1. Kidney transplantation in chronic kidney
disease
Chronic kidney disease (CKD) is a worldwide public
health problem. The constantly increasing prevalence of CKD
requires more research into new additional strategies in its
management. Impaired kidney function is accompanied by numerous
complications related to water and electrolyte balance disorders
and accumulation of uremic toxins which are physiologically
excreted in the urine, as well as increased risk of cardiovascular
events, thus affecting mortality, morbidity and the quality of life
of patients with CKD (1).
The prognosis of CKD patients is dependent on the
progression of renal dysfunction to its total function loss and
also, on the specific complications of their chronic disease
(2-4).
The optimal treatment for renal function impairment
is kidney transplantation, which ensures a higher quality of life
and longer survival than maintenance with dialysis, in patients
with end-stage renal disease (ESRD). Over the last few years the
prognosis of kidney transplanted patients has significantly
improved reaching a graft survival rate of over 92% per year
(5-7).
Currently, the attention is directed on prolonging long-term graft
survival as much as possible.
A narrative review was performed on articles
published between 1980 and 2020, which were identified on PubMed,
via specific mesh terms: ‘Perforin’, ‘kidney’, ‘granzyme’,
‘transplantation’, ‘graft dysfunction’. Furthermore, citation
tracking of the studies retrieved was used to identify additional
relevant articles. Only the articles written in English were
evaluated. A total of 93 references were introduced.
2. Brief immunology of renal
transplantation
Transplant rejection represents the rejection of a
transplant allograft or transplanted organ. This occurs because the
graft is accompanied by a series of antigens that the immune system
of the recipient perceives as non-self, and consequently an immune
response (host vs. graft reaction) is produced (8).
The mammalian immune system is an extremely complex
system developed over millions of years as evolved immune response
of vertebrates against microbial invasion and ensures the species
continuity. The system could be divided into adaptive and innate
immunity. Innate immunity represents a non-specific immune system,
and the first line of defense that involves recruitment and
participation of macrophages, neutrophils, natural killer (NK)
cells, cytokines, certain cell receptors and complement components
and precedes adaptive immunity functioning as secondary signals for
lymphocyte activation (8).
While the inherited immunity does not involve the
recognition of specific antigens, adaptive immunity involves the
recognition of a wide range of molecules, the identification of
different similar structures (high specificity to the pathogen
agent) and the immune memory (recognition of the aggressor at the
first contact and specific reaction by an accelerated and
protective response) (9). The
adaptive immune response is considered the most important hurdle in
organ transplantation.
The main target of the immune response to the graft
in organ transplantation are the major histocompatibility complex
molecules (MHC) expressed on the surface of the donor cells; this
feature is a form of adaptive immunity (9). The MHC is a complex of polymorphic
genes encoded in a locus situated on the short arm of human
chromosome 6. MHC protein products are expressed on the surfaces of
various cells. In humans these are called human leukocyte antigens
(HLA) and are analogous to the H-2 (in mice) and RT1 (in rats)
systems (9). Graft antigens that
serve as the main target of rejection are proteins encoded by the
MHC genes.
Graft rejection is the result of immune mechanism
activation due to antigenic differences between MHC I and II
molecules of the recipient and donor, the latter acting as major
antigens in the body of the recipient. These molecules present a
high polymorphism, in particular those of HLA class I A and B with
at least 200 and 250 alleles, respectively, which have been
described in the human population while HLA-C and HLA-DP have a
limited polymorphism and thus low significance (9,10).
In order to have moderate affinity for their own MHC
molecules, specifically selected T cells during thymus development
recognize portions of protein antigens that have been fragmented
into peptides bound to MHC class I and II molecules. T-cell
recognition of the antigen is the main event that initiates the
effect of immune response mechanisms followed by two discrete
signals. The first phase (signal 1) is the recognition of the
complex formed by the MHC class II molecule and the antigenic
peptide by the surface lymphocyte receptor (TCR-CD3) and Th (T
helper) fixing to the antigen-presenting cell (APC). The T-cell
receptor is a heterodimer consisting of an α polypeptide chain and
a β polypeptide chain, which associates on the surface of the T
cell, with the CD3 polypeptide complex. Signal 2 is received by the
CD28 accessory molecules that bind to the B7-1 (CD80) or B7-2
(CD86) molecule on the APC surface (11,12).
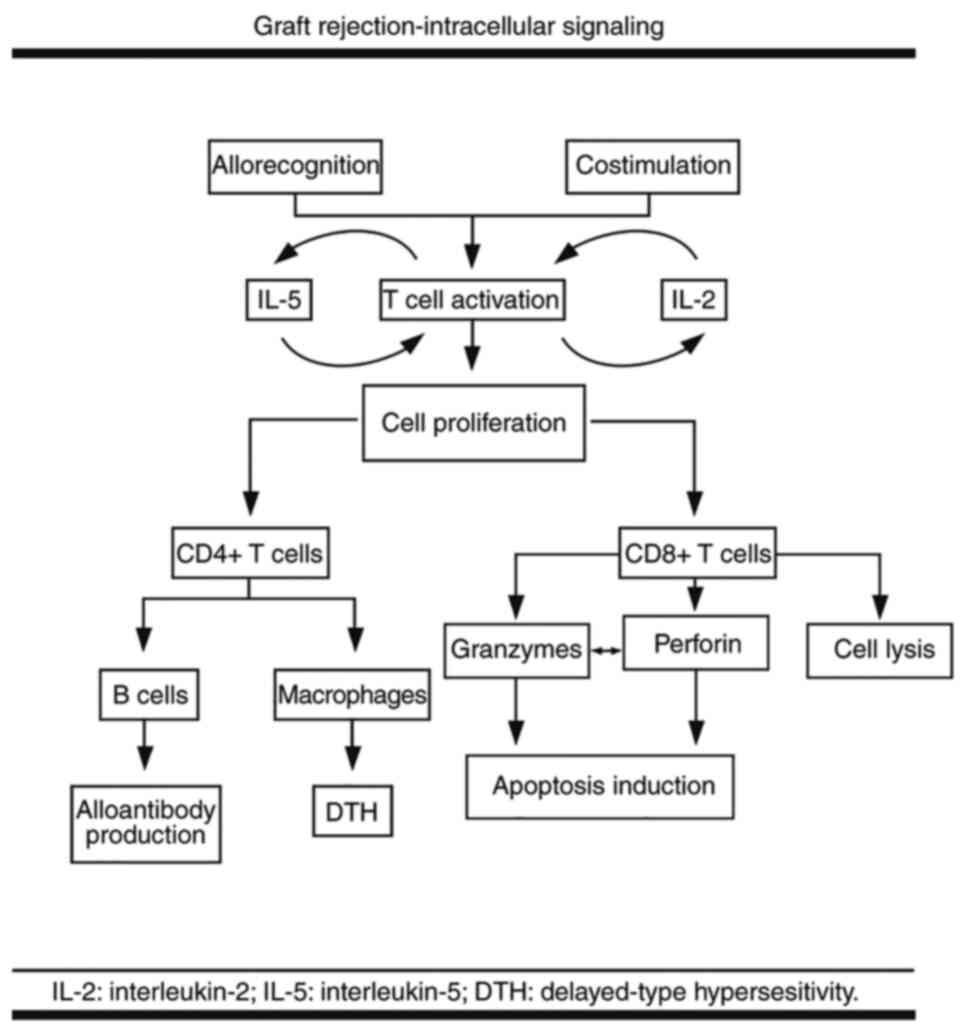
Once activated T cells undergo proliferation under
the influence of mitogenic growth and differentiation factors such
as interleukin (IL)-2 and IL-5, which activate the target of
rapamycin (TOR) paths; this process requires nucleotide synthesis.
Cell proliferation and differentiation induce cytotoxicity mediated
by the lymphocytes CD8+ T, activating B lymphocytes
(either directly or depending on Th lymphocytes) to produce
antibodies and determine macrophages to induce delayed
hypersensitivity responses (11,13)
(Fig. 1).
Detailed studies of these steps have led to the
development of targeted immunosuppressive therapies such as IL-2
receptor blockers (basiliximab), mTOR inhibitors (sirolimus and
everolimus), nucleotide synthesis inhibitors (mycophenolate) or
antimetabolites (azathioprine) (14).
In 2014, a new series of small molecules with
inhibiting PRF function was tested on male CD1 mice after
intravenous administration; the compounds exhibited microsomal
stability, which may lead to the development of a new
immunosuppressive therapy (15). In
contrast, the induction of PRF mRNA was partially blocked by the
immunosuppressive drug cyclosporine A, and therefore this therapy
has been recently avoided due to toxicity in favor of tacrolimus
administration (16).
3. PRF-granzyme pathway
The main mechanism used by lymphocytes for
eliminating infected or malignant cells in the host involves
granule exocytosis which contains PRF and a family of serine
esterases known under the name of granzymes. The fundamental basis
of the apoptotic pathway PRF-granzyme, is the synergy between its
components PRF and granzymes. These molecules have distinct roles.
PRF pores serve as entrance gates for the protease in the targeted
cell cytosol allowing granzymes to initiate various apoptotic
pathways. Although granzymes can internalize independent of PRF,
when seized into the endocytic vesicle lumen, they do not have
access to cytosolic substrates and remain harmless. Thus, the
expression of PRF is required for granule-mediated cytotoxicity,
ensuring the entry of granzymes into the targeted cells, the latter
to induce apoptosis (17-20)
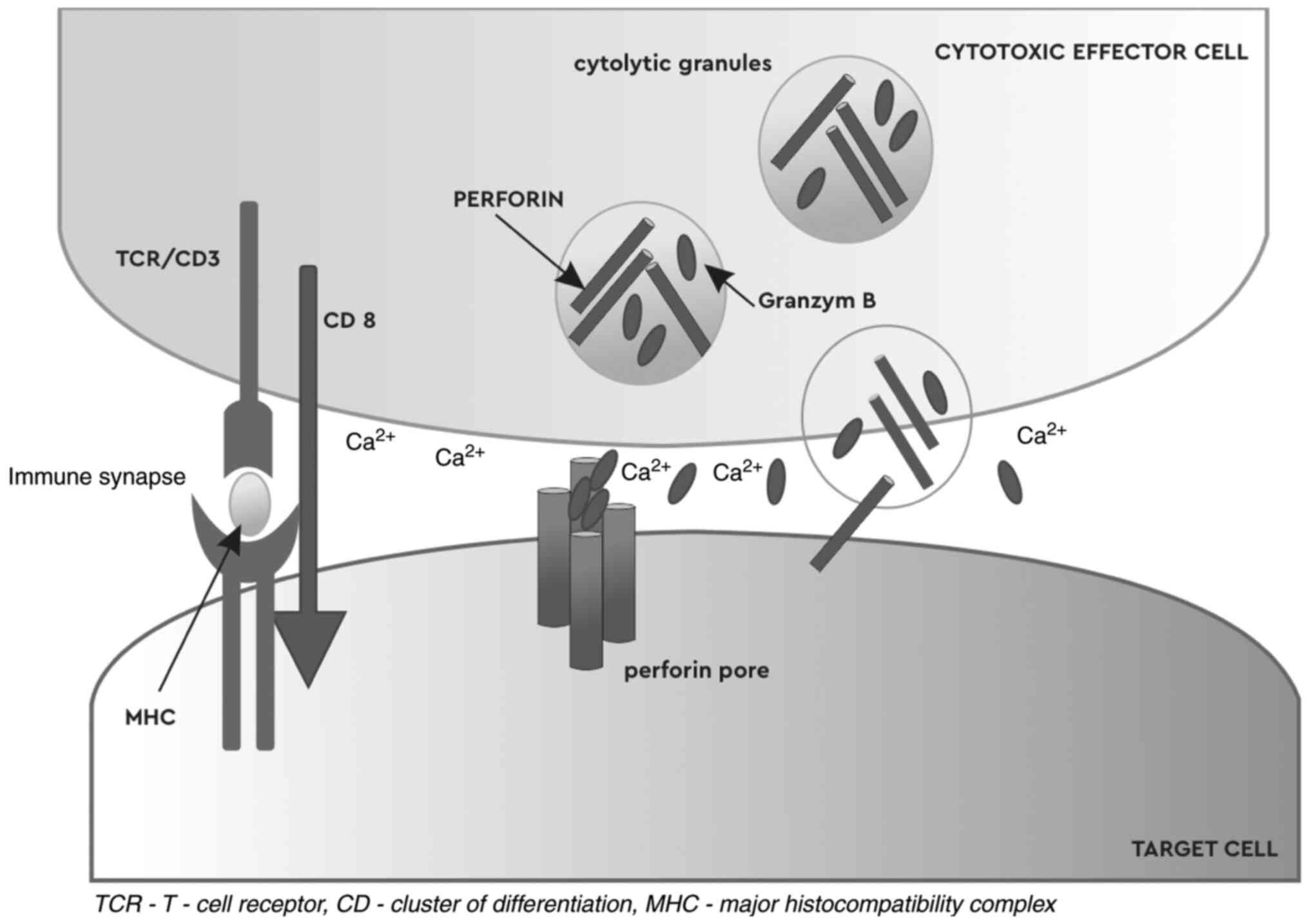
[Fig. 2, PRF-granzyme B pathway
adapted from (12); T-cell immune
response through the release of PRF and granzyme B, which attack
target cells, inducing apoptosis.].
PRF mRNA was identified in CD8+ cells
infiltrating the glomerulus of crescentic glomerulonephritis rats.
In human crescentic glomerulonephritis, both CD4- and CD8-positive
T lymphocytes are observed in glomeruli (21).
4. Chronic allograft dysfunction
Despite major advances in immunosuppression and
transplant management, acute and chronic rejection are the main
causes of kidney graft loss. It was revealed that acute rejection
is the strongest predictor of subsequent chronic rejection
(22).
CAN occurs due to repeated episodes of acute
rejection, HLA system lack of compatibility, improper
immunosuppression, ischemic injury (vascular occlusion caused by
arterial immune-mediated thickening and dysfunction) with secondary
fibrosis and late recovery of renal function (23).
Early allograft lesions by the PRF-granzyme pathway
could initiate the development of CAN in renal allografts (24). CD4+ Th lymphocytes
reactive to graft alloantigens produce cytokines which induce
endothelial and smooth muscle cell proliferation and cause
occlusion of the vascular lumen. It appears that cytotoxic T
lymphocytes infiltrated in the renal graft induce allogeneic
tubular epithelial cell death, via the native PRF pathway (25,26).
The diagnosis of chronic rejection is not always
made in a timely manner, required for the complete recuperation of
renal function. This problem occurs since kidney damage is detected
mainly as an increase in serum creatinine and the appearance of
proteinuria. However, an increase in serum creatinine is a late
sign of kidney damage, as the compensatory mechanisms in the
kidneys can maintain the glomerular filtration rate (GFR) despite
progressive structural damage. Although the long-term survival of
allografts is improving, late graft loss from CAN remains a
clinically significant problem and is the second most common cause
of late renal allograft loss, after death (27). Thus, the identification of early
markers associated with, or predicting CAN would be clinically
useful.
Recurrent or chronic inflammatory processes are
common in people with CKD and particularly in those with ESRD. This
is due to numerous underlying factors including the uremic
environment, high levels of circulating proinflammatory cytokines,
oxidative stress, carbonyl stress, waste of protein energy (PEW),
and increased incidence of infections (especially dialysis access)
to mention a few (28).
The acute-phase response is a major
pathophysiological phenomenon that accompanies inflammation. With
this reaction, normal homeostatic mechanisms are replaced by new
established factors that are likely to contribute to defensive or
adaptive capabilities (29,30).
In patients returning to a dialysis program after
acute or chronic rejection of the renal transplant but without
kidney transplant graftectomy performed, a chronic inflammatory
state was observed, that was reduced by the removal of the
non-functional graft (explant) (31).
5. Predictive markers of chronic allograft
dysfunction
To date, few studies have been performed on PRF in
the renal area. In an experimental study, using Wistar-Kyoto (WKY)
rats with antiglomerular basement membrane (GBM) crescentic
glomerulonephritis (GN), PRF protein and mRNA expression of PRF
were demonstrated in glomeruli by immunohistochemistry and in
situ hybridization. WKY rats treated with anti-PRF antibodies
revealed significantly reduced amounts of proteinuria and frequency
of crescentic glomeruli (21).
There is strong scientific evidence that
immunological non-invasive monitoring could be useful in the first
6 months after kidney transplantation in particular regarding
prediction of acute rejection episodes (32). It is less clear whether CAN is also
associated with consistent changes of peripheral blood or the
urinary cells. Several histological studies have demonstrated
enhanced expression of granzymes and PRF in numerous types of
transplanted grafts and their correlation with acute rejection
episodes (33-40).
It appears that the urinary mRNA levels of three markers including
PRF, granzyme B and FAS ligand appear to be correlated with acute
rejection and the increase of serum creatinine (41,42).
The molecular analysis of the expression of these three molecules
in renal biopsy samples revealed that only the expression of PRF
and FAS ligand were correlated with the acute rejection while the
expression of PRF and granzyme B could intensify at a longer time
after transplantation, possibly associated with chronic dysfunction
(43). In 1997, a concurrent
RT-qPCR assessment of PRF, granzyme B and Fas ligand revealed a
correlation with acute rejection even in cases of mild
infiltration, with 100% sensitivity and specificity. The combined
analysis of the expression of Fas ligand, PRF and granzyme B genes
by quantitative RT-PCR provided a reliable tool for the diagnosis
and management of acute renal rejection and antirejection therapy
leading to a rapid decrease in the expression of these genes
(44). Li et al confirmed
that the mRNA levels of PRF and granzyme B were increased in urine
samples from patients with acute rejection (42). To date, it has been demonstrated
that urinary mRNA levels of AGT, EGFR and TGF-β1 may be reliable
prediction markers of CAN (32,45-47).
The genetic expression of serum PRF has been
revealed to be more correlated with acute rejection in renal
transplanted patients in comparison with granzyme B and Fas ligand
thus supporting its use as a marker of acute rejection (48). The PRF-granzyme and the Fas ligand
are two major pathways by which cytotoxic T lymphocytes induce
apoptosis in target cells (49).
The expression of the message into the graft for these
immune-activating genes has been revealed to be markedly correlated
with graft rejection. In a retrospective pilot study, 140
fine-needle aspiration biopsy samples from 50 human renal
allografts were labeled using alkaline phosphatase/alkaline
anti-phosphatase immunocytochemistry incorporating monoclonal
antibodies to PRF, granzyme B, and Fas ligand. Positive labeling
levels for these markers were compared with the initial clinical
diagnosis of rejection. Only when all three antibodies yielded
positive labeling, was the association with the clinical rejection
status superior to conventional morphological cytology (50).
A recent study on humanized mice, revealed that
granzyme B expression was significantly increased in
CD8+ T cells in patients with graft rejection, while
surviving graft patients expressed less granzyme B, as they had an
increased level of HLA-G dimer which inhibited cytotoxicity of
CD8+ T cells (51). The
synergy between PRF and granzymes is already established.
Therefore, a high level of granzymes implies a high level of
PRF.
Recently, new categories of drugs, such as
inhibitors of PRF, have aroused the interest of researchers. Tampio
et al demonstrated that using L-type amino acid transporter
1 (LAT1)-utilizing prodrugs of PRF inhibitors for improved
administration of brain drug delivery, led to improved
pharmacological effects, decreased production of cellular apoptosis
mediators, decreased overall oxidative stress and inflammation in
the brain, and from the periphery, increasing cell survival
(52).
6. PRF ‘roots’
In the evolution of PRF there were complex models of
events of birth and death including duplication/pseudogenization to
mammals, multiple amplifications and losses in reptiles and fish as
well and a case of partial duplication with a new beginning codon
to fish. Approximately 500 million years ago, the primordial PRF
gene evolved, around the same time as T-cell receptor antigen
recognition based on the major histocompatibility complex. As it is
absent from primitive chordates and invertebrates, cytotoxic cells
from these lineages must have a different cytotoxic effector
molecule or mechanism. Orthologs and homologues of human PRF have
been identified in almost all species. Research has shown that in
species prior to Gnathostomata (Euteleostomi) the PRF gene did not
exist which suggests that cytotoxic cells of prior species have
another mechanism or different means for killing targeted cells. In
addition, there is evidence that PRF originated from the
duplication of the ancient gene MPEG1 and shares a common ancestor
with functionally related complement proteins (53,54).
7. PRF: Structure and genetics
PRF is a 67-kDa pore-forming protein, stored and
released from the secretory granules (SG) of the cytotoxic
lymphocytes which leads to osmotic lysis of the membrane of target
cells and subsequently allows proapoptotic granzymes (serine
proteases which split the peptide connections of proteins) with
broad specificity to enter the targeted cells and activate the cell
death program. PRF expression is increased in the activated
CD8+ cells, αγ T cells, in subpopulations of activated
CD4+ T cells, and NK cells (but with a high and stable
incorporation in NK cells) (17,19,55-58).
In addition, PRF expression may be stimulated in some activated
CD4+ cells (59,60).
In mammals, PRF is encoded by the PRF1 gene
expressed in cytotoxic lymphocytes and regulatory T cells. PRF1
transcription is the main mechanism that determines PRF expression
in cytotoxic T lymphocytes and NK cells. While PRF is uniformly
expressed by mature NK cells as a result of spontaneous stimulation
of constitutive gene transcription, its expression in peripheral T
cells requires gene activation (17,58,61-64).
Locus control position is essential for
PRF1-specific activity (NK and cytotoxic cell activation). A
heterochromatin-dependent regulation could allow certain exogenous
stimuli and certain endogenous controlling of the transcription
factors to induce PRF1 transcription in other types of cells
(59). In 2006, a study conducted
by Pipkin and Lichtenheld identified the locus control region for
perforin of 150 Kb of cis action sequences which leads to
the physiological PRF1 transcription, comprising 16 hypersensitive
DNase I (DHS) sites, four of them necessary for PRF expression
(17,65). PRF was identified for the first time
in 1983 and it was cloned from an expression library by a cross
reaction of the C9 antibody (59,66-71).
Fine-resolution comparisons by direct sequence comparisons have
revealed a similarity between the two proteins (C9 and PRF), that
contain in the middle part of their sequences a short region called
membrane-attack complex/PRF (MACPF) (59,72,73).
Both proteins polymerize in tubular complexes able to determine
lysis of the membrane insertion acting as large and
voltage-independent transmembrane channels. Initial studies have
revealed that while C9 polymerizes in physiological conditions
requiring the assembly of complex C5b-8 into the receptor, the
functional activity of PRF in the phospholipid membrane is
calcium-dependent (67,74,75).
After exocytosis, granules from the killer cells releasing PRF and
granzymes are exposed to immune synapses rich in calcium and
neutral pH (59,64). The PRF monomers bind to calcium by
its C2 domain acquiring the capacity of bounding lipids to the
targeted cell membrane and then to merge in transmembrane pores of
up to 100 Å, which allows granzymes access to the protein
substrates involved in apoptosis (59,76-78).
8. Perforinopathies
It is recognized that the residual function of the
PRF-dependent cytotoxic cells causes transplant rejection of
allogenic stem cells, allografts and solid organs. At the opposite
pole defects of the cytotoxic path and PRF deficiency (failure to
deliver PRF) lead to disorders called perforinopathies including
familial haemophagocytic lymphohistiocytosis (FHL), viral
infections and the predisposition to develop haemato-oncological
diseases (12,79-83). Voskoboinik et al have
proposed the term of perforinopathies in order to define a spectrum
of immune-mediated disease responses associated with monoallelic
mutations in genes related to FHL (84).
The complete absence of the PRF function results
into FHL, an immunoregulatory disease that appears in childhood and
is characterized by uncontrolled activation of CPA and
CD8+ T lymphocytes with secondary accumulation of T
cells. Recently it was discovered that the partial loss of PRF
function is strongly associated with FHL and a series of
hematological disorders that appear later in childhood or in
adolescence. In addition, PRF functionality is essential for
cytotoxic lymphocytes in humans since harmful mutations in PRF1
leads to FHL2 representing 30-60% of FHL cases (59,79,85).
PRF and CD107a tests are more sensitive and have a
similar specificity compared with NK cytotoxicity test and would be
able to enhance FHL screening (86).
Relative recent studies have revealed that UVB and
UVA radiations induce accumulation of granzyme B in human
keratinocytes. In addition, granzyme B secondary to UVB radiation
mediates cytotoxicity of keratinocytes, while in UVA irradiation it
increases the ability of the keratinocyte to degrade matrix
extracellular components; these observations could be the basis of
photoaging and photocarcinogenicity domain (87,88).
While cancer therapy has begun to use ‘suicide
genes’ to induce cell apoptosis, the role of vaccination with
apoptotic cells, either immune stimulatory or immune suppressive is
still debated. Recently a new technology called cytolytic DNA
technology has been developed, using a vaccine which encodes
truncated PRF incorporated in a bicistronic DNA vector that
activates dendritic cells thus stimulating the CD8+
T-cell response against HIV and HCV reducing the viral loads
similar to traditional vaccines (89-92).
PRF has gained the attention of cardiology
researchers, proving to date, that patients with left ventricular
dysfunction have PRF-positive infiltration of heart cells and that
PRF could be an adverse predictor of long-term mortality in
patients with inflammatory cardiomyopathy (93).
9. Conclusions
PRF is a pore-forming protein vital for cytotoxic
effector function and has an indispensable role in
granzyme-mediated apoptosis. It is responsible for endothelial
damage and plays a role in the pathogenesis of numerous
inflammatory diseases and targets cell apoptosis.
Over the last few years, the study of serum PRF and
its role in inflammatory and neoplastic diseases has captured the
attention of the medical world. To date, few studies have reported
the correlation between serum or urinary PRF, granzyme and ligand
FAS with acute transplant rejection. Further studies are required
to clarify the role of PRF as a potential early biomarker with a
predictive role in chronic allograft rejection.
Advances in immunosuppressive therapy, in order to
maintain kidney transplantation and avoid rejection, have led to
decreased rejection rates. However, these agents are not deprived
of side effects.
The residual function of PRF-dependent cytotoxic
cells causes transplant rejection of allogenic stem cells, the
allograft and solid organs. A deep understanding of the role of PRF
in inducing allograft rejection is necessary for the development of
new targeted post-transplant therapies. Highly specific inhibitors
of PRF function, are thus of interest as selective
immunosuppressive drugs.
The practical use of PRF expression has already been
demonstrated in various medical fields. Demonstrating the utility
of PRF as a predictive marker of CAN, such as with PRF inhibitors,
would have treatment implications that could mark the beginning of
a new era in immunosuppressive therapy.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the project
ANTREPRENORDOC, in the framework of Human Resources Development
Operational Program 2014-2020, financed from the European Social
Fund under the contract number 36355/23.05.2019 HRD
OP/380/6/13-SMIS Code: 123847.
Availability of data and materials
Not applicable.
Authors' contributions
AMPC conceived, wrote and edited the manuscript. EC
reviewed the manuscript for important intellectual content. LAT
revised the study before finally approving it for publication.
AMPC, EC and LAT confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Covic A, Mircescu G, Gluhovschi G and
Schiller A: Ghiduri de practica medicala. Boala cronica de rinichi.
1-109. 2007. Bucuresti AG de practica medicala. SR de N: No
Title.
|
|
2
|
Evans RW, Manninen DL, Garrison LP Jr,
Hart LG, Blagg CR, Gutman RA, Hull AR and Lowrie EG: The quality of
life of patients with end-stage renal disease. N Engl J Med.
312:553–559. 1985.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Laupacis A, Keown P, Pus N, Krueger H,
Ferguson B, Wong C and Muirhead N: A study of the quality of life
and cost-utility of renal transplantation. Kidney Int. 50:235–242.
1996.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Simmons RG and Abress L: Quality-of-life
issues for end-stage renal disease patients. Am J Kidney Dis.
15:201–208. 1990.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ojo AO, Hanson JA, Wolfe RA, Leichtman AB,
Agodoa LY and Port FK: Long-term survival in renal transplant
recipients with graft function. Kidney Int. 57:307–313.
2000.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Organ procurement and transplantation
network and scientific registry of transplant recipients 2010 data
report. Am J Transplant. 12 (Suppl 1):S1–S156. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Foroutan F, Friesen EL, Clark KE, Motaghi
S, Zyla R, Lee Y, Kamran R, Ali E, De Snoo M, Orchanian-Cheff A, et
al: Risk factors for 1-year graft loss after kidney transplantation
systematic review and meta-analysis. Clin J Am Soc Nephrol.
14:1642–1650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wyburn KR, Jose MD, Wu H, Atkins RC and
Chadban SJ: The role of macrophages in allograft rejection.
Transplantation. 80:1641–1647. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Williams TM: Human leukocyte antigen gene
polymorphism and the histocompatibility laboratory. J Mol
Diagnostics. 3:98–104. 2001.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Horton R, Wilming L, Rand V, Lovering RC,
Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW,
et al: Gene map of the extended human MHC. Nat Rev Genet.
5:889–899. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Sayegh MH and Turka LA: The role of T-cell
costimulatory activation pathways in transplant rejection. N Engl J
Med. 338:1813–1821. 1998.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Osińska I, Popko K and Demkow U: Perforin:
An important player in immune response. Cent Eur J Immunol.
39:109–115. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vella J: Transplantation immunobiology.
UpToDate, 2021.
|
|
14
|
Mukherjee S and Mukherjee U: A
comprehensive review of immunosuppression used for liver
transplantation. J Transplant. 2009(701464)2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bull MR, Spicer JA, Huttunen KM, Denny WA,
Ciccone A, Browne KA, Trapani JA and Helsby NA: The preclinical
pharmacokinetic disposition of a series of perforin-inhibitors as
potential immunosuppressive agents. Eur J Drug Metab Pharmacokinet.
40:417–425. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lu P, Garcia-Sanz JA, Lichtenheld MG and
Podack ER: Perforin expression in human peripheral blood
mononuclear cells: Definition of an IL-2-independent pathway of
perforin induction in CD8+ T cells. J Immunol. 148:3354–3360.
1992.PubMed/NCBI
|
|
17
|
Pipkin ME, Ljutic B, Cruz-Guilloty F,
Nouzova M, Rao A, Zúñiga-Pflücker JC and Lichtenheld MG: Chromosome
transfer activates and delineates a locus control region for
perforin. Immunity. 26:29–41. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Keefe D, Shi L, Feske S, Massol R, Navarro
F, Kirchhausen T and Lieberman J: Perforin triggers a plasma
membrane-repair response that facilitates CTL induction of
apoptosis. Immunity. 23:249–262. 2005.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kägi D, Ledermann B, Bürki K, Zinkernagel
RM and Hengartner H: Molecular mechanisms of lymphocyte-mediated
cytotoxicity and their role in immunological protection and
pathogenesis in vivo. Annu Rev Immunol. 14:207–232. 1996.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Henkart PA: Mechanism of
lymphocyte-mediated cytotoxicity. Annu Rev Immunol. 3:31–58.
1985.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Fujinaka H, Yamamoto T, Feng L, Nameta M,
Garcia G, Chen S, El-shemi AA, Ohshiro K, Katsuyama K, Yoshida Y,
et al: Anti-perforin antibody treatment ameliorates experimental
crescentic glomerulonephritis in WKY rats. Kidney Int. 72:823–830.
2007.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cecka JM, Cho YW and Terasaki PI: Analyses
of the UNOS scientific renal transplant registry at three
years-early events affecting transplant success. Transplantation.
53:59–63. 1992.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mitchell RN and Libby P: Vascular
remodeling in transplant vasculopathy. Circ Res. 100:967–978.
2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Almond PS, Matas A, Gillingham K, Dunn DL,
Payne WD, Gores P, Gruessner R and Najarian JS: Risk factors for
chronic rejection in renal allograft recipients. Transplantation.
55:752–756; discussion 756-7. 1993.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Miltenburg AM, Meijer-Paape ME, Daha MR,
van Bockel JH, Weening JJ, van Es LA and van*der*Woude FJ:
Donor-specific lysis of human kidney proximal tubular epithelial
cells by renal allograft-infiltrating lymphocytes. Transplantation.
48:296–302. 1989.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wever PC, Boonstra JG, Laterveer JC, Hack
CE, van Der Woude FJ, Daha MR and ten*Berge IJ: Mechanisms of
lymphocyte-mediated cytotoxicity in acute renal allograft reaction.
Transplantation. 66:259–264. 1998.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cecka JM: The UNOS scientific renal
transplant registry-2000. Clin Transpl. 1-18:2000.PubMed/NCBI
|
|
28
|
Owen WF and Lowrie EG: C-reactive protein
as an outcome predictor for maintenance hemodialysis patients.
Kidney Int. 54:627–636. 1998.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Guessous I, Ponte B, Marques-Vidal P,
Paccaud F, Gaspoz JM, Burnier M, Waeber G, Vollenweider P and
Bochud M: Clinical and biological determinants of kidney outcomes
in a population-based cohort study. Kidney Blood Press Res.
39:74–85. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Csaba P, Kovesdy*Kopple JD and
Kalantar-Zadeh K: Inflammation in renal insufficiency. UpToDate,
2011.
|
|
31
|
López-Gómez JM, Pérez-Flores I, Jofré R,
Carretero D, Rodríguez-Benitez P, Villaverde M, Pérez-García R,
Nassar GM, Niembro E and Ayus JC: Presence of a failed kidney
transplant in patients who are on hemodialysis is associated with
chronic inflammatory state and erythropoietin resistance. J Am Soc
Nephrol. 15:2494–2501. 2004.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mas VR, Mas LA, Archer KJ, Yanek K, King
AL, Gibney EM, Cotterell A, Fisher RA, Posner M and Maluf DG:
Evaluation of gene panel mrnas in urine samples of kidney
transplant recipients as a non-invasive tool of graft function. Mol
Med. 13:315–324. 2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Choy JC: Granzymes and perforin in solid
organ transplant rejection. Cell Death Differ. 17:567–576.
2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hameed A, Truong LD, Price V, Kruhenbuhl O
and Tschopp J: Immunohistochemical localization of granzyme B
antigen in cytotoxic cells in human tissues. Am J Pathol.
138:1069–1075. 1991.PubMed/NCBI
|
|
35
|
Griffiths GM, Namikawa R, Mueller C, Liu
CC, Young JD, Billingham M and Weissman I: Granzyme A and perforin
as markers for rejection in cardiac transplantation. Eur J Immunol.
21:687–692. 1991.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ciément MV, Haddad P, Soulié A, Benvenuti
C, Lichtenheld MG, Podack ER, Sigaux N and Sasportes M: Perform and
granzyme B as markers for acute rejection in heart transplantation.
Int Immunol. 3:1175–1181. 1991.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lipman ML, Stevens AC and Strom TB:
Heightened intragraft CTL gene expression in acutely rejecting
renal allografts. J Immunol. 152:5120–5127. 1994.PubMed/NCBI
|
|
38
|
Kummer JA, Wever PC, Kamp AM, ten Berge
IJ, Hack CE and Weening JJ: Expression of granzyme A and B proteins
by cytotoxic lymphocytes involved in acute renal allograft
rejection. Kidney Int. 47:70–77. 1995.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Krams SM, Villanueva JC, Quinn MB and
Martinez OM: Expression of the cytotoxic T cell mediator granzyme B
during liver allograft rejection. Transpl Immunol. 3:162–166.
1995.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Legros-Maïda S, Soulié A, Benvenuti C,
Wargnier A, Vallée N, Berthou C, Guillet J, Sasportes M and Sigaux
N: Granzyme B and perforin can be used as predictive markers of
acute rejection in heart transplantation. Eur J Immunol.
24:229–233. 1994.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yannaraki M, Rebibou JM, Ducloux D, Saas
P, Duperrier A, Felix S, Rifle G, Chalopin JM, Hervé P, Tiberghien
P and Ferrand C: Urinary cytotoxic molecular markers for a
noninvasive diagnosis in acute renal transplant rejection. Transpl
Int. 19:759–768. 2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li B, Hartono C, Ding R, Sharma VK,
Ramaswamy R, Qian B, Serur D, Mouradian J, Schwartz JE and
Suthanthiran M: Noninvasive diagnosis of renal-allograft rejection
by measurement of messenger RNA for perforin and granzyme B in
urine. N Engl J Med. 344:947–954. 2001.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Graziotto R, Del Prete D, Rigotti P,
Anglani F, Baldan N, Furian L, Valente M, Antonello A, Marchini F,
D'Angelo A and Gambaro G: Perforin, Granzyme B, and Fas ligand for
molecular diagnosis of acute renal-allograft rejection: Analyses on
serial biopsies suggest methodological issues. Transplantation.
81:1125–1132. 2006.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Strehlau J, Pavlakis M, Lipman M, Shapiro
M, Vasconcellos L, Harmon W and Strom TB: Quantitative detection of
immune activation transcripts as a diagnostic tool in kidney
transplantation. Proc Natl Acad Sci USA. 94:695–700.
1997.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Campistol JM, Iñigo P, Larios S, Bescos M
and Oppenheimer F: Role of transforming growth factor-beta1 in the
progression of chronic allograft nephropathy. Nephrol Dial
Transplant. 16 (Suppl 1):S114–S116. 2001.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Nocera A, Tagliamacco A, De Palma R, Del
Galdo F, Ferrante A, Fontana I, Barocci S, Ginevri F, Rolla D,
Ravetti JL and Valente U: Cytokine mRNA expression in chronically
rejected human renal allografts. Clin Transplant. 18:564–570.
2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Pribylova-Hribova P, Kotsch K, Lodererova
A, Viklicky O, Vitko S, Volk HD and Lacha J: TGF-beta1 mRNA
upregulation influences chronic renal allograft dysfunction. Kidney
Int. 69:1872–1879. 2006.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Shin GT, Kim SJ, Lee TS, Oh CK and Kim H:
Gene expression of perforin by peripheral blood lymphocytes as a
marker of acute rejection. Nephron Clin Pract. 100:c63–c70.
2005.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Trapani JA and Smyth MJ: Functional
significance of the perforin/granzyme cell death pathway. Nat Rev
Immunol. 2:735–747. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
50
|
Pascoe MD, Marshall SE, Welsh KI, Fulton
LM and Hughes DA: Increased accuracy of renal allograft rejection
diagnosis using combined perforin, granzyme B, and Fas ligand
fine-needle aspiration immunocytology. Transplantation.
69:2547–2553. 2000.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ajith A, Portik-Dobos V, Nguyen-Lefebvre
AT, Callaway C, Horuzsko DD, Kapoor R, Zayas C, Maenaka K, Mulloy
LL and Horuzsko A: HLA-G dimer targets Granzyme B pathway to
prolong human renal allograft survival. FASEB J. 33:5220–5236.
2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Tampio J, Huttunen J, Montaser A and
Huttunen KM: Targeting of perforin inhibitor into the brain
parenchyma via a prodrug approach can decrease oxidative stress and
neuroinflammation and improve cell survival. Mol Neurobiol.
57:4563–4577. 2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
D'Angelo ME, Dunstone MA, Whisstock JC,
Trapani JA and Bird PI: Perforin evolved from a gene duplication of
MPEG1, followed by a complex pattern of gene gain and loss within
Euteleostomi. BMC Evol Biol. 12(59)2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Araujo-Voces M and Quesada V: Frequent
birth-and-death events throughout perforin-1 evolution. BMC Evol
Biol. 20(135)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
De Rosa SC, Andrus JP, Perfetto SP,
Mantovani JJ, Herzenberg LA, Herzenberg LA and Roederer M: Ontogeny
of gamma delta T cells in humans. J Immunol. 172:1637–1645.
2004.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Grossman WJ, Verbsky JW, Barchet W,
Colonna M, Atkinson JP and Ley TJ: Human T regulatory cells can use
the perforin pathway to cause autologous target cell death.
Immunity. 21:589–601. 2004.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Gumperz JE, Miyake S, Yamamura T and
Brenner MB: Functionally distinct subsets of CD1d-restricted
natural killer T cells revealed by CD1d tetramer staining. J Exp
Med. 195:625–636. 2002.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Nakata M, Kawasaki A, Azuma M, Tsuji K,
Matsuda H, Shinkai Y, Yagita H and Okumura K: Expression of
perforin and cytolytic potential of human peripheral blood
lymphocyte subpopulations. Int Immunol. 4:1049–1054.
1992.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Brennan AJ, Chia J, Trapani JA and
Voskoboinik I: Perforin deficiency and susceptibility to cancer.
Cell Death Differ. 17:607–615. 2010.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Voskoboinik I, Smyth MJ and Trapani JA:
Perforin-mediated target-cell death and immune homeostasis. Nat Rev
Immunol. 6:940–952. 2006.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Salcedo TW, Azzoni L, Wolf SF and Perussia
B: Modulation of perforin and granzyme messenger RNA expression in
human natural killer cells. J Immunol. 151:2511–2520.
1993.PubMed/NCBI
|
|
62
|
Zhang J, Scordi I, Smyth MJ and
Lichtenheld MG: Interleukin 2 receptor signaling regulates the
perforin gene through signal transducer and activator of
transcription (Stat)5 activation of two enhancers. J Exp Med.
190:1297–1308. 1999.PubMed/NCBI View Article : Google Scholar
|
|
63
|
García-Sanz JA and Podack ER: Regulation
of perforin gene expression in a T cell hybrid with inducible
cytolytic activity. Eur J Immunol. 23:1877–1883. 1993.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Uellner R, Zvelebil MJ, Hopkins J, Jones
J, MacDougall LK, Morgan BP, Podack E, Waterfield MD and Griffiths
GM: Perforin is activated by a proteolytic cleavage during
biosynthesis which reveals a phospholipid-binding C2 domain. EMBO
J. 16:7287–7296. 1997.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Pipkin ME and Lichtenheld MG: A reliable
method to display authentic DNase I hypersensitive sites at
long-ranges in single-copy genes from large genomes. Nucleic Acids
Res. 34(e34)2006.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Podack ER and Dennert G: Assembly of two
types of tubules with putative cytolytic function by cloned natural
killer cells. Nature. 302:442–445. 1983.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Podack ER, Young JD and Cohn ZA: Isolation
and biochemical and functional characterization of perforin 1 from
cytolytic T-cell granules. Proc Natl Acad Sci USA. 82:8629–8633.
1985.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Young JD, Cohn ZA and Podack ER: The ninth
component of complement and the pore-forming protein (perforin 1)
from cytotoxic T cells: Structural, immunological, and functional
similarities. Science. 233:184–190. 1986.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Young JD, Hengartner H, Podack ER and Cohn
ZA: Purification and characterization of a cytolytic pore-forming
protein from granules of cloned lymphocytes with natural killer
activity. Cell. 44:849–859. 1986.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Lowrey DM, Aebischer T, Olsen K,
Lichtenheld M, Rupp F, Hengartner H and Podack ER: Cloning,
analysis, and expression of murine perforin 1 cDNA, a component of
cytolytic T-cell granules with homology to complement component C9.
Proc Natl Acad Sci USA. 86:247–251. 1989.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Shinkai Y, Takio K and Okumura K: Homology
of perforin to the ninth component of complement (C9). Nature.
334:525–527. 1988.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Tschopp J, Masson D and Stanley KK:
Structural/functional similarity between proteins involved in
complement- and cytotoxic T-lymphocyte-mediated cytolysis. Nature.
322:831–834. 1986.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Lichtenheld MG, Olsen KJ, Lu P, Lowrey DM,
Hameed A, Hengartner H and Podack ER: Structure and function of
human perforin. Nature. 335:448–451. 1988.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Blumenthal R, Millard PJ, Henkart MP,
Reynolds CW and Henkart PA: Liposomes as targets for granule
cytolysin from cytotoxic large granular lymphocyte tumors. Proc
Natl Acad Sci USA. 81:5551–5555. 1984.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Henkart PA, Yue CC, Yang J and Rosenberg
SA: Cytolytic and biochemical properties of cytoplasmic granules of
murine lymphokine-activated killer cells. J Immunol. 137:2611–2617.
1986.PubMed/NCBI
|
|
76
|
Müllbacher A, Waring P, Tha Hla R, Tran T,
Chin S, Stehle T, Museteanu C and Simon MM: Granzymes are the
essential downstream effector molecules for the control of primary
virus infections by cytolytic leukocytes. Proc Natl Acad Sci USA.
96:13950–13955. 1999.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Nakajima H, Park HL and Henkart PA:
Synergistic roles of granzymes A and B in mediating target cell
death by rat basophilic leukemia mast cell tumors also expressing
cytolysin/perforin. J Exp Med. 181:1037–1046. 1995.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Shi L, Mai S, Israels S, Browne K, Trapani
JA and Greenberg AH: Granzyme B (GraB) autonomously crosses the
cell membrane and perforin initiates apoptosis and GraB nuclear
localization. J Exp Med. 185:855–866. 1997.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Spicer BA, Conroy PJ, Law RHP, Voskoboinik
I and Whisstock JC: Perforin-A key (shaped) weapon in the
immunological arsenal. Semin Cell Dev Biol. 72:117–123.
2017.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Voskoboinik I, Whisstock JC and Trapani
JA: Perforin and granzymes: Function, dysfunction and human
pathology. Nat Rev Immunol. 15:388–400. 2015.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Jenkins MR, Rudd-Schmidt JA, Lopez JA,
Ramsbottom KM, Mannering SI, Andrews DM, Voskoboinik I and Trapani
JA: Failed CTL/NK cell killing and cytokine hypersecretion are
directly linked through prolonged synapse time. J Exp Med.
212:307–317. 2015.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Voskoboinik I, Thia MC, Fletcher J,
Ciccone A, Browne K, Smyth MJ and Trapani JA: Calcium-dependent
plasma membrane binding and cell lygis by perforin are mediated
through its C2 domain: A critical role for aspartate residues 429,
435, 483, and 485 but not 491. J Biol Chem. 280:8426–8434.
2005.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Taylor MA, Ward B, Schatzle JD and Bennett
M: Perforin- and Fas-dependent mechanisms of natural killer
cell-mediated rejection of incompatible bone marrow cell grafts.
Eur J Immunol. 32:793–799. 2002.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Voskoboinik I and Trapani JA:
Perforinopathy: A spectrum of human immune disease caused by
defective perforin delivery or function. Front Immunol.
4(441)2013.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Molleran Lee S, Villanueva J, Sumegi J,
Zhang K, Kogawa K, Davis J and Filipovich AH: Characterisation of
diverse PRF1 mutations leading to decreased natural killer cell
activity in North American families with haemophagocytic
lymphohistiocytosis. J Med Genet. 41:137–144. 2004.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Rubin TS, Zhang K, Gifford C, Lane A, Choo
S, Bleesing JJ and Marsh RA: Perforin and CD107a testing is
superior to NK cell function testing for screening patients for
genetic HLH. Blood. 129:2993–2999. 2017.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Hernandez-Pigeon H, Jean C, Charruyer A,
Haure MJ, Titeux M, Tonasso L, Quillet-Mary A, Baudouin C,
Charveron M and Laurent G: Human keratinocytes acquire cellular
cytotoxicity under UV-B irradiation. Implication of granzyme B and
perforin. J Biol Chem. 281:13525–13532. 2006.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Hernandez-Pigeon H, Jean C, Charruyer A,
Haure MJ, Baudouin C, Charveron M, Quillet-Mary A and Laurent G:
UVA Induces Granzyme B in Human Keratinocytes through MIF
implication in extracellular matrix remodeling. J Biol Chem.
282:8157–8164. 2007.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Shrestha AC, Wijesundara DK, Masavuli MG,
Mekonnen ZA, Gowans EJ and Grubor-Bauk B: Cytolytic perforin as an
adjuvant to enhance the immunogenicity of DNA vaccines. Vaccines
(Basel). 7(38)2019.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Gargett T, Grubor-Bauk B, Garrod TJ, Yu W,
Miller D, Major L, Wesselingh S, Suhrbier A and Gowans EJ:
Induction of antigen-positive cell death by the expression of
Perforin, but not DTa, from a DNA vaccine enhances the immune
response. Immunol Cell Biol. 92:359–367. 2014.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Gummow J, Li Y, Yu W, Garrod T,
Wijesundara D, Brennan AJ, Mullick R, Voskoboinik I, Grubor-Bauk B
and Gowans EJ: A Multiantigenic DNA vaccine that induces broad
hepatitis C Virus-specific T-cell responses in mice. J Virol.
89:7991–8002. 2015.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Wijesundara DK, Yu W, Quah BJC, Eldi P,
Hayball JD, Diener KR, Voskoboinik I, Gowans EJ and Grubor-Bauk B:
Cytolytic DNA vaccine encoding lytic perforin augments the
maturation of- and antigen presentation by-dendritic cells in a
time-dependent manner. Sci Rep. 7(8530)2017.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Escher F, Kühl U, Lassner D, Stroux A,
Gross U, Westermann D, Pieske B, Poller W and Schultheiss HP: High
perforin-positive cardiac cell infiltration and male sex predict
adverse long-term mortality in patients with inflammatory
cardiomyopathy. J Am Heart Assoc. 6(e005352)2017.PubMed/NCBI View Article : Google Scholar
|