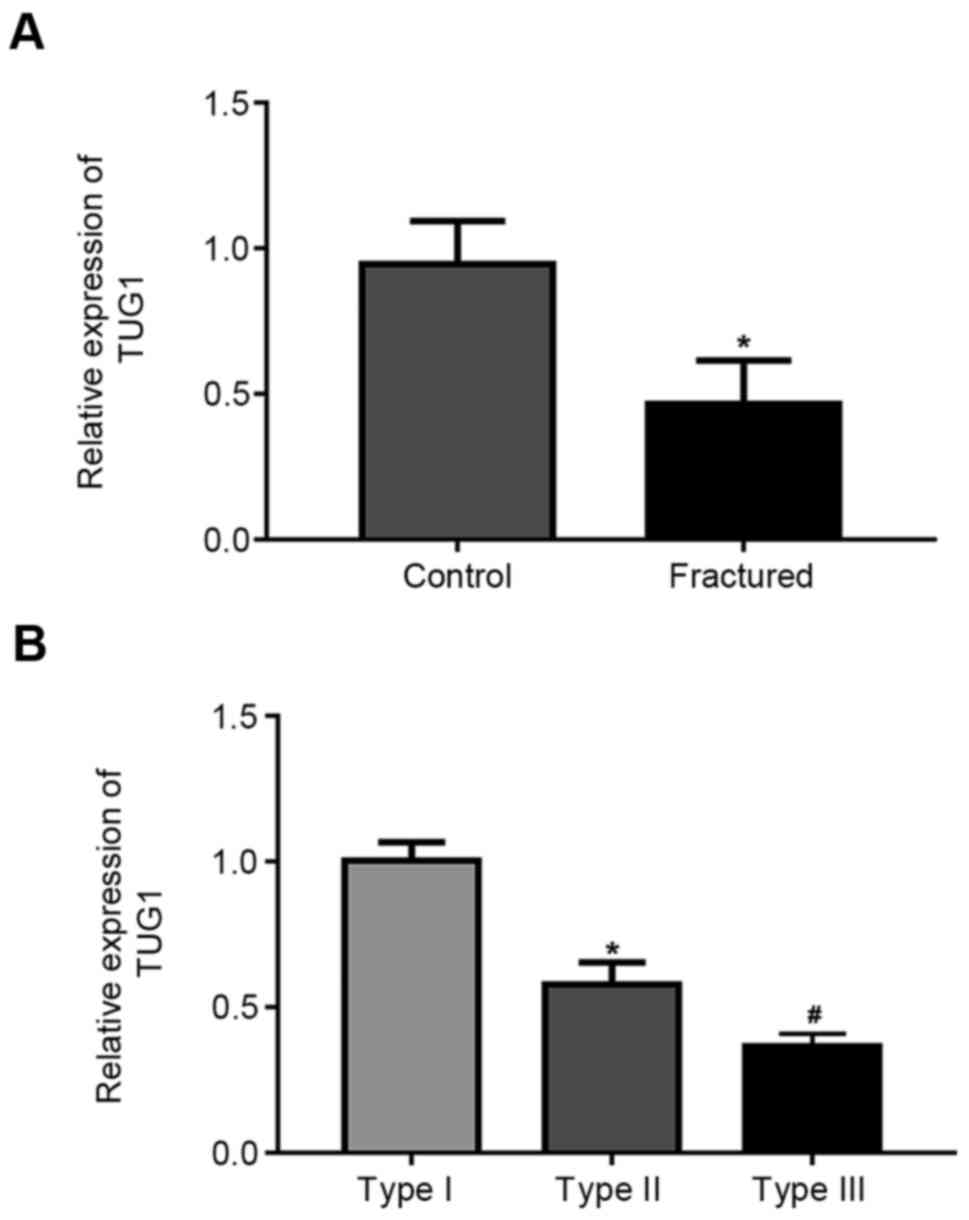
TUG1 expression is decreased in the
serum of patients with maxillary fractures
Results demonstrated that TUG1 expression was
significantly downregulated in patients compared with healthy
volunteers (Fig. 1A). Further, TUG1
expression significantly decreased with fracture severity (Fig. 1B).
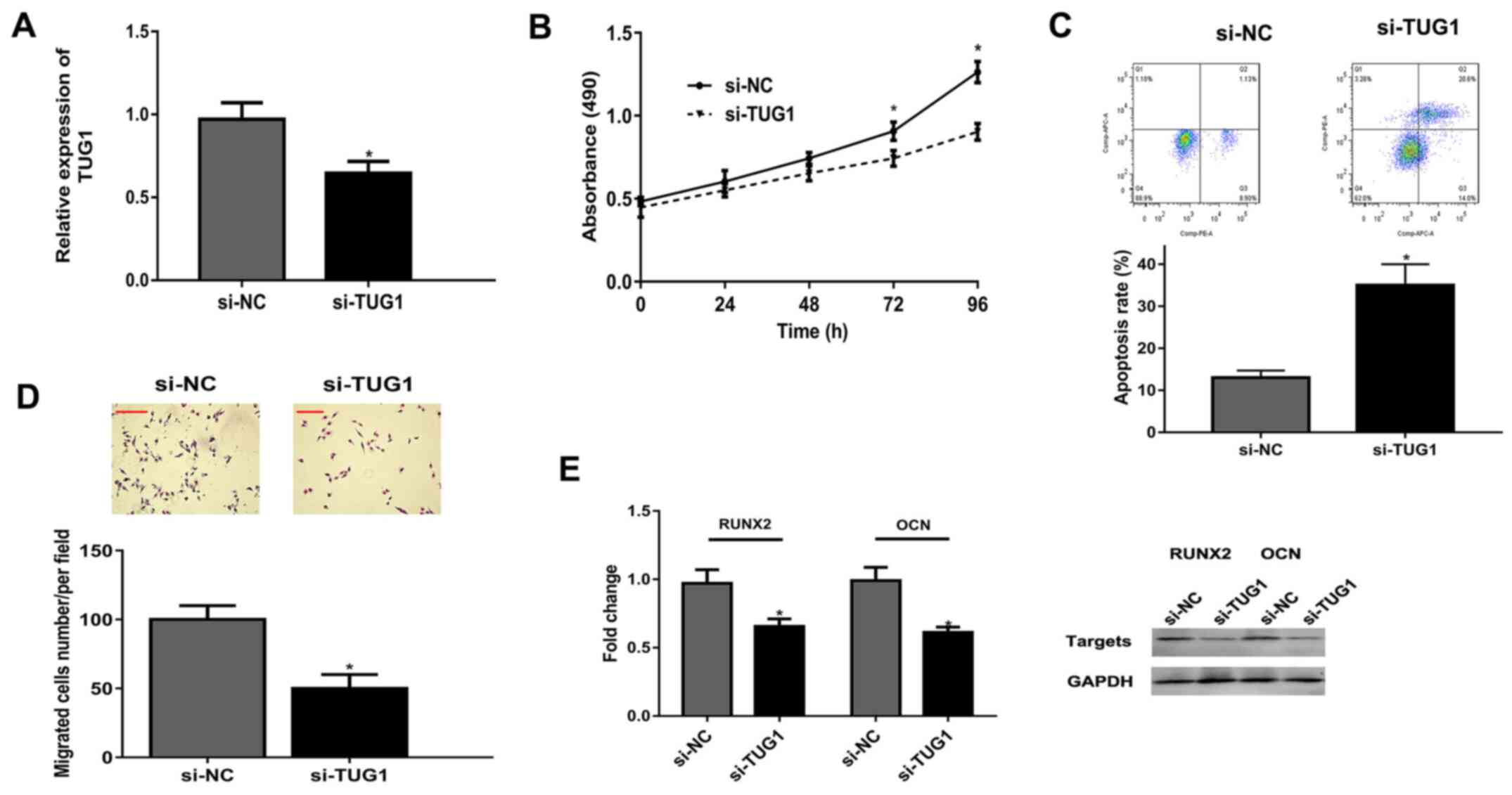
Knockdown of TUG1 represses viability,
migration and differentiation and induces apoptosis of
osteoblasts
Following transfection of osteoblasts with si-TUG1,
TUG1 expression was significantly decreased compared with the si-NC
group (Fig. 2A). MTT assay revealed
that TUG1 knockdown significantly suppressed osteoblast
proliferation at 72 and 96 h, compared with the si-NC group
(Fig. 2B). Flow cytometry revealed
knockdown of TUG1 significantly increased the apoptosis rate of
osteoblasts (Fig. 2C). Knockdown of
TUG1 inhibited osteoblast migration compared with si-NC (Fig. 2D). Finally, mRNAs and protein levels
of differentiation-related RUNX2 and OCN were lower in the si-TUG1
group than that in si-NC group (Fig.
2E).
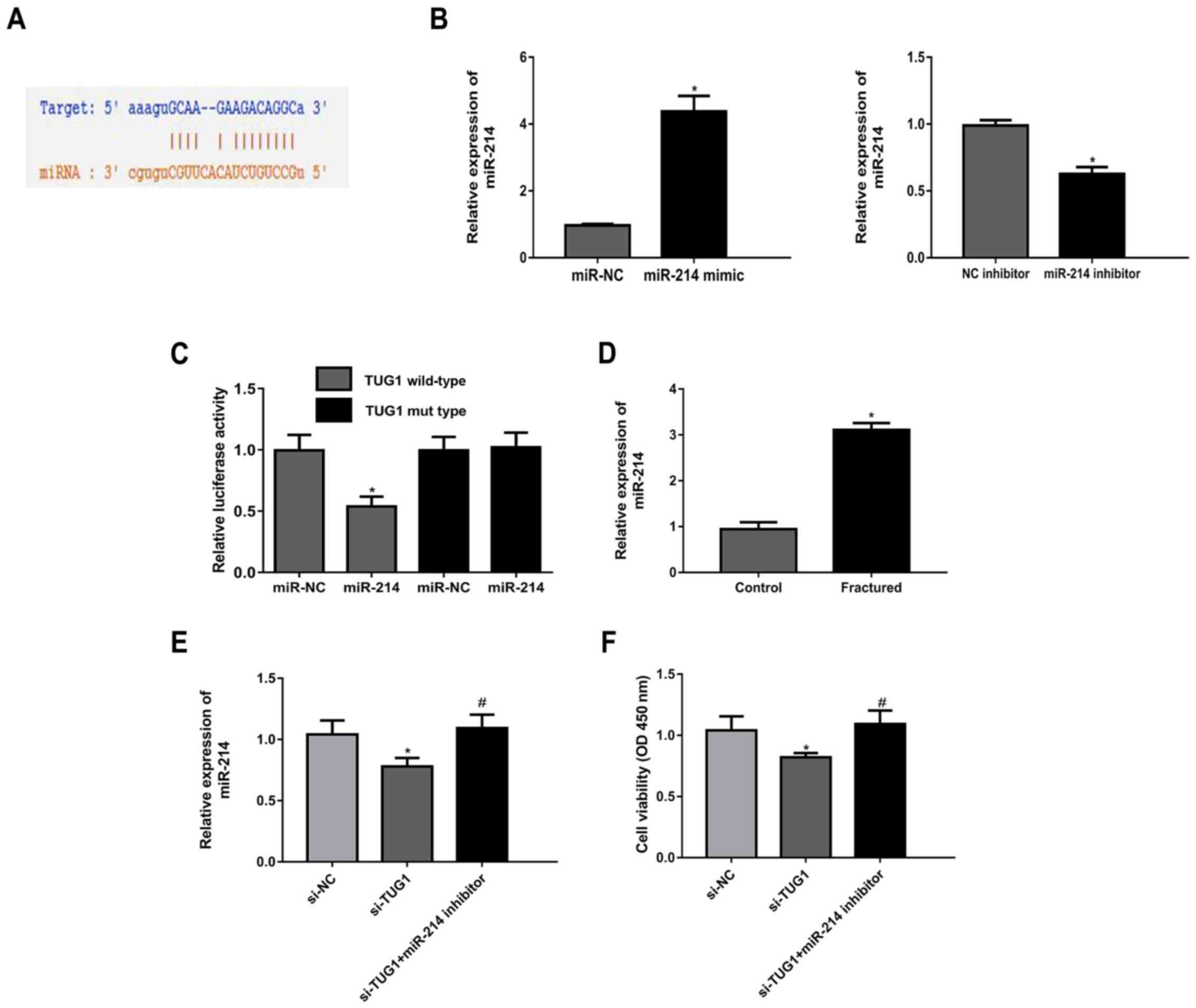
TUG1 acts as a sponge for miR-214
Most studies report that lncRNAs regulate gene
expression by sponging miRNAs (18). Given this, StarBase v2.0 was used to
predict the target microRNA binding with TUG1. It was demonstrated
that miR-214 had a potential binding site with TUG1 (Fig. 3A). The expression of miR-214 was
increased in cells transfected with miR-214 mimic compared with
miR-NC, while miR-214 expression was decreased in cells transfected
with miR-214 inhibitor compared with the NC inhibitor (Fig. 3B). Luciferase activity reporter
assays showed that miR-214 mimic inhibited the luciferase activity
of TUG1 WT compared with TUG1 MUT (Fig.
3C). Further, miR-214 expression level was significantly higher
in patients with maxillary fractures compared with controls
(Fig. 3D). TUG1 knockdown
upregulated miR-214 expression, which was reversed following
co-transfection of cells with a miR-214 inhibitor (Fig. 3E). The results of the MTT assay
indicated that si-TUG1 transfection resulted in a decrease in cell
viability, and this effect was abrogated following co-transfection
with miR-214 inhibitor (Fig.
3F).
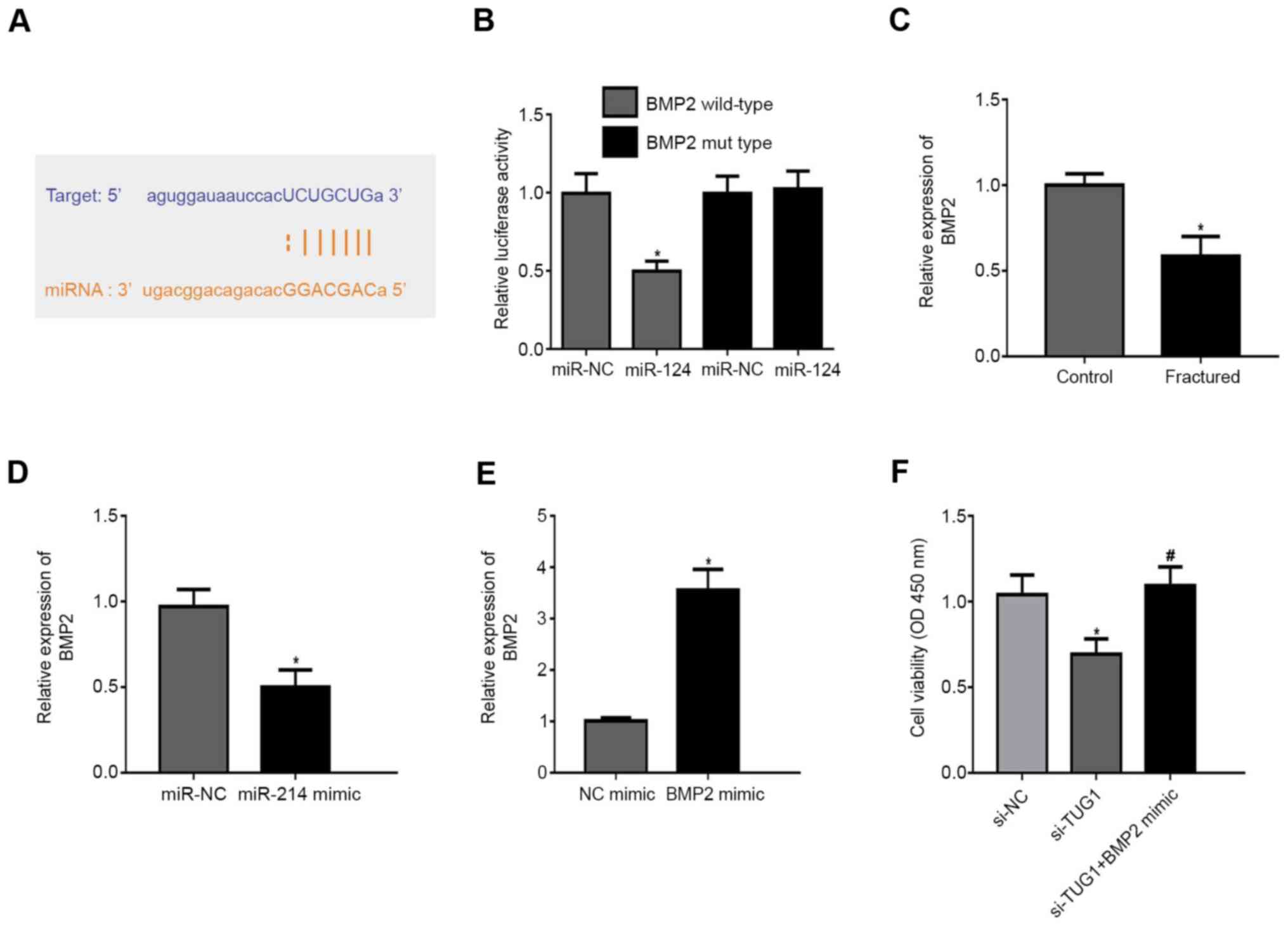
BMP2 is a direct target of
miR-214
TargetScan demonstrated 3'UTR of BMP2 was highly
conserved to bind with miR-214 (Fig.
4A). In BMP2 WT, miR-214 mimic repressed the fluorescence
(Fig. 4B). BMP2 expression was
lower in patients with maxillary fractures compared with healthy
controls (Fig. 4C). In addition,
miR-214 mimic inhibited BMP2 expression compared with the NC
(Fig. 4D). Transfection of cells
with a BMP2 mimic resulted in increased BMP2 expression levels
compared with the NC mimic (Fig.
4E). Notably, transfection of cells with si-TUG1 resulted in a
decrease in cell viability and this was reversed following
co-transfection with a BMP2 mimic (Fig.
4F).
Discussion
Due to its complex anatomical structure, the
diagnosis and treatment of maxillary bone injury is difficult.
Maxillary bone injury can cause dysfunction and morphological
deformities that severely impact quality of life. Given this,
effective diagnosis and treatment methods are needed to not only
improve patients' facial appearance, but to reshape the occlusal
relationship and peripheral structure (3,19). As
a novel long non-coding RNA, TUG1 has been reported to participate
in several types of orthopedic diseases including osteosarcoma
(20), osteoarthritis (21) and intervertebral disc degeneration
(22). In the present study, the
role of TUG1 in the development of maxillary fractures was
examined. It was revealed that TUG1 expression levels were
decreased in the serum of patients with maxillary fractures. Recent
studies have found TUG1 inhibition reduces osteoblast proliferation
and differentiation (23,24). Similarly, TUG1 knockdown attenuated
viability, migration and differentiation and induced apoptosis of
osteoblasts in the present study. These data suggest TUG1 knockdown
inhibits the activity of osteoblasts in fractures.
It is well known that lncRNAs play a regulatory role
in gene expression by sponging miRNAs. It was hypothesized that
miR-214 was the candidate target microRNA. Luciferase activity
reporter assays confirmed an inhibition of luciferase activity in
TUG1 WT. Other studies have found that miR-214 regulates bone
formation by weakening osteoblast proliferation and differentiation
(25,26). In the current study, increased
miR-214 expression was observed in patients with maxillary
fractures. Finally, miR-214 inhibitor reversed the repression of
osteoblast proliferation by si-TUG1.
Studies have shown miRNAs participate in the
regulation of gene expression through targeting 3'untranslated
region (3'-UTR) of mRNAs (27).
Subsequently, BMP2 was identified as the potential target gene of
miR-214 in the present study. BMP2 is critical in stimulating bone
formation during fracture healing (28,29).
It induces osteoblast proliferation and differentiation (30,31).
BMP2 expression was revealed to be lower in patients with maxillary
fractures and miR-214 mimic inhibited BMP2 expression in
osteoblasts. Finally, BMP2 mimic rescued the suppression of si-TUG1
in osteoblast proliferation.
The present study demonstrated that long noncoding
RNA TUG1 promoted proliferation and differentiation of MC3T3-E1
cells. It could be suggested that an elevation in TUG1 levels is
associated with later stages of maxillary fracture development, as
an increased level of TUG1 is required in maxillary fracture
repair. In the present study, the expression of TUG1 was only
examined at early stages of maxillary fractures. To hypothesize,
TUG1 expression would be expected to increase over time during the
recovery of maxillary fracture. Thus, further studies will be used
to explore the effect of TUG1 on the recovery of maxillary
fracture.
In conclusion, decreased TUG1 expression levels were
found in patients with maxillary fractures and TUG1 knockdown
repressed the biological process of osteoblasts by sponging
miR-214. Therefore, this study may provide some new information
about maxillary fracture development.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by Science Foundation of
Xinjiang Uygur Autonomous Region (grant no. 2014211C082).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZY and MT designed the experiments. ZY and WA
performed the experiments. AM analyzed the data. ZY and MT wrote
the manuscript. ZY and MT confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Medical Ethics
Committee of The First Affiliated Hospital of Xinjiang Medical
University (approval no. XJMU20171214; Urumqi, China). Written
informed consent was obtained from patients prior to
enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Motamedi MH, Dadgar E, Ebrahimi A, Shirani
G, Haghighat A and Jamalpour MR: Pattern of maxillofacial
fractures: A 5-year analysis of 8,818 patients. J Trauma Acute Care
Surg. 77:630–634. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shenoi S, Budhraja N and Badjate S: An
assessment of maxillofacial fractures: A two-year retrospective
study. J Emerg Trauma Shock. 5(205)2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhou HH, Ongodia D, Liu Q, Yang RT and Li
ZB: Dental trauma in patients with maxillofacial fractures. Dent
Traumatol. 29:285–290. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Feng Z, Chen R, Zhang Y, Yang M, Lin Y,
Tian W and Liu L: Outcome of postsurgical sequential functional
exercise of jaw fracture. J Craniofac Surg. 20:46–48.
2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Schindeler A, McDonald MM, Bokko P and
Little DG: Bone remodeling during fracture repair: The cellular
picture. Semin Cell Dev Biol. 19:459–466. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Giannoudis PV, Einhorn TA and Marsh D:
Fracture healing: The diamond concept. Injury. 38 (Suppl 4):S3–S6.
2007.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Claes L, Recknagel S and Ignatius A:
Fracture healing under healthy and inflammatory conditions. Nat Rev
Rheumatol. 8:133–143. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mattick JS and Rinn JL: Discovery and
annotation of long noncoding RNAs. Nat Struct Mol Biol. 22:5–7.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Li XG, Liu SC, Qiao XF, Kong Y, Liu JG,
Peng XM, Wang YX and Abdulkarim Mohammed Al-Mohana RA: LncRNA MEG3
promotes proliferation and differentiation of osteoblasts through
Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci.
23:4521–4529. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yin C, Tian Y, Yu Y, Wang H, Wu Z, Huang
Z, Zhang Y, Li D, Yang C, Wang X, et al: A novel long noncoding RNA
AK016739 inhibits osteoblast differentiation and bone formation. J
Cell Physiol. 234:11524–11536. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mulati M, Kobayashi Y, Takahashi A, Numata
H, Saito M, Hiraoka Y, Ochi H, Sato S, Ezura Y, Yuasa M, et al: The
long noncoding RNA Crnde regulates osteoblast proliferation through
the Wnt/β-catenin signaling pathway in mice. Bone.
130(115076)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512.
2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li Z, Shen J, Chan MT and Wu WK: TUG1: A
pivotal oncogenic long non-coding RNA of human cancers. Cell
Prolif. 49:471–475. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Katsushima K, Natsume A, Ohka F, Shinjo K,
Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, et
al: Targeting the Notch-regulated non-coding RNA TUG1 for glioma
treatment. Nat Commun. 7(13616)2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Manson PN: Some thoughts on the
classification and treatment of Le Fort fractures. Ann Plast Surg.
17:356–363. 1986.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Lim L and Sirichai P: Bone fractures:
Assessment and management. Aust Dent J. 61 (Suppl 1):S74–S81.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang Y, Yang T, Zhang Z, Lu M, Zhao W,
Zeng X and Zhang W: Long non-coding RNA TUG1 promotes migration and
invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells.
Cancer Sci. 108:859–867. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liang Z and Ren C: Emodin attenuates
apoptosis and inflammation induced by LPS through up-regulating
lncRNA TUG1 in murine chondrogenic ATDC5 cells. Biomed
Pharmacother. 103:897–902. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen J, Jia YS, Liu GZ, Sun Q, Zhang F, Ma
S and Wang YJ: Role of LncRNA TUG1 in intervertebral disc
degeneration and nucleus pulposus cells via regulating
Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun.
491:668–674. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu SC, Sun QZ, Qiao XF, Li XG, Yang JH,
Wang TQ, Xiao YJ and Qiao JM: LncRNA TUG1 influences osteoblast
proliferation and differentiation through the Wnt/β-catenin
signaling pathway. Eur Rev Med Pharmacol Sci. 23:4584–4590.
2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yu C, Li L, Xie F, Guo S, Liu F, Dong N
and Wang Y: LncRNA TUG1 sponges miR-204-5p to promote osteoblast
differentiation through upregulating Runx2 in aortic valve
calcification. Cardiovasc Res. 114:168–179. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu J, Li Y, Luo M and Yuan Z:
MicroRNA-214 inhibits the osteogenic differentiation of human
osteoblasts through the direct regulation of baculoviral IAP
repeat-containing 7. Exp Cell Res. 351:157–162. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li D, Liu J, Guo B, Liang C, Dang L, Lu C,
He X, Cheung HY, Xu L, Lu C, et al: Osteoclast-derived exosomal
miR-214-3p inhibits osteoblastic bone formation. Nat Commun.
7(10872)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Garrison KR, Shemilt I, Donell S, Ryder
JJ, Mugford M, Harvey I, Song F and Alt V: Bone morphogenetic
protein (BMP) for fracture healing in adults. Cochrane Database
Syst Rev. 2010(CD006950)2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Einhorn TA, Majeska RJ, Mohaideen A, Kagel
EM, Bouxsein ML, Turek TJ and Wozney JM: A single percutaneous
injection of recombinant human bone morphogenetic protein-2
accelerates fracture repair. J Bone Joint Surg Am. 85:1425–1435.
2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang RF, Liu JW, Yu SP, Sun D, Wang XH,
Fu JS and Xie Z: LncRNA UCA1 affects osteoblast proliferation and
differentiation by regulating BMP-2 expression. Eur Rev Med
Pharmacol Sci. 23:6774–6782. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Min HY, Kim KM, Wee G, Kim EJ and Jang WG:
Bmal1 induces osteoblast differentiation via regulation of BMP2
expression in MC3T3-E1 cells. Life Sci. 162:41–46. 2016.PubMed/NCBI View Article : Google Scholar
|