Introduction
Parkinson's disease (PD) is a neurodegenerative
disease that is characterized by the degeneration of dopaminergic
neurons in the substantia nigra compacta (SNc) area (1). The primary clinical manifestations of
PD are resting tremor, muscle rigidity and physical movement
disorders, such as motor retardation, abnormal posture and gait, as
well as non-motor symptoms, including depression and cognitive
impairment (2). The condition of
patients with PD progressively worsens, which seriously affects the
daily quality of life and sociability of patients (3). An article indicates that, among the
degenerative diseases of the neurodegenerative system, PD is the
most common disease in the middle-aged and elderly population
worldwide, second to Alzheimer's disease (4). The average incidence rate estimated
in developed countries is 14/100,000 people per year (5). Although the exact pathogenesis of PD
is not yet fully understood, various possible mechanisms have been
proposed, including oxidative stress, neuroinflammation,
mitochondrial dysfunction and ubiquitin proteasome system
dysfunction (6). At present, the
primary treatment for PD is to increase the concentration of
dopamine (DA) or directly stimulate drug-dependent DA receptors to
improve symptoms (7). However,
these treatments cannot prevent the progression of PD (8). Therefore, investigating the
pathogenesis and treatment of PD is of importance.
Tyrosine hydroxylase (TH), a type of monooxygenase,
is the rate-limiting enzyme that catalyzes the first step of the
reaction in the synthesis of DA (9). In the body, L-tyrosine is catalyzed
by TH to produce levodopa, which is then catalyzed by aromatic
decarboxylase to decarboxylate and finally forms DA. Due to the
important position of TH in the synthesis of DA, its absence or
reduced expression directly affects the synthesis and secretion of
DA, thus leading to PD (10). A
previous study revealed that forkhead box A1 (FOXA1) can maintain
dopaminergic properties, and the loss of FOXA1/2 can lead to the
downregulation of TH (11). FOXA1
is a member of the Fox transcription factor family and is expressed
in multiple human tissues, such as the digestive, urinary and
reproductive systems (12,13). It is also widely expressed in the
hippocampus, the region where the formation of new neurons occurs
in the brain (14). A previous
study has shown that FOXA1 can control the differentiation of
midbrain DA neurons by regulating the expression of genes that are
pivotal for neural differentiation (15). Furthermore, it was found that FOXA1
expression was significantly reduced in the serum samples of 30
Indian patients with PD (16).
To explore the potential role of FOXA1 in PD, the
present study collected blood samples from patients with PD to
determine the expression level of FOXA1. Mouse dopaminergic neuron
cells (MES23.5) were induced with 6-hydroxydopamine (6-OHDA) to
construct an in vitro PD model in order to study the effect
of FOXA1 on dopamine neuron damage, as well as the underlying
mechanism.
Materials and methods
Clinical samples
The present study was approved by the Ethics
Committee of Lianyungang Oriental Hospital Affiliated to Xuzhou
Medical University (approval no. KY-2020-003-01). All procedures
were performed in accordance with the 1964 Declaration of Helsinki
and its later amendments. The patients or their guardians (for
patients with reduced brain capacity) were informed of the project
and provided written informed consent. Blood samples were collected
from 15 patients with PD (nine male patients and six female; age,
63.4±7.76 years) and 15 healthy patients (controls; 11 male
patients and four female patients; age, 63.07±9.56 years) who were
admitted to the Lianyungang Oriental Hospital Affiliated to Xuzhou
Medical University (Lianyungang, China) between April 2020 and
August 2020. Inclusion criteria for patients with PD were as
follows: i) Clinical features of the patient were in line with the
clinical diagnostic criteria of the Parkinson's Society of England
Brain Bank; and ii) PD was clearly diagnosed by two physicians at
or above the deputy director level. The exclusion criteria for
patients with PD were as follows: i) Cerebrovascular disease; ii)
Parkinson's syndrome; iii) family-related PD; iv) Parkinson's
superimposed syndrome; and v) hepatolenticular degeneration caused
by external factors, such as drugs, encephalitis, and brain injury.
A total of 5 ml fasting venous blood was drawn from the study
subjects and placed into tubes containing anticoagulant.
Subsequently, total RNA was extracted from these samples, as
previously described (16).
Cell culture and transfection
MES23.5 cells were purchased from American Type
Culture Collection and cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% Penicillin-Streptomycin. Cells were grown
at 37˚C with a 5% CO2/95% air atmosphere. 6-OHDA (purity
≥97%; Merck & Co., Inc.) was dissolved in sterile deionized
water to prepare a 60 mmol/l stock solution, which was stored at
-20˚C in the dark. MES23.5 cells pretreated with 6-OHDA for 24 h
served as the 6-OHDA group, whereas untreated cells served as the
control group.
MES23.5 cells were plated into 10-cm dishes
(2x106 cells/dish) and then transfected with 25 nM
pcDNA3.1-FOXA1 vector plasmid [overexpression (Ov)-FOXA1; Hunan
Fenghui Biotechnology Co., Ltd.], empty vector plasmid (Ov-NC;
Hunan Fenghui Biotechnology Co., Ltd.), 100 nM small interfering
RNA (siRNA) targeted against trefoil factor 1 (TFF1; siRNA-TFF1-1,
5'-GGCCCAGGAAGAAACATGTAT-3'; siRNA-TFF1-2,
5'-GGCCATCGAGAACACTCAAGA-3'; Genepharm Biotech Corp.) or scrambled
siRNA (siRNA-NC, 5'-GAAGACATCCTGCGGAAGTAA-3'; Genepharm Biotech
Corp.) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37˚C according to the manufacturer's
instructions. Following 48 h of transfection, the expression levels
of FOXA1 and TFF1 were determined.
Reverse transcription-quantitative PCR
(RT-qPCR)
MES23.5 cells (2x106) were suspended and
homogenized in 0.75 ml TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) for 10 min on ice. The quantity of
total RNA was measured using a NanoDrop spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). Total RNA was
reverse transcribed into cDNA using a TaqMan™ RT kit
(cat. no. N8080234; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was performed using
SYBR Green PCR Master Mix (Beijing Solarbio Science &
Technology Co., Ltd.) on an ABI PRISM 7300 Sequence Detection
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 95˚C for 5 min; followed
by 38 cycles of 95˚C for 15 sec, 55˚C for 30 sec and 72˚C for 90
sec. The relative mRNA expression levels of each specific gene were
quantified using the 2-ΔΔCq method (17) and normalized to the internal
reference gene GAPDH. The sequences of the primers used for RT-qPCR
are listed in Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5'-3') |
|---|
| FOXA1 | F:
TGTGTATTCCAGACCCGTGC |
| | R:
AGGGGAAGGAGTGAAAGGGA |
| TNF-α | F:
AGGCACTCCCCCAAAAGATG |
| | R:
TGGTGGTTTGTGAGTGTGAGG |
| IL-1β | F:
TGCCACCTTTTGACAGTGATG |
| | R:
ATGTGCTGCTGCGAGATTTG |
| IL-6 | F:
GCCTTCTTGGGACTGATGCT |
| | R:
GTGACTCCAGCTTATCTCTTGGT |
| TFF1 | F:
CCATGGCCATCGAGAACACT |
| | R:
GGGGTTGAACTGTGTCACCA |
| TH | F:
TACTTTGTGCGCTTCGAGGT |
| | R:
TGGGTAGCATAGAGGCCCTT |
| GADPH | F:
GGGTCCCAGCTTAGGTTCAT |
| | R:
ATCCGTTCACACCGACCTTC |
Western blotting
MES23.5 cells (2x106) were lysed using
RIPA buffer (cat. no. P0013C; Beyotime Institute of Biotechnology)
for 10 min on ice, and total protein was quantified using a BCA
protein assay kit (cat. no. P0012; Beyotime Institute of
Biotechnology). Protein samples (25 µg/lane) were subjected to 10%
SDS-PAGE and then transferred onto PVDF membranes. After blocking
in 5% fat-free milk for 2 h at room temperature, the membranes were
incubated at 4˚C overnight with primary antibodies (all purchased
from Abcam) targeted against FOXA1 (cat. no. ab170933; 1:1,000),
TFF1 (cat. no. ab92377; 1:1,000), TH (cat. no. ab137869; 1:5,000),
Bcl-2 (cat. no. ab32124; 1:1,000), Bax (cat. no. ab32503; 1:1,000),
cleaved caspase 9 (cat. no. ab2324; 1:200), caspase 9 (cat. no.
ab202068; 1:2,000) and GAPDH (cat. no. ab9485; 1:2,500).
Subsequently, the membranes were incubated with an HRP-labelled
goat anti-rabbit secondary antibody (cat. no. ab6721; 1:5,000;
Abcam) for 2 h at room temperature. Protein bands were visualized
using an ECL kit (cat. no. P0018S; Beyotime Institute of
Biotechnology). Protein expression was semi-quantified using ImageJ
software (version 1.8; National Institutes of Health) with GAPDH as
the loading control.
Cell counting kit-8 (CCK-8) assay
MES23.5 cells (5x103 cells/well) were
seeded into a 96-well plate. Before the addition of 10 µl CCK-8
solution (Beyotime Institute of Biotechnology) to each well, the
cells were cultured for 24 h at 37˚C. Following a further
incubation for 2 h at 37˚C after the addition of CCK-8, the optical
density was measured at a wavelength of 450 nm using a microplate
reader (Molecular Devices, LLC).
Immunofluorescence
MES23.5 cells (2x104 cells/well) were
plated in 24-well plates, fixed with 4% paraformaldehyde (Macklin,
Inc.) for 1 h at room temperature, and blocked in 1% BSA (Beijing
Solarbio Science & Technology Co., Ltd.) for 0.5 h at room
temperature. Next, the cells were incubated in the presence of a
primary antibody against TH (cat. no. ab137869; 1:100; Abcam) at
4˚C overnight, followed by incubation with the Alexa
Fluor® 488-labelled goat anti-rabbit secondary antibody
(cat. no. ab150077; 1:200; Abcam) for 1 h at room temperature. The
nuclei were counterstained with DAPI for 5 min at room temperature.
The results were observed under a fluorescence microscope
(magnification, x200; Olympus Corporation).
Determination of oxidative stress
index
The level of reactive oxygen species (ROS; cat. no.
S0033S) and malondialdehyde (MDA; cat. no. S0131S), and the
activity of superoxidase dismutase (SOD; cat. no. S0101S) in
MES23.5 cells (2x106) were quantified using commercial
ELISA kits (all purchased from Beyotime Institute of Biotechnology)
according to the manufacturer's instructions. The optical density
was measured at a wavelength of 525 nm for ROS, 550 nm for SOD and
532 nm for MDA using a microplate reader (Molecular Devices,
LLC).
TUNEL assay
MES23.5 cells (2x104 cells/well) were
seeded into a 24-well plate. Subsequently, the TUNEL assay was
performed using a TUNEL assay kit (cat. no. C1086; Beyotime
Institute of Biotechnology) according to the manufacturer's
instructions. The results were observed under a fluorescence
microscope (magnification, x200; Olympus Corporation) at five
fields of view.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was performed with a ChIP assay kit (cat.
no. P2078; Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Briefly, MES23.5 cells
(1x106) were cross-linked with 1% formaldehyde followed
by centrifugation at 1,000 x g for 1 min at 4˚C. The precipitated
cells were lysed with SDS lysis buffer containing 1 mM PMSF for 10
min, and sonicated in an ice bath. Cell lysates (2 ml) were
incubated with 70 µl Protein A + G agarose for 30 min at 4˚C before
incubation with 60 µl Protein A + G beads coated with 1 µg
anti-FOXA1 antibody (cat. no. ab170933; Abcam) for 1 h at 4˚C.
Anti-rabbit IgG (cat. no. 172730; Abcam) served as a NC. After the
incubation, the sample was centrifuged at 1,000 x g for 1 min at
4˚C and the precipitate was collected. Cross-linked DNA released
from the precipitate was purified using a DNA purification kit
(cat. no. D0033; Beyotime Institute of Biotechnology) according to
the manufacturer's instructions. The eluted DNA was subjected to
RT-qPCR according to the aforementioned protocol.
Luciferase reporter assay
The TFF1 promoter region containing a FOXA1 putative
binding site [wild-type (WT), 5'-AGGTCACGGTGGCCAC-3') or mutant
(MUT) site (5'-TCCAGTGCCACCGGTG-3')] was cloned into the pGL3-based
vector (BioVector NTCC, Inc.). MES23.5 cells were seeded
(1x105 cells/well) into 24-well plates and
co-transfected with 0.5 µg aforementioned reporter vector and 0.5
µg Ov-FOXA1 or Ov-NC vectors using Lipofectamine® 2000
reagent (Thermo Fisher Scientific, Inc.) at room temperature.
Following 48 h of transfection, the relative luciferase activity
was normalized to Renilla luciferase and measured using a
dual-luciferase reporter assay system (Promega Corporation)
according to the manufacturer's instructions.
Statistical analysis
Data are presented as the mean ± SD from at least
three independent experiments. Statistical analyses were performed
using GraphPad Prism 8.0 software (GraphPad Software, Inc.).
Statistical differences between two groups were analyzed using an
unpaired Student's t-test. Comparisons among multiple groups were
analyzed using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
FOXA1 overexpression enhances
6-OHDA-induced MES23.5 cell viability and decreases TH
expression
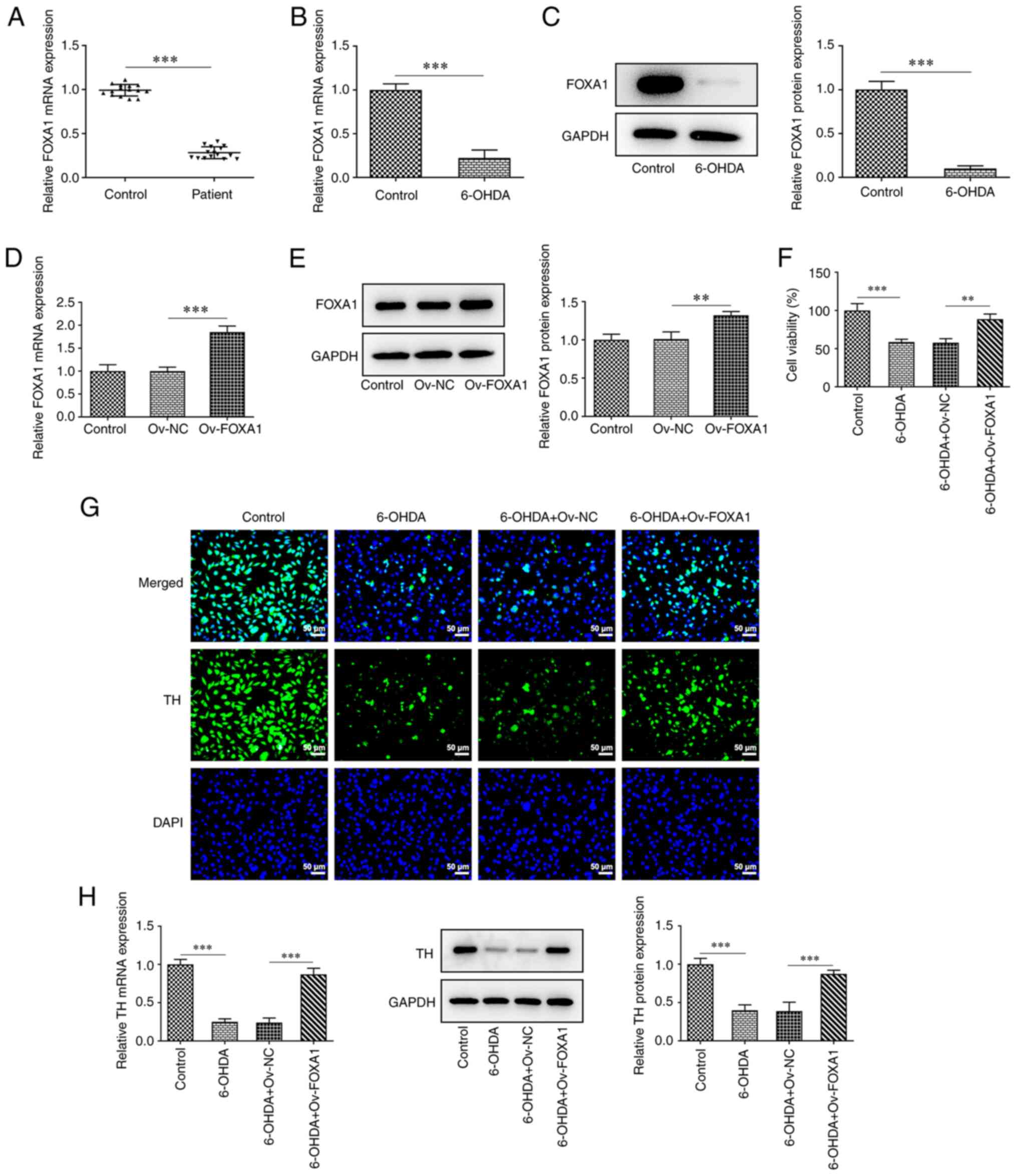
The mRNA expression levels of FOXA1 in the samples
from patients with PD and healthy subjects were determined using
RT-qPCR. The results revealed that the expression levels of FOXA1
in patients with PD were significantly lower compared with those in
the controls (Fig. 1A).
Subsequently, the expression levels of FOXA1 in
MES23.5 cells and pretreated cells (6-OHDA group) were determined
using RT-qPCR and western blotting. FOXA1 expression was
significantly downregulated in the 6-OHDA group compared with that
in the control group (Fig. 1B and
C).
A FOXA1 overexpression plasmid was constructed to
explore the role of FOXA1 in MES23.5 cells, and the transfection
efficiency was verified by RT-qPCR and western blotting (Fig. 1D and E). The expression level of FOXA1 in the
Ov-FOXA1 group was significantly upregulated compared with that in
the Ov-NC group. Subsequently, MES23.5 cells were divided into four
groups: i) Control; ii) 6-OHDA; iii) 6-OHDA + Ov-NC; and iv) 6-OHDA
+ Ov-FOXA1. Cell viability was assessed using the CCK-8 assay. The
results revealed that cell viability in the 6-OHDA group was
significantly downregulated compared with that in the control
group, whereas FOXA1 overexpression significantly attenuated this
decrease (Fig. 1F).
As aforementioned, the absence of TH can affect the
formation of DA (10); therefore,
the expression of TH in these groups was evaluated using
immunofluorescence, RT-qPCR and western blotting. The staining
results revealed that the fluorescence intensity of the 6-OHDA
group was markedly lower compared with that in the control group
(Fig. 1G). FOXA1 overexpression
notably increased the fluorescence intensity compared with that in
the 6-OHDA + Ov-NC group, suggesting that FOXA1 overexpression
could alleviate the downregulation of TH expression induced by
6-OHDA. In addition, the RT-qPCR and western blotting results also
indicated that FOXA1 overexpression could significantly inhibit
6-OHDA-induced downregulation of TH expression (Fig. 1H).
FOXA1 overexpression attenuates
6-OHDA-induced inflammation, oxidative stress and apoptosis in
MES23.5 cells
Cells were grouped as previously described, and the
effect of FOXA1 overexpression on cell inflammation and oxidative
stress was subsequently determined. The mRNA expression levels of
inflammatory factors, including TNF-α, IL-1β and IL-6, were
determined using RT-qPCR (Fig.
2A). The expression of these molecules was significantly
increased in the 6-OHDA group compared with that in the control
group, whereas FOXA1 overexpression significantly reversed these
effects. Furthermore, the levels of SOD, ROS and MDA were
determined using ELISA kits. The results demonstrated that the
level of SOD was significantly decreased in the 6-OHDA group
compared with that in the control group, but significantly
increased after FOXA1 overexpression (Fig. 2B). The levels of ROS and MDA were
significantly increased in the 6-OHDA group compared with those in
the control group, whereas FOXA1 overexpression significantly
decreased these levels. Furthermore, cell apoptosis was determined
using a TUNEL assay and western blotting. The fluorescence of
apoptotic cells was more intense in the 6-OHDA group compared with
that in the control group, and FOXA1 overexpression significantly
downregulated the fluorescence intensity (Fig. 2C). The western blotting results
showed that the protein expression levels of Bcl-2 were
significantly decreased in the 6-OHDA group compared with those in
the control group, which was accompanied by significantly increased
expression levels of Bax and cleaved caspase 9 (Fig. 2D). FOXA1 overexpression
significantly alleviated the effects of 6-OHDA on Bcl-2, Bax and
cleaved caspase 9 expression.
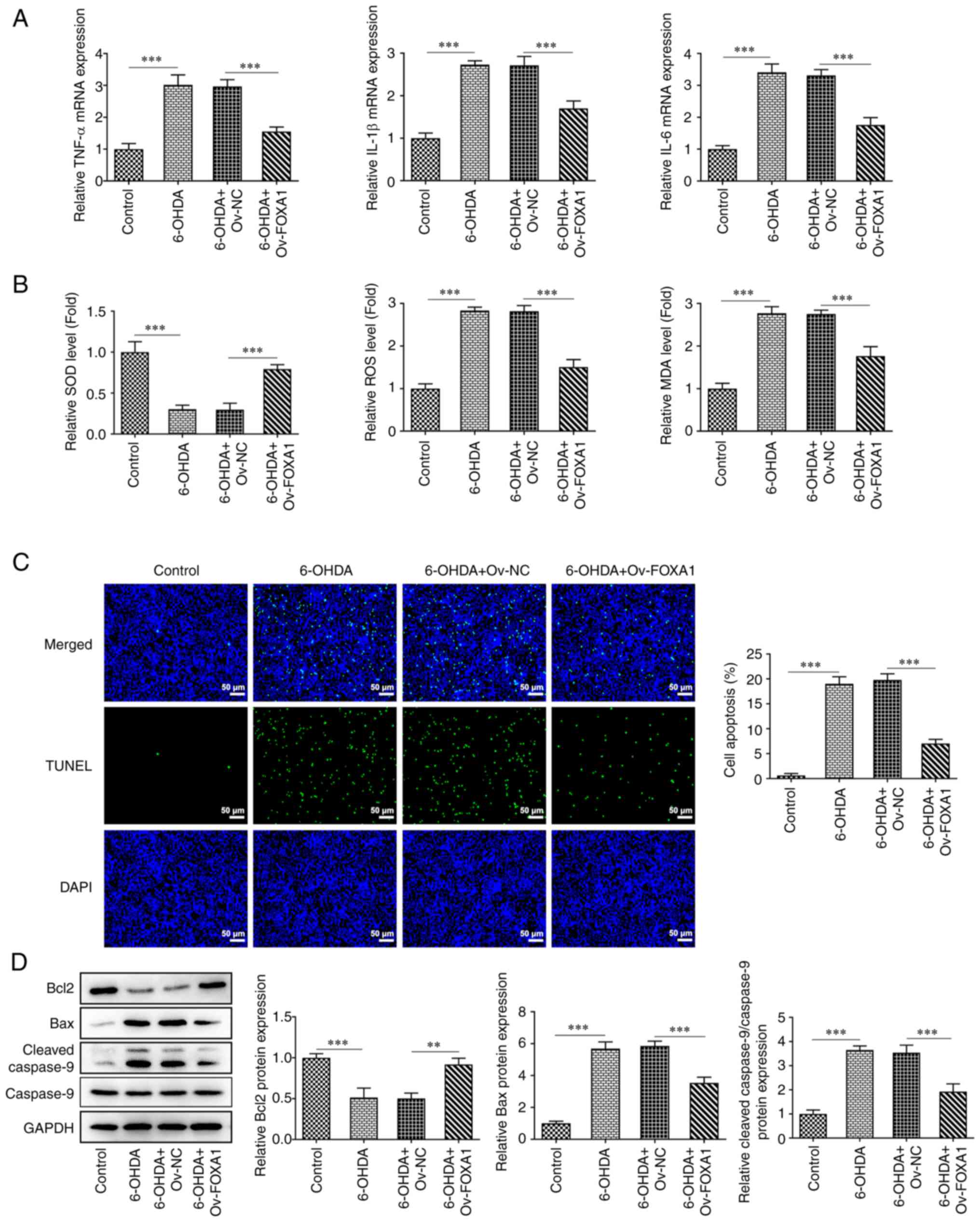 | Figure 2FOXA1 overexpression attenuates
6-OHDA-induced inflammation, oxidative stress and apoptosis in
MES23.5 cells. (A) mRNA expression levels of inflammatory factors,
including TNF-α, IL-1β and IL-6, were measured using reverse
transcription-quantitative PCR. (B) Expression levels of SOD, ROS
and MDA were determined using ELISA kits. Cell apoptosis was
determined using a (C) TUNEL assay (magnification, x200) and (D)
western blotting. **P<0.01 and
***P<0.001. SOD, superoxidase dismutase; ROS,
reactive oxygen species; MDA, malondialdehyde; Ov, overexpression;
NC, negative control; FOXA1, forkhead box A1; 6-OHDA,
6-hydroxydopamine. |
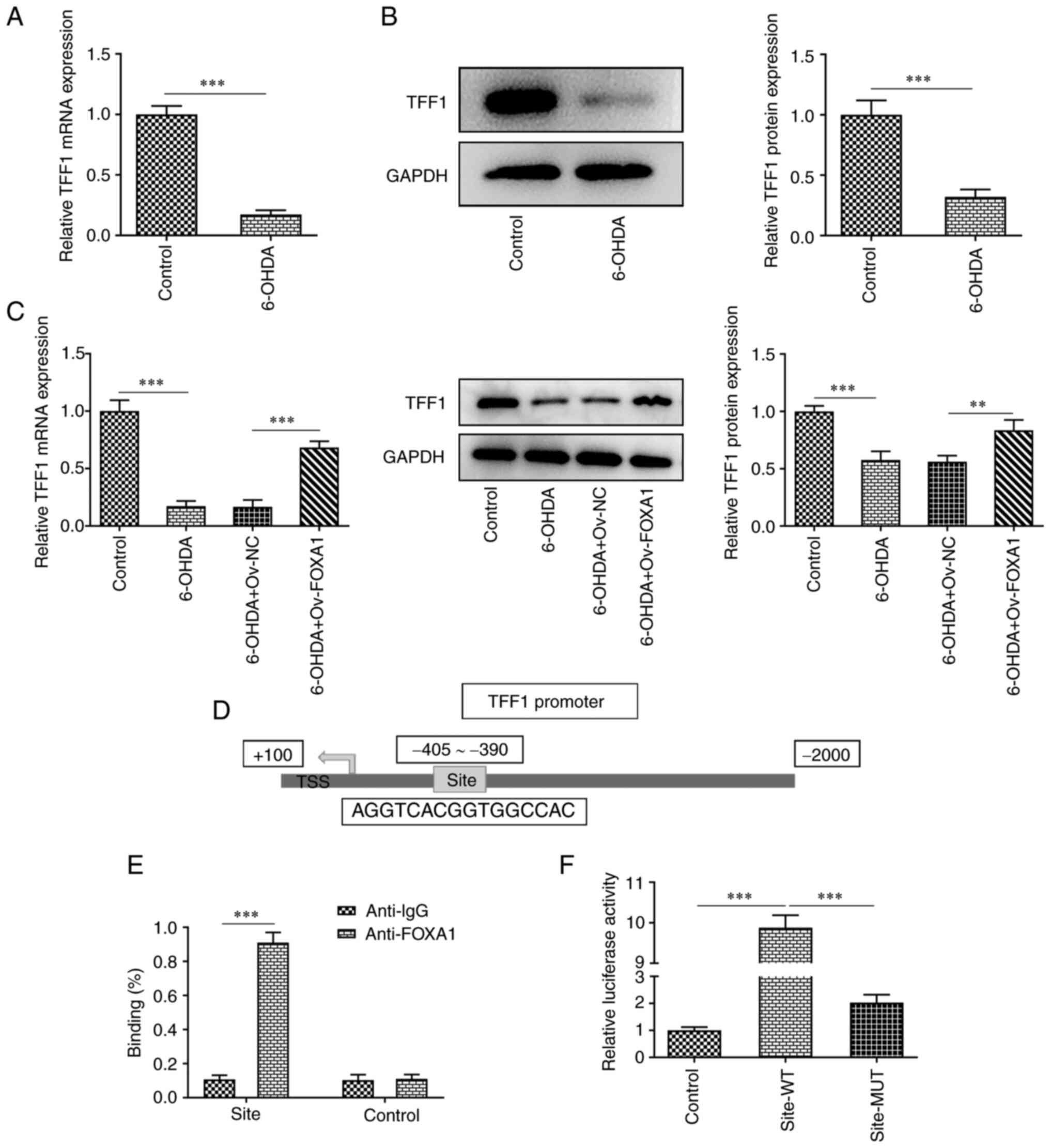
FOXA1 overexpression activates TFF1 in
6-OHDA-induced MES23.5 cells
It has been reported that TFF1 co-localizes with TH
in midbrain neurons in rats, and TFF1-positive neurons in a
6-OHDA-induced model were dead (18). Therefore, in the present study, the
expression levels of TFF1 in MES23.5 cells were determined using
RT-qPCR and western blotting. The results revealed that TFF1
expression was significantly decreased in the 6-OHDA group compared
with that in the control group (Fig.
3A and B). FOXA1
overexpression significantly increased the expression levels of
TFF1 in 6-OHDA-induced MES23.5 cells (Fig. 3C). In addition, FOXA1 was
considered to be able to bind to the site 405-390 bp of the TFF1
promoter (Fig. 3D). Therefore, a
ChIP assay was performed to verify the interaction of FOXA1 with
TFF1 promoter, and the results indicated that FOXA1 could bind to
the promoter of TFF1 (Fig. 3E).
Next, the binding site was mutated, and luciferase activities were
measured using a luciferase reporter assay. The activity of the WT
group was significantly higher compared with that of the MUT group,
indicating that FOXA1 did bind to the aforementioned site (Fig. 3F).
TFF1 mediates the mechanism of FOXA1
overexpression in 6-OHDA-induced MES23.5 cells
To study the role of TFF1 in MES23.5 cells, TFF1 was
silenced via siRNA, and its expression was determined using RT-qPCR
and western blotting. The results revealed that the expression
levels of the siRNA-TFF1-1 group were lower compared with those of
the siRNA-TFF1-2 group; thus, the siRNA-TFF1-1 group was selected
for subsequent experiments (Fig.
4A and B). MES23.5 cells were
divided into the following five groups: i) Control; ii) 6-OHDA;
iii) 6-OHDA + Ov-FOXA1; iv) 6-OHDA + Ov-FOXA1 + siRNA-NC; and v)
6-OHDA + Ov-FOXA1 + siRNA-TFF1. Cell viability of these groups was
assessed using a CCK-8 assay, and the results revealed that the
cell viability of the siRNA-TFF1 group was significantly reduced
compared with that of the siRNA-NC group, indicating that TFF1
silencing reversed the increase in cell viability induced by FOXA1
overexpression (Fig. 4C). Next,
the expression of TH in these groups was evaluated using
immunofluorescence, RT-qPCR and western blotting. The expression of
TH was significantly decreased after TFF1 silencing, which
indicated that TFF1 silencing could reverse the effect of FOXA1
overexpression on TH expression (Fig.
4D-F). Furthermore, the effect of TFF1 silencing on cell
inflammation was assessed by measuring the levels of TNF-α, IL-1β
and IL-6. The RT-qPCR results suggested that TFF1 silencing could
significantly elevate the levels of inflammatory cytokines in
6-OHDA-induced cells overexpressing FOXA1, which is equivalent to
blocking the inhibitory effect of FOXA1 overexpression on cell
inflammation (Fig. 5A). TFF1
silencing significantly reversed FOXA1 overexpression-induced
alterations in the activity of SOD, ROS and MDA, which indicated
that TFF1 could mediate oxidative stress (Fig. 5B). The effect of TFF1 silencing on
cell apoptosis was determined using a TUNEL assay and western
blotting. The fluorescence intensity in the siRNA-TFF1 group was
enhanced compared with that of the siRNA-NC group (Fig. 5C). Moreover, the expression levels
of Bax and cleaved caspase 9 were significantly increased and that
of Bcl2 was decreased in the siRNA-TFF1 group compared with those
in the siRNA-NC group (Fig. 5D).
These findings suggested that TFF1 silencing reversed the effect of
FOXA1 overexpression on cell apoptosis.
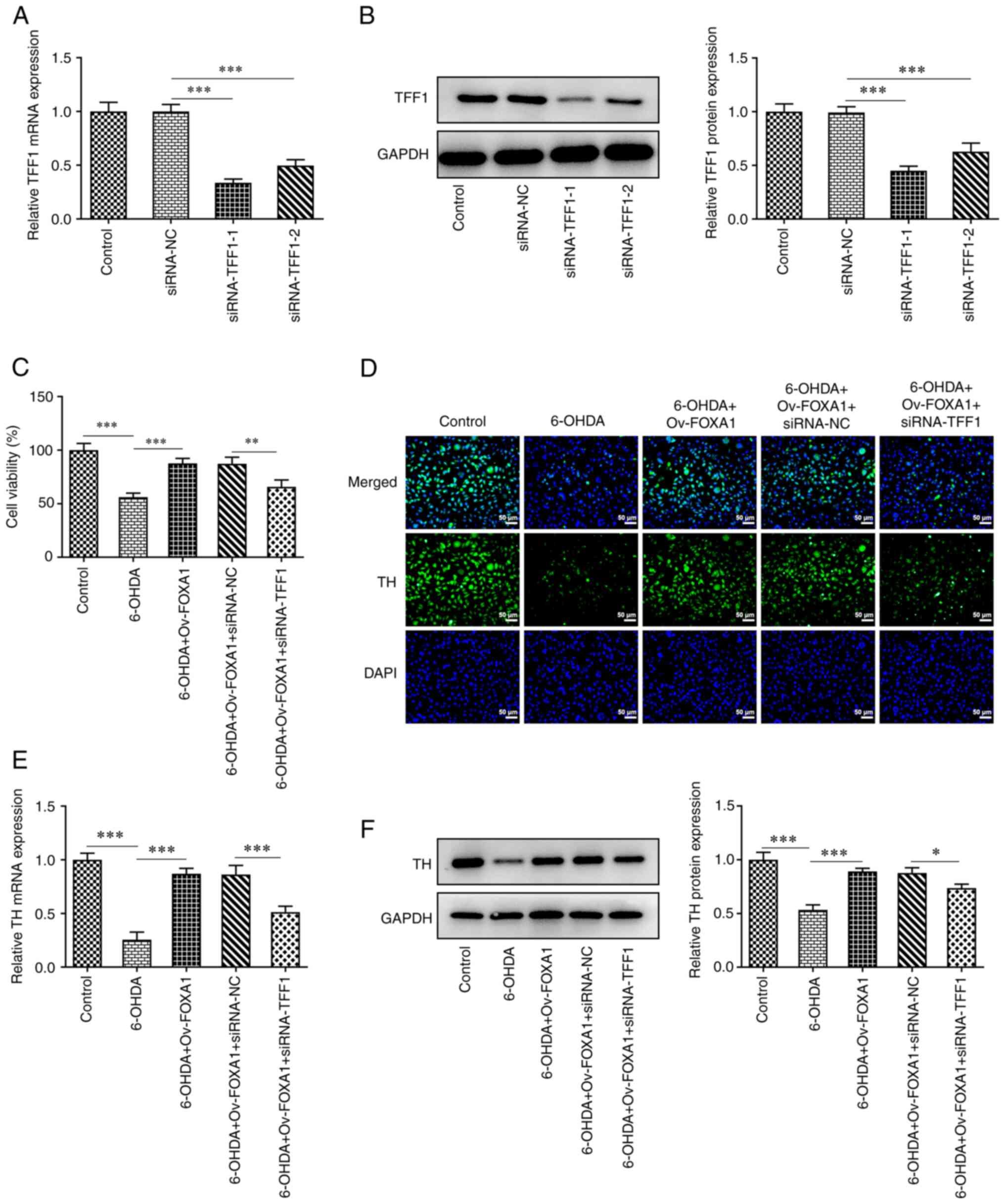 | Figure 4TFF1 mediates the mechanism of FOXA1
overexpression in 6-OHDA-induced MES23.5 cell viability and TH
expression. TFF1 silencing was verified using (A) RT-qPCR and (B)
western blotting. (C) Cell viability in MES23.5 cells was assessed
using a Cell Counting Kit-8 assay. Expression levels of TH in
MES23.5 cells was evaluated using (D) immunofluorescence
(magnification, x200), (E) RT-qPCR and (F) western blotting.
*P<0.05, **P<0.01 and
***P<0.001. TFF1, trefoil factor 1; RT-qPCR, reverse
transcription-quantitative PCR; TH, tyrosine hydroxylase; siRNA,
small interfering RNA; NC, negative control; FOXA1, forkhead box
A1; 6-OHDA, 6-hydroxydopamine. |
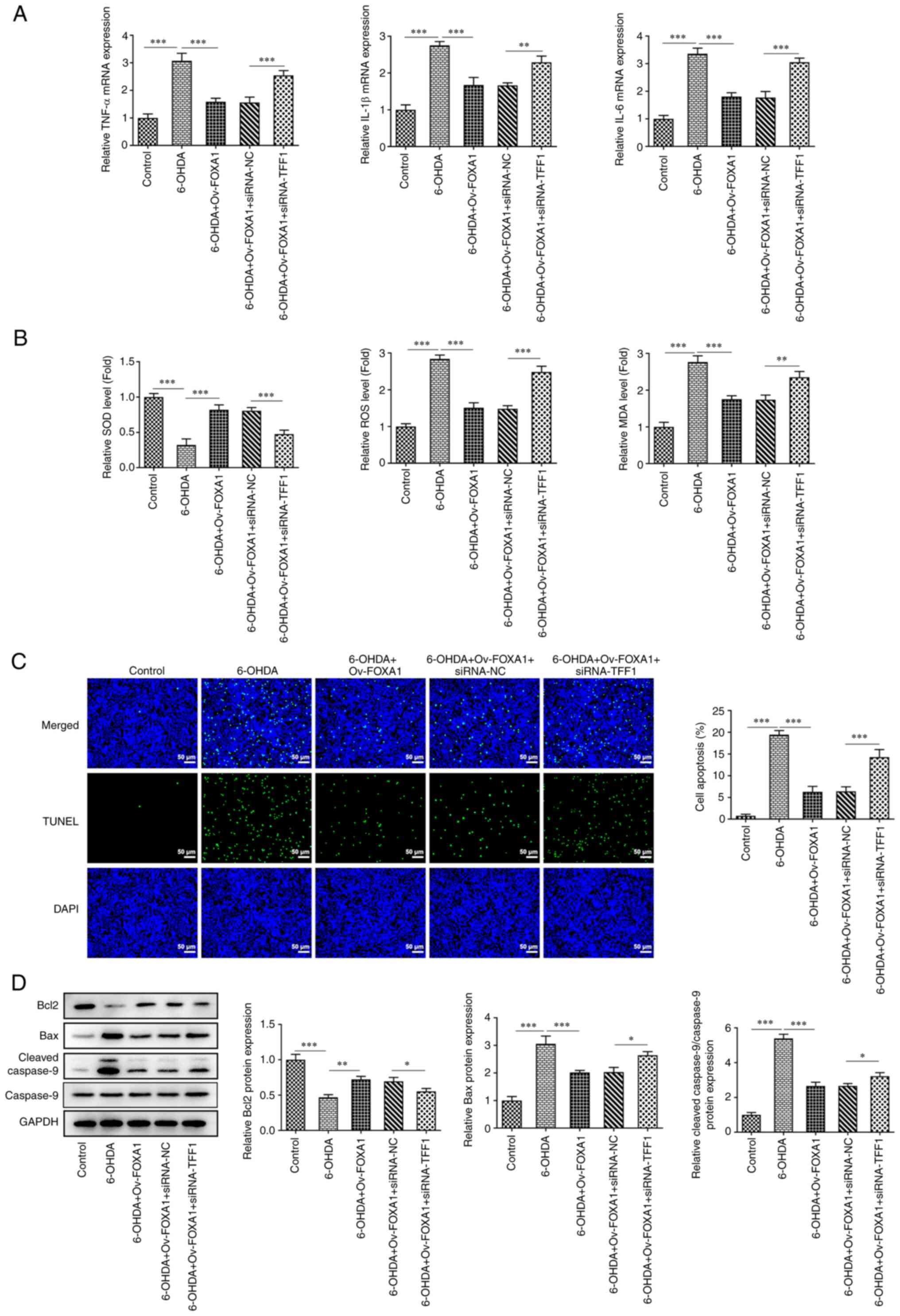 | Figure 5TFF1 mediates the mechanism of FOXA1
overexpression in inflammation, oxidative stress and apoptosis in
MES23.5 cells. (A) mRNA expression levels of inflammatory factors,
including TNF-α, IL-1β and IL-6, were measured using reverse
transcription-quantitative PCR. (B) Expression levels of SOD, ROS
and MDA were determined using ELISA kits. Cell apoptosis was
determined using a (C) TUNEL assay (magnification, x200) and (D)
western blotting. *P<0.05, **P<0.01 and
***P<0.001. SOD, superoxidase dismutase; ROS,
reactive oxygen species; MDA, malondialdehyde; Ov, overexpression;
FOXA1, forkhead box A1; siRNA, small interfering RNA; NC, negative
control; TFF1, trefoil factor 1; 6-OHDA, 6-hydroxydopamine. |
Discussion
Numerous studies on the pathogenesis of PD have
confirmed that neuroinflammation and oxidative stress participate
in, and promote the occurrence and progression of PD (19-21).
In the central nervous system, glial cells can be activated by
pathogen- and injury-related molecular patterns, leading to
persistent neuroinflammation (22). In addition, since the metabolic
activity of neurons is markedly high, the demand for energy and the
consumption of oxygen are high, which leads to the generation of
large quantities of ROS. By contrast, the content of antioxidants
in neurons is relatively low, which increases the risk of oxidative
stress (23). In the present
study, FOXA1 overexpression decreased the levels of inflammatory
factors in 6-OHDA-induced MES23.5 cells. The MES23.5 cell line is a
dopaminergic neuroblastoma, which displays the characteristics of
dopaminergic neurons, and its cultivation is easier than that of
midbrain nerve cells. Therefore, this cell line has been widely
used as a tool for studying neurodegenerative diseases (24). Considering that excessive ROS is
the cause of oxidative stress (25) and that MDA is a metabolite of
oxygen free radicals (26), the
levels of ROS and MDA were assessed in the present study.
Due to the development of novel experimental
techniques, the study of the regulatory role of transcription
factors in eukaryotes has attracted considerable attention. Through
studying the specific functions and mechanisms of various
transcription factors in transcriptional regulation, various drug
molecules have been designed, which possess the ability to inhibit
or activate transcription factors, thereby altering gene expression
patterns (27,28). This kind of strategy may have
important application prospects for the development of treatments.
In the present study, the role of the transcription factor FOXA1 in
DA nerve cells was evaluated. The results suggested that FOXA1
overexpression may reduce cell inflammation, oxidative stress and
apoptosis. Importantly, FOXA1 overexpression could increase the
expression of the marker TH, which implies that it can reduce the
loss of dopaminergic neurons. In previous studies, FOXA1/2 was also
considered to play an important role in the central nervous system.
It has been reported that FOXA1/2 participates in the early
formation, late development and function of dopaminergic neurons
after maturation, and it is necessary for maintaining the normal
neuronal firing activity of dopaminergic neurons in the SNc
(29,30).
TFF1 belongs to the trefoil family of proteins and
is primarily found to be stably expressed in the gastrointestinal
mucosa, which can protect the mucosa from damage, stabilize the
mucous layer and promote epithelial healing (31,32).
In the present study, it was considered that TFF1 was involved in
the FOXA1-mediated regulation of dopaminergic neuron cells. Upon
FOXA1 transcription, the expression of TFF1 is activated, which
reduces the damage of 6-OHDA to neurons. Notably, there are several
studies on TFF1 in human tumors; for example, TFF1 is considered to
inhibit the function of the IL-6/STAT3 proinflammatory signaling
axis in inhibiting gastric tumorigenesis (33). TFF1 is generally upregulated in the
serum of patients with breast cancer; it is considered to be
relevant to estrogen receptor level and it can serve as a
prognostic marker, particularly for non-triple-negative breast
cancer (34). However, it is worth
noting that TFF1 is reported to be a neuropeptide and is
co-expressed with TH in dopaminergic neurons (18). The present results further
indicated that TFF1 may play a role in neurons. Notably, the
present study exhibits certain limitations, including the fact that
experiments were only conducted at the cellular level; thus, future
in vivo studies in mice should be conducted. In addition,
the association between FOXA1 expression and individual disease
duration should be investigated in future studies.
In conclusion, the expression level of FOXA1 was
downregulated in patients with PD, and FOXA1 overexpression
attenuated 6-OHDA-induced inflammation, oxidative stress and
apoptosis in MES23.5 cells. Furthermore, FOXA1 overexpression
activated TFF1, indicating that TFF1 mediated the mechanism of
FOXA1 overexpression in MES23.5 cells. The present findings
suggested that FOXA1 may serve as a target for the treatment of PD
and contribute to the development of targeted drugs.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Key Project of Bengbu Medical College (grant no. 2020byzd389).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TL, PZ and HC designed the study. TL, PZ, XZ, XH and
LL performed the experiments. LL made considerable contributions to
the drafting of the manuscript. HC revised the manuscript for
important intellectual content. BH and LK collected the clinical
data and analyzed the data. All authors read and approved the final
manuscript. TL and LL confirm the authenticity of the raw data.
Ethics approval and consent to
participate
All procedures were performed in accordance with the
Declaration of the Institutional Research Committee's Ethical
standards, as well as the 1964 Declaration of Helsinki and its
later amendments. The present study was approved by the Ethics
Committee of Lianyungang Oriental Hospital Affiliated to Xuzhou
Medical University (approval no. KY-2020-003-01). All patients or
their parents/guardians provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Balestrino R and Schapira AHV: Parkinson
disease. Eur J Neurol. 27:27–42. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Salari S and Bagheri M: In vivo, in vitro
and pharmacologic models of Parkinson's disease. Physiol Res.
68:17–24. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Armstrong MJ and Okun MS: Diagnosis and
treatment of Parkinson disease: A review. JAMA. 323:548–560.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Aarsland D, Batzu L, Halliday GM, Geurtsen
GJ, Ballard C, Ray Chaudhuri K and Weintraub D: Parkinson
disease-associated cognitive impairment. Nat Rev Dis Primers.
7(47)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
López-Liria R, Parra-Egeda J, Vega-Ramírez
FA, Aguilar-Parra JM, Trigueros-Ramos R, Morales-Gázquez MJ and
Rocamora-Pérez P: Treatment of dysphagia in Parkinson's Disease: A
systematic review. Int J Environ Res Public Health.
17(4104)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sodhi RK, Bansal Y, Singh R, Saroj P,
Bhandari R, Kumar B and Kuhad A: IDO-1 inhibition protects against
neuroinflammation, oxidative stress and mitochondrial dysfunction
in 6-OHDA induced murine model of Parkinson's disease.
Neurotoxicology. 84:184–197. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
De Virgilio A, Greco A, Fabbrini G,
Inghilleri M, Rizzo MI, Gallo A, Conte M, Rosato C, Ciniglio
Appiani M and de Vincentiis M: Parkinson's disease: Autoimmunity
and neuroinflammation. Autoimmun Rev. 15:1005–1011. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Xu X, Fu Z and Le W: Exercise and
Parkinson's disease. Int Rev Neurobiol. 147:45–74. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Dunkley PR and Dickson PW: Tyrosine
hydroxylase phosphorylation in vivo. J Neurochem. 149:706–728.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang X, Zhu Z, Ding X, Wang X, Cui G, Hua
F and Xiang J: CaMKII inhibition ameliorated levodopa-induced
dyskinesia by downregulating tyrosine hydroxylase activity in an
experimental model of Parkinson's disease. Brain Res. 1687:66–73.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pristerà A, Lin W, Kaufmann AK,
Brimblecombe KR, Threlfell S, Dodson PD, Magill PJ, Fernandes C,
Cragg SJ and Ang SL: Transcription factors FOXA1 and FOXA2 maintain
dopaminergic neuronal properties and control feeding behavior in
adult mice. Proc Natl Acad Sci USA. 112:E4929–E4938.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Horisawa K, Udono M, Ueno K, Ohkawa Y,
Nagasaki M, Sekiya S and Suzuki A: The dynamics of transcriptional
activation by hepatic reprogramming factors. Mol Cell.
79:660–676.e8. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gredler ML, Patterson SE, Seifert AW and
Cohn MJ: Foxa1 and Foxa2 orchestrate development of the urethral
tube and division of the embryonic cloaca through an autoregulatory
loop with Shh. Dev Biol. 465:23–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ruiz i Altaba A, Palma V and Dahmane N:
Hedgehog-Gli signalling and the growth of the brain. Nat Rev
Neurosci. 3:24–33. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Yu Q, Chen J, Deng W, Cao X, Adu-Frimpong
M, Yu J and Xu X: Neural differentiation of fibroblasts induced by
intracellular co-delivery of Ascl1, Brn2 and FoxA1 via a non-viral
vector of cationic polysaccharide. Biomed Mater.
13(015022)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tippabathani J, Nellore J, Radhakrishnan
V, Banik S and Kapoor S: Identification of NURR1 (Exon 4) and FOXA1
(Exon 3) haplotypes associated with mRNA expression levels in
peripheral blood lymphocytes of Parkinson's patients in small
indian population. Parkinsons Dis. 2017(6025358)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jensen P, Heimberg M, Ducray AD, Widmer HR
and Meyer M: Expression of trefoil factor 1 in the developing and
adult rat ventral mesencephalon. PLoS One. 8(e76592)2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen Z and Trapp BD: Microglia and
neuroprotection. J Neurochem. 136 (Suppl 1):S10–S17.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Puspita L, Chung SY and Shim JW: Oxidative
stress and cellular pathologies in Parkinson's disease. Mol Brain.
10(53)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Vivekanantham S, Shah S, Dewji R, Dewji A,
Khatri C and Ologunde R: Neuroinflammation in Parkinson's disease:
Role in neurodegeneration and tissue repair. Int J Neurosci.
125:717–725. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pajares M, I Rojo A, Manda G, Boscá L and
Cuadrado A: Inflammation in Parkinson's disease: Mechanisms and
therapeutic implications. Cells. 9(1687)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Feuerstein D, Backes H, Gramer M, Takagaki
M, Gabel P, Kumagai T and Graf R: Regulation of cerebral metabolism
during cortical spreading depression. J Cereb Blood Flow Metab.
36:1965–1977. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Deng H, Liu S, Pan D, Jia Y and Ma ZG:
Myricetin reduces cytotoxicity by suppressing hepcidin expression
in MES23.5 cells. Neural Regen Res. 16:1105–1110. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Taysi S, Tascan AS, Ugur MG and Demir M:
Radicals, oxidative/nitrosative stress and preeclampsia. Mini Rev
Med Chem. 19:178–193. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Islam MT: Oxidative stress and
mitochondrial dysfunction-linked neurodegenerative disorders.
Neurol Res. 39:73–82. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Papavassiliou KA and Papavassiliou AG:
Transcription factor drug targets. J Cell Biochem. 117:2693–2696.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lambert M, Jambon S, Depauw S and
David-Cordonnier MH: Targeting transcription factors for cancer
treatment. Molecules. 23(1479)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lin W, Metzakopian E, Mavromatakis YE, Gao
N, Balaskas N, Sasaki H, Briscoe J, Whitsett JA, Goulding M,
Kaestner KH and Ang SL: Foxa1 and Foxa2 function both upstream of
and cooperatively with Lmx1a and Lmx1b in a feedforward loop
promoting mesodiencephalic dopaminergic neuron development. Dev
Biol. 333:386–396. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ang SL: Foxa1 and Foxa2 transcription
factors regulate differentiation of midbrain dopaminergic neurons.
Adv Exp Med Biol. 651:58–65. 2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Clyne M and May FEB: The interaction of
helicobacter pylori with TFF1 and its role in mediating the tropism
of the bacteria within the stomach. Int J Mol Sci.
20(4400)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hu J, Shi Y, Wang C, Wan H, Wu D, Wang H
and Peng X: Role of intestinal trefoil factor in protecting
intestinal epithelial cells from burn-induced injury. Sci Rep.
8(3201)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Soutto M, Chen Z, Bhat AA, Wang L, Zhu S,
Gomaa A, Bates A, Bhat NS, Peng D, Belkhiri A, et al: Activation of
STAT3 signaling is mediated by TFF1 silencing in gastric neoplasia.
Nat Commun. 10(3039)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yi J, Ren L, Li D, Wu J, Li W, Du G and
Wang J: Trefoil factor 1 (TFF1) is a potential prognostic biomarker
with functional significance in breast cancers. Biomed
Pharmacother. 124(109827)2020.PubMed/NCBI View Article : Google Scholar
|