Introduction
Adenomyosis is characterized as the abnormal
ingrowth and invagination of the basal endometrium into the
myometrium. Previous studies have documented an incidence rate of
30-50% (1). However, in infertile
women with endometriosis, the incidence rate of adenomyosis
increases to 70% (1). Therefore,
the detection of adenomyosis in patients with infertility is
important.
Extracellular vesicles (EVs) are secreted by most
cells and can be transported; they can exchange cargo between cells
as a means of intercellular communication at both the paracrine and
systemic levels (2). EVs can travel
systemically and target cells at very distant sites (3). EVs represent an information carrier
for the originating cell and transport a variety of molecules, such
as nucleic acids, cytokines, membrane-bound receptors and other
biologically active lipids and proteins (4). EV composition is largely determined by
the cell type and the physiological or pathological conditions of
the producing cell (5). Moreover,
the contents of EVs also reflect cell-specific pathological
processes with membrane protection, and EVs have great potential to
serve as circulating biomarkers that might improve current cancer
diagnosis (6). Recent research
suggests that EVs derived from adenomyosis endow endometrial
epithelial cells with an invasive phenotype via
epithelial-to-mesenchymal transition (EMT) (7). Therefore, adenomyosis-derived EVs
(AMEVs) may carry specific proteins that reveal the pathological
status of adenomyosis. Establishing these proteins as biological
biomarkers is thus pivotal for the early diagnosis of
adenomyosis.
A previous study confirmed that EVs exist in the
plasma of patients with endometriosis (8), and tissue-derived AMEVs (T-AMEVs) have
been isolated from adenomyosis lesion homogenates and blood-derived
AMEVs (B-AMEVs) from the peripheral blood of patients with
adenomyosis (7). Liquid
chromatography-mass spectrometry (LC-MS) has been used to assess
the proteins characteristics of T-AMEVs and B-AMEVs to more
accurately identify functional proteins and molecular biomarkers in
EVs for adenomyosis diagnosis (7).
The present study aimed to identify proteins in B-AMEVs as the
potential biomarkers for adenomyosis.
Materials and methods
Patients and samples
Adenomyotic lesions (n=31) were collected from
patients with adenomyosis undergoing hysterectomy in the Department
of Obstetrics and Gynecology of The First Affiliated Hospital of
Chongqing Medical University (Chongqing, China). Plasma samples of
patients with adenomyosis (n=25) and without adenomyosis (n=31,
control) were collected from The Affiliated Hospital of Zunyi
Medical University (Guizhou, China) during June 2018 to March 2021.
The patients with adenomyosis or without adenomyosis were diagnosed
by two gynecological pathologists via examining histologic sections
of surgical specimens. The recruited women were premenopausal with
regular menstrual cycles and at proliferative or secretory phase
during the procedure. Any recruited women with indication of
concomitant endometriosis, endometrial pathology or malignancy,
history of hormone therapy or intrauterine device placement within
3 months preoperatively were all excluded. The plasma samples of
patients with adenomyosis or without adenomyosis were collected
before hysterectomy and recruited as the surgical specimens of
uterus diagnosed by the gynaecological pathologist. The age
distribution of tissue group of adenomyosis were between 44 to 50
years old, and blood sample of adenomyosis group were 36 to 53
years, and blood sample of control group were 37 to 49 years
(Table I).
 | Table ICharacteristics of the patients with
adenomyosis. |
Table I
Characteristics of the patients with
adenomyosis.
|
Characteristics | Adenomyotic lesion
donors, n=31 | Blood donors,
n=25 | P-value |
|---|
| Age, years | 44.03±5.05 | 41.84±6.67 | 0.16 |
| Uterine volume,
cm3 | 546.57±373.23 | 409.30±189.17 | 0.10 |
| Age at menarche,
years | 13.45±1.29 | 13.96±1.27 | 0.15 |
| Age at first
marriage, years | 22.71±2.37 | 22.40±2.55 | 0.64 |
| Pregnancy | 3.45±1.67 | 3.60±1.85 | 0.75 |
| Parity | 1.42±0.62 | 1.60±1.12 | 0.45 |
| Dysmenorrhea, n
(%) | 21 (67.7) | 19 (76.0) | 0.42 |
| Hypermenorrhea, n
(%) | 15 (48.3) | 11 (44.0) | 0.42 |
Magnetic resonance imaging (MRI)
All patients underwent a pelvic enhanced MRI
examination using a standardized protocol prior to sample
collection (Magnetom symphony 1.5T MRI Tim system; Siemens Medical
Solutions), in order to define the size, volume and location of the
adenomyotic lesions following enhancement with gadolinium.
Haematoxylin and eosin (H&E)
staining
All collected adenomyotic tissue samples were fixed
in 10% neutral-buffered formalin overnight at room temperature,
embedded in paraffin and cut into 5-µm sections. Then, they were
stained in hematoxylin for 10 min at room temperature and
counterstained in 1% eosin solution for 4 min at room temperature
using a previously reported method (9). The H&E staining slides were
examined using a light microscope (BX51; Olympus Corporation).
Isolation of T-AMEVs and B-AMEVs from
patients with adenomyosis
T-AMEVs and B-AMEVs were isolated and purified using
a previously published protocol with some modifications (10,11).
Freshly collected adenomyosis lesions were washed in 4˚C cold PBS
(HyClone; Cytiva) and immediately homogenized between two pieces of
ground glass. The resulting cells were resuspended in 4˚C cold PBS.
The supernatants were centrifuged twice (800 x g for 10 min, then
1,000 x g for 10 min) at 4˚C and ultrafiltered through a 1.2-µm
Minisart Syringe Filter (Sartorius AG) to remove cells and cell
fragments.
Peripheral blood samples were collected into
vacutainer tubes and centrifuged twice at 4˚C, 2,000 x g for 10
min. The supernatant was ultrafiltered through a 1.2-µm Minisart
Syringe Filter (Sartorius AG) to remove cells and cell fragments.
The filtrate was stored at -80˚C until further differential
centrifugation.
The resulting tissue or peripheral blood supernatant
was subjected to differential centrifugation (50,000 x g for 90 min
followed by 100,000 x g for 90 min) at 4˚C. The precipitate was
resuspended in 1 ml PBS. The retained precipitates were further
purified by layering onto iodixanol density gradient medium
(Axis-Shield Diagnostics) using a top-down gradient of 5, 10, 20,
30, 40 and 50% with centrifugation at 4˚C, 100,000 x g for 90 min.
The purified EVs were centrifuged at 4˚C, 100,000 x g for 90 min,
and the pellets were resuspended in 1 ml PBS, then stored at -80˚C
until further processing. The protein content of the purified EVs
was quantified using an Enhanced BCA Protein Assay kit (Beyotime
Institute of Biotechnology).
Transmission electron microscopy (TEM)
with negative contrast staining
A 1-µl sample of purified EVs was mixed with 100 µl
PBS solution, dried on a newly discharged 300-mesh
Formvar/carbon-coated TEM grid (Ted Pella, Inc.) at room
temperature, then negatively stained with 2% potassium
phosphotungstate overnight at room temperature. All transmission
electron micrographs were obtained using an H7500 transmission
electron microscope (Hitachi, Ltd.) operating at 80 kV.
Low-vacuum scanning electron
microscopy (LVSEM)
Purified EVs (1 µl) were resuspended in 100 µl PBS,
then in 900 µl 2.5% glutaraldehyde solution (cat. no. P1126;
Beijing Solarbio Science & Technology Co., Ltd.) at 4˚C
overnight. Micrographs of AMEVs were acquired using a low-vacuum
scanning electron microscope (Nova Nano SEM 450; FEI; Thermo Fisher
Scientific, Inc.) operated at 5 kV.
Nanoparticle tracking analysis
(NTA)
Purified EVs (1 µl) were diluted in PBS (1 ml) and
quantified using NTA (Nanosight NS 3000; Malvern Instruments, Ltd.)
to determine the size distribution and particle concentration.
Liquid chromatography-mass
spectrometry (LC-MS)
LC-MS was performed according to a protocol
described in a previous study with some modifications (12). In positive ionization mode, A
Q-Exactive mass spectrometer (Thermo Fisher Scientific, Inc.) was
used as the detector. Protein identification was performed with
MASCOT software (version 2.3; Matrix Science, Inc.) by searching
the UniProt database (https://www.uniprot.org/taxonomy/9606).
Carbamidomethylation of cysteines was set as a fixed modification.
Trypsin was added at 1:50 trypsin:protein mass ratio for the first
digestion overnight and a 1:100 trypsin:protein mass ratio for a
second 4-h digestion. The maximal mass tolerance in MS mode was set
to 20 ppm for the first search, and the fragment mass tolerance was
set to 0.6 Da. The maximum false discovery rates (FDRs) for both
peptide and protein identification were set to 0.05.
Gene ontology (GO) annotation and
kyoto encyclopedia of genes and genomes (KEGG) pathway
analysis
Functional enrichment analyses of the GO terms and
KEGG pathways enriched in the proteins were conducted using the
STRING database (version 11.0) (https://string-db.org/), with the entire human genome
serving as the reference. GO analysis of enriched proteins was
conducted for cellular component, molecular function and biological
process annotation. The online Venn diagram tool FunRich3.1.3
(http://funrich.org/download) was used to
compare T-AMEV and B-AMEV protein data with proteins in the
Vesiclepedia database (12,13).
Western blot analysis
Protein expression was examined using previously
published protocols with some modifications (14). In brief, total protein was extracted
using Membrane and Cytosol Protein Extraction Kit (Beyotime
Institute of Biotechnology) and protein concentration was
determined using Enhanced BCA Protein Assay Kit (Beyotime Institute
of Biotechnology) according to the manufacturer's instructions.
Equal amounts of proteins (~30 µg per sample) were separated via
12% SDS-PAGE (Beyotime Institute of Biotechnology). The separated
proteins were transferred to a 0.45-µm PVDF membrane
(MilliporeSigma) and blocked in 0.5% bovine serum albumin solution
for 30 min at 37˚C, then incubated with anti-flotillin-2 (1:1,000;
cat. no. ab181988; Abcam), anti-CD9 (1:2,000; cat. no. ab92726;
Abcam), anti-CD63 (1:1,000; cat. no. ab134045; Abcam), anti-HSP90A
(1:500; cat. no. AF5368; Affinity Biosciences), anti-STIP1
(1:1,000; cat. no. ab126724; Abcam), anti-TAGLN-2 (1:500; cat. no.
AF12053; Affinity Biosciences) and anti-GAPDH (1:1,000; cat. no.
E021060-03; EarthOx Life Sciences) antibodies at 4˚C overnight.
After three washes with wash buffer [PBS (pH 7.2; HyClone; Cytiva)
+ 0.1% Tween 20 (Beijing Solarbio Science & Technology Co.,
Ltd.)], the membrane was incubated with a HRP-conjugated goat
polyclonal secondary antibody against rabbit IgG-H&L (cat. no.
E030130; EarthOx Life Sciences) for 1 h at 37˚C. After washing with
wash buffer, immunoreactive bands were visualized using ECL
detection reagent (cat. no. WBKLS0050; MilliporeSigma). The films
were scanned using an Azure c400 instrument (Azure Biosystems,
Inc.), and the labelled bands were semi-quantified using ImageJ
software (version 1.8.0; National Institutes of Health).
Statistical analysis
Data analysis was performed using SPSS software
(version 21; IBM Corp.). Normally distributed data are expressed as
the mean ± SD and were analysed using unpaired t-tests. The
incidence of hypermenorrhoea and dysmenorrhea are presented as
fractions and percentages and were statistically compared using
χ2 tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
MRI and pathological characteristics
of patients with adenomyosis
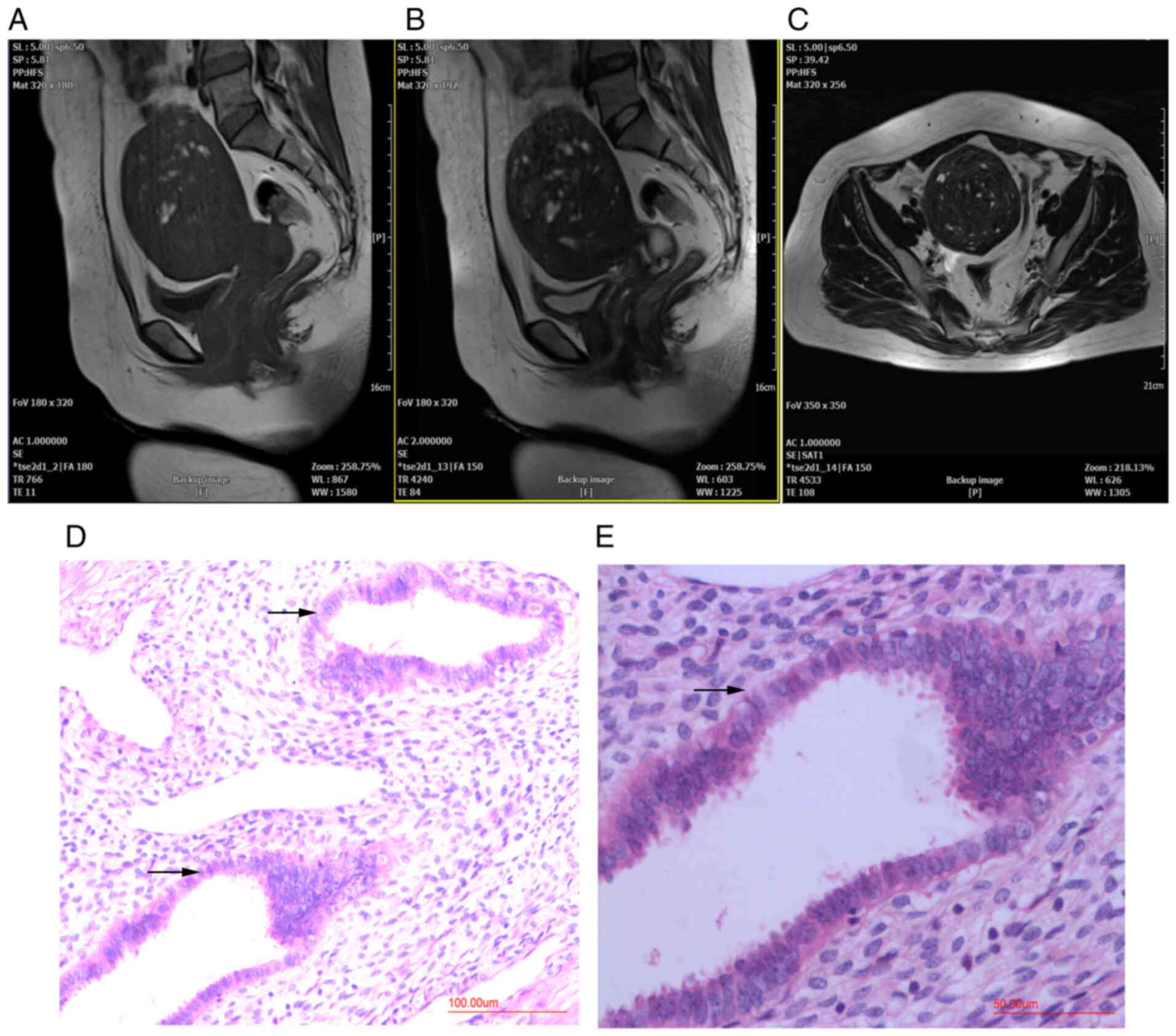
Patients with adenomyosis were diagnosed using MRI
before hysterectomy and histopathological examination after
hysterectomy. MRI indicated diffuse thickening of the myometrium,
an irregular junctional zone and the presence of localized lesions
in the posterior myometrium (Fig.
1A-C). H&E staining showed adenomyosis lesions in the
posterior wall of the uterus (Fig.
1D and E).
Characterization of T-AMEVs and
B-AMEVs
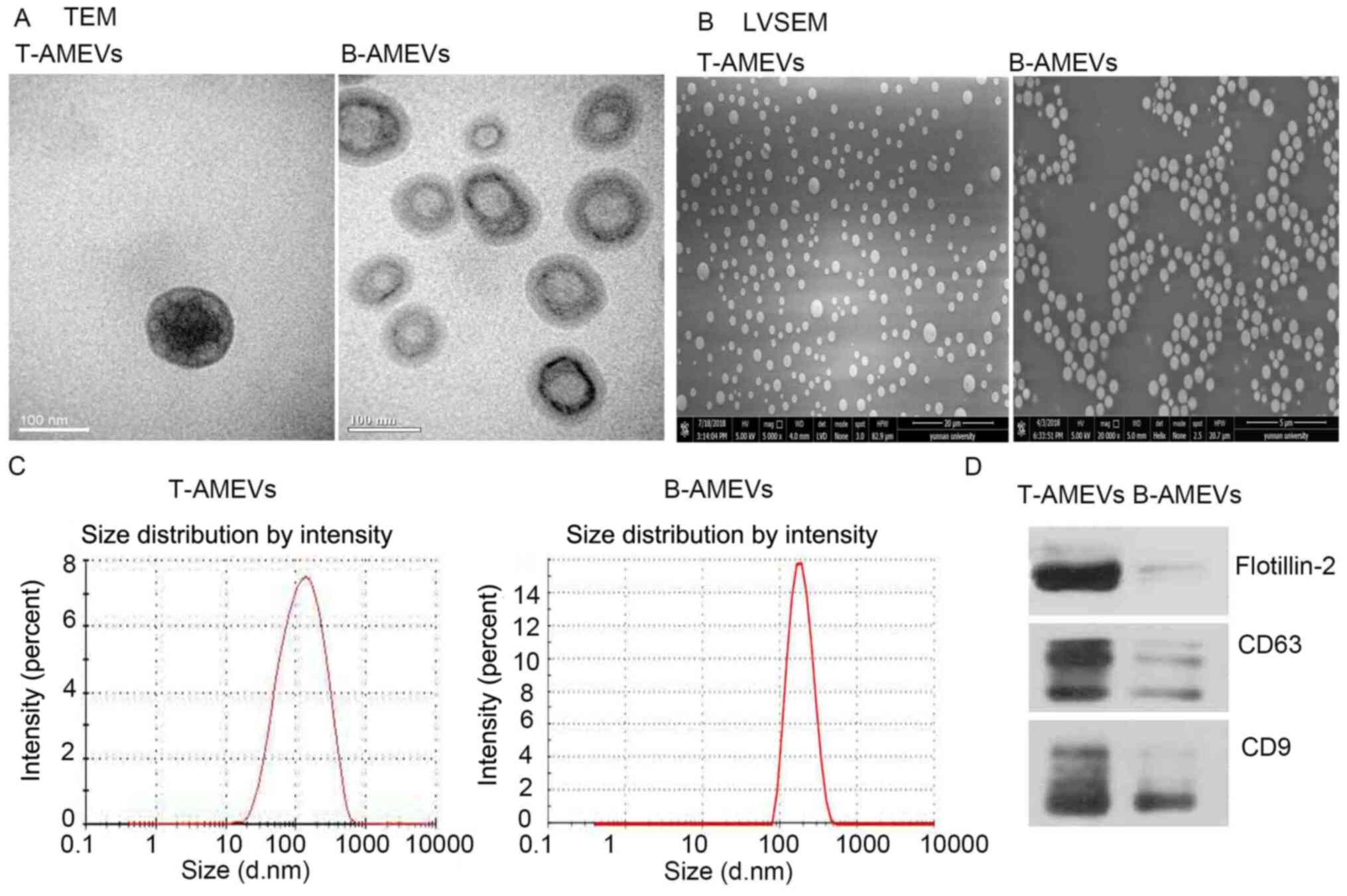
TEM, LVSEM and NTA were used to examine the
morphology and size distribution of T-AMEVs and B-AMEVs. The TEM
(Fig. 2A) and LVSEM (Fig. 2B) results suggested that the T-AMEVs
and B-AMEVs had a lipid bilayer membrane and spherical shape. NTA
showed that the average diameters of the T-AMEVs and B-AMEVs were
150.9±102.2 and 194.1±66.81 nm, respectively (Fig. 2C). Western blot analysis showed the
presence of CD9, CD81 and flotillin-2 in both T-AMEVs and B-AMEVs
(Fig. 2D).
Proteomic analysis of T-AMEVs and
B-AMEVs
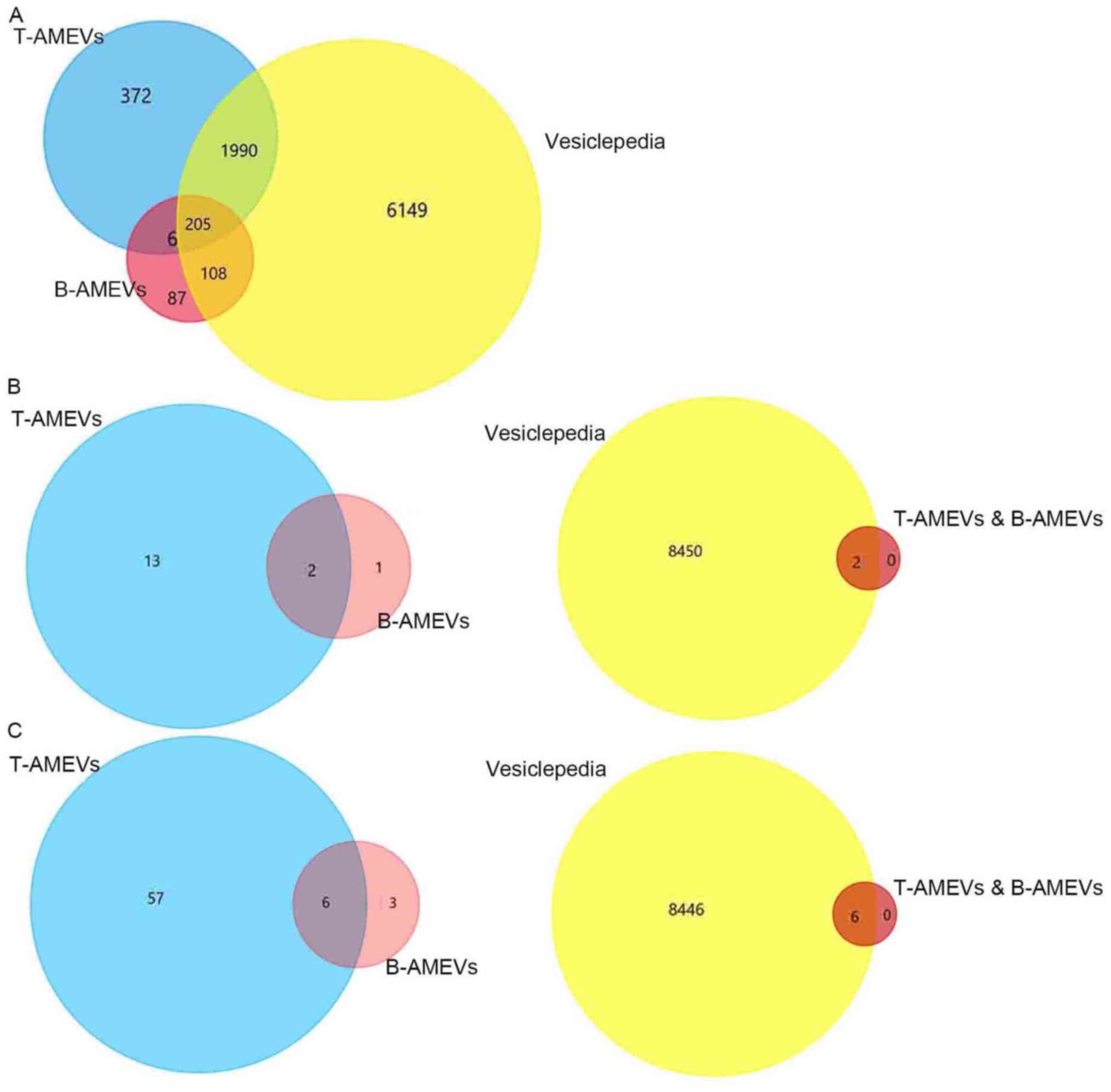
Mass spectrometric data for T-AMEVs and B-AMEVs were
compared to the EV protein data from the Vesiclepedia database. A
total of 2,195/2,573 proteins (85.30%) in T-AMEVs overlapped with
entries in Vesiclepedia. For B-AMEVs, 313/406 proteins (77.09%)
overlapped with Vesiclepedia (Fig.
3A). A total of 211 proteins were simultaneously expressed in
T-AMEVs and B-AMEVs (Table SI),
including 205 proteins that overlapped with entries in Vesiclepedia
and 6 proteins that were not in Vesiclepedia (Fig. 3A).
Further study of the proteomic data revealed that 15
proteins in T-AMEVs were related to the EMT process: Moesin, ezrin,
glycogen synthase kinase-3 β, 78-kDa glucose-regulated protein,
C-terminal-binding protein 1, C-terminal-binding protein 2, cell
cycle and apoptosis regulator protein 2, interleukin-like EMT
inducer, histidine triad nucleotide-binding protein 1 (HINT1),
stimulator of interferon genes protein, FK506-binding protein 1A,
junctional adhesion molecule 1, ubiquitin A-52 residue ribosomal
protein fusion product 1 (UBA52), melanoma
differentiation-associated protein 9 and transforming protein RhoA
(UniProt database) (Table SII).
Similarly, 3 proteins in B-AMEVs were related to the EMT process:
large tumour suppressor homolog 2, HINT1, and UBA52. Among the
EMT-related proteins, HINT1 and UBA52 were simultaneously expressed
in T-AMEVs and B-AMEVs (Fig. 3B;
Table SIII), which also overlapped
with the Vesiclepedia database (Fig.
3B).
A total of 63 proteins in T-AMEVs and 9 proteins in
B-AMEVs were found to promote the invasion of target cells in
adenomyosis (15). Of these, 6
proteins were shared between T-AMEVs and B-AMEVs and overlapped
with entries in Vesiclepedia: Fibulin-1, protein S100-A9,
plasminogen, fascin, heat shock cognate 71-kDa protein and Rho
GDP-dissociation inhibitor 1 (Fig.
3C; Table SIV).
GO and KEGG pathway analysis of
proteins simultaneously expressed in T-AMEVs and B-AMEVs
GO and KEGG pathway analyses of the proteins
simultaneously expressed in T-AMEVs and B-AMEVs were performed
using the STRING database. The biological processes of the 211
proteins primarily referred to the terms ‘regulation of actin
cytoskeleton organization’, ‘regulation of oxidative stress-induced
cell death’ and ‘regulation of cell morphogenesis’ (Fig. S1A). The cellular components of the
211 proteins were primarily related to the terms ‘cortical
cytoskeleton’, ‘cortical actin cytoskeleton’, ‘actin cytoskeleton’
and ‘intrinsic component of the cytoplasmic side of the plasma
membrane’ (Fig. S1B). The
molecular functions of the 211 proteins were primarily enriched in
the terms ‘actin filament binding’, protein-containing complex
binding’, ‘cytoskeletal protein binding’, ‘actin binding’ and
‘structural constituent of cytoskeleton’ (Fig. S1C).
KEGG pathway analysis revealed that the enriched
pathway terms of the 211 proteins with FDRs <0.05 were the
‘hypoxia-inducible factor (HIF)-1 signalling pathway’, ‘metabolic
pathways’, ‘central carbon metabolism in cancer’, ‘peroxisome
proliferator-activated receptor (PPAR) signalling pathway’ and
‘autophagy’ (Table SV). Among
these enriched pathways, the ‘PPAR signalling pathway’, ‘HIF-1
signalling pathway’ and ‘autophagy’ are related to the oliferation,
metastasis and autophagy of endometrial cells (Table SV).
HSP90A, STIP1 and TAGLN-2 in T-AMEVs
and B-AMEVs were identified as potential biomarkers for
adenomyosis
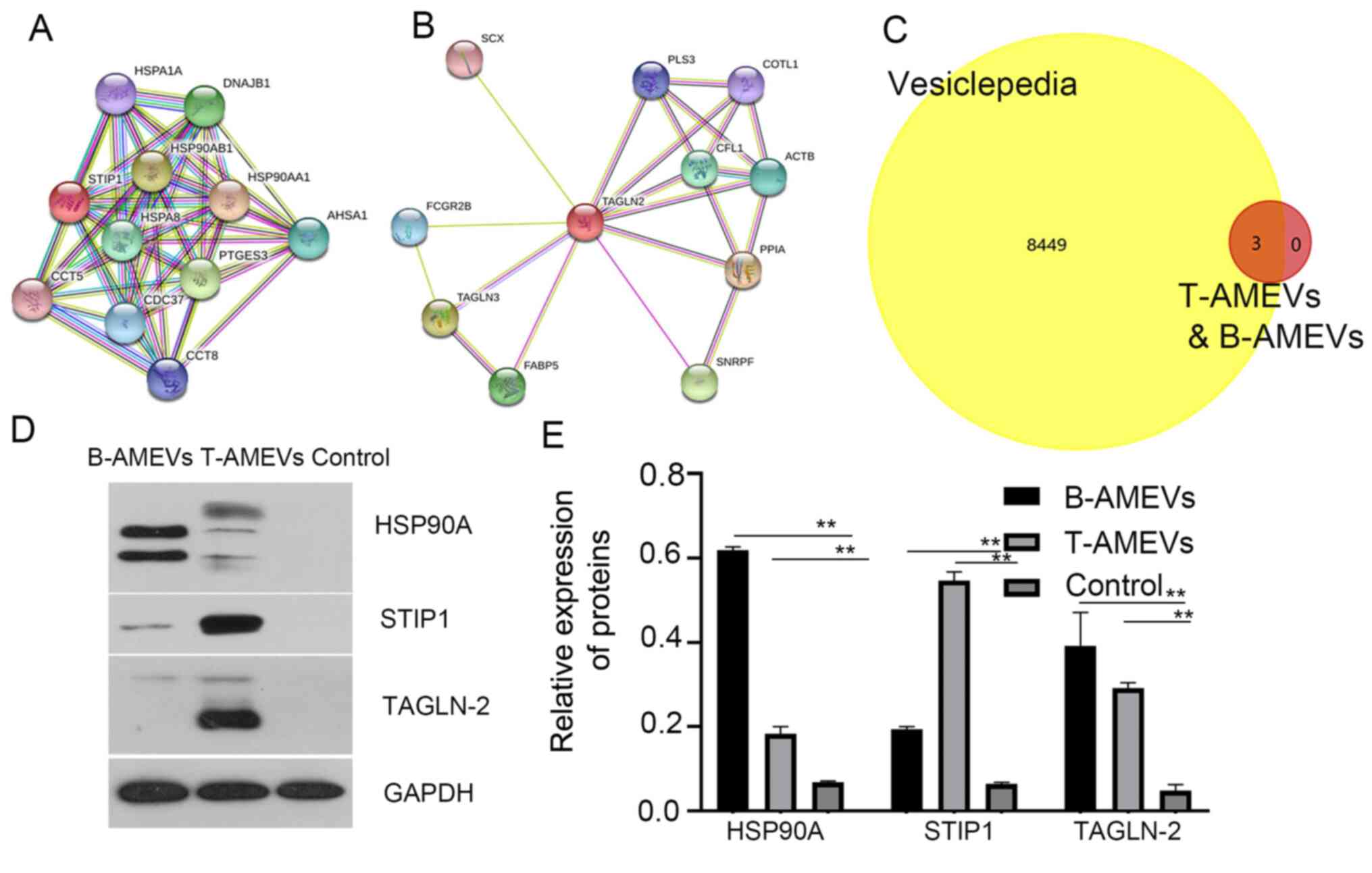
It has previously been suggested that STIP1 is
upregulated in women with adenomyosis compared with those without
adenomyosis (16). In the present
study, analysis in the STRING database showed that STIP1 and HSP90A
acted synergistically (17)
(Fig. 4A). TAGLN-2 is a homolog of
transgelin, which is an early marker of smooth muscle cell
differentiation (18), and STRING
database analysis showed that TAGLN-2 and TAGLN-3 act
synergistically (Fig. 4B).
Furthermore, 3 out of 211 proteins shared between
T-AMEVs and B-AMEVs, (HSP90A, STIP1 and TAGLN-2) were chosen as
potential biomarkers for adenomyosis. Research has reported that
STIP1 is over-expressed in adenomyosis. STIP1 is one of the
co-chaperone proteins that form the structural parts of the HSP90
machinery (16). TAGLN-2 is a
homolog of transgelin, which is an early marker of smooth muscle
cell differentiation (18). In
adenomyosis, myocytes exhibit cellular hypertrophy and show
differences in cytoplasmic organelles, nuclear structures and
intercellular junctions with normal myometrium (1). Therefore, the present study selected
the three proteins as potential biomarkers for adenomyosis.
FunRich3.1.3 software analysis demonstrated that these three
proteins overlapped with entries in the Vesiclepedia database,
indicating that they are EV-derived proteins (Fig. 4C).
To further determine whether HSP90A, STIP1 and
TAGLN-2 were expressed only in AMEVs and could be detected in EV
samples, plasma EVs were collected from women without
adenomyosis/endometriosis (n=31) as a control group (Table SVI; Fig. S2). Western blot analysis revealed
that HSP90A, STIP1 and TAGLN-2 were detectable in both T-AMEVs and
B-AMEVs, but not in EVs of the control group (Fig. 4D and E). STIP1 bands were detected at ~63 kDa
(predicted molecular weight, 63 kDa). Semi-quantitative analysis
showed that STIP1 was expressed in B-AMEVs and T-AMEVs. LC-MS
showed that the exponentially modified protein abundance index of
STIP1 in T-AMEVs and B-AMEVs was 0.58 and 0.11, respectively.
Discussion
Adenomyosis is a disease associated with endometrial
invasion. However, the factors involved in the invasive capacity of
the endometrium remain to be further studied. EVs are a new type of
intercellular communication mode. Our previous study demonstrated
EVs play an important role in modulating the disease process of
adenomyosis (7). Therefore, it is
hypothesized that it is necessary to investigate EVs in the
diagnosis and pathogenesis of adenomyosis. EVs have been isolated
from the serum of patients with endometriosis (8). Therefore, it is important to
investigate the role of EVs in the diagnosis and pathogenesis of
adenomyosis.
In the present study, EVs were first extracted from
lesion homogenates and peripheral blood of patients with
adenomyosis. The protein cargoes of AMEVs were analysed using
LC-MS, and potential biomarkers for adenomyosis were identified by
proteomic analysis. The proteomic results showed that 211 vesicular
proteins were simultaneously expressed in T-AMEVs and B-AMEVs.
Among the 211 proteins, 2 were associated with EMT, and 6 with
invasion. The present results are consistent with previously
reported results that EMT (19,20)
and the invasiveness (21) of
endometrial cells play critical roles in the pathogenesis and
development of adenomyosis. The 211 cargo proteins simultaneously
expressed in T-AMEVs and B-AMEVs might represent the molecular
characteristics of adenomyosis.
GO analysis revealed that proteins in T-AMEVs and
B-AMEVs were enriched in the ‘regulation of cellular component
organization’, ‘regulation of cytoskeleton organization’ and
‘regulation of cell morphogenesis’, indicating that the cargos of
T-AMEVs and B-AMEVs are closely related to EMT. This regulatory
characteristic indicated endometrial cells had a trend toward EMT
(Fig. S1A). The enrichment of
‘cytoplasmic part’ and ‘cortical cytoskeleton’ terms in the GO
cellular component analysis reflects the EV origin (Fig. S1B). In addition, the terms
‘cytoplasmic vesicle lumen’ and ‘secretory granule lumen’ indicate
that EV may originate from the cytoplasmic vesicle lumen and the
secretory granule lumens (Fig.
S1B). ‘Cytoskeletal protein binding’, ‘actin filament binding’
and ‘structural constituent of cytoskeleton’ were the most enriched
molecular function terms, which suggests that direct regulation of
the cytoskeleton might be the primary regulatory function of
T-AMEVs and B-AMEVs (Fig.
S1C).
KEGG pathway analysis revealed that the 211 proteins
of T-AMEVs and B-AMEVs are involved in several signalling pathways,
including ‘carbon metabolism’, ‘glycolysis’ and the ‘biosynthesis
of amino acids’ (Table SV), which
indicates that T-AMEVs and B-AMEVs had the capacity to transfer
information to other cells and thereby influence the recipient cell
function (5). Additionally, some
proteins were enriched in the HIF-1 signalling pathways, which are
related to the metastatic and proangiogenic capacity of adenomyotic
endometrial cells (22), and in the
PPAR signalling pathway, which is related to the proliferative
capacity of adenomyotic endometrial cells (23). These results demonstrated that
T-AMEVs and B-AMEVs carry specific proteins that represent the
pathological condition of adenomyosis and that these proteins are
candidate biological biomarkers for adenomyosis.
Currently, there is no specific diagnostic biomarker
for adenomyosis. The current non-invasive methods for the diagnosis
of adenomyosis are transvaginal ultrasound or MRI (2). EVs are increasingly important as
biomarkers for diseases (24). EVs
are nanoscale lipid bilayer particles that contain nucleic acid and
protein cargoes and can be secreted from cells under a variety of
normal and pathological conditions. EVs have recently attracted
widespread research interest due to their potential use as
circulating biomarkers for a variety of diseases, including
numerous types of cancers. EVs play a specific role in cancer
pathogenesis and exhibit potential for use in non-invasive disease
diagnosis and/or monitoring (25).
The presence of tumour-derived EVs in circulating
body fluids means that EVs represent a readily available source of
biomarkers. This observation suggests that EVs may have unique uses
in longitudinal disease monitoring and the early detection of
relapse (26). In addition, certain
EV-related proteins and nucleic acid species have been reported to
predict the response to treatment. Overall, accumulating evidence
indicates that EVs are a rich and easily accessible source of
cancer biomarkers.
From the 211 proteins simultaneously expressed in
T-AMEVs and B-AMEVs, we aimed to identify proteins as potential
EV-based biomarkers for adenomyosis. In vitro experiments
showed that patients with adenomyosis had significantly higher
preoperative serum STIP1 levels than women without adenomyosis.
Specifically, one study showed a positive correlation between STIP1
levels and adenomyosis (16). STIP1
is an adaptor protein that coordinates the functions of HSP70 and
HSP90 in protein folding. Increased STIP1 expression is related to
the pathogenesis of gynaecological malignancies, including ovarian
cancer and endometrial cancer (16). Evidence suggests that STIP1 can
stimulate DNA synthesis and enhance cell proliferation. Knockdown
of STIP1 expression in cancer cells has been shown to reduce tumour
invasiveness by downregulation of matrix metalloproteinases (MMPs)
(27). Through STRING database
analysis, we confirmed that STIP1 acts synergistically with
HSP90A.
HSP90 proteins are highly conserved molecules that
promote protein folding and block the aggregation of neosynthesized
or incorrectly folded proteins. HSP90 has been detected in exosomes
secreted from human cancer cells and is important in cancer
development because it regulates tumour growth, adhesion, invasion,
metastasis, angiogenesis, and apoptosis (28,29).
HSP90 is overexpressed in a variety of cancers, including
pancreatic, ovarian, breast, lung and endometrial cancers,
oropharyngeal squamous cell carcinoma (SCC), and multiple myeloma.
The high expression of HSP90 is considered a marker of poor
prognosis in lung cancer, oesophageal cancer, bladder cancer,
melanoma, and leukaemia (29).
HSP90A is the inducible form of HSP90.
In addition, studies have indicated ultrastructural
differences between smooth muscle cells from adenomyotic and normal
myometria (1). TAGLN-2 is a homolog
of transgelin, which is an early marker of smooth muscle cell
differentiation (18), and its
biological function involves the regulation of actin cytoskeleton
dynamics by stabilizing actin fibres. Transgelin also participates
in processes involving actin cytoskeleton remodelling, such as cell
proliferation, differentiation, migration, and apoptosis. TAGLN-2
is also considered to be correlated with tumorigenesis, tumour
metastasis and invasion, and it is highly expressed in uterine
cervical SCC tissues and endometrial cancer (30). TAGLN-2 and TAGLN-3 work
synergistically.
In our previous study, EVs isolated from adenomyotic
tissue sample highly expressed STIP1, HSP90A and TAGLN2(7). To further determine whether HSP90A,
STIP1 and TAGLN-2 were expressed only in AMEV and can be detected
in EV samples, plasmaEVs were isolated from women without
adenomyosis/endometriosis, and western blotting was used to measure
the protein expression levels of STIP1, HSP90A and TAGLN-2 in
T-AMEVs, B-AMEVs and serum EVs. The results suggested that STIP1,
HSP90A and TAGLN-2 were expressed in T-AMEVs and B-AMEVs but not in
EVs of control group This result further supports the use of STIP1,
HSP90A and TAGLN-2 as biomarkers for adenomyosis.
In conclusion, cargo proteins contained in EVs
derived from adenomyotic tissue and the peripheral blood of
patients with adenomyosis may induce endometrial cell invasion and
EMT. High expression of STIP1, HSP90A and TAGLN-2 may relate to
endometrial cell growth, invasion, EMT and smooth muscle cell
differentiation. The present study is limited by the sample size of
clinical patients. Therefore, further studies should increase the
volume of single blood samples by increasing EV output to
facilitate LC-MS detection of EV proteins in the blood of a single
patient, and further studies should be performed to verify the
three proteins' function in pathophysiology of adenomyosis using
experimental assays. STIP1, HSP90A and TAGLN-2 were expressed in
EVs derived from adenomyotic tissue and the peripheral blood of
patients with adenomyosis. These proteins could represent potential
biomarkers for the clinical diagnosis of adenomyosis.
Supplementary Material
Gene Ontology analysis of proteins
simultaneously expressed in T-AMEVs and B-AMEVs. (A) Biological
process analysis. (B) Cellular component analysis. (C) Molecular
function analysis. T-AMEV, tissue adenomyosis-derived extracellular
vesicle; B-AMEV, blood adenomyosis-derived extracellular vesicle.
Blue bars show background gene count, and the orange bars
show-log10 of false discovery rate.
Characterization of serum
extracellular vesicles in women without adenomyosis or
endometriosis (d, diameter; nm, nanometre). (A) Transmission
electron microscopy images with negative staining. (B) Nanoparticle
tracking analysis of serum extracellular vesicles from women
without adenomyosis/endometriosis.
A total of 211 protein markers of
T-AMEVs and B-AMEVs.
Epithelial-mesenchymal transition in T
AMEVs and uniport.
Epithelial to mesenchymal transition
associated proteins in T AMEVs and B AMEVs.
Invasion-associated proteins in
T-AMEVs and B-AMEVs.
Kyoto Encyclopedia of Genes and
Genomes pathway analysis.
Characteristics of blood samples from
women with or without adenomyosis/endometriosis.
Acknowledgements
Not applicable.
Funding
Funding: This work was partially supported by The Chongqing
Basic Science and Frontier Technology Research Project (grant no.
cstc2017jcyjAX0432).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW, LF, BY and DC conceived and designed the study
and analysed and interpreted the data. DC performed the experiments
and acquired the data with the assistance of LZ, HQ and YW for
in vitro work. DC, LZ and YX analysed the data and were
involved in drafting the manuscript. DC, BY and LF wrote and
revised the paper critically. All of the other authors were
involved in writing the manuscript. DC collected the blood and
tissue samples from adenomyosis patients. All authors read and
approved the final manuscript. LF and ZW confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The study protocol for human samples was approved by
the Ethics Committee of Chongqing Medical University (approval no.
2018014) and the Affiliated Hospital of Zunyi Medical University
(Guizhou, China; approval no. 20211-005). The study was carried out
in accordance with the Declaration of Helsinki Declaration of the
World Medical Association. All patients (including the control
group) provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Benagiano G, Habiba M and Brosens I: The
pathophysiology of uterine adenomyosis: An update. Fertil Steril.
98:572–579. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Xu R, Greening DW, Zhu HJ, Takahashi N and
Simpson RJ: Extracellular vesicle isolation and characterization:
Toward clinical application. J Clin Invest. 126:1152–1162.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Colombo M, Raposo G and Théry C:
Biogenesis, secretion, and intercellular interactions of exosomes
and other extracellular vesicles. Annu Rev Cell Dev Biol.
30:255–289. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Henderson MC and Azorsa DO: The genomic
and proteomic content of cancer cell-derived exosomes. Front Oncol.
2(38)2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles.
4(27066)2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Momen-Heravi F, Getting SJ and Moschos SA:
Extracellular vesicles and their nucleic acids for biomarker
discovery. Pharmacol Ther. 192:170–187. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chen D, Qiao H, Wang Y, Zhou L, Yin N,
Fang L and Wang Z: Adenomyosis-derived extracellular vesicles endow
endometrial epithelial cells with an invasive phenotype through
epithelial-mesenchymal transition. Genes Dis. 7:636–648.
2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Qiu JJ, Lin XJ, Zheng TT, Tang XY, Zhang Y
and Hua KQ: The exosomal long noncoding RNA aHIF is upregulated in
serum from patients with endometriosis and promotes angiogenesis in
endometriosis. Reprod Sci. 26:1590–1602. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Roeder HA, Cramer SF and Leppert PC: A
look at uterine wound healing through a histopathological study of
uterine scars. Reprod Sci. 19:463–473. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang GJ, Liu Y, Qin A, Shah SV, Deng ZB,
Xiang X, Cheng Z, Liu C, Wang J, Zhang L, et al: Thymus
exosomes-like particles induce regulatory T cells. J Immunol.
181:5242–5248. 2008.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Coumans FAW, Brisson AR, Buzas EI,
Dignat-George F, Drees EEE, El-Andaloussi S, Emanueli C, Gasecka A,
Hendrix A, Hill AF, et al: Methodological guidelines to study
extracellular vesicles. Circ Res. 120:1632–1648. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Pathan M, Keerthikumar S, Chisanga D,
Alessandro R, Ang CS, Askenase P, Batagov AO, Benito-Martin A,
Camussi G, Clayton A, et al: A novel community driven software for
functional enrichment analysis of extracellular vesicles data. J
Extracell Vesicles. 6(1321455)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CMJ, Williamson NJ, Mouradov D, Sieber OM, Simpson RJ,
Salim A, et al: FunRich: A standalone tool for functional
enrichment analysis. Proteomics. 15:2597–2601. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang DJ, Wang CM, Wang YT, Qiao H, Fang LQ
and Wang ZB: Lactation-related MicroRNA expression in microvesicles
of human umbilical cord blood. Med Sci Monit. 22:4542–4554.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Morgat A, Lombardot T, Coudert E, Axelsen
K, Neto TB, Gehant S, Bansal P, Bolleman J, Gasteiger E, de Castro
E, et al: Enzyme annotation in UniProtKB using Rhea.
Bioinformatics. 36:1896–1901. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang HS, Tsai CL, Chang PY, Chao A, Wu RC,
Chen SH, Wang CJ, Yen CF, Lee YS and Wang TH: Positive associations
between upregulated levels of stress-induced phosphoprotein 1 and
matrix metalloproteinase-9 in endometriosis/adenomyosis. PLoS One.
13(e0190573)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tsai CL, Lee YS, Chao A, Yen CF, Wang HS
and Wang TH: Associations between a single nucleotide polymorphism
of stress-induced phosphoprotein 1 and endometriosis/adenomyosis.
Taiwan J Obstet Gynecol. 57:270–275. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li L, Miano JM, Cserjesi P and Olson EN:
SM22 alpha, a marker of adult smooth muscle, is expressed in
multiple myogenic lineages during embryogenesis. Circ Res.
78:188–195. 1996.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Benagiano G, Brosens I and Habiba M:
Structural and molecular features of the endomyometrium in
endometriosis and adenomyosis. Hum Reprod Update. 20:386–402.
2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Syn N, Wang L, Sethi G, Thiery JP and Goh
BC: Exosome-mediated metastasis: From epithelial-mesenchymal
transition to escape from immunosurveillance. Trends Pharmacol Sci.
37:606–617. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gaetje R, Kotzian S, Herrmann G, Baumann R
and Starzinski-Powitz A: Invasiveness of endometriotic cells in
vitro. Lancet. 346:1463–1464. 1995.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhou S, Yi T, Liu R, Bian C, Qi X, He X,
Wang K, Li J, Zhao X, Huang C and Wei Y: Proteomics identification
of annexin A2 as a key mediator in the metastasis and
proangiogenesis of endometrial cells in human adenomyosis. Mol Cell
Proteomics. 11(M112.017988)2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wu Y and Guo SW: Peroxisome
proliferator-activated receptor-gamma and retinoid X receptor
agonists synergistically suppress proliferation of immortalized
endometrial stromal cells. Fertil Steril. 91 (Suppl 5):S2142–S2147.
2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Taleb RSZ, Moez P, Younan D, Eisenacher M,
Tenbusch M, Sitek B and Bracht T: Protein biomarker discovery using
human blood plasma microparticles. Methods Mol Biol. 1959:51–64.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lane RE, Korbie D, Hill MM and Trau M:
Extracellular vesicles as circulating cancer biomarkers:
Opportunities and challenges. Clin Trans Med. 7(14)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hornick NI, Huan J, Doron B, Goloviznina
NA, Lapidus J, Chang BH and Kurre P: Serum exosome microRNA as a
minimally-invasive early biomarker of AML. Sci Rep.
5(11295)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Walsh N, Larkin A, Swan N, Conlon K,
Dowling P, McDermott R and Clynes M: RNAi knockdown of Hop
(Hsp70/Hsp90 organising protein) decreases invasion via MMP-2 down
regulation. Cancer Lett. 306:180–189. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sims JD, McCready J and Jay DG:
Extracellular heat shock protein (Hsp)70 and Hsp90α assist in
matrix metalloproteinase-2 activation and breast cancer cell
migration and invasion. PLoS One. 6(e18848)2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu J, Liu T, Rios Z, Mei Q, Lin X and Cao
S: Heat shock proteins and cancer. Trends Pharmacol Sci.
38:226–256. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Meng T, Liu L, Hao R, Chen S and Dong Y:
Transgelin-2: A potential oncogenic factor. Tumour Biol.
39(1010428317702650)2017.PubMed/NCBI View Article : Google Scholar
|