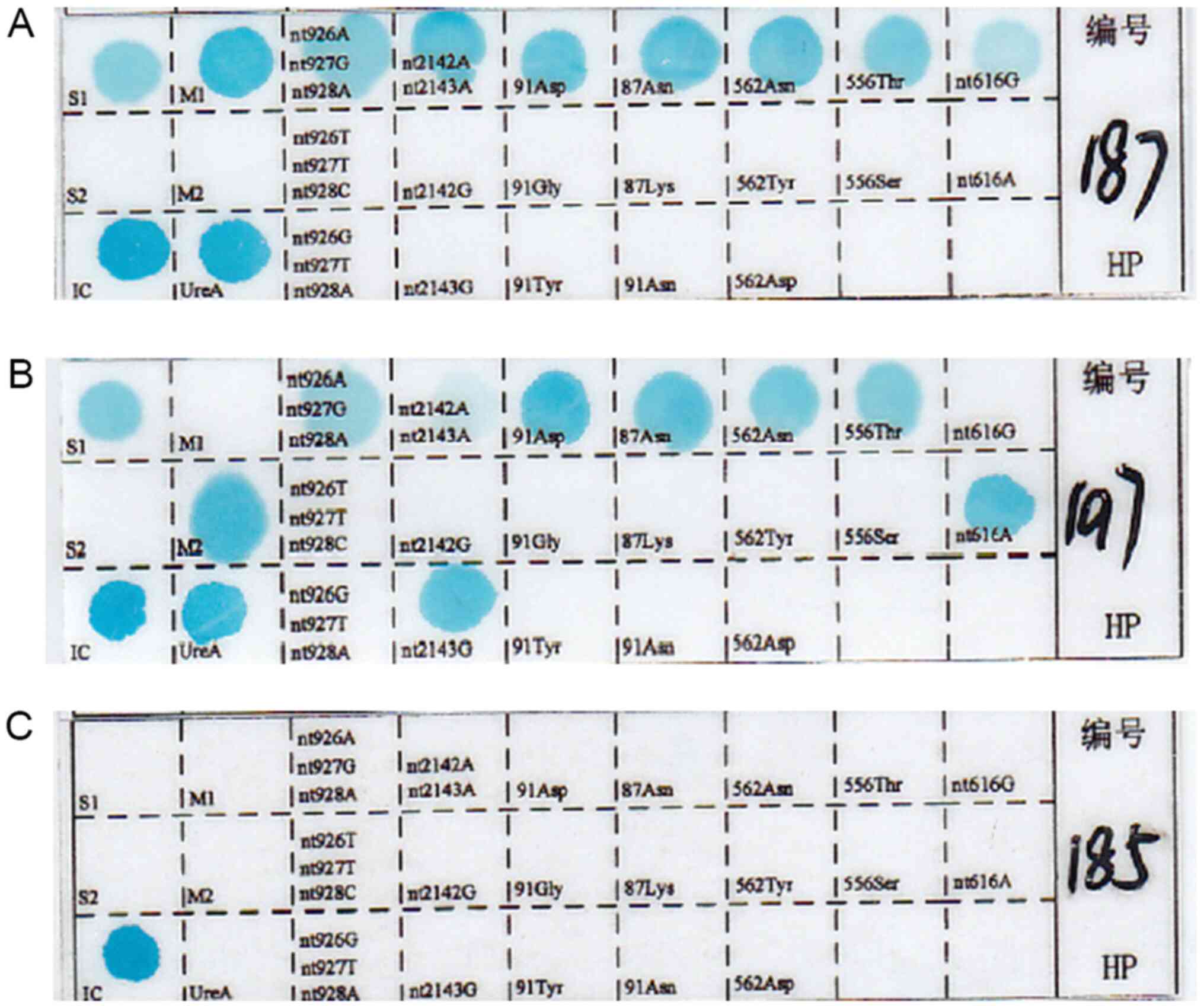
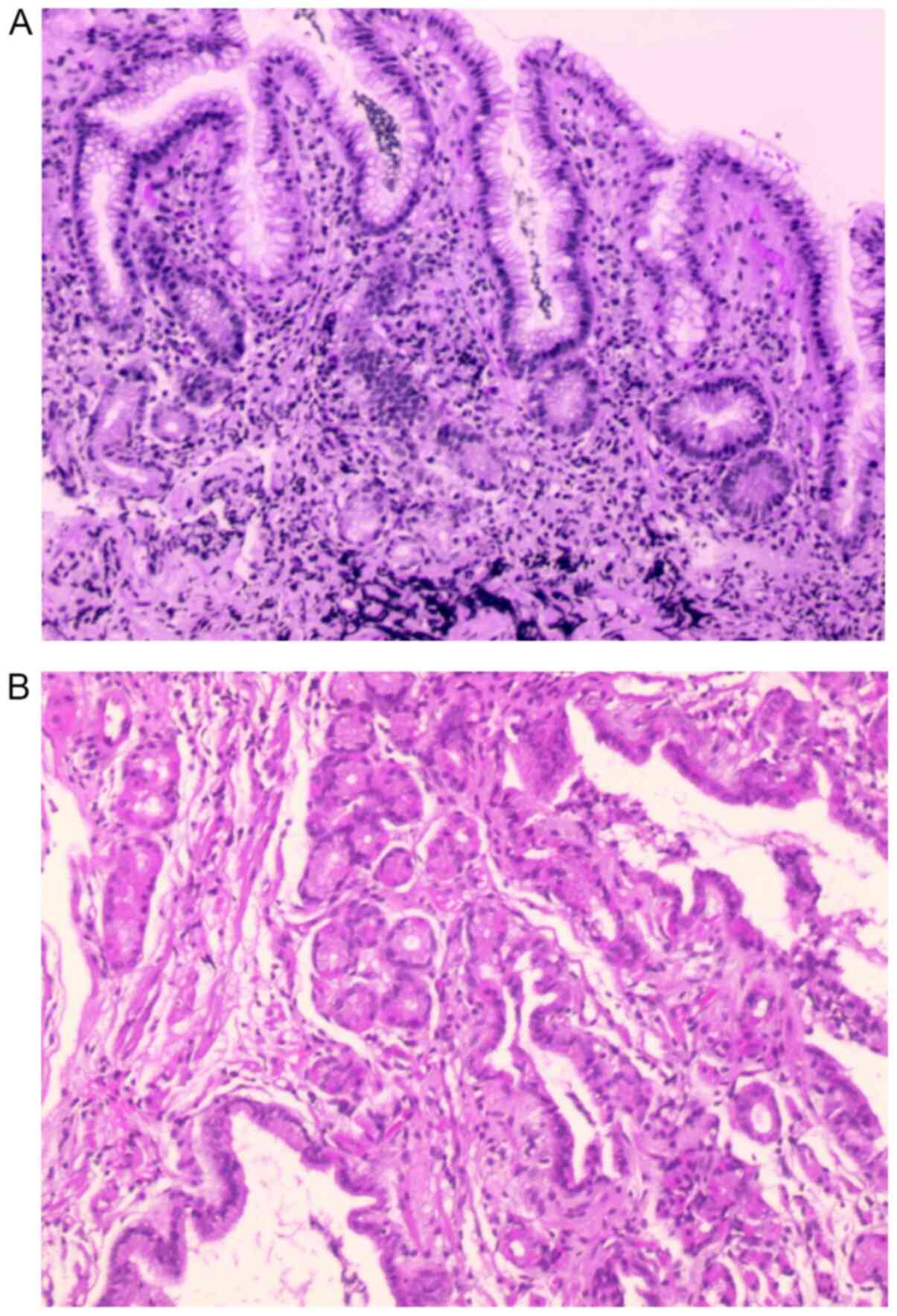
|
1
|
Marshall BJ and Warren JR: Unidentified
curved bacilli in the stomach of patients with gastritis and peptic
ulceration. Lancet. 1:1311–1315. 1984.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Schistosomes, liver flukes and
Helicobacter pylori. IARC Working Group on the evaluation of
carcinogenic risks to humans. Lyon, 7-14 June 1994. IARC Monogr
Eval Carcinog Risks Hum 61: 1-241, 1994.
|
|
3
|
Sugano K, Tack J, Kuipers EJ, Graham DY,
El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N and Malfertheiner
P: faculty members of Kyoto Global Consensus Conference. Kyoto
global consensus report on Helicobacter pylori gastritis.
Gut. 64:1353–1367. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cheng H, Hu F, Xie Y, Hu P, Wang J, Lv N,
Zhang J, Zhang G, Zhou Z, Wu K, et al: Prevalence of
Helicobacter pylori resistance to antibiotics and its
influence on the treatment outcome in china: A mulficenter clinical
study. Chin J Gastroenterol. 12:525–530. 2007.
|
|
5
|
Song Z, Zhang J, He L, Chen M, Hou X, Li Z
and Zhou L: Prospective multi-region study on primary antibiotic
resistance of Helicobacter pylori strains isolated from
Chinese patients. Dig Liver Dis. 46:1077–1081. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sun QJ, Liang X, Zheng Q, Gu WQ, Liu WZ,
Xiao SD and Lu H: Resistance of Helicobacter pylori to
antibiotics from 2000 to 2009 in Shanghai. World J Gastroenterol.
16:5118–5121. 2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu DS, Wang YH, Zeng ZR, Zhang ZY, Lu H,
Xu JM, Du YQ, Li Y, Wang JB, Xu SP, et al: Primary antibiotic
resistance of Helicobacter pylori in Chinese patients: A
multiregion prospective 7-year study. Clin Microbiol Infect.
24:780.e5–780.e8. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hu Y, Zhang M, Lu B and Dai J:
Helicobacter pylori and antibiotic resistance, a continuing
and intractable problem. Helicobacter. 21:349–363. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shao Y, Lu R, Yang Y, Xu Q, Wang B and Ye
G: Antibiotic resistance of Helicobacter pylori to 16
antibiotics in clinical patients. J Clin Lab Anal.
32(e22339)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Khoury J, Geffen Y, Shaul R, Sholy H,
Chowers Y and Saadi T: Secondary antibiotic resistance of
Helicobacter pylori isolates in Israeli children and adults.
J Glob Antimicrob Resist. 10:182–185. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Saniee P, Hosseini F, Kadkhodaei S,
Siavoshi F and Khalili-Samani S: Helicobacter pylori
multidrug resistance due to misuse of antibiotics in Iran. Arch
Iran Med. 21:283–288. 2018.PubMed/NCBI
|
|
12
|
Savoldi A, Carrara E, Graham DY, Conti M
and Tacconelli E: Prevalence of antibiotic resistance in
Helicobacter pylori: A systematic review and meta-analysis
in world health organization regions. Gastroenterology.
155:1372–1382.e17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pohl D, Keller PM, Bordier V and Wagner K:
Review of current diagnostic methods and advances in
Helicobacter pylori diagnostics in the era of next
generation sequencing. World J Gastroenterol. 25:4629–4660.
2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Thi Huyen Trang T, Thanh Binh T and
Yamaoka Y: Relationship between vacA types and development of
gastroduodenal diseases. Toxins (Basel). 8(182)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Karabiber H, Selimoglu MA, Otlu B,
Yildirim O and Ozer A: Virulence factors and antibiotic resistance
in children with Helicobacter pylori gastritis. J Pediatr
Gastroenterol Nutr. 58:608–612. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liou JM, Chang CY, Chen MJ L, et al: The
Primary Resistance of Helicobacter pyloriin Taiwan after the
National Policy to Restrict Antibiotic Consumption and Its Relation
to Virulence Factors-A Nationwide Study. PLoS One. 10(5,
e0124199)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yin G, Bie S, Gu H, Shu X, Zheng W, Peng
K, Zhao H, Li F, Chen B, Botchway BOA, et al: Application of gene
chip technology in the diagnostic and drug resistance detection of
Helicobacter pylori in children. J Gastroenterol Hepatol.
35:1331–1339. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Brisson-Noël A, Trieu-Cuot P and Courvalin
P: Mechanism of action of spiramycin and other macrolides. J
Antimicrob Chemother. 22 (Suppl B):S13–S23. 1988.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Seo SI, Do BJ, Kang JG, Kim HS, Jang MK,
Kim HY and Shin WG: Helicobacter pylori eradication
according to sequencing-based 23S ribosomal RNA point mutation
associated with clarithromycin resistance. J Clin Med.
9(54)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ong S, Kim SE, Kim JH, Yi NH, Kim TY, Jung
K, Park MI and Jung HY: Helicobacter pylori eradication
rates with concomitant and tailored therapy based on 23S rRNA point
mutation: A multicenter randomized controlled trial. Helicobacter.
24(e12654)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kim YJ, Chung WC and Kim DB: Efficacy of
bismuth added to standard triple therapy as the first-line
eradication regimen for Helicobacter pylori infection.
Helicobacter. 26(e12792)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hooper DC: Emerging mechanisms of
fluoroquinolone resistance. Emerg Infect Dis. 7:337–341.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liou JM, Chen CC, Chen MJ, Chang CY, Fang
YJ, Lee JY, Sheng WH, Wang HP, Wu MS and Lin JT: Empirical modified
sequential therapy containing levofloxacin and high-dose
esomeprazole in second-line therapy for Helicobacter pylori
infection: A multicentre clinical trial. J Antimicrob Chemother.
66:1847–1852. 2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Papastergiou V, Mathou N, Licousi S,
Evgenidi A, Paraskeva KD, Giannakopoulos A, Stavrou PZ, Platsouka E
and Karagiannis JA: Seven-day genotypic resistance-guided triple
Helicobacter pylori eradication therapy can be highly
effective. Ann Gastroenterol. 31:198–204. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Palmitessa V, Monno R, Panarese A, Cuppone
R, Burattini O, Marangi S, Curlo M, Fumarola L, Petrosillo A,
Parisi A, et al: Evaluation of antibiotic resistance of
Helicobacter pylori strains isolated in Bari, Southern
Italy, in 2017-2018 by phenotypic and genotyping methods. Microb
Drug Resist. 26:909–917. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Miyachi H, Miki I, Aoyama N, Shirasaka D,
Matsumoto Y, Toyoda M, Mitani T, Morita Y, Tamura T, Kinoshita S,
et al: Primary levofloxacin resistance and gyrA/B mutations among
Helicobacter pylori in Japan. Helicobacter. 11:243–249.
2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Di Caro S, Fini L, Daoud Y, Grizzi F,
Gasbarrini A, De Lorenzo A, Di Renzo L, McCartney S and Bloom S:
Levofloxacin/amoxicillin-based schemes vs quadruple therapy for
Helicobacter pylori eradication in second-line. World J
Gastroenterol. 18:5669–5678. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Debnath N and Yadav A: Indigenous
probiotic lactobacillus strains to combat gastric pathogen
Helicobacter pylori: Microbial interference therapy. Indu
pal Kaur, Sandip V. Pawar and Praveen rishi. Chandigarh, India. In:
Probiotic Research in Therapeutics, pp215-230, 2021.
|
|
29
|
Zamani M, Rahbar A and Shokri-Shirvani J:
Resistance of Helicobacter pylori to furazolidone and
levofloxacin: A viewpoint. World J Gastroenterol. 23:6920–6922.
2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xie C and Lv N: Helicobacter pylori
infection in China. Dis Surveill. 33:272–275. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lee SM, Kim N, Kwon YH, Nam RH, Kim JM,
Park JY, Lee YS and Lee DH: rdxA, frxA, and efflux pump in
metronidazole-resistant Helicobacter pylori: Their relation
to clinical outcomes. J Gastroenterol Hepatol. 33:681–688.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Jeong JY, Mukhopadhyay AK, Dailidiene D,
Wang Y, Velapatiño B, Gilman RH, Parkinson AJ, Nair GB, Wong BC,
Lam SK, et al: Sequential inactivation of rdxA (HP0954) and frxA
(HP0642) nitroreductase genes causes moderate and high-level
metronidazole resistance in Helicobacter pylori. J
Bacteriol. 182:5082–5090. 2000.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jeong JY and Berg DE: Mouse-colonizing
Helicobacter pylori SS1 is unusually susceptible to
metronidazole due to two complementary reductase activities.
Antimicrob Agents Chemother. 44:3127–3132. 2000.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kwon DH, El-Zaatari FA, Kato M, Osato MS,
Reddy R, Yamaoka Y and Graham DY: Analysis of rdxA and involvement
of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA)
and ferredoxin-like protein (FdxB) in metronidazole Helicobacter
pylori. Antimicrob Agents Chemother. 44:2133–2142.
2000.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Rasheed F, Campbell BJ, Alfizah H, Varro
A, Zahra R, Yamaoka Y and Pritchard DM: Analysis of Clinical
Isolates of Helicobacter pylori in Pakistan reveals high
degrees of pathogenicity and high frequencies of antibiotic
resistance. Helicobacter. 19:387–399. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang Y, Wen Y, Xiao Q, Zheng W, Long G,
Chen B, Shu X and Jiang M: Mutations in the antibiotic target genes
related to clarithromycin, metronidazole and levofloxacin
resistance in Helicobacter pylori strains from children in
China. Infect Drug Resist. 13:311–322. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang D, Guo Q, Yuan Y and Gong Y: The
antibiotic resistance of Helicobacter pylori to five
antibiotics and influencing factors in an area of China with a high
risk of gastric cancer. BMC Microbiol. 19(152)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bachir M, Allem R, Tifrit A, Medjekane M,
Drici AE, Diaf M and Douidi KT: Primary antibiotic resistance and
its relationship with cagA and vacA genes in Helicobacter
pylori isolates from Algerian patients. Braz J Microbiol.
49:544–551. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xu J and Xie GY: Relationship between cagA
and VacA genotypes of Helicobacter pylori and drug
resistance. Med J West China. 5:818–819, 821. 2010.(In
Chinese).
|
|
40
|
Schubert JP, Gehlert J, Rayner CK,
Roberts-Thomson IC, Costello S, Mangoni AA and Bryant RV:
Antibiotic resistance of Helicobacter pylori in Australia
and New Zealand: A systematic review and meta-analysis. J
Gastroenterol Hepatol. 36:1450–1456. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Giorgio F, Ierardi E, Sorrentino C,
Principi M, Barone M, Losurdo G, Iannone A, Giangaspero A, Monno R
and Di Leo A: Helicobacter pylori DNA isolation in the
stool: An essential pre-requisite for bacterial noninvasive
molecular analysis. Scand J Gastroenterol. 15:1429–1432.
2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen S, Li Y and Yu C: Oligonucleotide
microarray: A new rapid method for screening the 23S rRNA gene of
Helicobacter pylori for single nucleotide polymorphisms
associated with clarithromycin resistance. J Gastroenterol Hepatol.
23:126–131. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Xuan SH, Zhou YG, Shao B, Cui YL, Li J,
Yin HB, Song XP, Cong H, Jing FX, Jin QH, et al: Enzymic
colorimetry-based DNA chip: A rapid and accurate assay for
detecting mutations for clarithromycin resistance in the 23S rRNA
gene of Helicobacter pylori. J Med Microbiol. 58:1443–1448.
2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kaprou GD, Bergšpica I, Alexa EA,
Alvarez-Ordóñez A and Prieto M: Rapid methods for antimicrobial
resistance diagnostics. Antibiotics (Basel). 10:209–239.
2021.PubMed/NCBI View Article : Google Scholar
|