The diagnosis of prostate cancer (PCa) is usually
not a death sentence. Overall, the relative survival rate in five
years is almost 100 percent (1).
Even when the disease progresses, it usually has a relatively slow
clinical course. Prostate cancer is usually diagnosed in elderly
men who often have several comorbidities and reduced expected
lifespan. This suggests that if PCa patients were able to survive
15 or 20 years with the disease, prostate cancer-specific mortality
would greatly decrease.
Androgen deprivation therapy (ADT) is the mainstay
of therapy in early disseminated prostate cancer. Despite the
initial response to treatment, the disease eventually relapses into
an androgen independent state. Several attempts have been made to
therapeutically target the mechanisms of resistance to ADT
(2-4).
However, the responses to these novel treatments are generally
short lived (5). New candidate
treatment regimens or combinations that counter the effects of
resistance mechanisms are not always successful (6,7).
Similar to all biological systems, cancer cells have molecular
redundancies and it is rare that a viable combination of
therapeutic compounds can kill every single cancer cell while
sparing normal cells. In addition, tumors within individual
patients are highly heterogeneous, hence there is a marked
probability that at least a few tumor clones will survive a certain
treatment (8). Most research in
prostate cancer is following ‘down the rabbithole’ of discovering
and subsequently targeting the emerging resistance mechanisms in
order to eliminate every single cancer cell (3-7).
Not much research has been directed towards discovering novel ways
to slow the progression rate of the disease.
Early stage localized PCa can be successfully
treated with radical prostatectomy (RP) and/or radiation therapy
(9). The 5-year biochemical
disease-free survival in patients with Grade Group 1 through 5
disease after RP is 96, 88, 63, 48 and 26% respectively (10). Among patients with T0-2 clinical
stage disease, only 11% eventually die from prostate cancer
(11). However, the effects of RP
and radiotherapy are not so dramatic. These high survival rates
mostly result from the indolent clinical course of the tumors. In
early localized disease, excellent survival rates can be achieved
even without intervention. In a 2015 meta-analysis of active
surveillance studies, only 8 out of 7,627 patients eventually died
from prostate cancer, with a median follow up of 3.5 years (range
of 1.5-7.5 years) (12). In a
prospective randomized trial, radical prostatectomy did not
increase survival compared to watchful waiting in patients above 65
years old with early-stage prostate cancer (13). Longer follow up confirms the
relatively indolent course of most localized prostate cancers. The
15-year metastasis-free survival in patients with Gleason 6 or less
and PSA between 10-20 ng/ml is 94%. In Gleason 3+4 and PSA <20
ng/ml is 84%, while in Gleason 4+3 and PSA <20 ng/ml is 63%
(13).
Contrary to most other cancers, in prostate cancer,
even the diagnosis of locally advanced or metastatic disease is not
always a death sentence. A study conducted in 1997 found that
patients with locally advanced disease had a corrected 15-year
survival rate of 57% (11). Even
6% of patients discovered with initial metastatic disease did not
die from prostate cancer after 15 years (11). Several attempts have been made to
develop a genetic signature to inform us regarding which patients
will have a reduced survival. This has led to the development of
several experimental molecular assays, such as Decipher, and others
(14-16).
After RP, patients with low, intermediate or high risk Decipher
scores have 10-year cumulative metastasis rates of 5.5, 15 and
26.7% respectively (17). This
means that patients with the most aggressive genetic signatures
will potentially be 73.3% metastasis-free at 10 years based on this
method (17). Patients with
biochemical-only recurrence also have a mostly indolent disease
course. Post-RP biochemical relapse has a 37% likelihood of
metastatic disease in 5 years (18). Median time to clinical metastases
after PSA elevation is approximately 8 years (18). Even for patients who develop
biochemical relapse <1.2 years after RP, the ten-year
cancer-specific mortality rate is about 10 percent (19). Similarly, biochemical relapse after
prostate radiotherapy yields high survival rates (20). Post-RP salvage radiotherapy results
in 10-year prostate cancer specific survival rates of 86 percent
(21).
Even in the recurrent or metastatic setting, a
subset of patients can achieve remarkable and durable responses to
ADT and novel antiandrogen therapies (22). Among men with biochemical
recurrence who are placed on immediate ADT, the 5-year overall
survival rate is 91.2% (22).
Among patients who develop distant metastatic disease, the 5-year
prostate cancer-specific mortality is 57% (23). In the phase 3 AFFIRM study, there
was a group of long-term responders to enzalutamide after treatment
with docetaxel, who had a median survival of 7.9 years (24). In the STAMPEDE trial, almost half
of the patients with metastatic disease who received ADT plus
abiraterone acetate in the hormone-naive setting, were free from
disease progression after 4.5 years (25). While subsets of prostate cancers
can have a remarkably indolent course and show good response to
therapy, other subsets can be particularly aggressive and
refractory to treatments (26,27).
Hence, the fundamental question that arises is what is the
underlying cause of these differences? Is it an inherent property
of the tumors, dictated by their genetic and epigenetic signature?
Is it a matter of the tumor microenvironment, including the immune
microenvironment? In this review, we will attempt to summarize what
is already known regarding the underlying molecular mechanisms
related to the indolent course of subsets of prostate cancers. Our
hope is to provide evidence that the mechanisms that drive the
aggressive variants are reversible.
Several distinct genomic subsets of PCa exist.
Unsupervised clustering of molecular profiling (which include gene
mutations, fusions, copy number alterations, gene expression levels
and DNA methylation) indicate that 74% of all tumors can be
assigned in one out of seven classes based on oncogenic drivers:
fusions that involve 1) ERG, 2) ETV1, 3) ETV4, 4) FLI1, or
mutations in 5) SPOP, 6) FOXA1, 7) IDH1(28). The relative distribution of these
subgroups is similar in tumors derived from both primary and
metastatic sites. This molecular taxonomy cannot accurately predict
whether the tumors will be aggressive or indolent. The tumor
mutational burden in prostate cancer is relatively low (28). Overall, increased number of copy
number alterations (CNA) is associated with worse prognosis
(29,30). One of the most frequent events in
the prostate cancer genome with prognostic significance is a loss
in the short arm of chromosome 8 (31,32).
Loci frequently lost (>40%) include 8p21.2 and 8p23.2(31). The latter is associated with
advanced disease and is most commonly found in progressors vs. non
progressors (50 vs. 31%) (31).
The most frequent gains (>50%) include 11p15.4, 2p25.1, 13q34,
and 11q13.1. The latter is associated with biochemical recurrence
independent of tumor stage and grade. Genes that overlap with this
region include MEN1, MAP4K2, SF1, PPP2R5B and others (31). Fusions of androgen-regulated
promoters with members of the ETS family of transcription factors
are also very common. About 53% of prostate cancers have ETS-family
fusions (33-36).
TMPRSS2-ERG fusion analysis for CNAs reveals three important
regions of copy-number loss: Two regions spanning PTEN and TP53 and
the third spanning the region at 3p14, which likely contains FOXP1,
RYBP and SHQ1 genes (30).
Whole exome sequencing reveals that there is only a
small number of recurrent genes with alterations among various
subtypes (28). In primary tumors,
the most frequently altered gene is PTEN (17%), followed by TP53
(8%) (28). This suggests that no
specific gene alteration by itself is solely responsible for
carcinogenesis or disease aggressiveness. However, when distinct
pathways as a whole are analyzed, the hypothesis changes. Taylor
et al reported androgen receptor (AR) pathway gene
aberrations in 56 percent of primaries and 100% of metastases
(30). While AR gene
amplifications and mutations were almost exclusively found in
metastatic disease, it appeared that aberrations in NCOA2 and NCOR2
genes were important in primary tumors (30). PI3K signaling pathway was affected
in 42% of primary tumors and 100% of metastases. The RB signaling
pathway was affected in 34% of primary sites and 74% of metastases.
The RAS/RAF signaling pathway was affected in 43% of primary sites
and 90% of metastases (30). These
findings support the notion that genetic factors are associated
with the development and progression of prostate cancer. It is
unlikely that clinically significant prostate cancer is a result of
a single altered gene. The frequencies of altered critical pathways
in primary tumors (many of which are indolent) suggest that
clinically significant prostate cancers are unlikely to also result
from a single altered molecular pathway. On the contrary, the
evidence points towards combinations of genetic aberrations, which
result in several altered molecular pathways. Accumulation of
critical genomic aberrations over time and divergent clonal
evolution are also hallmarks of the disease progression towards a
more aggressive state (37)
(Table I). Identifying and
targeting key aberrant genes or pathways simultaneously may
theoretically ‘force’ the disease to regress into a more indolent
state. However, it is unknown whether the aggressive state is
actually reversible once it occurs.
The low mutation rate in PCa suggests that other
factors might also determine the clinical course of the disease. It
is now well established that epigenetic modifications play an
important role in prostate cancer (38-41)
(Table I). Epigenetics is the
study of heritable changes in gene expression, without the presence
of changes in the DNA sequence itself (42). While cells can alter their
epigenome as a response to various conditions, it is known that
epigenetic abnormalities frequently accumulate in cancer (43,44).
Epigenetic changes can predispose genes to mutations, while genes
that modify the epigenome are frequently mutated (45-47).
DNA methylation has been implicated in the lineage plasticity of
PCa (48). Several hypermethylated
cell cycle genes and growth suppressor genes have been linked to
worse prognosis (43). Aberrant
DNA hypomethylation has also been observed more frequently in late
stages of PCa (49). Epigenetic
reprogramming is associated with loss of luminal epithelial
identity and the transition from a typical prostate adenocarcinoma
towards an aggressive neuroendocrine PCa (NEPC) (37,50,51).
Neuroendocrine PCa cell lines possess a unique chromatin
accessibility profile, distinct from prostate adenocarcinoma
(52). Inactivation of TP53 and/or
RB1 leads to upregulation of DNA methyltransferase family member 1
(DNTM1) (53,54). DNA methylation is linked to the
activity of EZH2, which serves as a recruitment platform for DNMTs
(55). EZH2 is a central regulator
of neuroendocrine differentiation and the transition from an
androgen receptor (AR)-dependent disease towards an aggressive
state that is independent of AR signaling and indifferent to the
effects of antiandrogens (51,56-58).
The activity of AR can also be directly regulated by epigenetic
modifiers, such as histone deacetylases (HDAC) (59). Post-transcriptional mechanisms,
such as mRNA splicing or regulation by miRNA also play a role in
the progression of prostate cancer (60-62).
EZH2 can act both as a transcriptional activator or repressor,
depending on post-translational modifications of EZH2 (63-65).
Prostate tumors with ‘bad’ morphologic or genetic
features can still run an indolent clinical course. This suggests
that the cellular and secreted factors in the tumor immune
microenvironment (TIME) might play a role in the balance between
tumor clearance and tumor progression, as well as the response to
treatment. However, PCa in general has an immunologically ‘cold’
TIME (65). Overall mutation
rates, as well as DDR gene defects in PCa are low, especially in
the early disease setting (66).
Hence, neoantigen expression is diminished compared to many other
cancers. This results in a non-inflamed TIME, where tumor cells
proliferate freely and evade immune-mediated elimination. Although
the presence of cytotoxic and helper T-lymphocytes within tumor
margins have been associated with favorable prognosis across
several cancer types, PCa exhibits unique features (66-69).
Studies suggest that high density of stromal CD8+ tumor
infiltrating lymphocytes (TILs), and high PD-L1 expression are not
associated with better outcomes in PCa (66,70-72).
Some studies indicate that they might even be detrimental (73). Prostate cancer-infiltrating TILs
are frequently dysfunctional (71,74)
(Table I). However, high
proportions of Foxp3+ regulatory TILs are associated with worse
progression-free and overall survival in prostate cancer (75). In addition, high levels of M2
macrophages are pro-tumorigenic, suggesting that TGFβ might play a
role in immune exclusion in prostate cancer (76,77).
Epigenetic silencing of MHC Class I expression is common in
advanced prostate tumors (78,79).
Several signaling pathways, including the INF axis can also affect
the expression of MHC Class I and the subsequent activation and
expansion of CD8+ TILs within the invasive margins of a tumor
(80). PTEN loss and other DDR
defects also impact the TIME by modulating the activation of
cellular INF pathways (66,81-83).
It is known that the PTEN axis can confer sensitivity to
T-cell-based immunotherapies (84). The development of bone metastases
promotes the interaction between the tumor cells and the bone
microenvironment. This further decreases the immunogenicity of the
lesions (85). However, a few
prostate cancers are immunologically ‘hot’ tumors and show durable
responses when treated with immunotherapeutic agents (85-87).
This suggests that the TIME has the potential to affect the
clinical course of a patient who develops PCa, given the right
circumstances.
Cancer associated fibroblasts (CAFs) constitute the
most abundant cell population in the TME. They have been shown to
play a major role in prostate cancer progression (88). During tumorigenesis, stromal
fibroblasts crosstalk and likely coevolve with the epithelial
compartment and become CAFs. Experiments revealed that CAFs from
aggressive disease are sufficient to drive the progression of
prostate cancer cells with low tumorigenic potential (88). They can also contribute to
castration-resistance (89). On
the other hand, normal fibroblasts can slow the proliferation rate
of aggressive prostate cancer models (90). The amount of tumor-associated
stroma diminishes as prostate cancer becomes more aggressive
(89). Although it is hard to
prove causality, TME features that characterize aggressive disease
include a high proportion of CAFs, low proportion of smooth
muscles, high vimentin expression, asporin expression, increased
manifestation of matrix metalloproteinases, increased expression of
COL5A2, and decreased expression of COL4A6 (89,91-93).
The increased deposition of various collagen types, such as I or
III, contributes to matrix stiffness, which leads to increased
tumor invasiveness and metastatic potential (88). It was also suggested that
CAF-derived neuregulin 1 (NRG1) induces antiandrogen resistance,
via a NRG1-HER3 axis (94). CAFs
don't have the genetic alterations of the epithelial compartment.
On the contrary, CAFs from aggressive prostate cancers have
discrete methylation differences compared to CAFs from moderate
risk disease (95). For example,
epigenetic regulation of Ras activity in prostatic CAFs, was found
to regulate the metabolic and neuroendocrine activity in prostate
cancer that fails ADT (96).
Stromal AR expression also diminishes early in prostate
tumorigenesis and continues to gradually decrease as the disease
evolves into a more aggressive phenotype. It has been suggested
that stromal AR inhibits the growth of malignant epithelial cells
(97). Neural tissue is also an
active TME element in prostate cancer. Perineural invasion is a
known adverse prognostic factor (98). Several neural transmission
receptors are present in cancer cells (98). Moreover, it has been recently shown
that the abundance of neurotrophic growth factors in the patient's
urine may perform better than PSA to separate aggressive prostate
tumors from indolent ones (99).
There are currently no reliable molecular signatures
to identify prostate tumors destined to run an indolent clinical
course. Emerging evidence suggests that the upregulation of
pathways related to cellular aging and senescence can distinguish
between indolent and aggressive disease (100) (Table
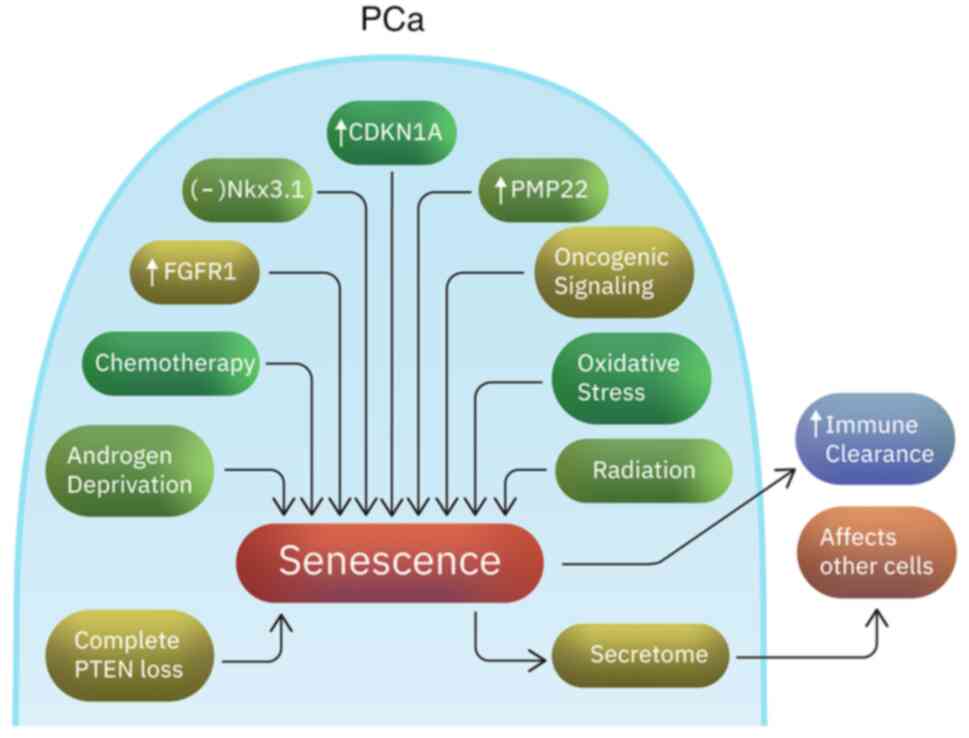
I). The ‘indolence signature’ includes inactivation of Nkx3.1,
and increased expression of CDKN1A (p21), FGFR1 and PMP22 genes
(100) (Fig. 1). CDKN1A is a cell regulatory gene
associated with senescence (101). FGFR1 is known to play a critical
role in prostate development and prostate tumorigenesis (102). This suggests a potentially
complex activity of FGFR1 in prostate cancer. FGF signaling plays
an important role in stem cell renewal, cellular aging and
senescence (103). Although PMP22
is a gene highly expressed in neurons, it has also been associated
with cellular proliferation regulation in other tissues and growth
arrest in fibroblasts (104,105). Increased senescence has also been
associated with reduced PSA recurrence rates (106). Senescent cells not only undergo
cell cycle arrest, but they also trigger an immune response that
can help the clearance of neoplastic cells (107). However, they are metabolically
active and their secretome can impact the surrounding non-senescent
cancer cells in ways that promote cancer progression and metastasis
(108,109). Senescence can also be caused by
long term oncogenic signaling or DNA damage and increased oxidative
stress as a result of anticancer agents or radiation (109,110). In addition, complete PTEN loss
triggers a p53-dependent cellular senescence response (The
overwhelming majority of patients have PTEN loss heterozygosity,
which results in tumor initiation and progression) (111-113).
Moreover, androgen deprivation frequently induces senescence in
prostate tumor cells (114,115). In conclusion, the induction of
senescent molecular signatures might contribute to the indolent
clinical course in some patients with PCa, before and after
treatment, especially antiandrogen therapy.
Data from histology and gene expression analysis can
provide useful prognostic information. Luminal B tumors carry the
worst outcome (69% overall survival at 10-years), followed by basal
and luminal A tumors (10-year overall survival at 80% and 82%
respectively) (116). It is well
known that the amount of Gleason 4 disease in the primary tumor is
strongly associated with clinical outcomes and disease
aggressiveness (117,118). High grade localized tumors are
marked by epigenetic loss of heterogeneity and common
trans-regulatory signatures. They exhibit enrichment for FOXA1,
CDX2 and HOXB13 transcription factor binding sites (119). A few studies compared the
differential gene expression between Gleason grade 3 and Gleason
grade 4 lesions (120,121). The genes exclusively expressed in
Gleason 4 tumors are those that are upregulated in neuronal,
embryonic and hematopoietic stem cells. Overexpression of EGFR and
HER2/neu are almost exclusively confined to Gleason 4 and above
cancers (120,121). These genes are associated with
independent tumor cell proliferation and enhanced cell motility
(117). Gleason 4 and 5 lesions
have lesser frequencies of cyclin D2 methylation, which results in
cyclin D2 activation and CDKN1B sequestration. This subsequently
results in cell cycle entry. CDKN1B immunostaining progressively
diminishes with increasing Gleason score (122-124).
Gleason score is also strongly associated with expression levels of
the anti-apoptotic genes DAD1 and BCL2 (125,126). Moreover, indolent cancers are
more capable of subverting a brake in replication. High Gleason
grade lesions also show decreased androgen signaling, suggesting
dedifferentiation. The downregulation of androgen responsive genes
in high grade tumors results in increased cell proliferation
(127,128). In addition, poor prognosis tumors
are related to increased VEGF production and microvessel density,
as well as irregularity in vessel diameter (129-131).
Microvessel pericyte density score is also associated with disease
aggressiveness (132). Higher
grade lesions overexpress elements that are permissive for tumor
migration. For example, the chemokine receptor CXCR4, which is
overexpressed in Gleason 4 lesions, poses a key role in the
development of lymph nodes and bone metastases (133-135).
Interestingly, its ligand CXCL12 is secreted in high concentrations
by lymph nodes and bone marrow stroma (117).
In PCa, metastatic disease may occur years after RP.
This suggests that in some cases cancer cells undergo a
long-lasting quiescent state (136). Quiescent cells are reversibly
suspended in the G0 phase, but they retain the ability to re-enter
the cell cycle and initiate symptomatic metastatic disease
(137) (Table I; Fig.
2). Tumor dormancy might be due to intrinsic factors or due to
conditions provided to the tumor cells by the surrounding
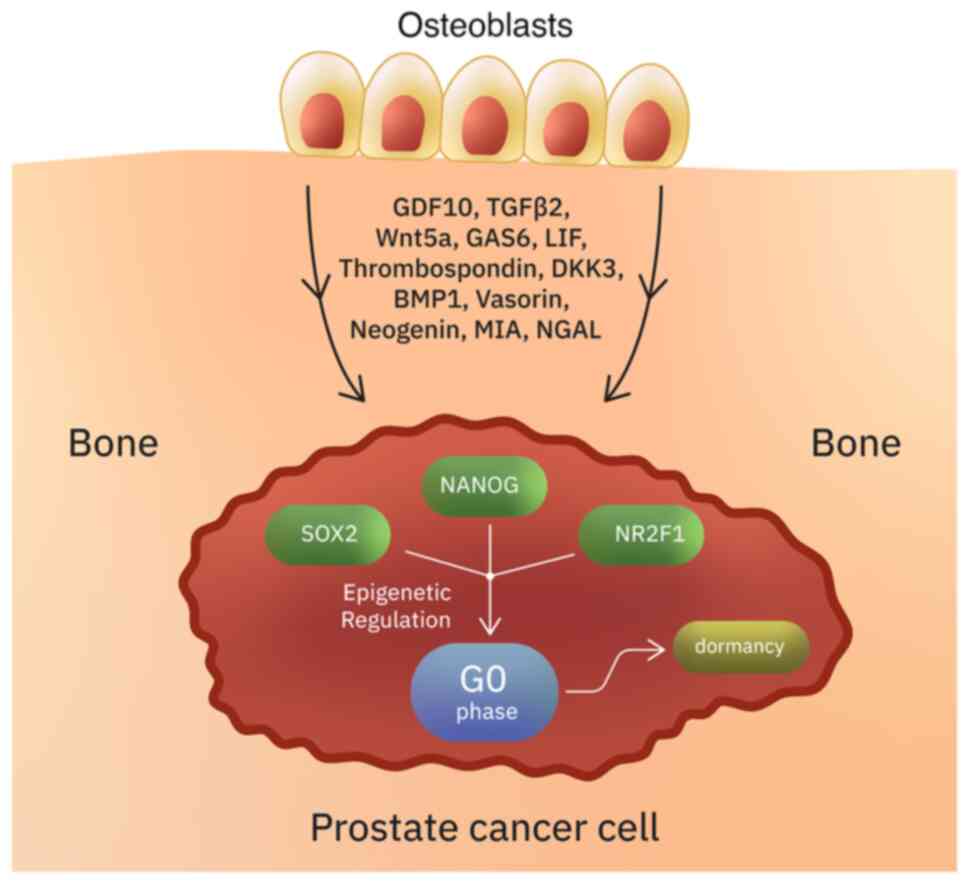
microenvironment (138,139). The bone is a major site of PCa
recurrence, suggesting that the bone microenvironment promotes a
state of dormancy (136). In
vitro studies have shown that two members of the TGFβ/BMP
family, GDF10 and TGFβ2 (which are secreted from differentiated
osteoblasts), induce quiescence in PCa cells (136,138). Several other osteoblast secreted
factors such as Wnt5a (maintains hematopoietic stem cells in
dormant state), GAS6, LIF, thrombospondin, DKK3, BMP1, vasorin,
neogenin, MIA and NGAL, have also been suggested to promote
dormancy in PCa (136,140-142).
For example, vasorin, neogenin and DKK3 induce dormancy via
activation of the p38MAPK signaling pathway (136). Besides osteoblasts, other bone
marrow stromal cells secrete dormancy-inducing factors (136). The transcription factors SOX2,
NANOG and the orphan receptor NR2F1 are important for the
maintenance of a quiescent phenotype through epigenetic regulation
(143-147).
Studies suggest that dormant cells at the metastatic sites continue
to acquire genomic changes (148). The receptor tyrosine kinases
TYRO3 and MERTK (TAM family) were shown to promote dormancy escape
(145). Norepinephrine was also
hypothesized to stimulate dormancy escape in PCa through the beta-2
adrenergic signaling (149).
The majority of prostate cancers follow an indolent
clinical course. It is unclear whether there is one or several
types of indolence in PCa. Several factors have been linked to
disease aggressiveness. However, it is still largely unknown which
of these factors actually have a causative role. High quality tumor
analyses suggest that complex genetic aberrations in multiple
critical pathways are associated with a worse phenotype. The escape
from indolence into an aggressive disease is associated with a
complex interplay in which genetic and epigenetic factors are
likely involved. The effect of the immediate immune tumor
microenvironment is also very important. Future studies will show
whether halting the stepwise tumor evolution is a feasible option.
Further research will also determine whether we can meaningfully
prolong the life of PCa patients by inducing senescence or
long-term tumor dormancy.
Not applicable.
Funding: No funding was received.
Not applicable.
MS conceptualized this review, reviewed the
literature and drafted and critically reviewed the final
manuscript. LJF reviewed the literature, and drafted and critically
reviewed the final version of the manuscript. SR reviewed the
literature, and drafted and critically reviewed the manuscript.
Data authentication is not applicable. All authors read and
approved the final manuscript.
Not applicable.
Not applicable.
All authors declare that they have no competing
interests.
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wadosky KM and Koochekpour S: Molecular
mechanisms underlying resistance to androgen deprivation therapy in
prostate cancer. Oncotarget. 7:64447–64470. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Varkaris A, Katsiampoura AD, Araujo JC,
Gallick GE and Corn PG: Src signaling pathways in prostate cancer.
Cancer Metastasis Rev. 33:595–606. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Koinis F, Corn P, Parikh N, Song J,
Vardaki I, Mourkioti I, Lin SH, Logothetis C, Panaretakis T and
Gallick G: Resistance to MET/VEGFR2 inhibition by cabozantinib is
mediated by YAP/TBX5-dependent induction of FGFR1 in
castration-resistant prostate cancer. Cancers (Basel).
12(244)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Smith M, De Bono J, Sternberg C, Le Moulec
S, Oudard S, De Giorgi U, Krainer M, Bergman A, Hoelzer W, De Wit
R, et al: Phase III study of cabozantinib in previously treated
metastatic castration-resistant prostate cancer: COMET-1. J Clin
Oncol. 34:3005–3013. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Michaelson MD, Oudard S, Ou YC, Sengeløv
L, Saad F, Houede N, Ostler P, Stenzl A, Daugaard G, Jones R, et
al: Randomized, placebo-controlled, phase III trial of sunitinib
plus prednisone versus prednisone alone in progressive, metastatic,
castration-resistant prostate cancer. J Clin Oncol. 32:76–82.
2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Spreafico A, Chi KN, Sridhar SS, Smith DC,
Carducci MA, Kavsak P, Wong TS, Wang L, Ivy SP, Mukherjee SD, et
al: A randomized phase II study of cediranib alone versus cediranib
in combination with dasatinib in docetaxel resistant, castration
resistant prostate cancer patients. Invest New Drugs. 32:1005–1016.
2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dagogo-Jack I and Shaw AT: Tumour
heterogeneity and resistance to cancer therapies. Nat Rev Clin
Oncol. 15:81–94. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Denmeade SR and Isaacs JT: A history of
prostate cancer treatment. Nat Rev Cancer. 2:389–396.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Epstein JI, Zelefsky MJ, Sjoberg DD,
Nelson JB, Egevad L, Magi-Galluzzi C, Vickers AJ, Parwani AV,
Reuter VE, Fine SW, et al: A contemporary prostate cancer grading
system: A validated alternative to the gleason score. Eur Urol.
69:428–435. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Johansson JE, Holmberg L, Johansson S,
Bergström R and Adami HO: Fifteen-year survival in prostate cancer.
A prospective, population-based study in Sweden. JAMA. 277:467–471.
1997.PubMed/NCBI
|
|
12
|
Simpkin AJ, Tilling K, Martin RM, Lane JA,
Hamdy FC, Holmberg L, Neal DE, Metcalfe C and Donovan JL:
Systematic review and meta-analysis of factors determining change
to radical treatment in active surveillance for localized prostate
cancer. Eur Urol. 67:993–1005. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Musunuru HB, Yamamoto T, Klotz L, Ghanem
G, Mamedov A, Sethukavalan P, Jethava V, Jain S, Zhang L, Vesprini
D and Loblaw A: Active surveillance for intermediate risk prostate
cancer: Survival outcomes in the sunnybrook experience. J Urol.
196:1651–1658. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Herlemann A, Huang HC, Alam R, Tosoian JJ,
Kim HL, Klein EA, Simko JP, Chan JM, Lane BR, Davis JW, et al:
Decipher identifies men with otherwise clinically
favorable-intermediate risk disease who may not be good candidates
for active surveillance. Prostate Cancer Prostatic Dis. 23:136–143.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kornberg Z, Cooperberg MR, Cowan JE, Chan
JM, Shinohara K, Simko JP, Tenggara I and Carroll PR: A 17-gene
genomic prostate score as a predictor of adverse pathology in men
on active surveillance. J Urol. 202:702–709. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Spratt DE, Zhang J, Santiago-Jiménez M,
Dess RT, Davis JW, Den RB, Dicker AP, Kane CJ, Pollack A, Stoyanova
R, et al: Development and validation of a novel integrated
clinical-genomic risk group classification for localized prostate
cancer. J Clin Oncol. 36:581–590. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Spratt DE, Yousefi K, Deheshi S, Ross AE,
Den RB, Schaeffer EM, Trock BJ, Zhang J, Glass AG, Dicker AP, et
al: Individual patient-level meta-analysis of the performance of
the decipher genomic classifier in high-risk men after
prostatectomy to predict development of metastatic disease. J Clin
Oncol. 35:1991–1998. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Pound CR, Partin AW, Eisenberger MA, Chan
DW, Pearson JD and Walsh PC: Natural history of progression after
PSA elevation following radical prostatectomy. JAMA. 281:1591–1597.
1999.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Boorjian SA, Thompson RH, Tollefson MK,
Rangel LJ, Bergstralh EJ, Blute ML and Karnes RJ: Long-term risk of
clinical progression after biochemical recurrence following radical
prostatectomy: The impact of time from surgery to recurrence. Eur
Urol. 59:893–899. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Martin NE, Chen MH, Beard CJ, Nguyen PL,
Loffredo MJ, Renshaw AA, Kantoff PW and D'Amico AV: Natural history
of untreated prostate specific antigen radiorecurrent prostate
cancer in men with favorable prognostic indicators. Prostate
Cancer. 2014(912943)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Trock BJ, Han M, Freedland SJ, Humphreys
EB, DeWeese TL, Partin AW and Walsh PC: Prostate cancer-specific
survival following salvage radiotherapy vs observation in men with
biochemical recurrence after radical prostatectomy. JAMA.
299:2760–2769. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Stone L: Prostate cancer: ADT after
radical prostatectomy-when and how? Nat Rev Urol.
13(367)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Muralidhar V, Mahal BA and Nguyen PL:
Conditional cancer-specific mortality in T4, N1, or M1 prostate
cancer: Implications for long-term prognosis. Radiat Oncol.
10(155)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Scher HI, Fizazi K, Saad F, Taplin ME,
Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, et
al: Increased survival with enzalutamide in prostate cancer after
chemotherapy. N Engl J Med. 367:1187–1197. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
James ND, de Bono JS, Spears MR, Clarke
NW, Mason MD, Dearnaley DP, Ritchie AWS, Amos CL, Gilson C, Jones
RJ, et al: Abiraterone for prostate cancer not previously treated
with hormone therapy. N Engl J Med. 377:338–351. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chandrasekar T, Yang JC, Gao AC and Evans
CP: Targeting molecular resistance in castration-resistant prostate
cancer. BMC Med. 13(206)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Fine SW: Neuroendocrine tumors of the
prostate. Mod Pathol. 31 (S1):S122–S132. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Cancer Genome Atlas Research Network. The
molecular taxonomy of primary prostate cancer. Cell. 163:1011–1025.
2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hieronymus H, Schultz N, Gopalan A, Carver
BS, Chang MT, Xiao Y, Heguy A, Huberman K, Bernstein M, Assel M, et
al: Copy number alteration burden predicts prostate cancer relapse.
Proc Natl Acad Sci USA. 111:11139–11144. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva
B, et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Paris PL, Andaya A, Fridlyand J, Jain AN,
Weinberg V, Kowbel D, Brebner JH, Simko J, Watson JE, Volik S, et
al: Whole genome scanning identifies genotypes associated with
recurrence and metastasis in prostate tumors. Hum Mol Genet.
13:1303–1313. 2004.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chu LW, Troncoso P, Johnston DA and Liang
JC: Genetic markers useful for distinguishing between
organ-confined and locally advanced prostate cancer. Genes
Chromosomes Cancer. 36:303–312. 2003.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648.
2005.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sboner A, Habegger L, Pflueger D, Terry S,
Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS,
et al: FusionSeq: A modular framework for finding gene fusions by
analyzing paired-end RNA-sequencing data. Genome Biol.
11(R104)2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang K, Singh D, Zeng Z, Coleman SJ, Huang
Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al:
MapSplice: Accurate mapping of RNA-seq reads for splice junction
discovery. Nucleic Acids Res. 38(e178)2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tomlins SA, Bjartell A, Chinnaiyan AM,
Jenster G, Nam RK, Rubin MA and Schalken JA: ETS gene fusions in
prostate cancer: From discovery to daily clinical practice. Eur
Urol. 56:275–286. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tzelepi V, Logotheti S, Efstathiou E,
Troncoso P, Aparicio A, Sakellakis M, Hoang A, Perimenis P,
Melachrinou M, Logothetis C and Zolota V: Epigenetics and prostate
cancer: Defining the timing of DNA methyltransferase deregulation
during prostate cancer progression. Pathology. 52:218–227.
2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Macedo-Silva C, Benedetti R, Ciardiello F,
Cappabianca S, Jerónimo C and Altucci L: Epigenetic mechanisms
underlying prostate cancer radioresistance. Clin Epigenetics.
13(125)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ngollo M, Dagdemir A, Karsli-Ceppioglu S,
Judes G, Pajon A, Penault-Llorca F, Boiteux JP, Bignon YJ, Guy L
and Bernard-Gallon DJ: Epigenetic modifications in prostate cancer.
Epigenomics. 6:415–426. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kumaraswamy A, Welker Leng KR, Westbrook
TC, Yates JA, Zhao SG, Evans CP, Feng FY, Morgan TM and Alumkal JJ:
Recent advances in epigenetic biomarkers and epigenetic targeting
in prostate cancer. Eur Urol. 80:71–81. 2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Weinhold B: Epigenetics: The science of
change. Environ Health Perspect. 114:A160–A167. 2006.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Sugiura M, Sato H, Kanesaka M, Imamura Y,
Sakamoto S, Ichikawa T and Kaneda A: Epigenetic modifications in
prostate cancer. Int J Urol. 28:140–149. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Dawson MA: The cancer epigenome: Concepts,
challenges, and therapeutic opportunities. Science. 355:1147–1152.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Herman JG, Umar A, Polyak K, Graff JR,
Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW,
et al: Incidence and functional consequences of hMLH1 promoter
hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA.
95:6870–6875. 1998.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kane MF, Loda M, Gaida GM, Lipman J,
Mishra R, Goldman H, Jessup JM and Kolodner R: Methylation of the
hMLH1 promoter correlates with lack of expression of hMLH1 in
sporadic colon tumors and mismatch repair-defective human tumor
cell lines. Cancer Res. 57:808–811. 1997.PubMed/NCBI
|
|
47
|
Shih AH, Abdel-Wahab O, Patel JP and
Levine RL: The role of mutations in epigenetic regulators in
myeloid malignancies. Nat Rev Cancer. 12:599–612. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ge R, Wang Z, Montironi R, Jiang Z, Cheng
M, Santoni M, Huang K, Massari F, Lu X, Cimadamore A, et al:
Epigenetic modulations and lineage plasticity in advanced prostate
cancer. Ann Oncol. 31:470–479. 2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yegnasubramanian S, Haffner MC, Zhang Y,
Gurel B, Cornish TC, Wu Z, Irizarry RA, Morgan J, Hicks J, DeWeese
TL, et al: DNA hypomethylation arises later in prostate cancer
progression than CpG island hypermethylation and contributes to
metastatic tumor heterogeneity. Cancer Res. 68:8954–8967.
2008.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Aggarwal R, Huang J, Alumkal JJ, Zhang L,
Feng FY, Thomas GV, Weinstein AS, Friedl V, Zhang C, Witte ON, et
al: Clinical and genomic characterization of treatment-emergent
small-cell neuroendocrine prostate cancer: A multi-institutional
prospective study. J Clin Oncol. 36:2492–2503. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Davies A, Zoubeidi A and Selth LA: The
epigenetic and transcriptional landscape of neuroendocrine prostate
cancer. Endocr Relat Cancer. 27:R35–R50. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Park JW, Lee JK, Sheu KM, Wang L, Balanis
NG, Nguyen K, Smith BA, Cheng C, Tsai BL, Cheng D, et al:
Reprogramming normal human epithelial tissues to a common, lethal
neuroendocrine cancer lineage. Science. 362:91–95. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
McCabe MT, Davis JN and Day ML: Regulation
of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res.
65:3624–3632. 2005.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS,
Chen CY, Lu YY, Tang YA, Yang YC, Yang PC and Wang YC:
Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1
overexpression in lung cancer. Cancer Res. 70:5807–5817.
2010.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Viré E, Brenner C, Deplus R, Blanchon L,
Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden
JM, et al: The Polycomb group protein EZH2 directly controls DNA
methylation. Nature. 439:871–874. 2006.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Shan J, Al-Muftah MA, Al-Kowari MK,
Abuaqel SWJ, Al-Rumaihi K, Al-Bozom I, Li P and Chouchane L:
Targeting Wnt/EZH2/microRNA-708 signaling pathway inhibits
neuroendocrine differentiation in prostate cancer. Cell Death
Discov. 5(139)2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Dardenne E, Beltran H, Benelli M, Gayvert
K, Berger A, Puca L, Cyrta J, Sboner A, Noorzad Z, MacDonald T, et
al: N-Myc induces an EZH2-mediated transcriptional program driving
neuroendocrine prostate cancer. Cancer Cell. 30:563–577.
2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Shah N, Wang P, Wongvipat J, Karthaus WR,
Abida W, Armenia J, Rockowitz S, Drier Y, Bernstein BE, Long HW, et
al: Regulation of the glucocorticoid receptor via a BET-dependent
enhancer drives antiandrogen resistance in prostate cancer. Elife.
6(e27861)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu
X, Powell MJ, Yang T, Gu W, Avantaggiati ML, et al: Hormonal
control of androgen receptor function through SIRT1. Mol Cell Biol.
26:8122–8135. 2006.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zhang X, Coleman IM, Brown LG, True LD,
Kollath L, Lucas JM, Lam HM, Dumpit R, Corey E, Chéry L, et al:
SRRM4 expression and the loss of REST activity may promote the
emergence of the neuroendocrine phenotype in castration-resistant
prostate cancer. Clin Cancer Res. 21:4698–4708. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Li Y, Donmez N, Sahinalp C, Xie N, Wang Y,
Xue H, Mo F, Beltran H, Gleave M, Wang Y, et al: SRRM4 drives
neuroendocrine transdifferentiation of prostate adenocarcinoma
under androgen receptor pathway inhibition. Eur Urol. 71:68–78.
2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Nam RK, Benatar T, Amemiya Y, Wallis CJD,
Romero JM, Tsagaris M, Sherman C, Sugar L and Seth A: MicroRNA-652
induces NED in LNCaP and EMT in PC3 prostate cancer cells.
Oncotarget. 9:19159–19176. 2018.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis
RT, Wu X, Stack EC, Loda M, Liu T, et al: EZH2 oncogenic activity
in castration-resistant prostate cancer cells is
Polycomb-independent. Science. 338:1465–1469. 2012.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Kim J, Lee Y, Lu X, Song B, Fong KW, Cao
Q, Licht JD, Zhao JC and Yu J: Polycomb- and
methylation-independent roles of EZH2 as a transcription activator.
Cell Rep. 25:2808–2820.e4. 2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Bilusic M, Madan RA and Gulley JL:
Immunotherapy of prostate cancer: Facts and hopes. Clin Cancer Res.
23:6764–6770. 2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Vitkin N, Nersesian S, Siemens DR and Koti
M: The tumor immune contexture of prostate cancer. Front Immunol.
10(603)2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Fridman WH, Zitvogel L, Sautès-Fridman C
and Kroemer G: The immune contexture in cancer prognosis and
treatment. Nat Rev Clin Oncol. 14:717–734. 2017.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Mellman I, Coukos G and Dranoff G: Cancer
immunotherapy comes of age. Nature. 480:480–489. 2011.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Topalian SL, Taube JM, Anders RA and
Pardoll DM: Mechanism-driven biomarkers to guide immune checkpoint
blockade in cancer therapy. Nat Rev Cancer. 16:275–287.
2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Leclerc BG, Charlebois R, Chouinard G,
Allard B, Pommey S, Saad F and Stagg J: CD73 expression is an
independent prognostic factor in prostate cancer. Clin Cancer Res.
22:158–166. 2016.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ness N, Andersen S, Valkov A, Nordby Y,
Donnem T, Al-Saad S, Busund LT, Bremnes RM and Richardsen E:
Infiltration of CD8+ lymphocytes is an independent prognostic
factor of biochemical failure-free survival in prostate cancer.
Prostate. 74:1452–1461. 2014.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Petitprez F, Fossati N, Vano Y, Freschi M,
Becht E, Lucianò R, Calderaro J, Guédet T, Lacroix L, Rancoita PMV,
et al: PD-L1 expression and CD8+ T-cell infiltrate are
associated with clinical progression in patients with node-positive
prostate cancer. Eur Urol Focus. 5:192–196. 2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Zhao SG, Lehrer J, Chang SL, Das R, Erho
N, Liu Y, Sjöström M, Den RB, Freedland SJ, Klein EA, et al: The
immune landscape of prostate cancer and nomination of PD-L2 as a
potential therapeutic target. J Natl Cancer Inst. 111:301–310.
2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Sharma P, Hu-Lieskovan S, Wargo JA and
Ribas A: Primary, adaptive, and acquired resistance to cancer
immunotherapy. Cell. 168:707–723. 2017.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Nardone V, Botta C, Caraglia M, Martino
EC, Ambrosio MR, Carfagno T, Tini P, Semeraro L, Misso G, Grimaldi
A, et al: Tumor infiltrating T lymphocytes expressing FoxP3, CCR7
or PD-1 predict the outcome of prostate cancer patients subjected
to salvage radiotherapy after biochemical relapse. Cancer Biol
Ther. 17:1213–1220. 2016.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Lundholm M, Hägglöf C, Wikberg ML, Stattin
P, Egevad L, Bergh A, Wikström P, Palmqvist R and Edin S: Secreted
factors from colorectal and prostate cancer cells skew the immune
response in opposite directions. Sci Rep. 5(15651)2015.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Mariathasan S, Turley SJ, Nickles D,
Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita
JL, Cubas R, et al: TGFβ attenuates tumour response to PD-L1
blockade by contributing to exclusion of T cells. Nature.
554:544–548. 2018.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Heninger E, Krueger TE, Thiede SM, Sperger
JM, Byers BL, Kircher MR, Kosoff D, Yang B, Jarrard DF, McNeel DG
and Lang JM: Inducible expression of cancer-testis antigens in
human prostate cancer. Oncotarget. 7:84359–84374. 2016.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Sanda MG, Restifo NP, Walsh JC, Kawakami
Y, Nelson WG, Pardoll DM and Simons JW: Molecular characterization
of defective antigen processing in human prostate cancer. J Natl
Cancer Inst. 87:280–285. 1995.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Martini M, Testi MG, Pasetto M, Picchio
MC, Innamorati G, Mazzocco M, Ugel S, Cingarlini S, Bronte V,
Zanovello P, et al: IFN-gamma-mediated upmodulation of MHC class I
expression activates tumor-specific immune response in a mouse
model of prostate cancer. Vaccine. 28:3548–3557. 2010.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Chen L and Guo D: The functions of tumor
suppressor PTEN in innate and adaptive immunity. Cell Mol Immunol.
14:581–589. 2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Ivashkiv LB and Donlin LT: Regulation of
type I interferon responses. Nat Rev Immunol. 14:36–49.
2014.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Pencik J, Schlederer M, Gruber W, Unger C,
Walker SM, Chalaris A, Marié IJ, Hassler MR, Javaheri T, Aksoy O,
et al: STAT3 regulated ARF expression suppresses prostate cancer
metastasis. Nat Commun. 6(7736)2015.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Peng W, Chen JQ, Liu C, Malu S, Creasy C,
Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al: Loss of
PTEN promotes resistance to T cell-mediated immunotherapy. Cancer
Discov. 6:202–216. 2016.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Jiao S, Subudhi SK, Aparicio A, Ge Z, Guan
B, Miura Y and Sharma P: Differences in tumor microenvironment
dictate T helper lineage polarization and response to immune
checkpoint therapy. Cell. 179:1177–1190.e13. 2019.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Stultz J and Fong L: How to turn up the
heat on the cold immune microenvironment of metastatic prostate
cancer. Prostate Cancer Prostatic Dis. 24:697–717. 2021.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Kwon ED, Drake CG, Scher HI, Fizazi K,
Bossi A, van den Eertwegh AJ, Krainer M, Houede N, Santos R,
Mahammedi H, et al: Ipilimumab versus placebo after radiotherapy in
patients with metastatic castration-resistant prostate cancer that
had progressed after docetaxel chemotherapy (CA184-043): A
multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol.
15:700–712. 2014.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Bonollo F, Thalmann GN, Kruithof-de Julio
M and Karkampouna S: The role of cancer-associated fibroblasts in
prostate cancer tumorigenesis. Cancers (Basel).
12(1887)2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Blom S, Erickson A, Östman A, Rannikko A,
Mirtti T, Kallioniemi O and Pellinen T: Fibroblast as a critical
stromal cell type determining prognosis in prostate cancer.
Prostate. 79:1505–1513. 2019.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Suhovskih AV, Kashuba VI, Klein G and
Grigorieva EV: Prostate cancer cells specifically reorganize
epithelial cell-fibroblast communication through proteoglycan and
junction pathways. Cell Adh Migr. 11:39–53. 2017.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Ma JB, Bai JY, Zhang HB, Gu L, He D and
Guo P: Downregulation of collagen COL4A6 is associated with
prostate cancer progression and metastasis. Genet Test Mol
Biomarkers. 24:399–408. 2020.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Ren X, Chen X, Fang K, Zhang X, Wei X,
Zhang T, Li G, Lu Z, Song N, Wang S and Qin C: COL5A2 promotes
proliferation and invasion in prostate cancer and is one of seven
gleason-related genes that predict recurrence-free survival. Front
Oncol. 11(583083)2021.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Bahmad HF, Jalloul M, Azar J, Moubarak MM,
Samad TA, Mukherji D, Al-Sayegh M and Abou-Kheir W: Tumor
microenvironment in prostate cancer: Toward identification of novel
molecular biomarkers for diagnosis, prognosis, and therapy
development. Front Genet. 12(652747)2021.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Zhang Z, Karthaus WR, Lee YS, Gao VR, Wu
C, Russo JW, Liu M, Mota JM, Abida W, Linton E, et al: Tumor
microenvironment-derived NRG1 promotes antiandrogen resistance in
prostate cancer. Cancer Cell. 38:279–296.e9. 2020.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Lawrence MG, Pidsley R, Niranjan B,
Papargiris M, Pereira BA, Richards M, Teng L, Norden S, Ryan A,
Frydenberg M, et al: Alterations in the methylome of the stromal
tumour microenvironment signal the presence and severity of
prostate cancer. Clin Epigenetics. 12(48)2020.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Mishra R, Haldar S, Placencio V, Madhav A,
Rohena-Rivera K, Agarwal P, Duong F, Angara B, Tripathi M, Liu Z,
et al: Stromal epigenetic alterations drive metabolic and
neuroendocrine prostate cancer reprogramming. J Clin Invest.
128:4472–4484. 2018.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Singh M, Jha R, Melamed J, Shapiro E,
Hayward SW and Lee P: Stromal androgen receptor in prostate
development and cancer. Am J Pathol. 184:2598–2607. 2014.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Sejda A, Sigorski D, Gulczyński J,
Wesołowski W, Kitlińska J and Iżycka-Świeszewska E: Complexity of
neural component of tumor microenvironment in prostate cancer.
Pathobiology. 87:87–99. 2020.PubMed/NCBI View Article : Google Scholar
|
|
99
|
March B, Lockhart KR, Faulkner S, Smolny
M, Rush R and Hondermarck H: ELISA-based quantification of
neurotrophic growth factors in urine from prostate cancer patients.
FASEB Bioadv. 3:888–896. 2021.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Irshad S, Bansal M, Castillo-Martin M,
Zheng T, Aytes A, Wenske S, Le Magnen C, Guarnieri P, Sumazin P,
Benson MC, et al: A molecular signature predictive of indolent
prostate cancer. Sci Transl Med. 5(202ra122)2013.PubMed/NCBI View Article : Google Scholar
|
|
101
|
López-Domínguez JA, Rodríguez-López S,
Ahumada-Castro U, Desprez PY, Konovalenko M, Laberge RM, Cárdenas
C, Villalba JM and Campisi J: Cdkn1a transcript variant 2 is a
marker of aging and cellular senescence. Aging (Albany NY).
13:13380–13392. 2021.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Yang F, Zhang Y, Ressler SJ, Ittmann MM,
Ayala GE, Dang TD, Wang F and Rowley DR: FGFR1 is essential for
prostate cancer progression and metastasis. Cancer Res.
73:3716–3724. 2013.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Coutu DL and Galipeau J: Roles of FGF
signaling in stem cell self-renewal, senescence and aging. Aging
(Albany NY). 3:920–33. 2011.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Adlkofer K, Martini R, Aguzzi A, Zielasek
J, Toyka KV and Suter U: Hypermyelination and demyelinating
peripheral neuropathy in Pmp22-deficient mice. Nat Genet.
11:274–280. 1995.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Suter U and Snipes GJ: Peripheral myelin
protein 22: Facts and hypotheses. J Neurosci Res. 40:145–151.
1995.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Wagner J, Damaschke N, Yang B, Truong M,
Guenther C, McCormick J, Huang W and Jarrard D: Overexpression of
the novel senescence marker β-galactosidase (GLB1) in prostate
cancer predicts reduced PSA recurrence. PLoS One.
10(e0124366)2015.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Prata LGPL, Ovsyannikova IG, Tchkonia T
and Kirkland JL: Senescent cell clearance by the immune system:
Emerging therapeutic opportunities. Semin Immunol.
40(101275)2018.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Wang B, Kohli J and Demaria M: Senescent
cells in cancer therapy: Friends or foes? Trends Cancer. 6:838–857.
2020.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Hwang HJ, Jung SH, Lee HC, Han NK, Bae IH,
Lee M, Han YH, Kang YS, Lee SJ, Park HJ, et al: Identification of
novel therapeutic targets in the secretome of ionizing
radiation-nduced senescent tumor cells. Oncol Rep. 35:841–850.
2016.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Ewald JA, Desotelle JA, Wilding G and
Jarrard DF: Therapy-induced senescence in cancer. J Natl Cancer
Inst. 102:1536–1546. 2010.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Parisotto M, Grelet E, El Bizri R and
Metzger D: Senescence controls prostatic neoplasia driven by Pten
loss. Mol Cell Oncol. 6(1511205)2018.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Jung SH, Hwang HJ, Kang D, Park HA, Lee
HC, Jeong D, Lee K, Park HJ, Ko YG and Lee JS: mTOR kinase leads to
PTEN-loss-induced cellular senescence by phosphorylating p53.
Oncogene. 38:1639–1650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Chen Z, Trotman LC, Shaffer D, Lin HK,
Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al:
Crucial role of p53-dependent cellular senescence in suppression of
Pten-deficient tumorigenesis. Nature. 436:725–730. 2005.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Blute ML Jr, Damaschke N, Wagner J, Yang
B, Gleave M, Fazli L, Shi F, Abel EJ, Downs TM, Huang W and Jarrard
DF: Persistence of senescent prostate cancer cells following
prolonged neoadjuvant androgen deprivation therapy. PLoS One.
12(e0172048)2017.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Pernicová Z, Slabáková E, Kharaishvili G,
Bouchal J, Král M, Kunická Z, Machala M, Kozubík A and Souček K:
Androgen depletion induces senescence in prostate cancer cells
through down-regulation of Skp2. Neoplasia. 13:526–536.
2011.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Zhao SG, Chang SL, Erho N, Yu M, Lehrer J,
Alshalalfa M, Speers C, Cooperberg MR, Kim W, Ryan CJ, et al:
Associations of luminal and basal subtyping of prostate cancer with
prognosis and response to androgen deprivation therapy. JAMA Oncol.
3:1663–1672. 2017.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Ahmed HU, Arya M, Freeman A and Emberton
M: Do low-grade and low-volume prostate cancers bear the hallmarks
of malignancy? Lancet Oncol. 13:e509–e517. 2012.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Sharma M and Miyamoto H: Percent Gleason
pattern 4 in stratifying the prognosis of patients with
intermediate-risk prostate cancer. Transl Androl Urol. 7 (Suppl
4):S484–S489. 2018.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Eksi SE, Chitsazan A, Sayar Z, Thomas GV,
Fields AJ, Kopp RP, Spellman PT and Adey AC: Epigenetic loss of
heterogeneity from low to high grade localized prostate tumours.
Nat Commun. 12(7292)2021.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Ross AE, Marchionni L, Vuica-Ross M,
Cheadle C, Fan J, Berman DM and Schaeffer EM: Gene expression
pathways of high grade localized prostate cancer. Prostate.
71:1568–1577. 2011.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Skacel M, Ormsby AH, Pettay JD, Tsiftsakis
EK, Liou LS, Klein EA, Levin HS, Zippe CD and Tubbs RR: Aneusomy of
chromosomes 7, 8, and 17 and amplification of HER-2/neu and
epidermal growth factor receptor in Gleason score 7 prostate
carcinoma: A differential fluorescent in situ hybridization study
of Gleason pattern 3 and 4 using tissue microarray. Hum Pathol.
32:1392–1397. 2001.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Susaki E and Nakayama KI: Multiple
mechanisms for p27(Kip1) translocation and degradation. Cell Cycle.
6:3015–3020. 2007.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Padar A, Sathyanarayana UG, Suzuki M,
Maruyama R, Hsieh JT, Frenkel EP, Minna JD and Gazdar AF:
Inactivation of cyclin D2 gene in prostate cancers by aberrant
promoter methylation. Clin Cancer Res. 9:4730–4734. 2003.PubMed/NCBI
|
|
124
|
Guo Y, Sklar GN, Borkowski A and Kyprianou
N: Loss of the cyclin-dependent kinase inhibitor p27(Kip1) protein
in human prostate cancer correlates with tumor grade. Clin Cancer
Res. 3:2269–2274. 1997.PubMed/NCBI
|
|
125
|
True L, Coleman I, Hawley S, Huang CY,
Gifford D, Coleman R, Beer TM, Gelmann E, Datta M, Mostaghel E, et
al: A molecular correlate to the Gleason grading system for
prostate adenocarcinoma. Proc Natl Acad Sci USA. 103:10991–10996.
2006.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Fleischmann A, Huland H, Mirlacher M,
Wilczak W, Simon R, Erbersdobler A, Sauter G and Schlomm T:
Prognostic relevance of Bcl-2 overexpression in surgically treated
prostate cancer is not caused by increased copy number or
translocation of the gene. Prostate. 72:991–997. 2012.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Tomlins SA, Mehra R, Rhodes DR, Cao X,
Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA,
Pienta KJ, et al: Integrative molecular concept modeling of
prostate cancer progression. Nat Genet. 39:41–51. 2007.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Hendriksen PJ, Dits NF, Kokame K,
Veldhoven A, van Weerden WM, Bangma CH, Trapman J and Jenster G:
Evolution of the androgen receptor pathway during progression of
prostate cancer. Cancer Res. 66:5012–5020. 2006.PubMed/NCBI View Article : Google Scholar
|
|
129
|
West AF, O'Donnell M, Charlton RG, Neal DE
and Leung HY: Correlation of vascular endothelial growth factor
expression with fibroblast growth factor-8 expression and
clinico-pathologic parameters in human prostate cancer. Br J
Cancer. 85:576–583. 2001.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Erbersdobler A, Isbarn H, Dix K, Steiner
I, Schlomm T, Mirlacher M, Sauter G and Haese A: Prognostic value
of microvessel density in prostate cancer: A tissue microarray
study. World J Urol. 28:687–692. 2010.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Mucci LA, Powolny A, Giovannucci E, Liao
Z, Kenfield SA, Shen R, Stampfer MJ and Clinton SK: Prospective
study of prostate tumor angiogenesis and cancer-specific mortality
in the health professionals follow-up study. J Clin Oncol.
27:5627–5633. 2009.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Killingsworth MC and Wu X: Vascular
pericyte density and angiogenesis associated with adenocarcinoma of
the prostate. Pathobiology. 78:24–34. 2011.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Lin D, Bayani J and Wang Y, Sadar MD,
Yoshimoto M, Gout PW, Squire JA and Wang Y: Development of
metastatic and non-metastatic tumor lines from a patient's prostate
cancer specimen-identification of a small subpopulation with
metastatic potential in the primary tumor. Prostate. 70:1636–1644.
2010.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Schioppa T, Uranchimeg B, Saccani A,
Biswas SK, Doni A, Rapisarda A, Bernasconi S, Saccani S, Nebuloni
M, Vago L, et al: Regulation of the chemokine receptor CXCR4 by
hypoxia. J Exp Med. 198:1391–1402. 2003.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Staller P, Sulitkova J, Lisztwan J, Moch
H, Oakeley EJ and Krek W: Chemokine receptor CXCR4 downregulated by
von Hippel-Lindau tumour suppressor pVHL. Nature. 425:307–311.
2003.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Yu-Lee LY, Lee YC, Pan J, Lin SC, Pan T,
Yu G, Hawke DH, Pan BF and Lin SH: Bone secreted factors induce
cellular quiescence in prostate cancer cells. Sci Rep.
9(18635)2019.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Phan TG and Croucher PI: The dormant
cancer cell life cycle. Nat Rev Cancer. 20:398–411. 2020.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Yu-Lee LY, Yu G, Lee YC, Lin SC, Pan J,
Pan T, Yu KJ, Liu B, Creighton CJ, Rodriguez-Canales J, et al:
Osteoblast-secreted factors mediate dormancy of metastatic prostate
cancer in the bone via activation of the
TGFβRIII-p38MAPK-pS249/T252RB pathway. Cancer Res. 78:2911–2924.
2018.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Esposito M, Guise T and Kang Y: The
biology of bone metastasis. Cold Spring Harb Perspect Med.
8(a031252)2018.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Ren D, Dai Y, Yang Q, Zhang X, Guo W, Ye
L, Huang S, Chen X, Lai Y, Du H, et al: Wnt5a induces and maintains
prostate cancer cells dormancy in bone. J Exp Med. 216:428–449.
2019.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Shiozawa Y, Pedersen EA, Patel LR, Ziegler
AM, Havens AM, Jung Y, Wang J, Zalucha S, Loberg RD, Pienta KJ and
Taichman RS: GAS6/AXL axis regulates prostate cancer invasion,
proliferation, and survival in the bone marrow niche. Neoplasia.
12:116–127. 2010.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Singh DK, Patel VG, Oh WK and
Aguirre-Ghiso JA: Prostate cancer dormancy and reactivation in bone
marrow. J Clin Med. 10(2648)2021.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Sosa MS: Dormancy programs as emerging
antimetastasis therapeutic alternatives. Mol Cell Oncol.
3(e1029062)2015.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Cackowski FC and Heath EI: Prostate cancer
dormancy and recurrence. Cancer Lett. 524:103–108. 2022.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Cackowski FC, Eber MR, Rhee J, Decker AM,
Yumoto K, Berry JE, Lee E, Shiozawa Y, Jung Y, Aguirre-Ghiso JA and
Taichman RS: Mer tyrosine kinase regulates disseminated prostate
cancer cellular dormancy. J Cell Biochem. 118:891–902.
2017.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Zhang J, Si J, Gan L, Di C, Xie Y, Sun C,
Li H, Guo M and Zhang H: Research progress on therapeutic targeting
of quiescent cancer cells. Artif Cells Nanomed Biotechnol.
47:2810–2820. 2019.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Sosa MS, Parikh F, Maia AG, Estrada Y,
Bosch A, Bragado P, Ekpin E, George A, Zheng Y, Lam HM, et al:
NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven
quiescence programmes. Nat Commun. 6(6170)2015.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Recasens A and Munoz L: Targeting cancer
cell dormancy. Trends Pharmacol Sci. 40:128–141. 2019.PubMed/NCBI View Article : Google Scholar
|
|
149
|
Decker AM, Jung Y, Cackowski FC, Yumoto K,
Wang J and Taichman RS: Sympathetic signaling reactivates quiescent
disseminated prostate cancer cells in the bone marrow. Mol Cancer
Res. 15:1644–1655. 2017.PubMed/NCBI View Article : Google Scholar
|