Introduction
Hypertrophic scar is a type of fibrotic skin disease
caused by abnormal healing of skin injuries including skin burns
(1), which is characterized by
excessive proliferation of fibroblasts, epidermal interstitial
transformation and collagen deposition (2). Hypertrophic scars commonly occur in
injured skin areas and cause pain, itching and other symptoms,
which cause great psychological effects to the patients. At
present, the main clinical treatment methods for scars include
surgical excision and steroid therapy, but the pathological
molecular mechanism of scar formation remains to be elucidated.
Therefore, the present study on the pathological mechanism of scar
formation may reveal a novel therapeutic target for the treatment
of hypertrophic scars.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs that contain >200 nucleotides. Studies have
demonstrated that lncRNAs significantly affect complicated
pathological processes of various diseases, e.g., cardiovascular
diseases (3), cerebral ischemic
diseases and carcinomas (4).
lncRNAs have no protein-coding capability, but they bind to micro
(mi)RNA as competitive endogenous RNAs and regulate the expression
of downstream target genes, thus serving a critical role in various
biological cellular processes and malignant diseases. LncRNA
nuclear-enriched transcripts 1 (NEAT1) is a tumor growth regulator
that plays an essential role in different types of cancer (5,6)
including breast (7), gastric
(8) and lung (9) cancer. It has been demonstrated that
lncRNA NEAT1 sponges miRNA (miR)-129 to regulate the
epithelial-mesenchymal transition (EMT) and inflammatory response
of renal fibrosis through regulation of collagen (COL)-I (10). Nonetheless, the molecular
regulation mechanism of lncRNA NEAT1 in hypertrophic scar formation
remains unclear.
The role of miRNA in the progression of malignant
diseases has attracted extensive attention (11). miRNA can competitively block the
translation of downstream target genes and negatively regulate the
expression level of target genes, thus serving a key part in the
treatment of hypertrophic scars (12). Wang et al (13) found that miR-31-5p participates in
the formation of hypertrophic scars (HSs) by inhibiting FIH and
regulating the expression of HIF-1α. Bi et al (14) found that miR-98 inhibits the
proliferation of hypertrophic scar fibroblasts by targeting COL1A1.
In addition, Li et al (15)
found that lncRNA8975-1 regulates the expression of COL3A1, COL1A1
and α-smooth muscle actin (α-SMA) and inhibits the proliferation of
fibroblasts in hypertrophic scars. COL3A1 has been identified as a
marker for fibroblast differentiation in hypertrophic scar
formation, implying that the expression level of COL3A1 is
associated with the pathological process of hypertrophic scars.
Given the abnormal proliferation of fibroblasts, it was
hypothesized that COL3A1 is involved in the proliferation of
fibroblasts during scar formation. The present study aimed to
investigate the inhibitive effects of lncRNA NEAT1 on cell
proliferation of hypertrophic scars and elucidate the molecular
mechanism of the miR-488-3p/COL3A1 axis in regulating scar
fibroblast proliferation.
Materials and methods
Cell culture and treatments
Scar fibroblasts purchased from American Type
Culture Collection were incubated in Dulbecco's Modified Eagle's
Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin-streptomycin in 5% CO2 at 37˚C.
The groups of the experiment included the control group (21%
O2), the 10% hypoxia group (10% O2), the 5%
hypoxia group (5% O2) and the 1% hypoxia group (1%
O2). After 48 h of culture, the experiments were
started.
Bioinformatics analysis
Bioinformatics analysis was performed to predict the
downstream miRNA and mRNA of lncRNA NEAT1 using Starbase
(https://starbase.sysu.edu.cn/).
Cell transfection
Scar fibroblasts underwent transfection with 20 nM
miR-488-3p mimic or the corresponding negative control (Shanghai
GenePharma Co., Ltd.) by using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at 37˚C. For
COL3A1 overexpression, the recombinant sense expression vector
plasmid Cytomegalovirus promoter DNA 3.1 for COL3A1 (1 µg/µl
pcDNA3.1-COL3A1; Invitrogen; Thermo Fisher Scientific, Inc.) was
constructed by subcloning the cDNA fragment of COL3A1 containing
the complete coding sequence between KpnI and BamHI.
Short hairpin RNA (shRNA) targeting NEAT1 (sh-NEAT1) and their
negative control (sh-NC) were purchased from Shanghai GenePharma
Co., Ltd. Sequences were cloned in the pEGFP plasmid. miR-488-3p
mimic (miR-488-3p; 5'-UUGAAAGGCUAUUUCUUGGUC-3') and mimic control
(miR-NC; 5'-UUCUCCGAACGUGUCACGUTT-3' and
5'-ACGUGACACGUUCGGAGAATT-3'), miR-488-3p inhibitor
(anti-miR-488-3p; 5'-CUGUUCCUGCUGAACUGAGCCA-3') and inhibitor
control (anti-miR-NC; 5'-CAGUACUUUUGUGUAGUACAA-3') were also
purchased from Shanghai GenePharma Co., Ltd. Scar fibroblasts were
seeded into 24-well plates at a density of 2.0x104
cells/well, following which 50 nM synthetic oligonucleotides or 2
µg vectors were transfected into the cells using
Lipofectamine® 2000 for 24 h at 37˚C (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Following transfection, cells were incubated in fresh
DMEM at 37˚C for 24 h and finally collected for the subsequent
experiments.
CCK-8 assay
Scar fibroblasts transfected with designated vectors
for 72 h were seeded into 96-well plates at a density of
1x103 cells/well. Cell viability was detected by Cell
Counting Kit-8 assay (CCK-8 assay; Beyotime Institute of
Biotechnology) at 48 h following the manufacturer's instructions.
The optical density at 450 nm of each well was determined by using
a microplate reader.
Luciferase reporter assay
lncRNA NEAT1 with or without miR-488-3p putative
target sites were synthesized and obtained from Shanghai GenePharma
Co., Ltd. and cloned into the Xho I/Not I sites of a psiCHECK
vector (Promega Corporation). For transfection assays, cells were
seeded into 24-well plates and transfected with miR-488-3p mimics
or miR-488-3p inhibitor using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. After further incubation for 24 h, the
transfected cells were harvested, lysed and centrifuged (10,000 x
g, 10 min, 4˚C) to obtain the supernatant for the luciferase assay
by Dual-Luciferase Reporter Assay kit (Promega Corporation)
according to the manufacturer's instructions. Luciferase activity
was normalized to Renilla luciferase activity.
Immunofluorescence staining
Transfected cells were fixed with 4%
paraformaldehyde for 15 min and permeabilized by 0.1% Triton X-100
(Beyotime Institute of Biotechnology) for 30 min at room
temperature. After being blocked in normal goat serum (Beijing
Solarbio Science & Technology Co., Ltd.) for 15 min at room
temperature, the cells were incubated with Ki-67 primary antibody
at a 1:200 dilution (Abcam) in a wet box at 4˚C overnight. Then,
Cy3-labeled goat anti-rabbit IgG secondary antibody at a 1:200
dilution (Beyotime Institute of Biotechnology) was added to
incubate the cells at room temperature for 60 min. Next, the cells
were rinsed with PBS and the nucleus counterstained with DAPI for
10 min at room temperature (Beyotime Institute of Biotechnology).
Finally, the cells were sealed with the mounting medium (Beijing
Solarbio Science & Technology Co., Ltd.) and fluorescence was
observed under a fluorescent microscope (Olympus Corporation;
magnification, x400) by a pathology experimenter blinded to the
experimental or control group.
Reverse transcription-quantitative
(RT-q)PCR
A total of 1 µg RNA was extracted from scar
fibroblasts by using TRIzol® (Thermo Fisher Scientific,
Inc.). Reverse transcription was performed using RT Reagent kit
(cat. no. RR037A; Takara, Bio, Inc.). qPCR (cat. no. RR820A;
Takara, Bio, Inc.) was performed using SYBR Green mix (Takara Bio,
Inc.) with primers specific to miR-488-3p (Guangzhou RiboBio Co.,
Ltd.). The PCR conditions were as follows: 95˚C for 10 min,
followed by 40 cycles of 95˚C for 30 sec, 60˚C for 30 sec and 72˚C
for 1 min. Relative quantification of the miRNA expression was
calculated through the 2-ΔΔCq method (16). Primers: miR-488-3p forward,
5'-ACACTCCAGCTGGGTTGAAAGGCTATTTC-3' and reverse,
5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGACCAAGA-3'; NEAT1 forward,
5'-GGAGAGGGTTGGTTAGAGAT-3' and reverse, 5'-CCTTCAACCTGCATTTCCTA-3';
U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse,
5'-AACGCTTCACGAATTTGCGT-3'; and GAPDH forward,
5'-GCACCGTCAAGGCTGAGAAC-3' and reverse 5'-GGATCTCGCTCCTGGAAGATG-3'.
U6 and GAPDH were selected as the housekeeping gene to normalize
the expression of miRNA and mRNA.
Western blotting
Total protein lysates were generated using RIPA
lysis buffer supplemented with protease and phosphatase inhibitor
mixtures (cat. no. KC-440; Shanghai KangChen Biological Technology
Co. Ltd.). Nuclear proteins were extracted by using the NE-PER
nuclear and cytoplasmic extraction reagents (Pierce; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
concentration of the protein in cells lysates was detected using a
BCA kit (Beijing Solarbio Science & Technology Co., Ltd.).
Proteins (40 µg) were loaded onto a 5-10% polyacrylamide gel,
separated by electrophoresis and transferred onto a polyvinylidene
difluoride (PVDF) membrane. Then, the PVDF membrane was blocked
with a 5% solution of non-fat milk at room temperature for 3 h and
incubated with rabbit polyclonal antibodies to COL-I (1:1,000 cat.
no. ab260043; Abcam) and COL-III (1:1,000; cat. no. ab7778; Abcam),
α-SMA (1:1,000; cat. no. 19245s; Cell Signaling Technology, Inc.)
and GAPDH (1:1,000; cat. no. 5174s; Cell Signaling Technology,
Inc.) at 4˚C overnight. Horseradish peroxidase-conjugated goat
anti-rabbit (cat. no. BA1054) or anti-mouse (cat. no. BA1050)
antibody (1:15,000) (Wuhan Boster Biological Technology, Ltd.) was
utilized as a secondary antibody, incubated at 37˚C for 1 h.
Proteins were visualized by an enhanced chemiluminescence system
using the FluorChem FC system (ProteinSimple). ImageJ software
V.1.4 (National Institutes of Health) was used to measure the gray
values of the bands and analyze the changes in relative protein
expression levels.
Statistical analyses
Statistical analyses were performed using SPSS 13.0
software (SPSS Inc., Chicago, IL, USA). The data are shown as mean
± standard error of the mean (SEM) from three independent
experiments. Statistical analyses were conducted using Student's
t-test or ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
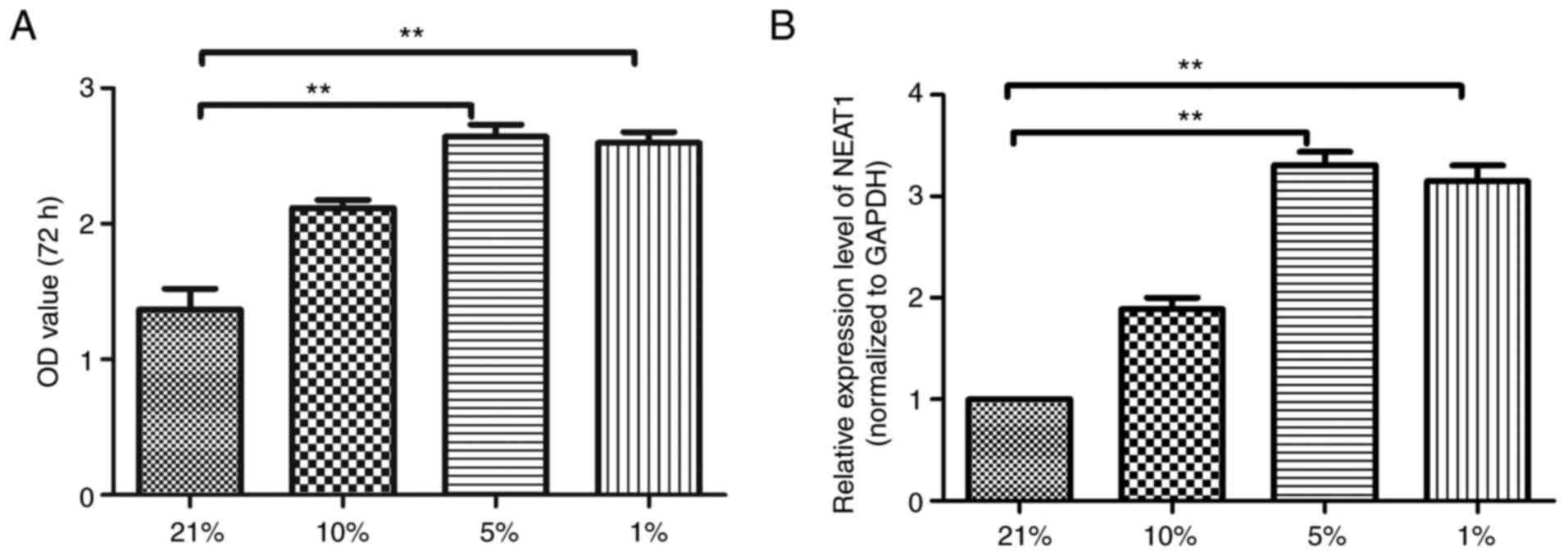
Upregulated lncRNA-NEAT1 expression in
scar fibroblasts under different hypoxic conditions
First, the viability of scar fibroblasts under
different hypoxic conditions was detected by CCK-8 kit. As shown in
Fig. 1A, the CCK-8 results
indicated that with the decrease of oxygen concentration, the
viability of scar fibroblasts was significantly enhanced; the
viability of scar fibroblasts in the 5% group was the highest. The
expression of lncRNA NEAT1 in scar fibroblasts under different
hypoxic conditions was detected. The RT-qPCR analysis results
(Fig. 1B) implied that compared
with the 21% group, the expression of lncRNA NEAT1 in scar
fibroblasts in the 5% group and the 1% group were markedly
increased (P<0.001).
Effects of lncRNA NEAT1 on
hypoxia-induced scar fibroblast proliferation
RT-qPCR analysis was performed to detect the
expression of lncRNA NEAT1 in different groups. As shown in
Fig. 2A, compared with the NC
group, the expression of lncRNA NEAT1 in the lncRNA NEAT1 group and
the hypoxia (5%) + lncRNA NEAT1 group was significantly reduced,
whereas that in the hypoxia (5%) + NC group was increased. To
investigate the effect of lncRNA NEAT1, the hypoxia-induced scar
fibroblasts proliferation was detected by CCK-8 kit. The results
(Fig. 2B) demonstrated that
compared with the NC group, the viability of scar fibroblasts in
the hypoxia (5%) + NC group was substantially enhanced while that
in the hypoxia (5%) + lncRNA NEAT1 group was inhibited. Ki-67
protein can be used as a proliferation marker for hypertrophic scar
fibroblasts, hence RT-qPCR analysis and immunofluorescence staining
were performed to detect the expression of Ki-67 in scar
fibroblasts. The results (Fig. 2C)
indicated that compared with the NC group, the expression of Ki-67
in the hypoxia (5%) + NC group was significantly increased while
that in the hypoxia (5%) + lncRNA NEAT1 group was markedly reduced,
which is consistent with the immunofluorescence staining results
(Fig. 2D). Therefore, lncRNA NEAT1
can inhibit the proliferation of hypoxia-induced scar
fibroblasts.
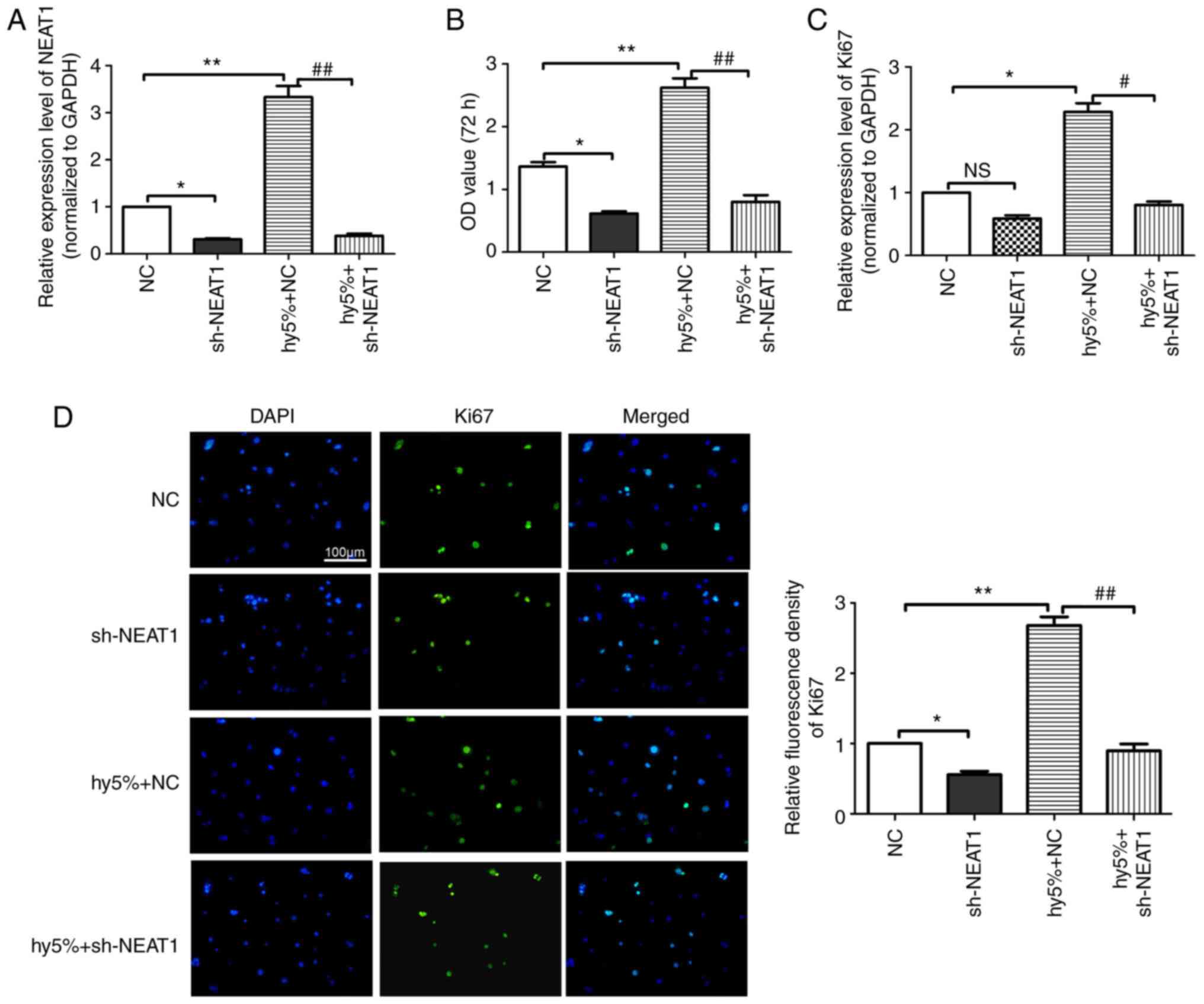 | Figure 2Effects of lncRNA NEAT1 on
hypoxia-induced scar fibroblast proliferation. (A) The mRNA
expression level of NEAT1 in different groups detected by RT-qPCR
analysis. (B) The ability of proliferation in hypoxia-induced scar
fibroblasts measured by CCK-8 kit. (C) The mRNA expression of Ki-67
in hypoxia-induced scar fibroblasts assessed by RT-qPCR. (D) The
positive rate of Ki-67 in fibroblasts detected by
immunofluorescence staining: the representative staining results
(left) and the statistical results (right). The data are presented
as mean ± standard error of the mean, vs. NC group,
*P<0.05, **P<0.01; vs. hy 5% + lncRNA
NEAT1 group, #P<0.05, ##P<0.01, n=6.
lncRNA, long non-coding RNA; NEAT1, nuclear-enriched transcripts 1;
RT-qPCR, reverse transcription-quantitative PCR; NC, negative
control; sh, short hairpin; hy, hypoxia. |
Effects of lncRNA NEAT1 on
steady-state protein levels of COL-I, COL-III and α-SMA in scar
fibroblasts
To investigate the regulatory mechanism of lncRNA
NEAT1 in scar fibroblasts, RT-qPCR analysis and western blotting
were performed to detect the mRNA and protein expression levels of
COL-I, COL-III and α-SMA in scar fibroblasts. The RT-qPCR results
(Fig. 3A-C) indicated that
compared with the NC group, the mRNA expression of COL-I, COL-III
and α-SMA in the Hy + NC group was substantially upregulated while
that in the Hy + lncRNA NEAT1 group was downregulated, which is
consistent with the western blotting results (Fig. 3D). In summary, the RT-qPCR results
and the western blotting results demonstrated that lncRNA NEAT1 can
downregulate the expression levels of COL-I, COL-III and α-SMA in
hypoxia-induced scar fibroblasts.
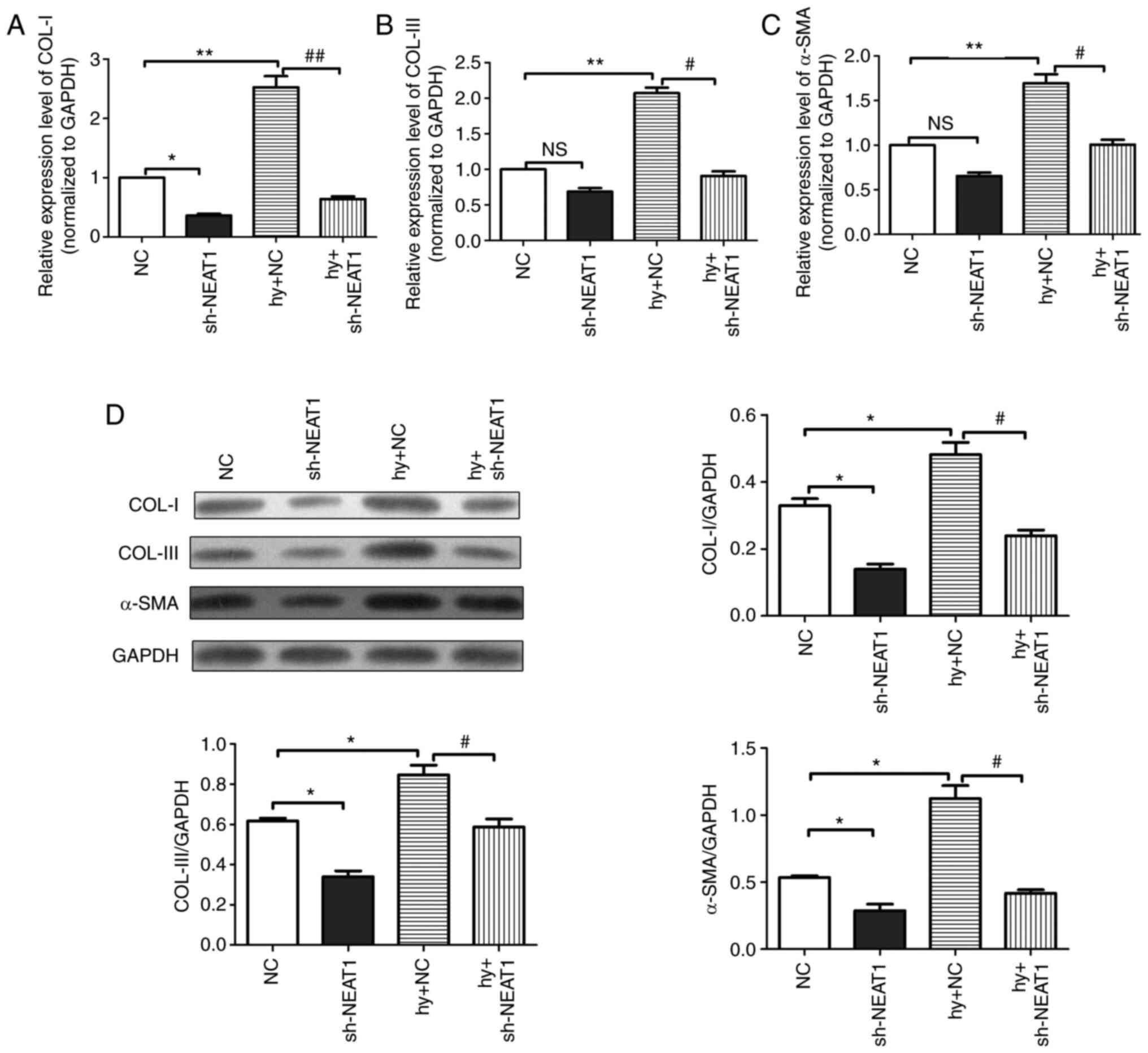 | Figure 3Effects of lncRNA NEAT1 on the
steady-state protein levels of COL-I, COL-III and α-SMA in scar
fibroblasts. The mRNA expression levels of (A) COL-I, (B) COL-III
and (C) α-SMA in scar fibroblasts detected by reverse
transcription-quantitative PCR. (D) The protein expression levels
of COL-I, COL-III and α-SMA in scar fibroblasts detected by the
western blotting assay. All protein expression levels were
normalized to GAPDH. The data are presented as mean ± standard
error of the mean, vs. NC group, *P<0.05,
**P<0.01; vs. Hy + NC group, #P<0.05,
##P<0.01, n=6. long non-coding RNA; NEAT1,
nuclear-enriched transcripts 1; COL, collagen; α-SMA, α-smooth
muscle actin; sh, short hairpin; Hy, hypoxia; NC, negative control;
NS, not significant. |
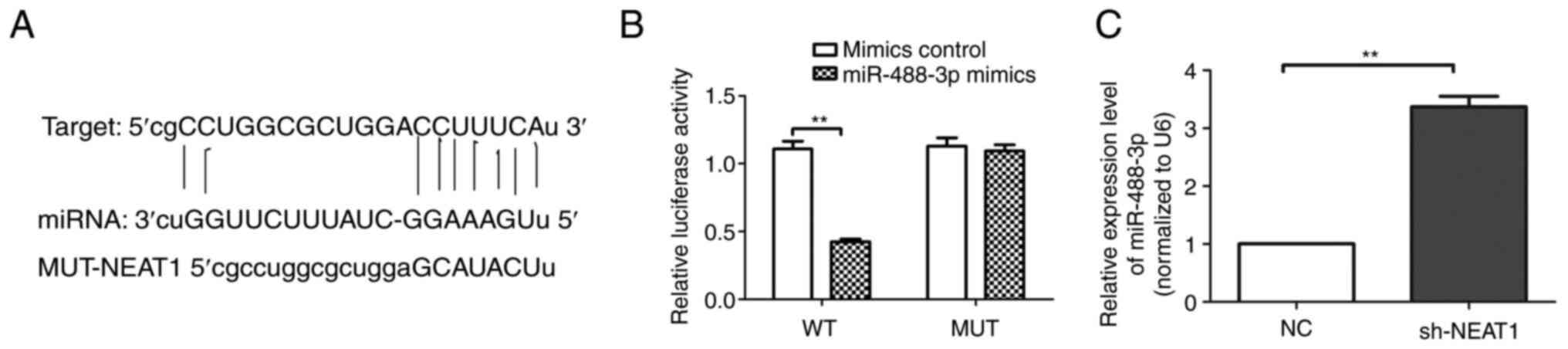
lncRNA NEAT1 directly targeted
miR-488-3p
Luciferase activity of scar fibroblasts (Fig. 4A and B) transfected with miR-488-3p mimics was
markedly reduced compared with the mimics control group
(P<0.01). In addition, the RT-qPCR results (Fig. 4C) demonstrated that the expression
of miR-488-3p in the lncRNA NEAT1 group was significantly increased
compared with the NC group. Therefore, it was confirmed that lncRNA
NEAT1 directly targets miR-488-3p.
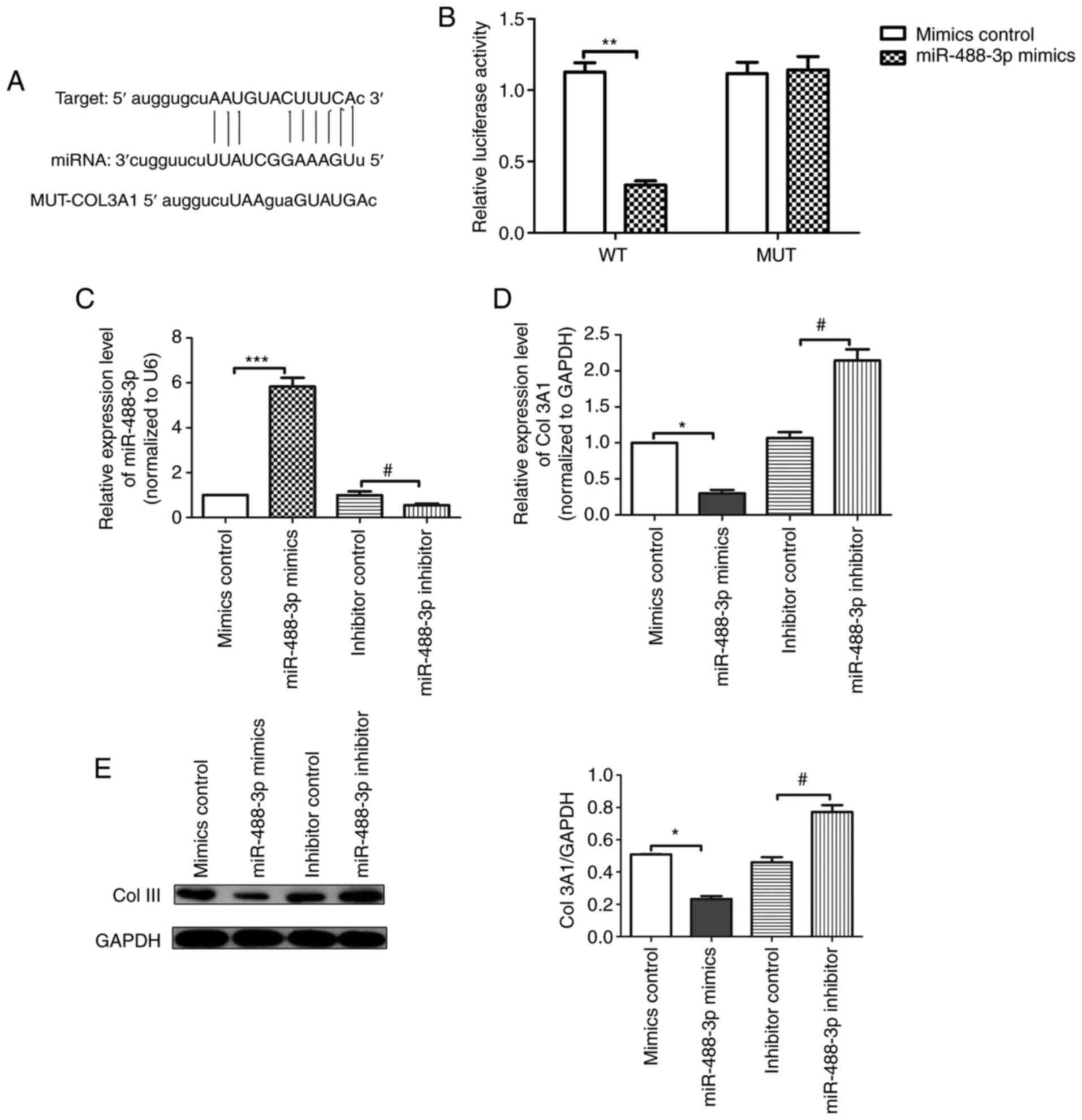
miR-488-3p directly targets
COL3A1
As indicated by the results of the luciferase
activity assay (Fig. 5A and
B), luciferase activity of scar
fibroblasts transfected with miR-488-3p mimics was substantially
reduced compared with the mimics control group (P<0.01). In
addition, compared with the mimics control group, the expression
level of miR-488-3p (Fig. 5C) in
the miR-488-3p mimics group was significantly increased. Compared
with the inhibitor control group, the expression level of
miR-488-3p was significantly reduced in the miR-488-3p inhibitor
group. Furthermore, the RT-qPCR analysis results (Fig. 5D) indicated that the mRNA
expression level of COL3A1 in the miR-488-3p mimics group was
substantially downregulated compared with the mimics control group,
whereas that in the miR-488-3p inhibitor group was increased
compared with the inhibitor control group. The western blotting
results (Fig. 5E) were consistent
with the RT-qPCR results. Given the above results, it was concluded
that miR-488-3p directly targets COL3A1 in scar fibroblasts.
lncRNA NEAT1 inhibited hypoxia-induced
scar fibroblasts proliferation through regulation of
miR-488-3p/COL3A1 axis
To detect the mRNA expression level of COL3A1 in
scar fibroblasts after transfection, RT-qPCR analysis was
performed. The results (Fig. 6A)
demonstrated that compared with the pcDNA3.1 group, the mRNA
expression level of COL3A1 in the pcDNA3.1-COL3A1 group was
significantly increased. The western blotting results (Fig. 6B) indicated that compared with the
Hy + NC group, the protein expression level of COL-III in the Hy +
lncRNA NEAT1 group was substantially reduced; compared with the Hy
+ lncRNA NEAT1 + inhibitor control group, the protein expression
level of COL-III in the Hy + lncRNA NEAT1 + miR-488-3p inhibitor
group was substantially increased; compared with the Hy + lncRNA
NEAT1 + pcDNA3.1 group, the protein expression level of COL-III in
the Hy + lncRNA NEAT1 + pcDNA3.1-COL3A1 group was markedly
increased. The proliferation of scar fibroblasts was detected by
CCK-8 kit. The results (Fig. 6C)
suggested that compared with the Hy + NC group, the viability of
scar fibroblasts in the Hy + lncRNA NEAT1 inhibitor group was
substantially inhibited; compared with the Hy + lncRNA NEAT1 +
inhibitor control group, the viability of scar fibroblasts in the
Hy + lncRNA NEAT1 + miR-488-3p inhibitor group was significantly
enhanced; compared with the Hy + lncRNA NEAT1 + pcDNA3.1 group, the
viability in the Hy + lncRNA NEAT1 + pcDNA3.1-COL3A1 group was
increased with statistic difference (P<0.05). Meanwhile, the
expression of Ki-67 protein in scar fibroblasts was detected
through RT-qPCR analysis and the results (Fig. 6D) were consistent with the CCK-8
assay results. Given the above, lncRNA NEAT1 inhibited
hypoxia-induced scar fibroblast proliferation through regulation of
miR-488-3p/COL3A1 axis.
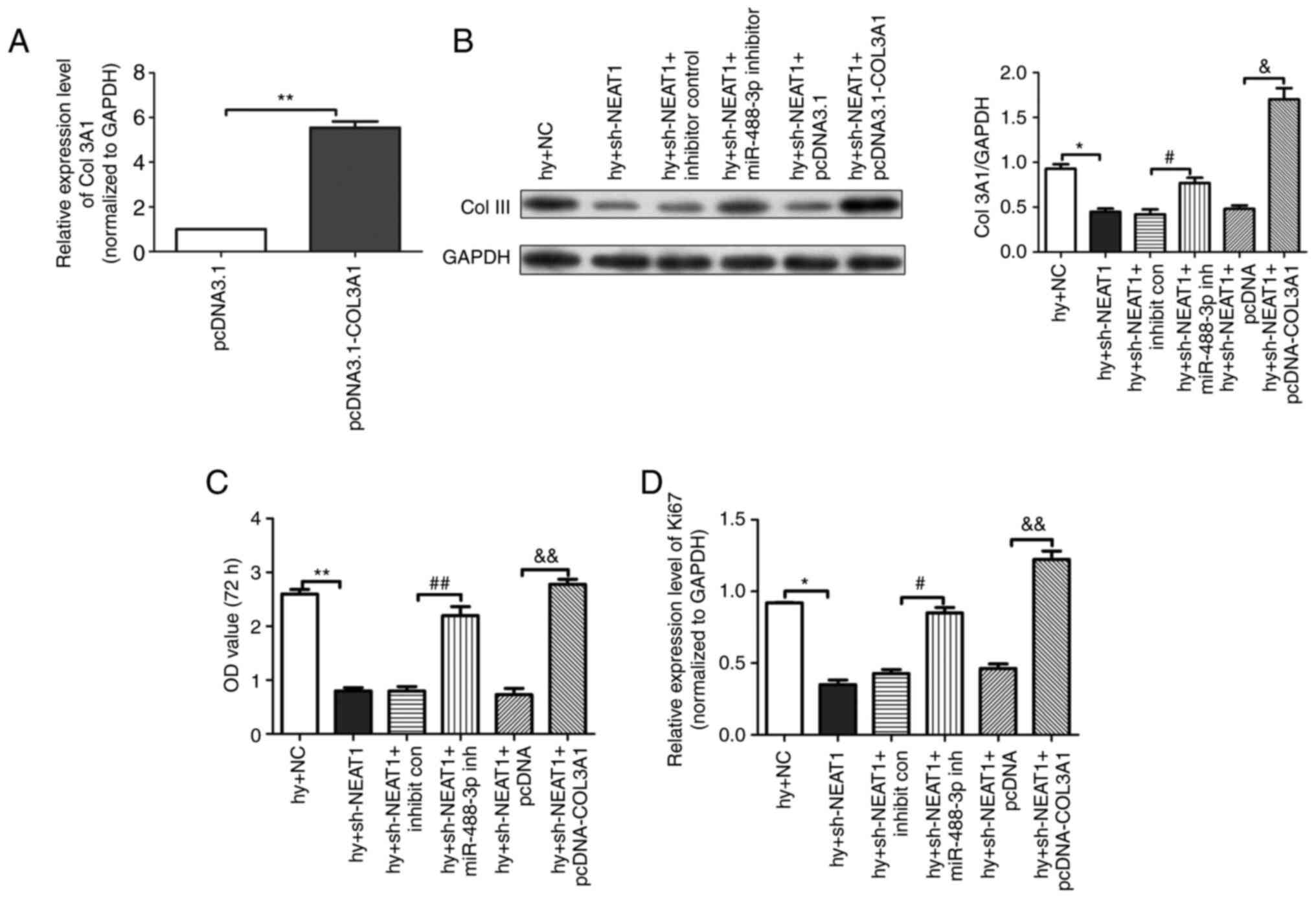 | Figure 6lncRNA NEAT1 inhibited hypoxia-induced
scar fibroblasts proliferation through regulation of
miR-488-3p/COL3A1 axis. (A) The mRNA expression level of COL3A1 in
scar fibroblasts detected by RT-qPCR analysis. (B) The protein
expression of COL-III in fibroblasts measured by the western
blotting assay. (C) The ability of cell proliferation assessed by
CCK-8 kit. (D) The mRNA expression of Ki-67 detected by RT-qPCR
analysis. The data are presented as mean ± standard error of the
mean, vs. pcDNA3.1 & Hy + NC group, *P<0.05,
**P<0.01; vs. Hy + lncRNA NEAT1 + inhibitor control
group, #P<0.05, ##P<0.01; vs. Hy +
lncRNA NEAT1 + pcDNA3.1 group, &P<0.05 and
&&P<0.01. n=3. LncRNA; long non-coding RNA;
NEAT1, nuclear-enriched transcripts 1; miR/miRNA, microRNA; COL,
collagen; RT-qPCR, reverse transcription-quantitative PCR; Hy,
hypoxia; NC, negative control; NS, not significant; sh, short
hairpin. |
Discussion
Hypertrophic scar is a common proliferative disease
associated with abnormal wound healing responses (17), so clarifying its pathological
mechanism is conducive to determining appropriate treatment
strategies. Abnormal proliferation and apoptosis of scar
fibroblasts directly or indirectly affect their collagen deposition
and scar formation (18). With the
development of biomedical science, increasing evidences have
demonstrated that lncRNAs play a regulatory role in the
pathogenesis of various diseases including cancer, myocardial
infarction, pulmonary fibrosis and hypertrophic scars by regulating
key proteins with competing endogenous (ce) RNAs (19-21).
LncRNA NEAT1 regulates FRS2 by targeting miR-29-3p in hypertrophic
scar fibroblasts, thereby exacerbating the pathological process of
scar formation (22). The results
of the present study indicated that lncRNA NEAT1 serves an
important role in hypertrophic scars by mediating miR-488-3p/COL3A1
axis. Therefore, it was hypothesized that lncRNA NEAT1 acts as a
mediator in the progression of hypertrophic scars.
Hypertrophic scar formation is a complicated
pathological process characterized by inflammation, collagen
deposition and fibroblast dysfunction. Activated fibroblasts are
the main effector cells in this fibrosis process (23). The abnormal proliferation of scar
fibroblasts and the inflammation-mediated fibrosis directly affect
scar formation. Bai et al (24) found that loureirin B suppresses
scar fibroblasts proliferation and fibrosis induced by TGF-β1 by
downregulating the expression of fibrosis-related molecules by
regulating MMPs. Liu et al (25) found that miR-6836-3p promotes the
proliferation of scar fibroblasts by upregulating the expression of
connective tissue growth factor, hence miR-6836-3p may be a
potential target in the treatment of hypertrophic scars. The
present study investigated the effects of lncRNA NEAT1 on the
function of hypoxia-induced scar fibroblasts. The results
demonstrated that there was a significant decrease in cell
viability and the expression level of Ki-67 protein in the lncRNA
NEAT1 group compared with the sh-NC group; silencing lncRNA NEAT1
inhibited the proliferation of fibroblasts and the pathological
progression of hypertrophic scars. Nonetheless, the underlying
molecular mechanism of the effects of lncRNA NEAT1 on proliferation
in hypertrophic scar formation remains to be investigated in future
research.
As the main participants of scar formation in wound
healing, scar fibroblasts are involved in biological processes
including collagen synthesis, extracellular matrix (ECM) formation
and deposition (26) and skin
fibrosis. TGF-β1 recruits macrophages to release inflammatory
factors, promotes the chemotaxis of fibroblasts and smooth muscle
and regulates the collagen gene expression in fibrosis (27). Collagens are known to regulate the
migration, proliferation and gene expression of cells (28). In hypertrophic scars, fibroblasts
synthesize excessive ECM proteins, among which the deposition of
collagens, especially COL-I and COL-III, is significantly increased
(29). Consistent with previous
studies, the results of the present study confirmed that collagen
deposition is significantly increased in hypoxia-induced
hypertrophic scars. The present study found that silencing lncRNA
NEAT1 targeted miR-488-3p to downregulate the expression levels of
COL-I, COL-III and α-SMA in hypoxia-induced fibroblasts under
hypoxic pathological conditions. Zhang et al (30) concluded that Ft1 activates the
PI3K/Akt/mTOR signaling pathway and promotes the expression levels
of COL1A1 and COL3A1, thereby stimulating fibroblast proliferation
and myofibroblast differentiation and accelerating wound healing.
The present study found that silencing lncRNA NEAT1 mediated
miR-488-3p/COL3A1 axis, downregulated collagen expression levels
and attenuated the process of hypertrophic scarring.
In summary, the knockdown of lncRNA NEAT1 expression
inhibited scar fibroblast proliferation through regulation of the
miR-488-3p axis and regulated a series of collagens including
COL3A1 to serve protective roles in hypertrophic scar formation.
The results demonstrated that lncRNA NEAT1 may be a novel
therapeutic target for the treatment of hypertrophic scars.
Nonetheless, the limitation of this study lies in the fact that the
underlying regulatory mechanism of lncRNA NEAT1 in hypertrophic
scar formation remains unconfirmed in vivo experiments.
Acknowledgements
Not applicable.
Funding
Funding: The study was supported by the hospital level project
of Children's Hospital of Shanxi (grant no. 202028)
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HX wrote the manuscript, designed experiments and
analyzed the data. XG and YT participated in experiments and data
analysis. JW participated in experiments and literature review. HX
and XG confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen L and Li J, Li Q, Yan H, Zhou B, Gao
Y and Li J: Non-coding RNAs: The new insight on hypertrophic scar.
J Cell Biochem. 118:1965–1968. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lee HJ and Jang YJ: Recent understandings
of biology, prophylaxis and treatment strategies for hypertrophic
scars and keloids. Int J Mol Sci. 19(711)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhang J, Yu L, Xu Y, Liu Y, Li Z, Xue X,
Wan S and Wang H: Long noncoding RNA upregulated in hypothermia
treated cardiomyocytes protects against myocardial infarction
through improving mitochondrial function. Int J Cardiol.
266:213–217. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shan G, Tang T, Xia Y and Qian HJ: Long
non-coding RNA NEAT1 promotes bladder progression through
regulating miR-410 mediated HMGB1. Biomed Pharmacother.
121(109248)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang X, Guan MX, Jiang QH, Li S, Zhang
HY, Wu ZG, Cong HL and Qi XH: NEAT1 knockdown suppresses
endothelial cell proliferation and induces apoptosis by regulating
miR-638/AKT/mTOR signaling in atherosclerosis. Oncol Rep.
44:115–125. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
lncRNA NEAT1 facilitates cell
proliferation, invasion and migration by regulating CBX7 and RTCB
in breast cancer (retraction). Onco Targets Ther.
13(7807)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang J, Zhao B, Chen X, Wang Z, Xu H and
Huang B: Silence of long noncoding RNA NEAT1 inhibits malignant
biological behaviors and chemotherapy resistance in gastric cancer.
Pathol Oncol Res. 24:109–113. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gu G, Hu C, Hui K, Chen T, Zhang H and
Jiang X: NEAT 1 knockdown enhances the sensitivity of human
non-small-cell lung cancer cells to anlotinib. Aging (Albany NY).
13:13941–13953. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li C, Liu YF, Huang C, Chen YX, Xu CY and
Chen Y: Long noncoding RNA NEAT1 sponges miR-129 to modulate renal
fibrosis by regulation of collagen type I. Am J Physiol Renal
Physiol. 319:F93–F105. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen X, Xie D, Zhao Q and You ZH:
MicroRNAs and complex diseases: From experimental results to
computational models. Brief Bioinform. 20:515–539. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wu X, Li J, Yang X, Bai X, Shi J, Gao J,
Li Y, Han S, Zhang Y, Han F, et al: miR-155 inhibits the formation
of hypertrophic scar fibroblasts by targeting HIF-1α via PI3K/AKT
pathway. J Mol Histol. 49:377–387. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang X, Zhang Y, Jiang BH, Zhang Q, Zhou
RP, Zhang L and Wang C: Study on the role of Hsa-miR-31-5p in
hypertrophic scar formation and the mechanism. Exp Cell Res.
361:201–209. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bi S, Chai L, Yuan X, Cao C and Li S:
MicroRNA-98 inhibits the cell proliferation of human hypertrophic
scar fibroblasts via targeting Col1A1. Biol Res.
50(22)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li J, Chen L, Cao C, Yan H, Zhou B, Gao Y,
Li Q and Li J: The long non-coding RNA LncRNA8975-1 is upregulated
in hypertrophic scar fibroblasts and controls collagen expression.
Cell Physiol Biochem. 40:326–334. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kirkpatrick LD, Shupp JW, Smith RD,
Alkhalil A, Moffatt LT and Carney BC: Galectin-1 production is
elevated in hypertrophic scar. Wound Repair Regen. 29:117–128.
2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tian S, Zheng Y, Xiao S, Luo P, Sun R, Liu
J and Xia Z: Ivermectin inhibits cell proliferation and the
expression levels of type I collagen, α-SMA and CCN2 in
hypertrophic scar fibroblasts. Mol Med Rep. 24(488)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20(5758)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Peng Q, Li L and Bi X: Long noncoding RNA
small nuclear RNA host gene 7 knockdown protects mouse cardiac
fibroblasts against myocardial infarction by regulating
miR-455-3p/platelet-activating factor receptor axis. J Cardiovasc
Pharmacol. 77:796–804. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tu L, Huang Q, Fu S and Liu D: Aberrantly
expressed long noncoding RNAs in hypertrophic scar fibroblasts
in vitro: A microarray study. Int J Mol Med. 41:1917–1930.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu Q, Chen J, Tan Z, Wang D, Zhou J, Li D
and Cen Y: Long non-coding RNA (lncRNA) nuclear enriched abundant
transcript 1 (NEAT1) regulates fibroblast growth factor receptor
substrate 2 (FRS2) by targeting microRNA (miR)-29-3p in
hypertrophic scar fibroblasts. Bioengineered. 12:5210–5219.
2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang J, Dodd C, Shankowsky HA, Scott PG
and Tredget EE: Wound Healing Research Group. Deep dermal
fibroblasts contribute to hypertrophic scarring. Lab Invest.
88:1278–1290. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bai X, He T, Liu J, Wang Y, Fan L, Tao K,
Shi J, Tang C, Su L and Hu D: Loureirin B inhibits fibroblast
proliferation and extracellular matrix deposition in hypertrophic
scar via TGF-β/Smad pathway. Exp Dermatol. 24:355–360.
2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu F, Chen WW, Li Y, Zhang JQ and Zheng
QB: MiR-6836-3p promotes proliferation of hypertrophic scar
fibroblasts by targeting CTGF. Eur Rev Med Pharmacol Sci.
22:4069–4074. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Deng J, Shi Y, Gao Z, Zhang W, Wu X, Cao W
and Liu W: Inhibition of pathological phenotype of hypertrophic
scar fibroblasts via coculture with adipose-derived stem cells.
Tissue Eng Part A. 24:382–393. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hamed S, Ullmann Y, Egozi D, Daod E,
Hellou E, Ashkar M, Gilhar A and Teot L: Fibronectin potentiates
topical erythropoietin-induced wound repair in diabetic mice. J
Invest Dermatol. 131:1365–1374. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang X, Song Z, Hu B, Chen Z, Chen F and
Cao C: MicroRNA-642a-5p inhibits colon cancer cell migration and
invasion by targeting collagen type I α1. Oncol Rep. 45:933–944.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang Q, Peng Z, Xiao S, Geng S, Yuan J and
Li Z: RNAi-mediated inhibition of COL1A1 and COL3A1 in human skin
fibroblasts. Exp Dermatol. 16:611–617. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang E, Gao B, Yang L, Wu X and Wang Z:
Notoginsenoside Ft1 promotes fibroblast proliferation via
PI3K/Akt/mTOR signaling pathway and benefits wound healing in
genetically diabetic mice. J Pharmacol Exp Ther. 356:324–332.
2016.PubMed/NCBI View Article : Google Scholar
|