Introduction
Ovarian cancer is the second most common cause of
gynecological cancer-related mortality among women around the world
(1). There were 295,000 new cases
and 185,000 deaths reported worldwide in 2018, with increasing
trends predicted (1,2). Despite marked efforts in surgery and
chemotherapy, clinical outcomes of OC patients remain unfavorable
(2). Although numerous studies have
indicated that the International Federation of Gynecology and
Obstetrics (FIGO) stage and lymph node metastasis are independent
prognostic factors for survival of patients with OC, they may not
be able to accurately estimate prognosis due to heterogeneities in
the patient population. Therefore, it is important to perform
experimental and clinical studies to identify novel biomarkers for
early diagnosis and prognosis of OC.
Long non-coding RNAs (lncRNA) are molecules
consisting of >200 nucleotides that do not encode for proteins
(3). Accumulating studies indicated
that lncRNAs regulate cancer cellular activities, including
proliferation, apoptosis and migration (4). Certain lncRNAs may be potential
markers for diagnosing and targets for treating cancer (5,6). To
date, the emerging functional roles of lncRNAs in OC remain to be
fully elucidated.
A previous microarray analysis performed by our
group confirmed a significant number of aberrantly expressed
lncRNAs and mRNAs in OC compared to normal ovarian tissues. Among
these significantly differentially expressed lncRNAs, BC041954 was
the most upregulated lncRNA (7).
BC041954 is a novel lncRNA that is located on chromosome 3 near the
zinc finger of the cerebellum 4 (ZIC4) gene. The aim of the present
study was to investigate the association between BC041954
expression and clinicopathological characteristics and further
explore the clinical significance of BC041954 in OC.
Patients and methods
Clinical specimens
The present study included 103 primary tumor samples
from patients with OC who underwent surgery at Shengjing Hospital
affiliated to China Medical University between September 2006 and
September 2015 (Shengjing, China). None of the patients had
received any preoperative chemotherapy or radiation treatments. A
total of 60 non-tumor tissue specimens (NT), including normal
ovarian and fallopian tube tissues, were collected from female
patients (mean age, 58.27±6.79 years) of the same age receiving
hysterectomy for non-malignant conditions (mean age 57.45±8.51
years). There is no significant difference in the age in the two
groups. The present study was approved by the Ethics Committee of
Shengjing Hospital, China Medical University (Shengjing, China;
ethical approval no. 2015PS158K). All cases of OC were
histologically diagnosed and classified in accordance with the
World Health Organization criteria (8,9). All
clinical data were collected by physicians and the investigators
who performed the experiments with the samples were blinded to the
clinical data. The clinicopathological characteristics of the
patients are listed in Table I.
 | Table IAssociations between BC041954
expression and clinicopathological characteristics in ovarian
cancer. |
Table I
Associations between BC041954
expression and clinicopathological characteristics in ovarian
cancer.
| | BC041954
expression | |
|---|
| Characteristic | Cases (n) | High (n=52) | Low (n=51) | P-value |
|---|
| Age (years) | | | | 0.279 |
|
≤55 | 43 | 19 | 24 | |
|
>55 | 60 | 33 | 27 | |
| Histological
subtypes | | | | 0.737 |
|
Serous | 62 | 32 | 30 | |
|
Mucinous | 19 | 7 | 12 | |
|
Endometrioid | 9 | 5 | 4 | |
|
Clear
cell | 8 | 5 | 3 | |
|
Others | 5 | 3 | 2 | |
| FIGO stage | | | | 0.018 |
|
I+II | 35 | 12 | 23 | |
|
III+IV | 68 | 40 | 28 | |
| Grade | | | | 0.387 |
|
G1 | 44 | 20 | 24 | |
|
G2 + G3 | 59 | 32 | 27 | |
| Distant
metastasis | | | | 0.031 |
|
Yes | 39 | 25 | 14 | |
|
No | 64 | 27 | 37 | |
RNA extraction
Tissue samples were collected, immediately
snap-frozen in liquid nitrogen and carefully stored at -80˚C until
use. TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) was used
to extract total RNA. RNA quantity and quality were evaluated using
NanoDrop® 2000 (Thermo Fisher Scientific, Inc.)
technology and agarose gel electrophoresis was used to detect the
RNA integrity (10).
Reverse transcription-quantitative
(RT-q)PCR
According to the manufacturer's protocols, RT of
total RNA into complementary DNA was performed using the
SuperScript™ III Reverse Transcriptase Kit (Invitrogen; Thermo
Fisher Scientific, Inc.). The full temperature protocol was 50˚C
for 60 min and 70˚C for 15 min. The ViiA 7 Real-time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was utilized
for RT-qPCR amplification with the Power SYBR Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling protocol for qPCR was as follows: Initial
denaturation at 95˚C for 10 min, followed by 40 cycles of 95˚C for
10 sec and 60˚C for 60 sec. GAPDH expression was monitored as the
endogenous control. The specific primer sequences for the lncRNA
BC041954 were as follows: Forward, 5'-TCTGTAGTTCGTTGTTGGTCGTG-3'
and reverse, 5'-GCGGTCCTGATTCATTAGCG-3'. The specific primer
sequences for GAPDH were as follows: Forward,
5'-GGGAAACTGTGGCGTGAT-3' and reverse, 5'-GAGTGGGTGTCGCTGTTGA-3'.
mRNA levels were normalized to GAPDH and fold changes in expression
were calculated using the 2-∆∆Cq method (11).
Construction of the
lncRNA-micro(mi)RNA-mRNA competing endogenous (ce)RNA network
miRNA response elements are the foundation of the
ceRNA hypothesis of RNA transcript crosstalk. Algorithms from
Targetscan and miRanda were used to determine putative miRNA-lncRNA
interactions. The miRNA-lncRNA and miRNA-mRNA pairs were used to
construct the lncRNA-miRNA-mRNA ceRNA network with an in-house Perl
script. All interaction information was imported into Cytoscape
software version 3.1.1 (http://www.cytoscape.org) to generate the regulatory
network (12).
Statistical analysis
lncRNA expression was calculated using fold-change
filtering and the independent-samples t-test. The association
between BC041954 and clinicopathological variables was evaluated
using the Chi-square test. Kaplan-Meier analysis was used to
determine the influence of BC041954 on OS. Furthermore, the Cox
proportional hazards regression model was used for univariate and
multivariate analyses. All experiments were performed in
triplicate. P<0.05 was set as the criterion for statistical
significance. SPSS software (version 19.0; IBM Corp.) was utilized
for statistical analysis.
Results
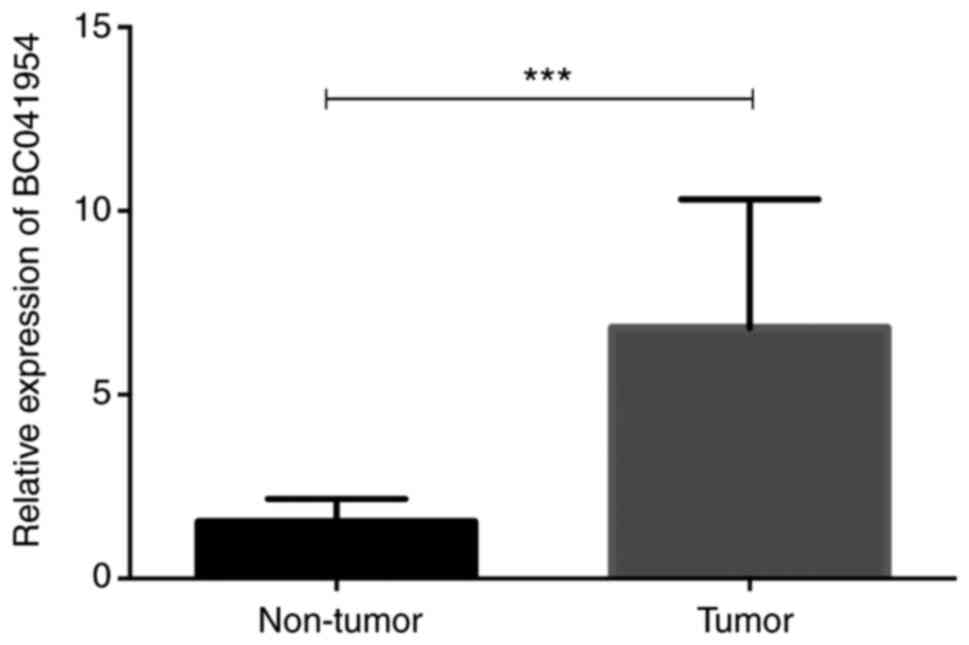
BC041954 is significantly increased in
OC tissues
First, BC041954 expression in OC and NT tissues was
detected using RT-qPCR. It was revealed that BC041954 was
significantly increased in OC compared to NT tissues (P<0.001;
Fig. 1).
Association between BC041954 and
clinicopathological parameters in patients with OC
To further explore the clinical significance of
BC041954 in OC, the association between lncRNA expression and
clinicopathological parameters was evaluated in the 103 patients
with OC. According to the median expression level of BC041954 as
the cutoff value, patients were divided into two groups: A high
BC041954 group and a low BC041954 group. As presented in Table I, high BC041954 expression was
negatively associated with the FIGO stage (P=0.018). In addition,
higher expression of BC041954 was more frequent in patients with
distant metastasis (P=0.031). There was no significant association
between BC041954 and any of the other clinicopathological
characteristics, including histological subtype and grade.
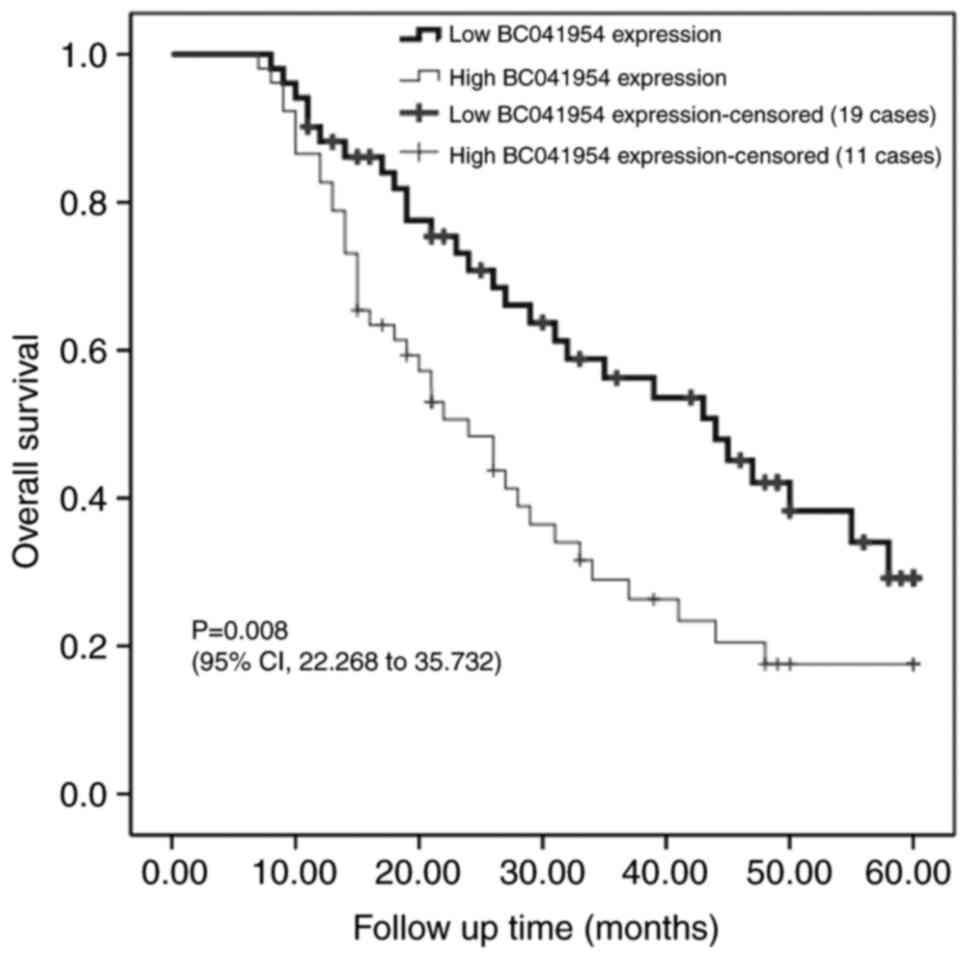
Influence of BC041954 on the prognosis
of patients with OC
The OS of patients with OC was evaluated using
Kaplan-Meier analysis. As presented in Fig. 2, OS was significantly lower in
patients with high BC041954 expression (P=0.008). Univariate
analysis indicated that BC041954 expression (P=0.010), FIGO stage
(P=0.001) and distant metastasis (P=0.006) were closely associated
with OS of patients with OC. Furthermore, multivariate analysis
with the Cox proportional hazards model was performed, according to
which the FIGO stage (P=0.001), distant metastasis (P=0.044) and
BC041954 expression (P=0.040) were independent prognostic factors
in OC (Table II).
 | Table IIUnivariate and multivariate analyses
for overall survival using the Cox regression model. |
Table II
Univariate and multivariate analyses
for overall survival using the Cox regression model.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Variable | Risk ratio | 95%CI | P-value | Risk ratio | 95%CI | P-value |
|---|
| Age (≤55 years vs.
>55 years) | 0.987 | 0.605-1.610 | 0.959 | | | |
| Histological subtype
[serous vs. (mucinous + endometrioid + clear cell + others)] | 1.057 | 0.640-1.747 | 0.828 | | | |
| FIGO stage (I + II
vs. III + IV) | 0.359 | 0.199-0.646 | 0.001 | 0.371 | 0.203-0.677 | 0.001 |
| Grade (G1 vs. G2 +
G3) | 1.237 | 0.762-2.008 | 0.390 | | | |
| Distant metastasis
(yes vs. no) | 2.124 | 1.247-3.616 | 0.006 | 1.755 | 1.061-3.033 | 0.044 |
| lncRNA BC041954
(high vs. low) | 0.522 | 0.319-0.856 | 0.010 | 0.582 | 0.348-0.975 | 0.040 |
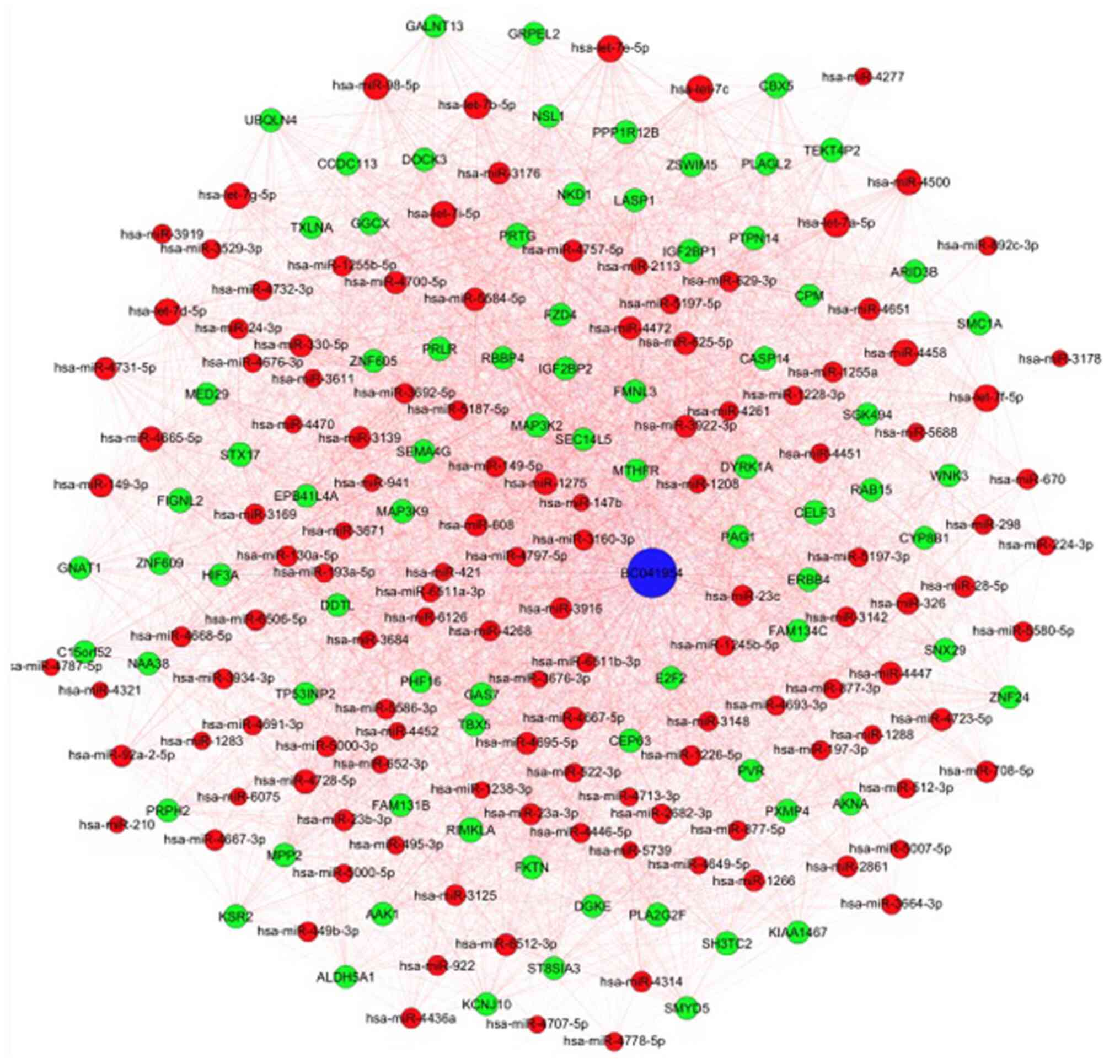
Functional predictions of the
lncRNA/miRNA/mRNA interactions
A ceRNA analysis was performed to provide an
overview of the potential lncRNA/miRNA/mRNA interactions (Fig. 3). The potential binding miRNAs of
lncRNA BC041954 are depicted. Blue nodes represent lncRNAs
including BC041954, red nodes represent miRNAs and green nodes
represent mRNAs. These networks were utilized to create functional
annotations of the predicted target mRNAs by searching the Gene
database. According to the ceRNA theory, lncRNAs may act as natural
miRNA sponges to inhibit miRNA function. The expression of
lncRNA-miRNA and miRNA-mRNA was negatively correlated. Thus, the
ceRNA modulation network of these lncRNAs, miRNAs and mRNAs may
provide molecular mechanisms involved in the initiation and
progression of OC, providing novel clues for the discovery of
clinical diagnostic markers and therapeutic targets. The ceRNA
analysis provided an overview of potential BC041954/miRNA/mRNA
interactions, indicating that numerous miRNAs interact with
BC041954, such as hsa-miR-197-3p, hsa-miR-23b-3p, hsa-miR-149-3p
and miR-193a. From the network constructed, 115 miRNAs that
potentially interact with BC041954 were identified. BC041954 may
serve as a miRNA sponge to inhibit miRNA function. Specifically,
miR-193a may directly interact with lncRNA BC041954. Based on the
identification of these miRNAs, the miRNA-target interactions
database was used to predict mRNA targets. The analysis indicated
that miRNAs may interact with receptor protein-tyrosine kinase 4
(ERBB4), γ-glutamyl carboxylase (GGCX), insulin-like growth factor
2 mRNA-binding protein 2 (IGF2BP2) and growth arrest specific 7
(GAS7) as targets. Furthermore, a previous study confirmed that
miR-193a is able to modulate ERBB4 via MAPK/ERK signaling to
enhance the progression of OC (13). Thus, it may be speculated that the
lncRNA BC041954/miR-193a/ERBB4 interaction has an important role in
OC pathogenesis.
Discussion
Numerous differentially expressed lncRNAs have been
implicated in tumorigenesis and the progression of various cancer
types (6,14). The functional roles of lncRNAs are
being increasingly recognized, which has led to a better
understanding of the biological processes of different cancer
types, such as breast (15),
hepatocellular (16), ovarian
(1,2,17) and
colorectal cancer (18). Various
lncRNAs may serve as therapeutic targets as well as biomarkers for
early diagnosis and prognosis of human cancers.
It has been reported that lncRNAs have prognostic
potential and have critical roles in OC progression (17). Qiu et al (19) reported that overexpression of hox
transcript antisense intergenic RNA was associated with poor
prognosis and facilitated tumor metastasis in patients with OC.
Metastasis-associated lung adenocarcinoma transcript 1 was
increased in OC tissues and significantly associated with
metastasis and tumor size in OC (20). Patients with high colon
cancer-associated transcript 2 (CCAT2) had shorter OS and CCAT2 was
positively associated with the tumor grade, FIGO stage and
metastasis in patients with OC (21). Reduced growth arrest specific 5
(GAS5) was detected in OC tissues and overexpression of GAS5
suppressed aggressive behaviors of OC cells (22). Taken together, lncRNAs may be a
novel prognostic factor and a potential therapeutic target for
OC.
A previous study by our group profiled
differentially expressed lncRNAs and mRNAs in OC vs. normal tissues
and detected 2,870 dysregulated lncRNAs. There were 2,658
differentially expressed mRNAs in OC (1,014 were upregulated and
1,644 were downregulated compared with those in NT). BC041954 was
selected from the obviously upregulated lncRNA for further
validation using RT-qPCR in 25 OC and 15 NT samples and the results
confirmed the alterations of the lncRNA expression in OC (7). However, the role of BC041954 in OC has
remained elusive. BC041954, located on chromosome 3 near the ZIC4
gene, is a novel lncRNA that was identified by microarray analysis.
In the present study, the relative expression of BC041954 OC was
detected in patients using RT-qPCR to estimate the clinical
significance of BC041954 in OC. First, it was confirmed that
BC041954 expression was markedly increased in OC tissues compared
with that in NTs. Furthermore, it was indicated that abnormal
BC041954 expression may be associated with OC progression. Elevated
BC041954 was closely associated with the FIGO stage and distant
metastasis. In addition, patients with high BC041954 expression had
significantly shorter OS, as determined using Kaplan-Meier
analysis. Multivariate analysis demonstrated that the FIGO stage,
distant metastasis and BC041954 expression were independent
prognostic factors for patients with OC. These results reveal that
BC041954 is a potential predictor of OC progression. The ceRNA
analysis provided an overview of potential lncRNA/miRNA/mRNA
interactions, indicating that numerous miRNAs interact with
BC041954. Among these miRNAs, miR-193a was identified to directly
interact with BC041954. It was previously reported that miR-193a
silencing contributes to a dynamic process of cell growth
suppression and migratory/invasive abilities of OC cells (13).
Based on the present results, an miRNA-target
interactions database was used to predict miRNA targets. The miRNAs
may interact with ERBB4, GGCX, IGF2BP2 and GAS7 as targets, most of
which are cancer-associated genes, including ERBB4(23), IGF2BP2(24) and GAS7(25), which have important roles in tumor
cell apoptosis, proliferation and metastasis.
Accumulating evidence has indicated that ErbB family
members are overexpressed and mutated in OC, and further that ErbB
family members are considered important therapeutic targets. The
ErbB receptor family (including ERBB4) and its downstream pathways
have been reported to be involved in the regulation of
epithelial-mesenchymal transition, migration and tumor invasion
(26). It has been indicated that
the ERBB4 rs1836724 polymorphism is associated with OS of patients
with OC (27). miR-193a is able to
modulate ERBB4 via the MAPK/ERK signaling pathway to increase the
oncogenic properties of OC (13).
The present results suggested that ERBB4 may also directly interact
with BC041954. Based on the present results, in conjunction with
those of previous studies, it may be hypothesized that the
BC041954/miR-193a/ERBB4 interaction has an important role in the
pathogenesis of OC. Therefore, further functional analyses of
BC041954 as a biomarker for tumor diagnosis and prognosis in OC are
required. The present study indicated that targeting the lncRNA
BC041954 may provide benefits for treating OC.
In conclusion, the present study indicated for the
first time that upregulation of lncRNA BC041954 is a frequent event
in OC and BC041954 may be a novel biomarker for predicting poor
prognosis in patients with OC. However, the molecular mechanisms by
which BC041954 is upregulated in OC requires to be further
investigated.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported in part by grants from The
Natural Science Foundation of Liaoning Province (grant nos.
2015020462 and 2019-ZD-0790).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YML contributed to the study design, writing of the
manuscript and adjustment of the experimental design. YRG collected
and analyzed the data. MYZ performed the experiments and analyzed
the data. YW generated the lncRNA/miRNA/mRNA regulatory network and
associated data analysis. All authors read and approved the final
manuscript. YML and YRG confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
This study was approved by our Ethics Committee in
Shengjing Hospital, China Medical University (Shengjing, China).
Written informed consent was obtained from the patient at the time
of recruitment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
R, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cortez AJ, Tudrej P, Kujawa KA and
Lisowska KM: Advances in ovarian cancer therapy. Cancer Chemother
Pharmacol. 81:17–38. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mattick JS: The genetic signatures of
noncoding RNAs. PLoS Genet. 5(e1000459)2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kawasaki N, Miwa T, Hokari S, Sakurai T,
Ohmori K, Miyauchi K, Miyazono K and Koinuma D: Long noncoding RNA
NORAD regulates transforming growth factor-β signaling and
epithelial-to-mesenchymal transition-like phenotype. Cancer Sci.
109:2211–2220. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tripathi MK, Doxtater K, Keramatnia F,
Zacheaus C, Yallapu MM, Jaggi M and Chauhan SC: Role of lncRNAs in
ovarian cancer: Defining new biomarkers for therapeutic purposes.
Drug Discov Today. 203:1635–1643. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lu YM, Wang Y, Liu SQ, Zhou MY and Guo YR:
Profile and validation of dysregulated long noncoding RNAs and
mRNAs in ovarian cancer. Oncol Rep. 40:2964–2976. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Barber HR, Sommers SC, Synder R and Kwon
TH: Histologic and nuclear grading and stromal reactions as indices
for prognosis in ovarian cancer. Am J Obstet Gynecol. 121:795–807.
1975.PubMed/NCBI
|
|
9
|
Szafron LM, Balcerak A, Grzybowska EA,
Pienkowska-Grela B, Podgorska A, Zub R, Olbryt M, Pamula-Pilat J,
Lisowska KM, Grzybowska E, et al: The putative oncogene, CRNDE, is
a negative prognostic factor in ovarian cancer patients.
Oncotarget. 6:43897–43910. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xu Q, Deng F, Xing Z, Wu Z, Cen B, Xu S,
Zhao Z, Nepomuceno R, Bhuiyan MI, Sun D, et al: Long non-coding RNA
C2dat1 regulates CaMKIIδ expression to promote neuronal survival
through the NF-κB signaling pathway following cerebral ischemia.
Cell Death Dis. 7(e2173)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen K, Liu MX, Mak CS, Yung MM, Leung TH,
Xu D, Ngu SF, Chan KK, Yang H, Ngan HY and Chan DW:
Methylation-associated silencing of miR-193a-3p promotes ovarian
cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways.
Theranostics. 8:423–436. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chan JJ and Tay Y: Noncoding RNA: RNA
regulatory networks in cancer. Int J Mol Sci.
19(1310)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mansoori Y, Tabei MB, Askari A, Izadi P,
Daraei A, Bastami M, Naghizadeh MM, Nariman-Saleh-Fam Z, Mansoori B
and Tavakkoly-Bazzaz J: Expression levels of breast cancer-related
GAS5 and LSINCT5 lncRNAs in cancer-free breast tissue: Molecular
associations with age at menarche and obesity. Breast J.
24:876–882. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang BG, Lv Z, Ding HX, Fang XX, Wen J, Xu
Q and Yuan Y: The association of lncRNA-HULC polymorphisms with
hepatocellular cancer risk and prognosis. Gene. 670:148–154.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Worku T, Bhattarai D, Ayers D, Wang K,
Wang C, Rehman ZU, Talpur HS and Yang L: Long non-coding RNAs: The
new horizon of gene regulation in ovarian cancer. Cell Physiol
Biochem. 44:948–966. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ding D, Li C, Zhao T, Li D, Yang L and
Zhang B: lncRNA H19/miR-29b-3p/PGRN axis promoted
epithelial-mesenchymal transition of colorectal cancer cells by
acting on Wnt signaling. Mol Cells. 41:423–435. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Qiu H, Wang X, Guo R, Liu Q, Wang Y, Yuan
Z, Li J and Shi H: HOTAIR rs920778 polymorphism is associated with
ovarian cancer susceptibility and poor prognosis in a Chinese
population. Future Oncol. 13:347–355. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zou A, Liu R and Wu X: Long non-coding RNA
MALAT1 is up-regulated in ovarian cancer tissue and promotes
SK-OV-3 cell proliferation and invasion. Neoplasma. 63:865–872.
2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Huang S, Qing C, Huang Z and Zhu Y: The
long non-coding RNA CCAT2 is up-regulated in ovarian cancer and
associated with poor prognosis. Diagn Pathol. 11(49)2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li J, Huang H, Li Y, Li L, Hou W and You
Z: Decreased expression of long non-coding RNA GAS5 promotes cell
proliferation, migration and invasion, and indicates a poor
prognosis in ovarian cancer. Oncol Rep. 36:3241–3250.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang H, Sun W, Sun M, Fu Z, Zhou C, Wang
C, Zuo D, Zhou Z, Wang G, Zhang T, et al: HER4 promotes cell
survival and chemoresistance in osteosarcoma via interaction with
NDRG1. Biochim Biophys Acta Mol Basis Dis. 1864 (5 Pt A):1839–1849.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu XL, Lu RY, Wang LK, Wang YY, Dai YJ,
Wang CY, Yang YJ, Guo F, Xue J and Yang DD: Long noncoding RNA
HOTAIR silencing inhibits invasion and proliferation of human colon
cancer LoVo cells via regulating IGF2BP2. J Cell Biochem Oct 18,
2018 (Online ahead of print).
|
|
25
|
Chang JW, Kuo WH, Lin CM, Chen WL, Chan
SH, Chiu MF, Chang IS, Jiang SS, Tsai FY, Chen CH, et al: Wild-type
p53 upregulates an early onset breast cancer-associated gene GAS7
to suppress metastasis via GAS7-CYFIP1-mediated signaling pathway.
Oncogene. 37:4137–4150. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lu YM, Rong ML, Shang C, Wang N, Li X,
Zhao YY and Zhang SL: Suppression of HER-2 via siRNA interference
promotes apoptosis and decreases metastatic potential of SKOV3
human ovarian carcinoma cells. Oncol Rep. 29:1133–1139.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wei P, Li L, Zhang Z, Zhang W, Liu M and
Sheng X: A genetic variant of miR-335 binding site in the ERBB4
3'-UTR is associated with prognosis of ovary cancer. J Cell
Biochem. 119:5135–5142. 2018.PubMed/NCBI View Article : Google Scholar
|