Introduction
Type 2 diabetes mellitus (T2DM) is one of the most
common metabolic disorders worldwide and is primarily caused by
defective insulin secretion (1).
Over the past 30 years, the number of individuals with T2DM and
prediabetes has increased two-fold globally, indicating T2DM as a
rapidly growing public health challenge. However, T2DM is a
multifactorial disease that slowly progresses over several years
(2). Environmental factors
including obesity, aging, an unhealthy diet, a lack of physical
activity, smoking, as well as genetic factors and epigenetic
modifications all contribute to the accelerating diabetes epidemic
in China (3). However, genetic
variation accounts for only a small ratio of risk of T2DM
development and environmental factors play a pivotal role in
driving the progression of T2DM. In addition, several studies have
reported increased hypertension rates among T2DM subjects (4-6).
It is estimated that the incidence of hypertension is increased
~two-fold in patients with T2DM compared with those without T2DM
(7). A previous study indicated
that moderate consumption of alcohol has been associated with a
reduced risk of T2DM (8). However,
moderate drinking needs to be monitored cautiously under a
culturally appropriate context, particularly considering the stable
increase in alcohol consumption in several Asian countries
(9) and the excess increase in
alcohol consumption in European countries (10).
Recent studies reported the occurrence of gut
microbiota (GM) dysbiosis in obese patients with T2DM and indicated
that the gut microflora may be a major environmental factor
involved in the onset and progression of diabetes. Additionally,
intestinal microbiome changes were also associated with the onset
of type 1 DM and gestational DM (11,12).
Therefore, it is necessary to develop a reliable early method for
detecting T2DM that could lead to earlier interventions and
treatments for T2DM.
The human gut microbiome has been demonstrated to
possess 500-1,000 bacterial species, which are estimated to
encompass ~2,000,000 genes. Surprisingly, the bacterial genes
possess 100 times more genes than the human genes (13). The GM is a very diversified
ecosystem and its function is dependent on several factors, such as
host genetics, species, sex, age, body mass index (BMI), diet,
smoking and drugs (14,15). GM may be key to the management of
T2DM development. The aim of the present study was to develop a
rapid machine learning-based method to predict the risk of
T2DM.
Materials and methods
Sample and clinical data
collection
A total of 118 newly diagnosed patients with T2DM
and 89 controls (non-T2DM) were randomly recruited between January
2019 and October 2020 from the affiliated Hospital of Chengde
Medical University (Chengde, China). The inclusion criteria for
T2DM were as follows: i) Patients with T2DM were enrolled in
accordance with the 1999 WHO diagnostic criteria as previously
described (16); and ii) were aged
>18 years. The exclusion criteria were as follows: i) Acute
infection, trauma or surgery within the past month; ii) use of
antibiotics, glucocorticoids or other immune regulators within the
past month; iii) severe coronary heart disease, stroke or malignant
disease; iv) pregnancy or lactation; v) autoimmune diseases, such
as hyperthyroidism; and vi) other types of diabetes (17). The exclusion criteria for the
controls were the same as those aforementioned.
All procedures were performed and approved (approval
no. CYFYLL2021171) in accordance with the ethical standards of the
Clinical Research Ethics Committee of the affiliated Hospital of
Chengde Medical University (Chengde, China), and written informed
consent was obtained from all participants included in the
study.
Fecal sample collection and DNA
extraction
A total of 207 fresh fecal samples were collected in
sterile collection tubes (Thermo Fisher Scientific, Inc.). All
samples were stored at -20˚C for temporary preservation and then
transferred to -80˚C for longer term storage. Specifically, 200 mg
fecal sample was added to 1 ml PBS in a 1.5 ml tube, vortexed at
maximum speed for 3 min and centrifuged at 167.7 x g for 5 min at
4˚C and then the supernatant was collected and transferred to a
2-ml tube. A total of ~800 µl supernatant was centrifuged at 1,677
x g for 5 min at 4˚C and then the supernatant was removed.
Subsequently, the microbial DNA was extracted from the fecal
samples using a nucleic acid extraction kit (cat. no. T221S)
according to the manufacturer's protocol (Xi'an Tianlong Science
& Technology Co., Ltd,). Finally, ~60 µl DNA was obtained for
downstream experiments.
Primers and PCR amplification
A total of 10 microbial oligonucleotide primers were
synthesized and purified by General Biol (generalbiol.com/) (Table
SI). Quantitative PCR (qPCR) was performed using an ABI-7500
real-time PCR system (Thermo Fisher Scientific, Inc.). The
thermocycling conditions were: Pre-denaturation at 95˚C for 10 min;
followed by 45 cycles of denaturation at 95˚C for 15 sec and
annealing at 60˚C for 45 sec. Following amplification, melting
temperature analysis of PCR products was performed to determine the
specificity of the PCR amplification. The melting curves were
obtained by heating from 60 to 95˚C at a rate of 0.3˚C/sec, with
continuous fluorescence measurement. Differences in threshold
cycles between the positive control (universal 16S rDNA) and each
bacteria were quantified using the 2-ΔCq method as
previously described (18), where
ΔCq was the differences in Cq values for each bacteria and
universal 16S rDNA and the relative abundance of each bacteria was
calculated.
Construction of the prediction
models
In the present study, three machine learning tools
were established to predict T2DM development, including an
artificial neural network of the multilayer perceptron (MLP) model,
an XGboost model and a support vector machine (SVM) model and
combined 6 clinical features and 10 bacterial species. The SVM
model has been reported to predict chronic kidney disease in
clinical applications (19), the
XGBoost model exhibits improved performance in predicting patients
with postoperative sepsis (20),
and the MLP model performed well when applied to computed
tomography for coronary artery disease and myocardial perfusion
(21). K-fold is a common cross
validation approach, particularly when the datasets are limited
(22). Therefore, k-fold (k=5) was
used to train, construct and compare the three predictive models.
Additionally, the parameters of the three predictive models were
tuned for the optimization of the equations in Python (Table SII).
A total of 207 participants (118 patients with T2DM
and 89 controls) were randomly allocated into a training set (80%)
and a test set (20%). In the training set, k=5 was used and various
parameter combinations were exhausted using grid search. For each
model, the confusion matrix, area under the receiver operating
characteristic (ROC) curve (AUC), accuracy, sensitivity (recall),
specificity, positive predictive value [ppv (precision)] and
negative predictive value (npv) and were used to evaluate and
compare the comprehensive performance of feature selection as
previously described (23).
Statistical analysis
The three models were used to predict the risk of
T2DM and evaluated using Python (version 3.6.12; Python Software
Foundation) and incorporated including 6 clinical features and 10
bacterial species. The diagnostic values of the three models were
assessed using ROC analysis. After preprocessing the data with
pandas and sklearn, XGboost was used to analyze the importance of
features and evaluated by Python as previously described (24). Categorical variables were presented
by numbers or proportions, and differences in distribution between
the two groups were analyzed using a χ2 test in SPSS
(version 19.0; IBM Corp.) Continuous variables are presented as the
median and range. Continuous variables between the two groups were
compared using a nonparametric Mann-Whitney U test (for two groups)
or nonparametric Kruskal-Wallis test followed by Dunn's post hoc
test (for more than two groups). Statistical calculations were
performed in GraphPad Prism (version 8.0; GraphPad Software, Inc.).
P≤0.05 was considered to indicate a statistically significant
difference.
Results
Clinical characteristics of the
participants
The clinical characteristics of the patients with
T2DM and controls are shown in Table
SIII. There were no significant differences in age (P=0.502),
sex (P=0.683), BMI (P=0.230), smoking (P=0.146), alcohol
consumption (P=0.220) and hypertension status (P=0.055) between
patients with T2DM and controls.
Comparison of the 10 bacteria between
patients with T2DM and controls
A total of 10 bacteria, including
Veillonellaceae, Clostridium leptum, Roseburia
inulinivorans, Bacteroides, Prevotella,
Bifidobacterium, Lactobacillus, Faecalibacterium
prausnitzii, Enterococcus and Eubacterium rectale
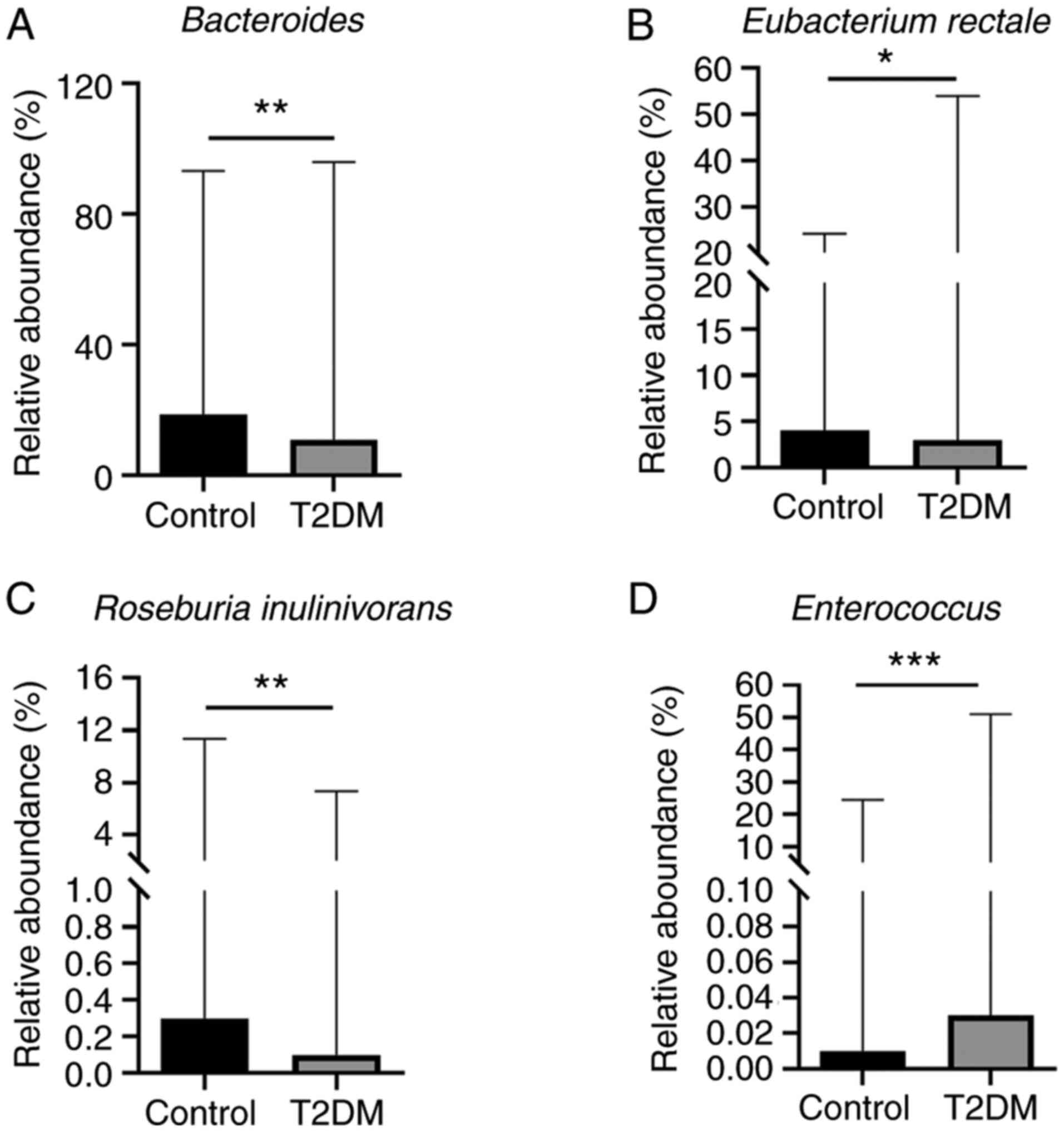
were detected by qPCR. The abundance of Bacteroides
(P=0.0055), Eubacterium rectale (P=0.0432) and Roseburia
inulinivorans (P=0.0019) was significantly lower in the T2DM
group than in the control group (Fig.
1A-C). In addition, the abundance of Enterococcus
(P=0.0002) was significantly higher in the T2DM group than in the
control group (Fig. 1D). However,
there were no significant differences in Prevotella
(P=0.164), Bifidobacterium (P=0.103), Veillonellaceae
(P=0.642), Faecalibacterium prausnitzii (P=0.157),
Lactobacillus (P=0.078) and Clostridium leptum
(P=0.493) between the two groups (Fig. S1).
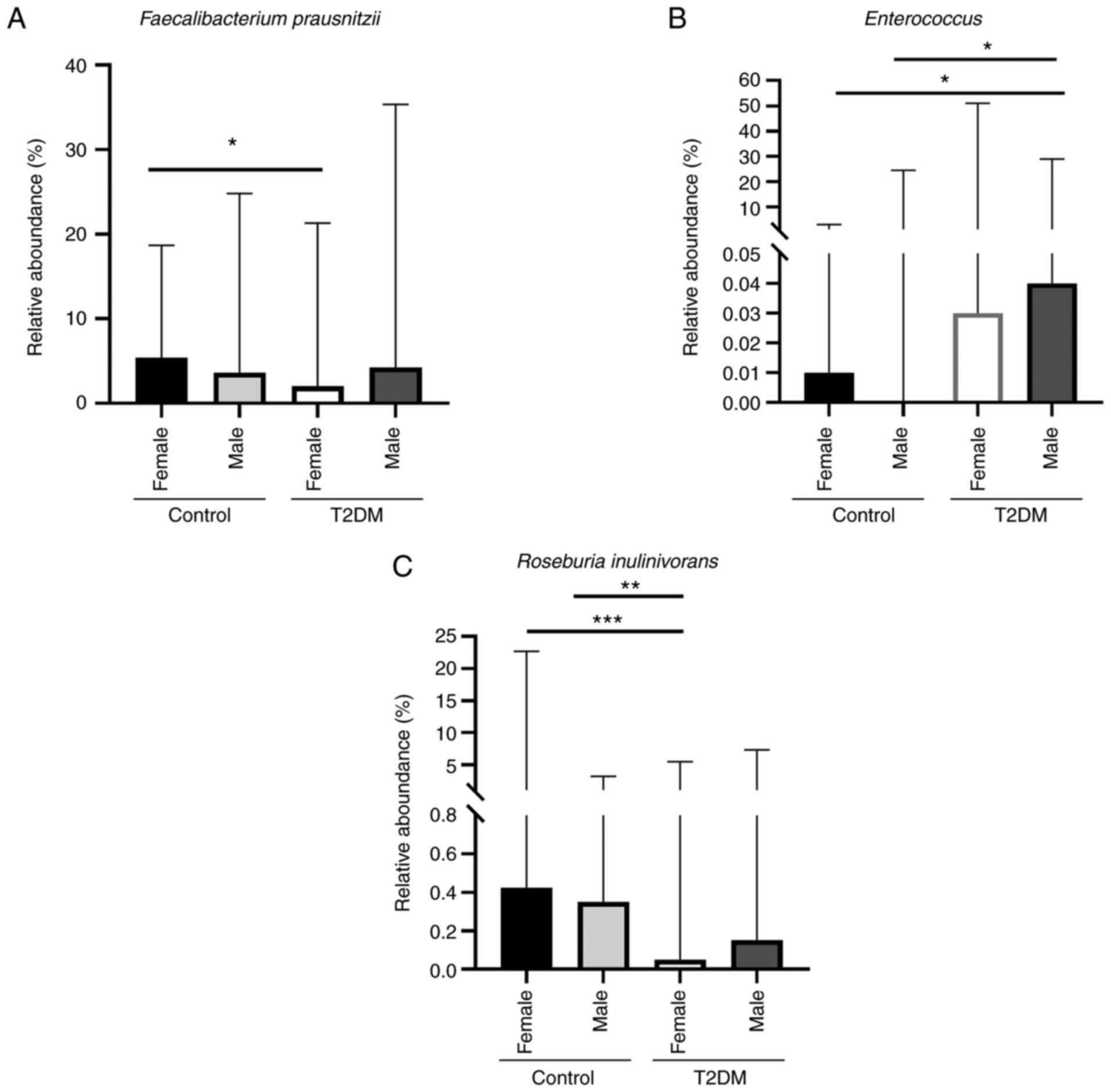
In addition, Faecalibacterium prausnitzii was
significantly higher in the control female subgroup than in the
T2DM female subgroup (P=0.032; Fig.
2A). Furthermore, the abundance of Enterococcus was
higher in the T2DM male subgroup than in both control female
(P=0.025) and male (P=0.0121) subgroups (Fig. 2B). Roseburia inulinivorans
was significantly higher in both control female and male subgroups
than in the T2DM female subgroup (P=0.0008 and P=0.0026,
respectively) (Fig. 2C). However,
there were no significant differences in the abundance of the
Bacteroides (P=0.0477), Prevotella (P=0.468),
Bifidobacterium (P=0.35), Lactobacillus (P=0.326),
Eubacterium rectale (P=0.118), Veillonellaceae
(P=0.124) and Clostridium leptum (P=0.178) between each
subgroup (Fig. S2).
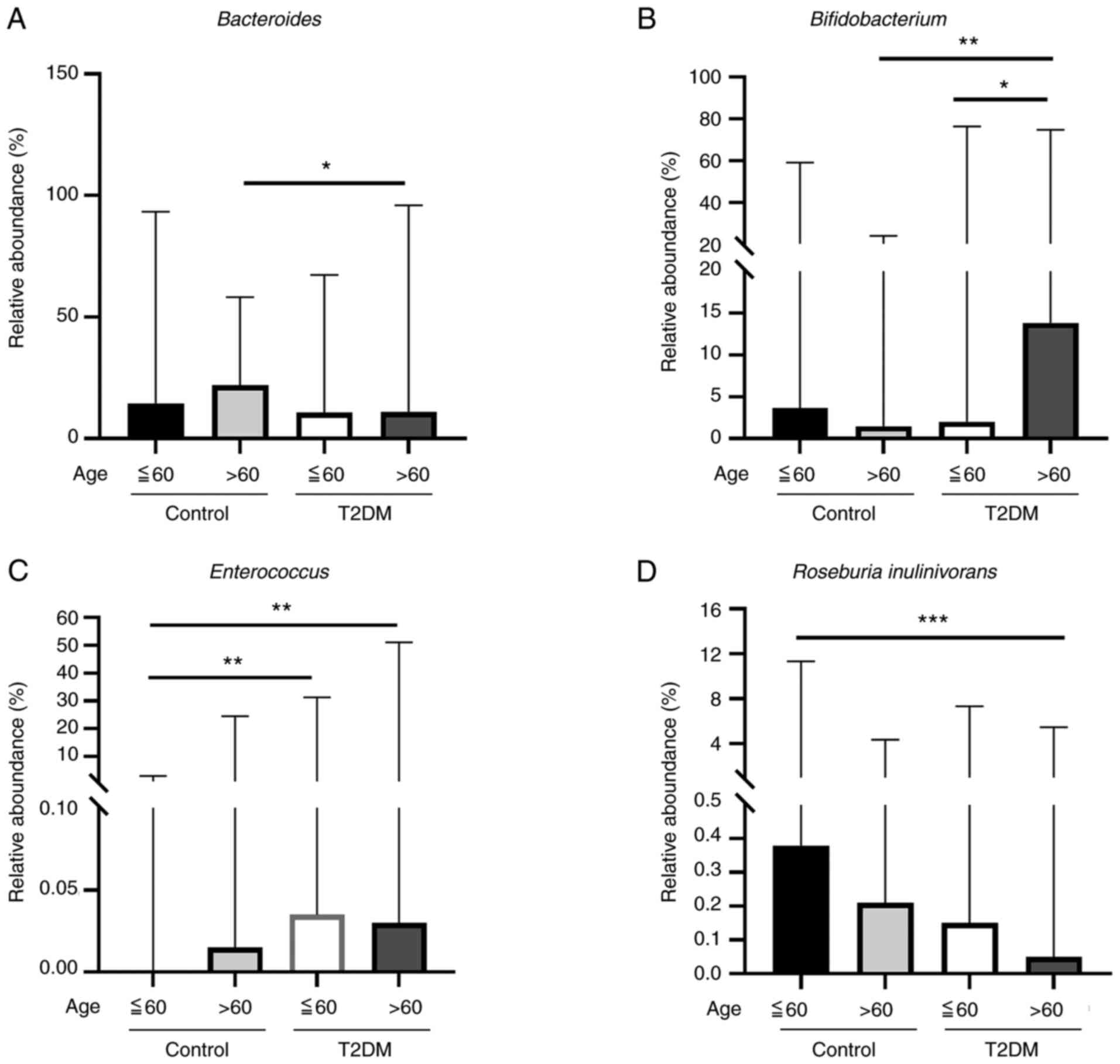
The abundance of Bacteroides in the control
older age (>60 years old) subgroup was higher than that in the
T2DM older age subgroup (P=0.0208; Fig. 3A). The abundance of
Bifidobacterium in the T2DM older age subgroup (>60 years
old) was higher than that in the T2DM younger age (≤60 years old)
subgroup (P=0.0343) and the control older age subgroup (P=0.0041;
Fig. 3B). The abundance of
Enterococcus was significantly higher in both the T2DM older
age and younger age subgroups (P=0.0012 and P=0.0012, respectively;
Fig. 3C), compared with control
participants less than 60 years old. Furthermore, Roseburia
inulinivorans was significantly higher in the control younger
age subgroup than in the T2DM older age subgroup (P=0.0007;
Fig. 3D). However, there were no
significant differences in the abundance of Prevotella
(P=0.0975), Lactobacillus (P=0.0697), Eubacterium
rectale (P=0.102), Faecalibacterium prausnitzii
(P=0.455), Veillonellaceae (P=0.507) and Clostridium
leptum (P=0.904) between each subgroup (Fig. S3).
Comparison of the 3 machine learning
models
SVM, XGboost and MLP models were used to predict the
risk of T2DM by incorporating 6 clinical features and 10 bacterial
species. A total of 207 samples were randomly divided into a
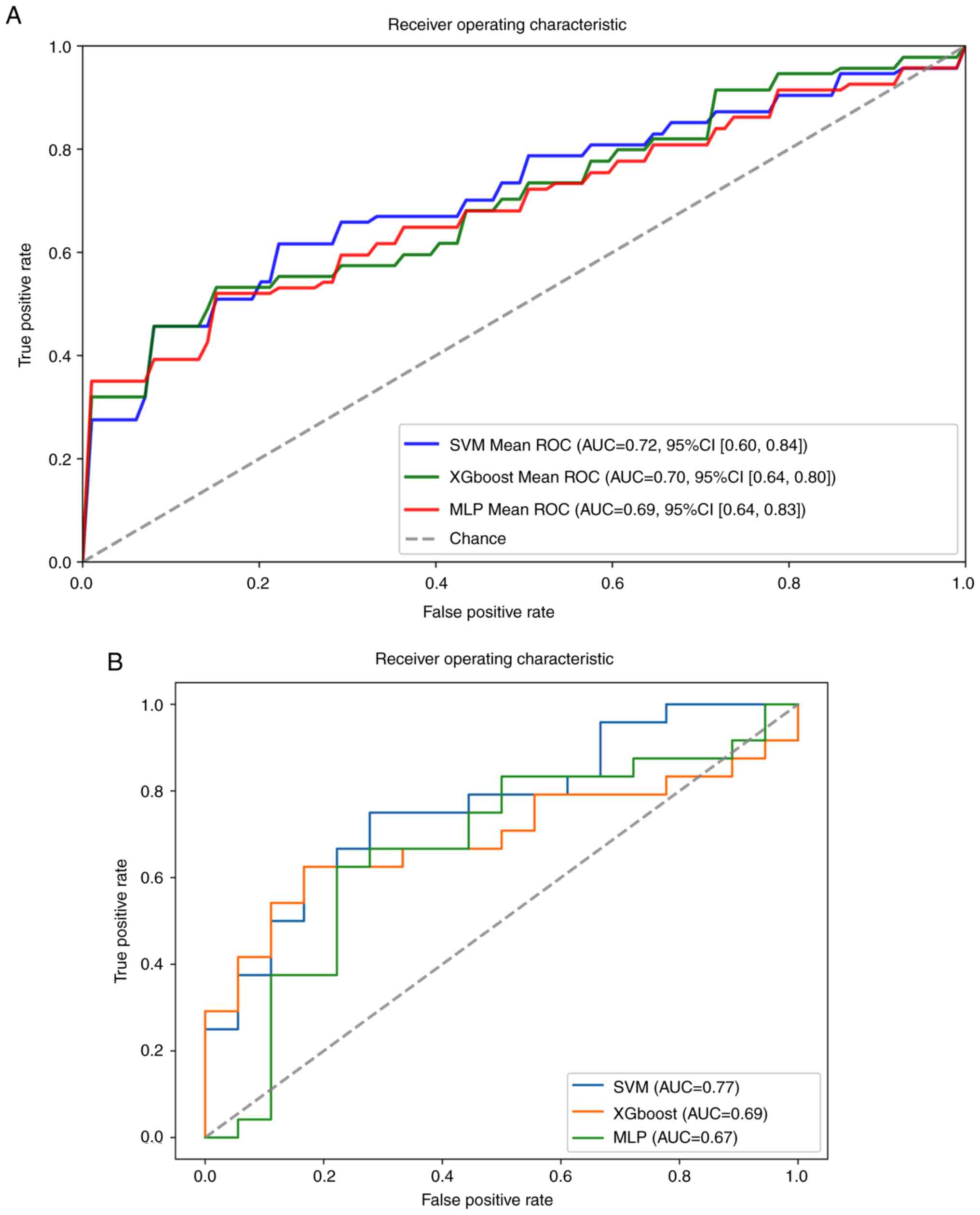
training set (80%) and test set (20%). The ROC curve is widely used
to validate the performance of prediction models, and the average
AUC and 95% CI are shown in Fig.
4A. In the training set, the results indicated that the AUC
values of SVM, XGboost and MLP models were 0.72, 0.70 and 0.69,
respectively. Furthermore, the accuracy, ppv (precision) and
sensitivity (recall) were >0.61 in all models (Table SIV). However, specificity and npv
were poor in the three models. In the test set, the results showed
that the SVM obtained the highest AUC value (0.77); the XGboost and
MLP model AUC values were 0.69 and 0.67, respectively (Fig. 4B). The accuracy was >0.67 in the
three models and the specificity and precision were >0.72
(Table SV). However, recall and
npv did not perform well in all the models. Furthermore, the
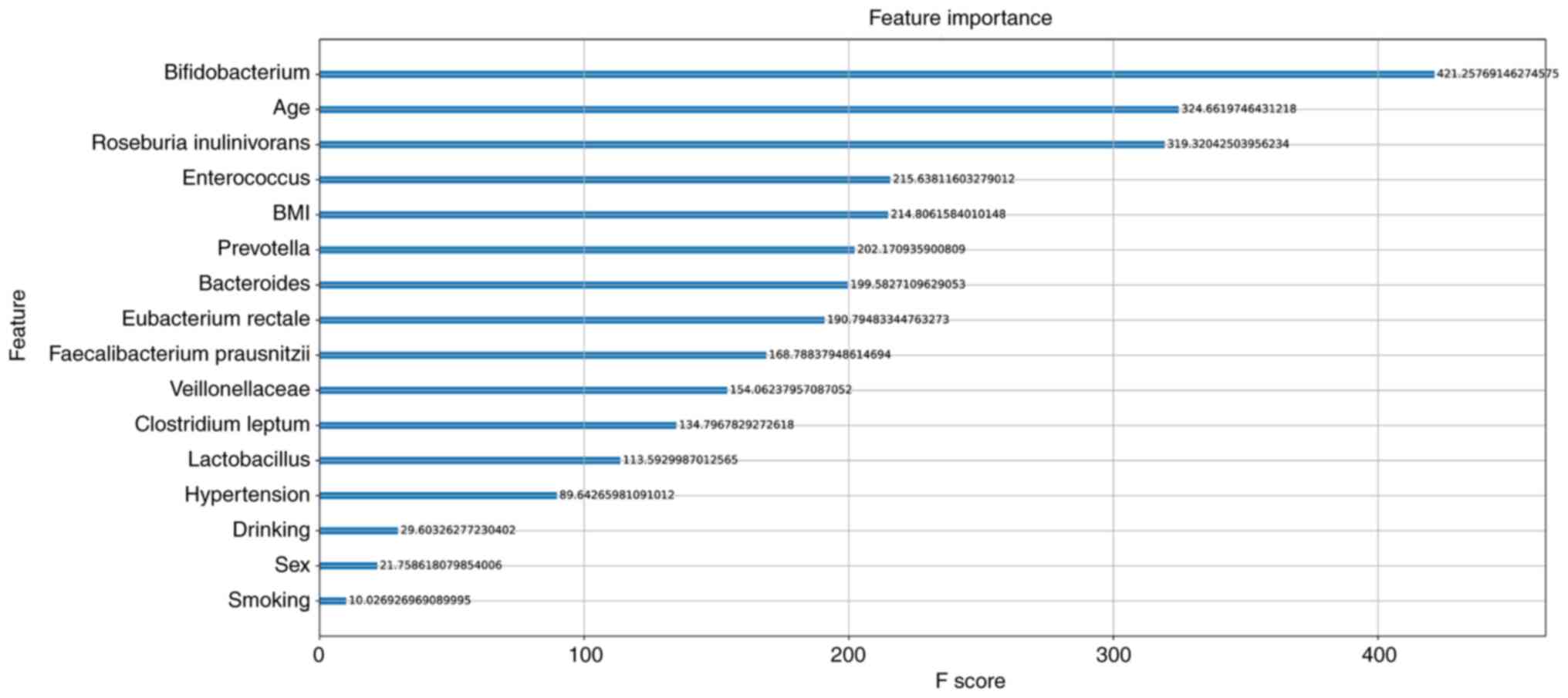
XGboost model was used to analyze the importance of the 16
features, and then the feature score rankings were measured
(Fig. 5). The results showed that
Bifidobacterium, age and Roseburia inulinivorans were
the top three features in the model.
Discussion
The prevalence of T2DM has become a major public
concern, with its continuingly increasing incidence worldwide. Gut
dysbiosis in patients with T2DM is caused by not only environmental
factors but also the host genetics. Studies have suggested that the
composition of the intestinal microbiota can trigger T2DM (25-27).
Therefore, further research is required to elucidate the connection
between GM and T2DM.
T2DM is a systemic disease which is characterized by
hyperglycemia, hyperlipidemia and organismic insulin resistance
(28). In the present study, six
clinical data including age, sex, BMI, smoking, alcohol consumption
and hypertension status which were associated with T2DM development
were collected. Additionally, numerous studies have shown that
non-alcoholic fatty liver disease (NAFLD) is commonly observed in
patients with T2DM (29,30). However, whether NAFLD is a cause or
consequence of the diabetic pathology remains a source of debate
(28). Thus, this relationship is
bidirectional, since T2DM substantially predicts the development of
these metabolic disorders (3).
Therefore, the NAFLD status, hyperlipidemia and disorders of
glucose and lipid metabolism status were not collected to predict
the risk of T2DM in the present study.
According to previous
studies, 10 bacteria including Veillonellaceae,
Clostridium leptum, Roseburia inulinivorans,
Bacteroides, Prevotella, Bifidobacterium,
Lactobacillus, Faecalibacterium prausnitzii,
Enterococcus and Eubacterium rectale were associated
with T2DM (31,32). The abundance of Bacteroides
was significantly lower in the T2DM group, which is consistent with
a previous study in animals, which revealed that after
administration of metformin, the relative abundance of
Bacteroides was increased in mice and rats treated with
metformin (33). Roseburia
inulinivorans was more abundant in the control group in the
present study, which is similar to a study which showed that the
abundance of Roseburia inulinivorans was increased in
patients after diabetic remission, achieved by both laparoscopic
Roux-en-Y gastric bypass or sleeve gastrectomy surgery (34). Interestingly, Roseburia
inulinivorans was significantly higher in both control female
and male subgroups than in the T2DM female subgroup. Moreover,
Roseburia inulinivorans was significantly higher in the
control younger age subgroup than in the T2DM older age subgroup.
In the present study, the abundance of Eubacterium rectale
was significantly higher in the control group than in the T2DM
group, which is consistent with a metagenome-wide association study
which revealed that the relative abundance of Eubacterium
rectale was higher in the control group than in the patients
with T2DM (31). A previous study
found that Enterococcus was positively correlated with
obesity (35). Enterococcus
was significantly higher in the patients with T2DM in the present
study, which is in accordance with a study that showed that
Enterococcus was more enriched in the DM group than in the
control group (36).
Interestingly, the abundance of Enterococcus was higher in
both T2DM younger and older age subgroups than in the control
younger age subgroup. Additionally, Enterococcus was higher
in the T2DM male subgroup than in both control female and male
subgroups.
Additionally, several studies have shown that
Bifidobacterium was negatively associated with T2DM
(37,38). Conversely, Sasaki et al
(39) reported that
Bifidobacterium was significantly increased in the patients
with T2DM when compared with the healthy controls. However, in the
present study, the abundance of Bifidobacterium did not
significantly differ between the control group and the T2DM group.
Interestingly, Bifidobacterium exhibited the higher
abundance in the T2DM older age subgroup than in the T2DM younger
age subgroup and the control older age subgroup.
Faecalibacterium prausnitzii was found to be negatively
associated with T2DM (32,40). Interestingly, Faecalibacterium
prausnitzii was significantly higher in the control female
subgroup than in the T2DM female subgroup; although there was no
significant difference in Faecalibacterium prausnitzii
abundance between the control group and T2DM group. Penckofer et
al (41) reported that
Lactobacillus was more abundant in women with T2DM than in
the controls. Conversely, previous studies have demonstrated the
beneficial effects of Lactobacillus for human health,
including improving T2DM, exhibiting anti-inflammatory effects and
reducing body weight (42-44).
Human gut Lactobacillus can reduce blood glucose responses
in vivo (45). The
aforementioned studies indicated that Lactobacillus shows
the most discrepant results among studies. Furthermore,
Prevotella was significantly correlated with lipid
metabolites, such as lysophosphatidylglycerol and
phosphatidylinositol-3, resembling obese and diabetic phenotypes
(46). The abundance of
Clostridium leptum in the probiotic group was significantly
higher than in the control group who did not take probiotics than
in Japanese patients with T2DM (47). In a previous study, it was
demonstrated that Veillonellaceae was significantly higher
in the acarbose group than in the placebo group (48). However, in the present study, the
abundance of Lactobacillus, Prevotella,
Clostridium leptum and Veillonellaceae did not
significantly differ between the control and T2DM groups.
In order to improve earlier warnings in patients
with T2DM, the SVM, XGboost and MLP models were used, incorporating
6 clinical features and 10 bacterial species to predict the risk of
T2DM. A total of 207 samples were randomly divided into a training
set (80%) and test set (20%). Among the three models, SVM and
XGboost models obtained AUC values of 0.72 and 0.70, respectively,
in the training set, and the accuracy, precision and recall were
>0.61. While in the test set, only the SVM model obtained an AUC
value of 0.77, the precision and specificity were >0.77, and the
accuracy, recall, and npv were >0.60. Previous studies reported
that if the model AUC is >0.70, the model has high accuracy
(49,50). Although the SVM model had the
highest overall predictive power, the sample size in the training
and test set were small. Thus, large samples are required to verify
this result.
In addition, the XGboost model was used to analyze
the importance of the 16 features, including 6 clinical features
and 10 bacterial species. The results revealed that
Bifidobacterium, age and Roseburia inulinivorans
played major roles in the model, while alcohol consumption, smoking
status and sex were less important. Bifidobacterium
represents beneficial genera, most frequently reported in studies
of T2DM, and appears to be the most consistent genus supported by
the literature, exhibiting potentially protective effects against
T2DM (51). Roseburia
inulinivorans is also the most consistently reported to exhibit
a negative association with T2DM (51). Therefore, the results indicated
that the gut microbiome can be a potential marker for predicting
the risk of T2DM. Yang et al (52) reported that the meta-analysis of
the prevalence T2DM rate at the age of 55-74 years was six- to
seven-fold higher than that of individuals aged 20-34 years, in
China. Thus, age may be a major factor in the risk of T2DM. In the
present study, age was ranked as the second most important factor
in the model. Additionally, Bifidobacterium, Roseburia
inulinivorans and Enterococcus were associated with an
older age. Bifidobacterium, Roseburia inulinivorans
and Enterococcus were ranked as the top 5 important features
in the model. In addition, a previous study showed that a high BMI
was the single strongest risk factor for T2DM (53), and was associated with several
metabolic abnormalities that result in insulin resistance (54). According to a series of nationwide
surveys reported in China (55),
the prevalence of being overweight (23 kg/m2 ≤ BMI
<27.5 kg/m2) in Chinese adults aged 20-59 years old
increased from 37.4% in 2000 to 39.2% in 2005, 40.7% in 2010, and
41.2% in 2014. The prevalence of obesity (BMI ≥27.5
kg/m2) increased from 8.6% in 2000 to 10.3% in 2005,
12.2% in 2010 and 12.9% in 2014. Notably, T2DM develops at a
considerably lower BMI in the Chinese population than in European
populations. The relatively high risk of diabetes at a lower BMI
could be partially attributed to the tendency towards visceral
adiposity in East Asian populations, including the Chinese
population (56). Therefore, BMI
may not play an important role in developing T2DM in the Chinese
population. In the present study, BMI ranked as the fifth most
important feature in the model. In addition, smoking has shown to
induce insulin resistance and compensatory insulin-secretion
responses (57), which may explain
the increased risk of T2DM in individuals who smoke. On the one
hand, moderate consumption of alcohol has been associated with a
reduced risk of T2DM (8). On the
other hand, it may be due to the public education campaigns to
reduce the prevalence of smoking in China in recent years. A
meta-analysis indicated that the prevalence of T2DM was 9.9% for
men and 11.6% for women in China (2000-2014) (52). It appears that the effect of sex on
the prevalence of T2DM amongst the Chinese is equal. Thus, alcohol
consumption, smoking and sex are less important in the model
ranking. Meanwhile, the abundance of Faecalibacterium
prausnitzii, Veillonellaceae, Clostridium leptum
and Lactobacillus did not differ between the control and
T2DM groups, which may explain why they were ranked lower.
There are several limitations in the present study.
First, the sample size used was relatively small and the total
cohort of patients with T2DM and cohort of controls was unbalanced.
Second, only 6 clinical features and 10 bacterial species were used
to establish the models between the two groups. The 16S rRNA gene
is a promising method for detecting GM, but in the present study,
the abundance of the 10 bacterial species between the two groups
was assessed by qPCR instead. Third, although the SVM model
obtained an AUC value of 0.77 in the test set, larger cohorts are
required to validate in the model before the model can be assessed
in the clinic for detection of early stage T2DM.
In conclusion, three machine learning models were
constructed and compared to predict the risk of T2DM, revealing
that the SVM model exhibited the highest overall predictive power.
In addition, Bifidobacterium, age and Roseburia
inulinivorans had important impacts in predicting early stage
T2DM. Therefore, SVM machine learning may have potential to aid in
the early prediction and treatment of patients with T2DM in the
near feature.
Supplementary Material
Comparison of the abundance of the 6
bacterial species between the patients with T2DM and controls. No
significant difference was observed in the abundance of (A)
Prevotella, (B) Bifidobacterium, (C)
Veillonellaceae, (D) Faecalibacter iumprausnitzii,
(E) Lactobacillus and (F) Clostridium leptum between
the two groups. Results represent the median and range. T2DM, type
2 diabetes mellitus.
Comparison of the abundance of the 7
bacterial species between the control female and male subgroups,
and the T2DM female and male subgroups. No significant difference
was observed in the abundance of (A) Bacteroides, (B)
Prevotella, (C) Bifidobacterium, (D)
Lactobacillus, (E) Eubacterium rectale, (F)
Veillonellaceae and (G) Clostridium leptum between
the four subgroups. Results represent the median and range. T2DM,
type 2 diabetes mellitus.
Comparison of the abundance of the 6
bacterial species between the control older age and younger age
subgroups, and the T2DM older age and younger age subgroups. No
significant difference was observed in the abundance of (A)
Prevotella, (B) Lactobacillus, (C) Eubacterium
rectale, (D) Faecalibacter iumprausnitzii, (E)
Veillonellaceae and (F) Clostridium leptum between
the four subgroups. Results represent the median and range. T2DM,
type 2 diabetes mellitus.
Sequences of the primers used in the
present study.
Selection of parameters in the
predictive models.
Clinical characteristics of patients
with T2DM and control subjects.
Summary of the efficacy of the SVM,
MLP and XGboost models in the training set.
Summary of the efficacy of the SVM,
MLP and XGboost models in the test set.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
XG, HT, JZ and YG designed the experiments. AZ, LL,
QS, JH and YW collected the samples and performed the experiments.
XG, RT, YX, JZ and YP analyzed the data. XG, HT, JZ and YG confirm
the authenticity of all the raw data. XG, HT, YX and YG wrote the
manuscript. JZ, YX, HT and YG revised the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and informed consent
The present study was approved (approval no.
CYFYLL2021171) by the Ethics Committee of the affiliated Hospital
of Chengde Medical University (Chengde, China). Written informed
consent was obtained from all participants involved in the present
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Roden M and Shulman GI: The integrative
biology of type 2 diabetes. Nature. 576:51–60. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Massey W and Brown JM: The gut microbial
endocrine organ in type 2 diabetes. Endocrinology.
162(bqaa235)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hu C and Jia W: Diabetes in China:
Epidemiology and genetic risk factors and their clinical utility in
personalized medication. Diabetes. 67:3–11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Pavlou DI, Paschou SA, Anagnostis P,
Spartalis M, Spartalis E, Vryonidou A, Tentolouris N and Siasos G:
Hypertension in patients with type 2 diabetes mellitus: Targets and
management. Maturitas. 112:71–77. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Colosia AD, Palencia R and Khan S:
Prevalence of hypertension and obesity in patients with type 2
diabetes mellitus in observational studies: A systematic literature
review. Diabetes Metab Syndr Obes. 6:327–338. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sabuncu T, Sonmez A, Eren MA, Sahin I,
Çorapçioğlu D, Üçler R, Akin Ş, Haymana C, Demirci İ, Atmaca A, et
al: Characteristics of patients with hypertension in a population
with type 2 diabetes mellitus. Results from the Turkish Nationwide
SurvEy of Glycemic and other metabolic parameters of patients with
diabetes mellitus (TEMD Hypertension Study). Prim Care Diabetes.
15:332–339. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
National high blood pressure education
program working group report on hypertension in diabetes.
Hypertension. 23:145–158; discussion 159-160. 1994.PubMed/NCBI
|
|
8
|
Baliunas DO, Taylor BJ, Irving H, Roerecke
M, Patra J, Mohapatra S and Rehm J: Alcohol as a risk factor for
type 2 diabetes: A systematic review and meta-analysis. Diabetes
Care. 32:2123–2132. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ezzati M and Riboli E: Behavioral and
dietary risk factors for noncommunicable diseases. N Engl J Med.
369:954–964. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Powles JW, Zatonski W, Vander Hoorn S and
Ezzati M: The contribution of leading diseases and risk factors to
excess losses of healthy life in Eastern Europe: Burden of disease
study. BMC Public Health. 5(116)2005.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhou H, Sun L, Zhang S, Zhao X, Gang X and
Wang G: Evaluating the causal role of gut microbiota in type 1
diabetes and its possible pathogenic mechanisms. Front Endocrinol
(Lausanne). 11(125)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hasain Z, Mokhtar NM, Kamaruddin NA,
Mohamed Ismail NA, Razalli NH, Gnanou JV and Raja Ali RA: Gut
microbiota and gestational diabetes mellitus: A review of host-gut
microbiota interactions and their therapeutic potential. Front Cell
Infect Microbiol. 10(188)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gilbert JA, Blaser MJ, Caporaso JG,
Jansson JK, Lynch SV and Knight R: Current understanding of the
human microbiome. Nat Med. 24:392–400. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Takagi T, Naito Y, Inoue R, Kashiwagi S,
Uchiyama K, Mizushima K, Tsuchiya S, Dohi O, Yoshida N, Kamada K,
et al: Differences in gut microbiota associated with age, sex, and
stool consistency in healthy Japanese subjects. J Gastroenterol.
54:53–63. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pasolli E, Asnicar F, Manara S, Zolfo M,
Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, et
al: Extensive unexplored human microbiome diversity revealed by
over 150,000 genomes from metagenomes spanning age, geography, and
lifestyle. Cell. 176:649–662.e620. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Alberti KG and Zimmet PZ: Definition,
diagnosis and classification of diabetes mellitus and its
complications. Part 1: Diagnosis and classification of diabetes
mellitus provisional report of a WHO consultation. Diabet Med.
15:539–553. 1998.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lin Q, Zhou W, Wang Y, Huang J, Hui X,
Zhou Z and Xiao Y: Abnormal peripheral neutrophil transcriptome in
newly diagnosed type 2 diabetes patients. J Diabetes Res.
2020(9519072)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xiao J, Ding R, Xu X, Guan H, Feng X, Sun
T, Zhu S and Ye Z: Comparison and development of machine learning
tools in the prediction of chronic kidney disease progression. J
Transl Med. 17(119)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yao RQ, Jin X, Wang GW, Yu Y, Wu GS, Zhu
YB, Li L, Li YX, Zhao PY, Zhu SY, et al: A machine learning-based
prediction of hospital mortality in patients with postoperative
sepsis. Front Med (Lausanne). 7(445)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Souza Filho JB, Sanchez M, Seixas JM,
Maidantchik C, Galliez R, Moreira AD, da Costa PA, Oliveira MM,
Harries AD and Kritski AL: Screening for active pulmonary
tuberculosis: Development and applicability of artificial neural
network models. Tuberculosis (Edinb). 111:94–101. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Vabalas A, Gowen E, Poliakoff E and Casson
AJ: Machine learning algorithm validation with a limited sample
size. PLoS One. 14(e0224365)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ma X, Wu Y, Zhang L, Yuan W, Yan L, Fan S,
Lian Y, Zhu X, Gao J, Zhao J, et al: Comparison and development of
machine learning tools for the prediction of chronic obstructive
pulmonary disease in the Chinese population. J Transl Med.
18(146)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu D, Yang Q, Su B, Hao J, Ma H, Yuan W,
Gao J, Ding F, Xu Y, Wang H, et al: Low-density lipoprotein
cholesterol 4: The notable risk factor of coronary artery disease
development. Front Cardiovasc Med. 8(619386)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sircana A, Framarin L, Leone N, Berrutti
M, Castellino F, Parente R, De Michieli F, Paschetta E and Musso G:
Altered gut microbiota in type 2 diabetes: Just a coincidence? Curr
Diab Rep. 18(98)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Salgaco MK, Oliveira LG, Costa GN, Bianchi
F and Sivieri K: Relationship between gut microbiota, probiotics,
and type 2 diabetes mellitus. Appl Microbiol Biotechnol.
103:9229–9238. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu Q, Wu S, Cheng Y, Zhang Z, Mao G, Li S,
Yang Y, Zhang X, Wu M and Tong H: Sargassum fusiforme fucoidan
modifies gut microbiota and intestinal metabolites during
alleviation of hyperglycemia in type 2 diabetic mice. Food Funct.
12:3572–3585. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pinti MV, Fink GK, Hathaway QA, Durr AJ,
Kunovac A and Hollander JM: Mitochondrial dysfunction in type 2
diabetes mellitus: An organ-based analysis. Am J Physiol Endocrinol
Metab. 316:E268–E285. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Masuoka HC and Chalasani N: Nonalcoholic
fatty liver disease: An emerging threat to obese and diabetic
individuals. Ann N Y Acad Sci. 1281:106–122. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Targher G and Byrne CD: Clinical review:
Nonalcoholic fatty liver disease: A novel cardiometabolic risk
factor for type 2 diabetes and its complications. J Clin Endocrinol
Metab. 98:483–495. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F,
Liang S, Zhang W, Guan Y, Shen D, et al: A metagenome-wide
association study of gut microbiota in type 2 diabetes. Nature.
490:55–60. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Karlsson FH, Tremaroli V, Nookaew I,
Bergström G, Behre CJ, Fagerberg B, Nielsen J and Bäckhed F: Gut
metagenome in European women with normal, impaired and diabetic
glucose control. Nature. 498:99–103. 2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang Q and Hu N: Effects of metformin on
the gut microbiota in obesity and type 2 diabetes mellitus.
Diabetes Metab Syndr Obes. 13:5003–5014. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Murphy R, Tsai P, Jüllig M, Liu A, Plank L
and Booth M: Differential changes in gut microbiota after gastric
bypass and sleeve gastrectomy bariatric surgery vary according to
diabetes remission. Obes Surg. 27:917–925. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Qiao Y, Sun J, Ding Y, Le G and Shi Y:
Alterations of the gut microbiota in high-fat diet mice is strongly
linked to oxidative stress. Appl Microbiol Biotechnol.
97:1689–1697. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhao X, Zhang Y, Guo R, Yu W, Zhang F, Wu
F and Shang J: The alteration in composition and function of gut
microbiome in patients with type 2 diabetes. J Diabetes Res.
2020(8842651)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Gao R, Zhu C, Li H, Yin M, Pan C, Huang L,
Kong C, Wang X, Zhang Y, Qu S and Qin H: Dysbiosis signatures of
gut microbiota along the sequence from healthy, young patients to
those with overweight and obesity. Obesity (Silver Spring).
26:351–361. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sedighi M, Razavi S, Navab-Moghadam F,
Khamseh ME, Alaei-Shahmiri F, Mehrtash A and Amirmozafari N:
Comparison of gut microbiota in adult patients with type 2 diabetes
and healthy individuals. Microb Pathog. 111:362–369.
2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Sasaki M, Ogasawara N, Funaki Y, Mizuno M,
Iida A, Goto C, Koikeda S, Kasugai K and Joh T: Transglucosidase
improves the gut microbiota profile of type 2 diabetes mellitus
patients: A randomized double-blind, placebo-controlled study. BMC
Gastroenterol. 13(81)2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang X, Shen D, Fang Z, Jie Z, Qiu X,
Zhang C, Chen Y and Ji L: Human gut microbiota changes reveal the
progression of glucose intolerance. PLoS One.
8(e71108)2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Penckofer S, Limeira R, Joyce C, Grzesiak
M, Thomas-White K and Wolfe AJ: Characteristics of the microbiota
in the urine of women with type 2 diabetes. J Diabetes
Complications. 34(107561)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yadav H, Jain S and Sinha PR: Antidiabetic
effect of probiotic dahi containing Lactobacillus acidophilus and
Lactobacillus casei in high fructose fed rats. Nutrition. 23:62–68.
2007.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Naito E, Yoshida Y, Makino K, Kounoshi Y,
Kunihiro S, Takahashi R, Matsuzaki T, Miyazaki K and Ishikawa F:
Beneficial effect of oral administration of Lactobacillus casei
strain Shirota on insulin resistance in diet-induced obesity mice.
J Appl Microbiol. 110:650–657. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kang JH, Yun SI and Park HO: Effects of
Lactobacillus gasseri BNR17 on body weight and adipose tissue mass
in diet-induced overweight rats. J Microbiol. 48:712–714.
2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Panwar H, Calderwood D, Grant IR, Grover S
and Green BD: Lactobacillus strains isolated from infant faeces
possess potent inhibitory activity against intestinal alpha- and
beta-glucosidases suggesting anti-diabetic potential. Eur J Nutr.
53:1465–1474. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu H, Pan LL, Lv S, Yang Q, Zhang H, Chen
W, Lv Z and Sun J: Alterations of gut microbiota and blood lipidome
in gestational diabetes mellitus with hyperlipidemia. Front
Physiol. 10(1015)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Sato J, Kanazawa A, Azuma K, Ikeda F, Goto
H, Komiya K, Kanno R, Tamura Y, Asahara T, Takahashi T, et al:
Probiotic reduces bacterial translocation in type 2 diabetes
mellitus: A randomised controlled study. Sci Rep.
7(12115)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang X, Fang Z, Zhang C, Xia H, Jie Z,
Han X, Chen Y and Ji L: Effects of acarbose on the gut microbiota
of prediabetic patients: A randomized, double-blind, controlled
crossover trial. Diabetes Ther. 8:293–307. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Luo X, Lin F, Zhu S, Yu M, Zhang Z, Meng L
and Peng J: Mine landslide susceptibility assessment using IVM, ANN
and SVM models considering the contribution of affecting factors.
PLoS One. 14(e0215134)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Hao S, Bai J, Liu H, Wang L, Liu T, Lin C,
Luo X, Gao J, Zhao J, Li H and Tang H: Comparison of machine
learning tools for the prediction of AMD based on genetic, age, and
diabetes-related variables in the Chinese population. Regen Ther.
15:180–186. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Gurung M, Li Z, You H, Rodrigues R, Jump
DB, Morgun A and Shulzhenko N: Role of gut microbiota in type 2
diabetes pathophysiology. EBioMedicine. 51(102590)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yang L, Shao J, Bian Y, Wu H, Shi L, Zeng
L, Li W and Dong J: Prevalence of type 2 diabetes mellitus among
inland residents in China (2000-2014): A meta-analysis. J Diabetes
Investig. 7:845–852. 2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Hu FB, Manson JE, Stampfer MJ, Colditz G,
Liu S, Solomon CG and Willett WC: Diet, lifestyle, and the risk of
type 2 diabetes mellitus in women. N Engl J Med. 345:790–797.
2001.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sinha R, Dufour S, Petersen KF, LeBon V,
Enoksson S, Ma YZ, Savoye M, Rothman DL, Shulman GI and Caprio S:
Assessment of skeletal muscle triglyceride content by (1)H nuclear
magnetic resonance spectroscopy in lean and obese adolescents:
Relationships to insulin sensitivity, total body fat, and central
adiposity. Diabetes. 51:1022–1027. 2002.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Tian Y, Jiang C, Wang M, Cai R, Zhang Y,
He Z, Wang H, Wu D, Wang F, Liu X, et al: BMI, leisure-time
physical activity, and physical fitness in adults in China: Results
from a series of national surveys, 2000-14. Lancet Diabetes
Endocrinol. 4:487–497. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Nazare JA, Smith JD, Borel AL, Haffner SM,
Balkau B, Ross R, Massien C, Alméras N and Després JP: Ethnic
influences on the relations between abdominal subcutaneous and
visceral adiposity, liver fat, and cardiometabolic risk profile:
The international study of prediction of intra-abdominal adiposity
and its relationship with cardiometabolic risk/intra-abdominal
adiposity. Am J Clin Nutr. 96:714–726. 2012.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Reaven G and Tsao PS: Insulin resistance
and compensatory hyperinsulinemia: The key player between cigarette
smoking and cardiovascular disease? J Am Coll Cardiol.
41:1044–1047. 2003.PubMed/NCBI View Article : Google Scholar
|