Introduction
Head and neck squamous cell carcinoma (HNSCC) is one
of the most lethal malignancies with >800,000 new-onset cases
reported per year. Mortality rates are increased with the stage of
the disease and rates >50% have been identified in critically
ill patients (1). HNSCC originates
from epithelial cells of the upper respiratory tract and esophageal
mucosa; of note, HNSCC exhibits high heterogeneity due to the
complex anatomy and genetic variations in that area. Over the past
few years, research on the correlation between ferroptosis and
tumors has become a major research hotspot. Several studies
confirmed that the expression levels of ferroptosis proteins
exhibited a wide variation in HNSCC, thereby significantly
regulating cell proliferation (2)
and epithelial-mesenchymal transformation (3) and even affecting cisplatin resistance
(4). However, systematic studies
focusing on ferroptosis and HNSCC have been continuously lacking
thus far and research on novel precision therapy targets remains in
its infancy.
Over the past decade, research on the significance
of ferroptosis in cancer has gained momentum. Ferroptosis is
another type of programmed necrotic cell death involving
iron-dependent lipid peroxidation and oxidative stress is thought
to be a principal cause of ferroptosis (5). Due to cancer cells' high iron levels
and their increased sensitivity to ferroptosis induction,
ferroptosis has been proposed to be promising for cancer
therapeutics. A growing number of studies indicated that for
numerous types of solid tumor, positive treatment effects were
achieved by targeting ferroptosis-related genes.
Besides ferroptosis agonists and inhibitors,
multiple drugs and Traditional Chinese Medicine (TCM) monomers were
confirmed to effectively affect the level of ferroptosis and exert
antitumor effects. Kong et al (6) proved that baicalin exerted its
anticancer activity via ferritin heavy chain 1 (FTH1) in both 5637
and KU19-19 cells. Zhu et al (7) proposed that artemisinin and its
derivatives may be used in the future as cancer therapies with
broader applications due to their induction of ferroptosis.
Tanshinone IIA (TanIIA) has long been confirmed to have a pivotal
role in HNSCC treatment. Studies in other organs indicate that
TanIIA exerts a significant impact on ferroptosis. For example, it
has been proved that Tan IIA induced ferroptosis in BGC-823 and
NCI-H87 gastric cancer cells via upregulating p53 expression and
downregulating xCT expression in gastric cancer (8). However, the association between
TanIIA and ferroptosis in HNSCC has remained elusive.
TanIIA, a pharmacologically active component
isolated from the rhizome of the Chinese herb Salvia
miltiorrhiza Bunge (Danshen), has been reported to possess
pleiotropic effects, such as anti-inflammatory effects, antioxidant
and anti-atherogenic effects. Recent studies have suggested that
TanIIA exhibits highly potent antitumor activity by inducing
apoptosis and triggering autophagy in tumor cells (9). A previous study by our group
confirmed TanIIA has a significant effect to promote FaDu cell
apoptosis by causing cell cycle arrest at the S phase, which
resulted in observably higher apoptotic cell fractions and
downregulation of survivin protein expression (10). In addition, TanIIA was confirmed to
exert its protective effect on endothelial cells by inhibiting
ferroptosis via activation of NRF2(11). However, the exact target and
mechanisms by which TanIIA regulates ferroptosis remain to be
elucidated.
In the present study, data acquired from FerrDb were
mined to comprehensively analyze the correlations between
ferroptosis marker genes and HNSCC. Cell survival, invasion and
expression status were evaluated after treatment with TanIIA, with
the goal of uncovering the function of TanIIA.
Materials and methods
Data sources
The RNA-sequencing expression (level 3) profiles and
corresponding clinical information for HNSCC were downloaded from
The Cancer Genome Atlas (TCGA) dataset (portal.gdc.com), including 502 tumor samples and 44
normal controls. Ferroptosis-related genes were acquired from the
FerrDb database [Zhou and Bao (12), 2020] which includes 125 ferroptosis
marker genes, of which 9 genes have been verified.
Differentially expressed gene (DEG)
analysis
The limma package in R (version 3.4.2; http://www.bioconductor.org/packages/release/bioc/html/limma.html)
was used to analyze the differentially expressed mRNAs among
samples. ‘Adjusted P<0.05 and Log2 (Fold Change) >1 or
Log2(Fold Change) <-1’ were defined as the threshold for the
differential expression of mRNAs. Spearman's correlation analysis
was used to describe the correlation between the 9 marker genes.
P<0.05 was considered to indicate statistical significance
(Table SI). The DEG-related
protein-protein interaction (PPI) network was established using the
Search Tool for the Retrieval of Interacting Genes and proteins
(STRING) database (string-db.org/) (13)
(Table SII), followed by
visualization using Cytoscape software (version 3.8.2; manual.cytoscape.org) (14).
To further confirm the underlying function of
potential targets, the data were analyzed by functional enrichment.
To better understand the role of these mRNAs in carcinogenesis, the
ClusterProfiler package (version: 3.18.0) in R was employed to
analyze the Gene Ontology function of potential targets and the
Kyoto Encyclopedia of Genes and Genomes pathway. The R software
ggplot2 package was used to draw the boxplot, while the R software
pheatmap package was used to draw the heatmap.
Validated ferroptosis marker genes and
HNSCC correlation analysis
The multi-gene correlation pheatmap was displayed by
the R software package. Spearman's correlation analysis was used to
determine the correlation between the 9 validated ferroptosis
markers. A Venn diagram analysis was carried out between genes
encoding ferroptosis markers and validated markers that are
associated with DEGs using the Interacti-Venn website (interactivenn.net/; Table SIII). By the above means, FTH1,
transferrin (TF) and TF receptor (TFRC) were screened out. The
Human Protein Atlas (http://www.proteinatlas.org) was used to validate FTH1
expression in the alimentary system (which includes the larynx and
hypopharynx).
The expression distributions of FTH1, TFRC and TF
genes in tumor tissues and normal tissues were downloaded from the
TCGA dataset. The current-release (V8) GTEx datasets were obtained
from the GTEx data portal (https://www.gtexportal.org/home/datasets) website.
Data were analyzed using R software v4.0.3 (R Foundation for
Statistical Computing). P<0.05 was considered to indicate
statistical significance.
Analyses of prognosis
Correlations between the expression levels of FTH1,
TFRC and TF genes and HNSCC overall survival were separately
analyzed through the Kaplan-Meier method and the log-rank test.
Time-receiver operating characteristic (ROC; v 0.4) (15) analysis was used to compare the
predictive accuracy of mRNA. All the analytic methods and R
packages were implemented by R (foundation for statistical
computing 2020) version 4.0.3. Furthermore, images of protein
immunohistochemistry (IHC) stains for FTH1, TFRC and TF in tumor
tissues were obtained from the Human Protein Atlas (16).
Cell acquisition
FaDu and human hypopharyngeal cell lines were
purchased from Procell Life Science & Technology Co., Ltd. The
obtained cells were cultured and passaged as primary cells and then
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1% double antibody
(penicillin-streptomycin mixture) (MilliporeSigma) in an incubator
with 5% CO2 at 37˚C. The cells were subcultured to the
third passage.
IHC method
FTH1 expression was visualized using the IHC method.
FaDu cells were seeded on to cover slips at 5x104/ml
density and then fixed in 4% paraformaldehyde (Beyotime Institute
of Biotechnology) for 15 min at 37˚C, permeabilized using 0.1%
Triton X-100 (Beyotime Institute of Biotechnology) for 20 min
blocked using 5% bovine serum albumin (MilliporeSigma) for 1 h at
37˚C Heat-mediated antigen retrieval was performed with Tris/EDTA
buffer (Gibco; Thermo Fisher Scientific, Inc.). A total of 100 µl
reaction enhancer was added to the samples and incubated for 20 min
at 37˚C before IHC staining (cat. no. PV-6000; Zhongshanjinqiao).
The samples were then incubated with HRP-labeled secondary antibody
(cat. no. A18781; 1:1,000 dilution; Thermo Fisher Scientific, Inc.)
for 30 min at 37˚C, after which the nuclei were counterstained with
diaminobenzidine (1:1,000; Beyotime Institute of Biotechnology,
Inc.) for 5-8 min. Following washing with tap water, samples were
stained with hematoxylin (Beyotime Institute of Biotechnology,
Inc.) for 20 sec, then dehydrated with different concentrations of
ethanol and clarified using dimethyl benzene (MilliporeSigma)
solution. Images were captured with an inverted phase-contrast
microscope (CH30; Olympus Corp.). A total of 5 images were acquired
from different fields of view of each slide and the staining of
yellow particles was regarded as positive expression.
Immunofluorescence (IF) method
IF assays were conducted to determine the cellular
location of FTH1 protein expression. The pre-treatment procedure
was the same as that used for IHC. IF was performed with FTH1 (5
µg/ml; cat. no. ab65080; Abcam) antibody for 12 h at 4˚C in the
dark, followed by incubation with DyLight 488 conjugated, goat
anti-human IgG (cat. no. ab96907; 1:1,000; Abcam.). The nuclei were
stained with 4',6-diamidino-2-phenylindole (DAPI; Beijing Solarbio
Science & Technology, Co., Ltd.). A total of five fluorescence
images were captured using a fluorescence microscope (Leica DMi8;
Leica Microsystems GmbH) with different excitation wavelengths for
the same field (dylight488 maximum emission is 518 nm; DAPI maximum
emission is 454 nm).
Cell survival
A Cell Counting Kit-8 assay (CCK-8; MedChemExpress)
(17) was adopted to detect cell
survival. TanIIA (cat. no. HY-N0135; MedChemExpress) was dissolved
with DMSO using ultrasonication, followed by treatment for 24, 48
and 72 h at individual concentrations of 0.25, 0.5 or 1.0 mg/l as
the treatment group. Cells were grown in 96-well plates and cell
survival was measured after 24 h following the manufacturer's
protocol. CCK-8 stain and serum-free medium were mixed at a volume
ratio of 1:10 and incubated in 5% CO2 at 37˚C for 1 h.
The absorbance at 450 nm wavelength (optical density value) was
then measured with a microplate reader (Multiskan-MK3; Thermo
Fisher Scientific, Inc.).
Tumor invasive ability
The tumor cell invasion capacity was analyzed using
a Transwell chamber (8.0 µm pore size; Corning, Inc.). BD Matrigel
(cat. no. 354248; BD Biosciences), which had been frozen in a -80˚C
freezer, was kept at 4 degrees overnight for 24 h, after which 300
µl serum-free medium was added to 60 µl Matrigel® to
prepare a mixed solution and the upper chambers were coated with it
in 24-well plates. FaDu cells (1x105) were then seeded
into five 96-well plates (1x103 cells/well), with five
parallel wells for each cell group. Furthermore, 500 µl DMEM
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS was
added to the lower chamber. The cells were then incubated for 24 h
at 37˚C in a 5% CO2 incubator. Transwell chambers were
removed and cells that had transgressed through the membrane to the
lower side were washed 2 times with PBS and fixed with 5%
glutaraldehyde (Beijing Solarbio Science & Technology, Co.,
Ltd.) at 4˚C. The attached cells were stained with crystal violet
(0.5% crystal violet in 20% methanol) for 5-10 min and then washed
two times with PBS. Images were acquired using an Axiovert 40 CFL
inverted microscope (Zeiss AG) and invaded cells were counted and
recorded. Five fields per slide were used in the analysis and each
experiment was performed in triplicate.
Western-blot analysis
FaDu and normal mucosa cells were seeded in 6-well
plates at 5x104/ml density (8 ml) and incubated
overnight in a 5% CO2 incubator at 37˚C. Cells were
first starved in serum-free medium for 48 h, then treated with
complete medium with 10% FBS for 12 h and treated with TanIIA at
0.5 mg/l for another 12 h. Cells cultured in a drug-free medium
were used as the control group. Total proteins were extracted using
One Step Animal Tissue/Cell Active Protein Extraction buffer (RIPA;
Thermo Fisher Scientific, Inc.) Protein concentration was
determined via BCA assay (cat. no. 23227; Thermo Fisher Scientific,
Inc.). Equal amounts of total protein (40 µg) were loaded in each
protein lane. Samples were separated by 10% SDS-PAGE (Beijing
Solarbio Science & Technology, Co., Ltd.) using a Bio-Rad
Electrophoresis System (Bio-Rad Laboratories, Inc.). The proteins
were transferred to a nitrocellulose membrane (cat. no. IFPL00010;
Merck KGaA) for gel electrophoresis separation, followed by
blocking with 5% skimmed milk (cat. no. 232100; BD Difco) for 1 h.
Tris-buffered saline with Tween-20 (Beijing Solarbio Science &
Technology, Co., Ltd.) was used to wash the membranes, and
subsequently, they were incubated at 4˚C overnight with the
following primary antibodies: FTH1 (1 µg/ml; 21 kDa; cat. no.
ab65080; Abcam); GAPDH (1:500 dilution; 36 kDa; cat. no. ab8245;
Abcam). Subsequently, membranes were incubated with a 1:10,000
dilution of HRP-labeled secondary antibody (cat. no. 7076s; Cell
Signaling Technology, Inc.) at room temperature for 1 h. GAPDH was
used as a normalization control. The membrane was visualized using
the ChemiDoc™ MP Imaging System (Bio-Rad Laboratories,
Inc.) and analysis was performed using Image Lab software (v4.0;
Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analyses were performed using SPSS
version 21.0 (IBM Corporation) and GraphPad Prism 8 (GraphPad
Software, Inc.). Each experiment was performed in triplicate and
the mean ± SD. The normality of distribution of data was assessed
using Shapiro-Wilk test. Data comparisons between two groups were
performed using an unpaired t-test. Comparisons among multiple
groups were performed using one-way ANOVA, followed by Tukey's
multiple-comparisons test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Correlation between HNSCC and
ferroptosis marker genes
The gene expression data and clinical
characteristics of 502 HNSCC and 44 normal tissue samples from the
TCGA database were included in the present study (Table SIV). The limma package in the R
software was used to study the differentially expressed mRNAs. A
total of 1,457 genes were significantly upregulated and 715
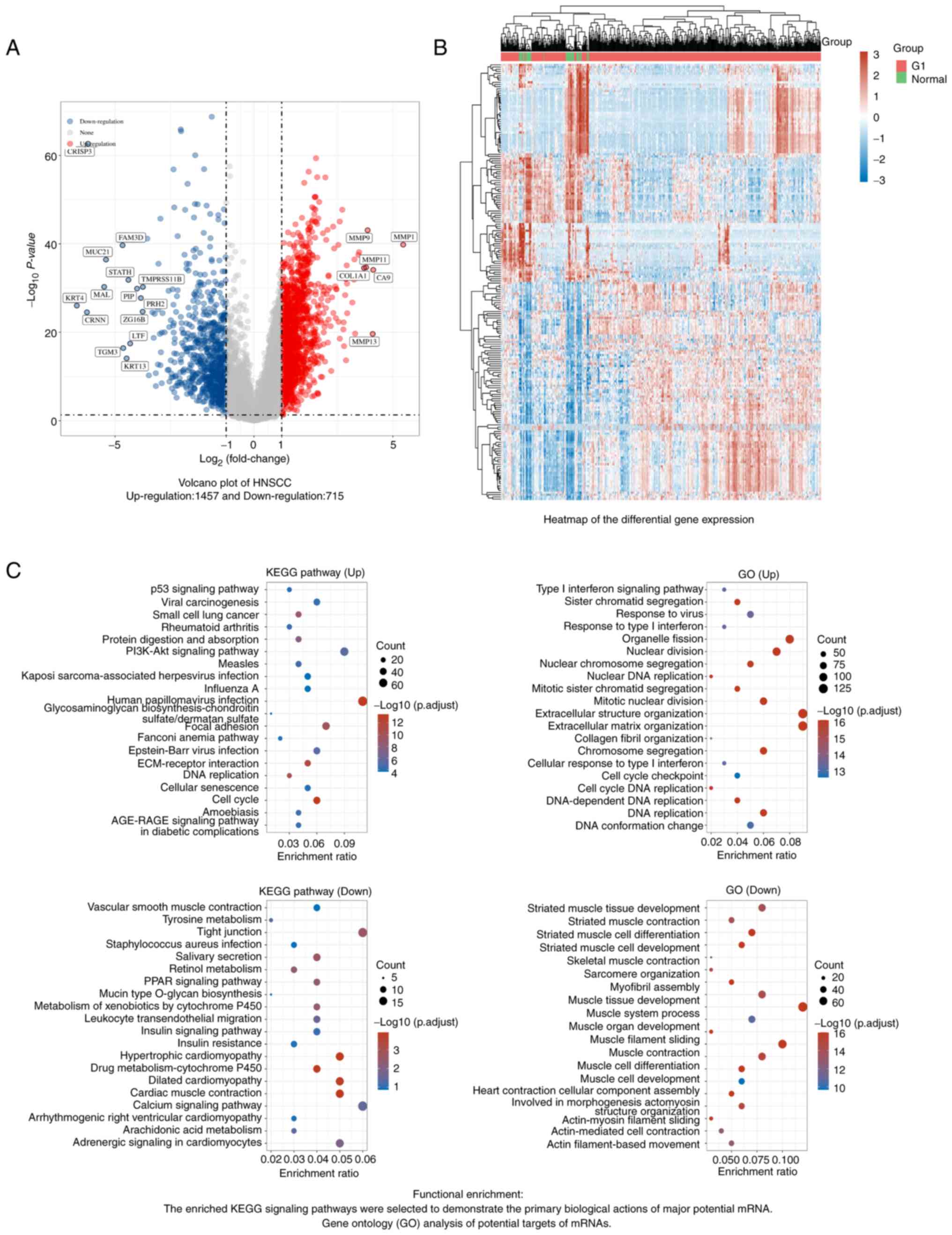
downregulated (Fig. 1). Enrichment
analysis indicated that the DEGs were enriched in ‘cell cycle’.
Next, 125 ferroptosis genes were downloaded from the FerrDb
database, among which nine were validated ferroptosis-related
markers (PTGS2, CHAC1, FTH1, SLC40A1, TF, TFRC, GPX4, HSPB1 and
NFE2L2). Pearson correlation analysis was conducted to assess the
correlations of expressions between each pair of genes of the above
9 ferroptosis marker genes (Fig.
2A) and the PPI network of these genes was further mapped
(Fig. 2B). The results indicated
that the expression of most of the gene pairs exhibited a
statistical correlation (Table
SI). The gene annotations and scores are presented in Table SII.
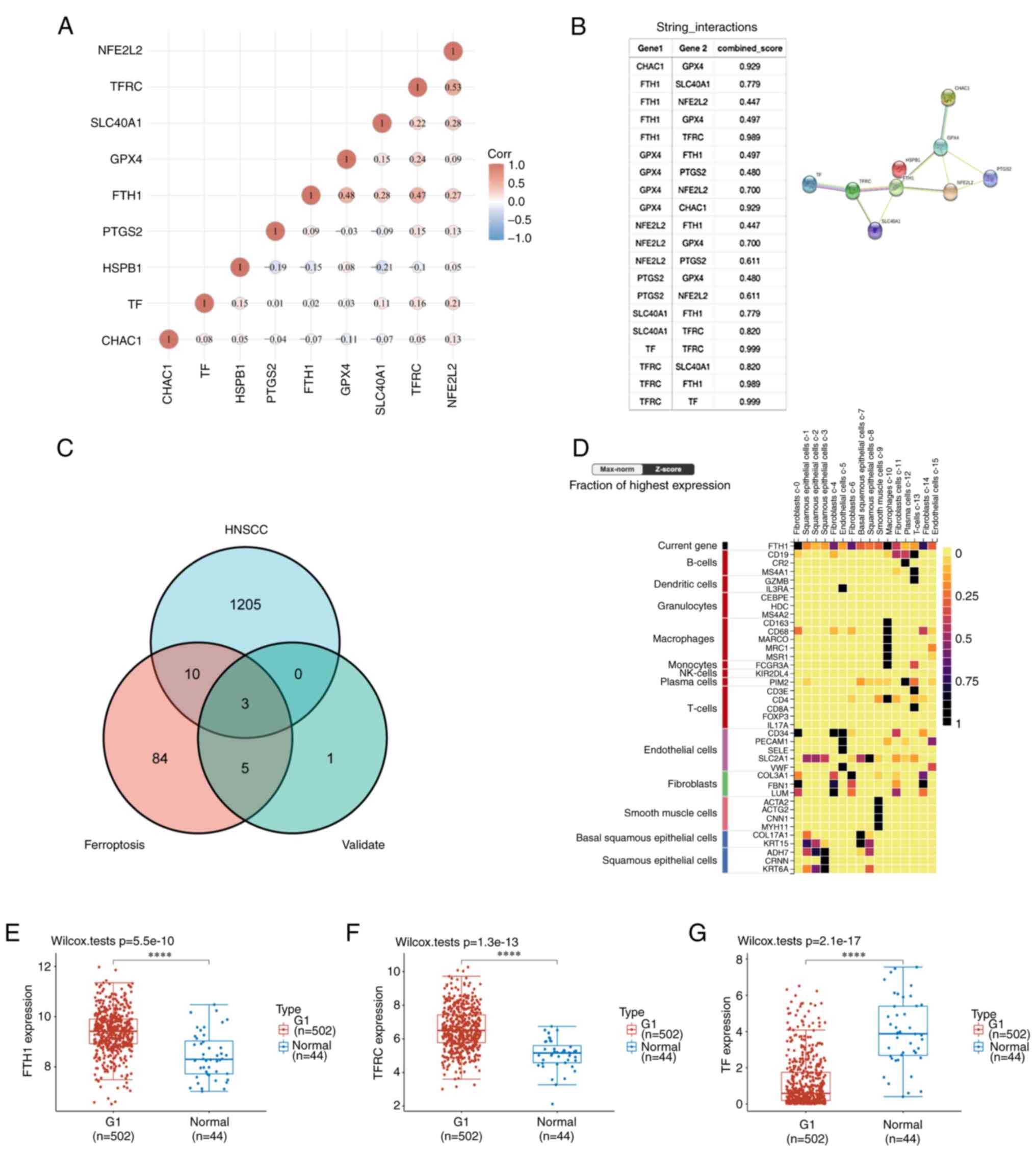 | Figure 2Screened out significantly expressed
ferroptosis genes in HNSCC. (A) The correlations among 9 validated
ferroptosis genes: A heatmap of the correlation between these 9
genes is provided. The abscissa and ordinate represent genes,
different colors represent different correlation coefficients (blue
represents a positive correlation whereas red represents a negative
correlation), with a darker color indicating a stronger
correlation. (B) Annotation of ferroptosis-related differentially
expressed marker proteins and their co-expression scores (STRING
analysis). (C) Venn diagram of common genes encoding ferroptosis
genes that were validated by PCR and are DEGs in HNSCC (R package
analysis). (D) Cell type marker heatmap of FTH1 in the alimentary
system; FTH1 is mainly expressed in fibroblasts and squamous
epithelial cells. Comparison of (E) FTH1, (F) TFRC and (G) TF
expression between HNSCC and normal mucosa by the Mann-Whitney
U-test (Wilcoxon rank-sum test). Data were acquired from The Cancer
Genome Atlas (https://portal.gdc.cancer.gov/) HNSCC database;
RNA-seq data in the level 3 HTSeq-FPKM format.
****P<0.0001. HNSCC, head and neck squamous
carcinoma; DEG, differentially expressed gene; Corr, correlation
coefficient; STRING, Search Tool for the Retrieval of Interacting
Genes and Proteins; FTH1, ferritin heavy chain 1; TF, transferrin;
TFRC, TF receptor; Seq, sequencing. |
FTH1, TFRC and TF expression in
HNSCC
Venn diagram analysis was used and FTH1, TFRC and TF
were screened out as common genes expressed in HNSCC and validated
ferroptosis markers (Fig. 2C).
Subsequently, the expression distributions of these genes in tumor
tissues and normal tissues were analyzed and the results indicated
that all the three genes were significantly differentially
expressed in HNSCC (FTH1, P=5.5x10-10; TFRC,
P=1.3x10-13; and TF, P=2.1x10-17; Fig. 2E-G).
FTH1 is an independent prognostic
factor in HNSCC
The associations between ferroptosis marker-related
DEGs and survival were then analyzed. The log-rank test was used to
compare differences in survival between groups with high and low
expression of the genes FTH1 [hazard ratio (HR): 1.562, 95%CI:
1.191-2.051, P=0.00129], TFRC (HR: 1.228, 95%CI: 0.94-1.606,
P=0.132) and TF (HR: 0.854, 95%CI: 0.653-1.115, P=0.246; Fig. 3A). Time-receiver ROC analysis was
used to compare the predictive accuracy for FTH1 [3-year area under
the ROC curve (AUC)=0.585, 95%CI: 0.531-0.639], TFRC (3-year
AUC=0.525, 95%CI: 0.470-0.480) and TF (3-year AUC=0.514, 95%CI:
0.460-0.569; Fig. 3B).
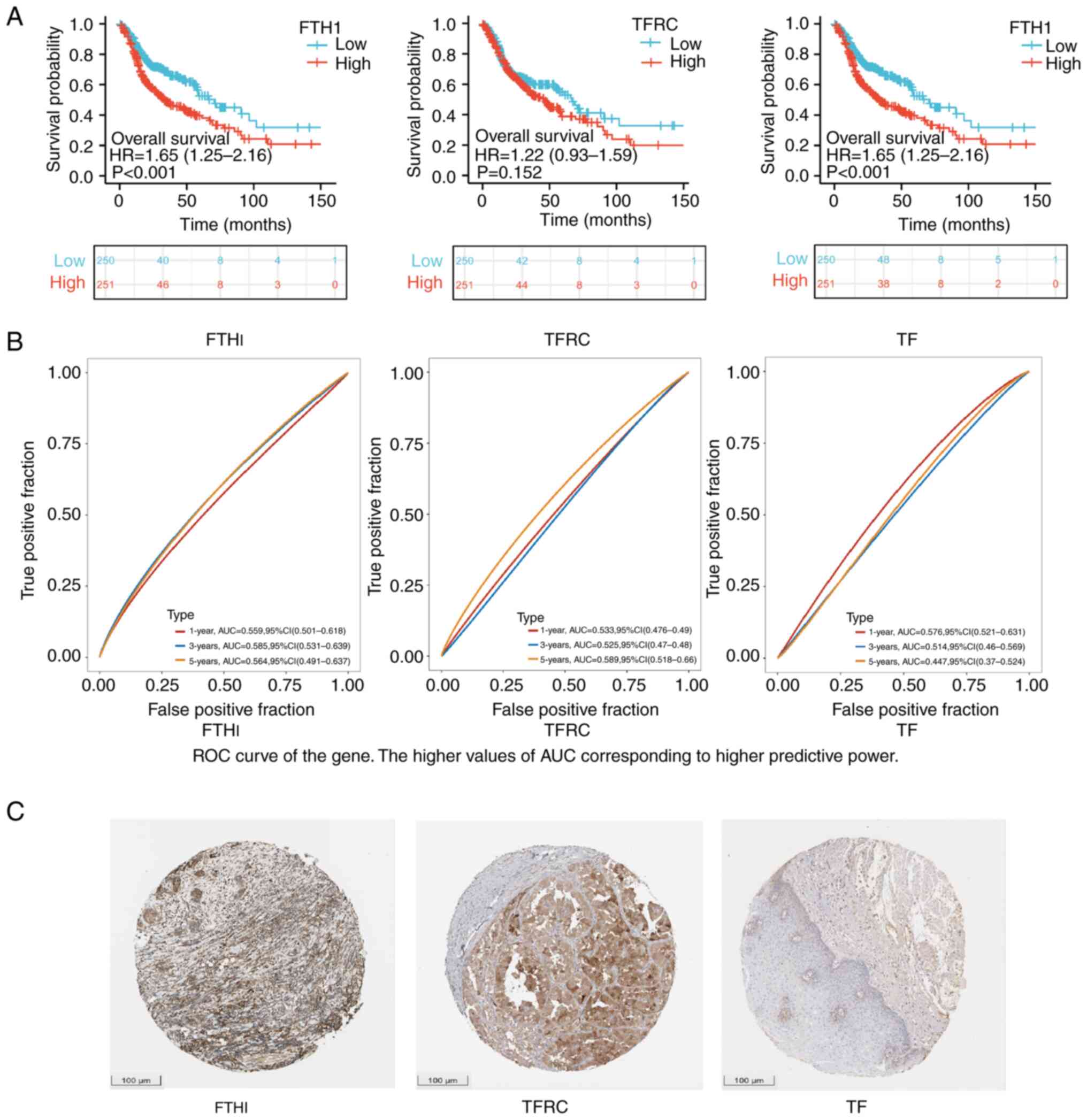 | Figure 3Validated ferroptosis markers and
HNSCC. (A) Kaplan-Meier survival analysis of FTH1, TFRC and TF in
The Cancer Genome Atlas dataset. Comparison among different groups
was performed using the log-rank test. Adjusted HR was computed and
95%CI for HR and median survival time were calculated. (B) ROC
curves of FTH1, TFRC and TF at 1,3,5 years. The median values were
used as cutoff. A higher AUC corresponds to a higher predictive
power. (C) Identification of FTH1, TFRC and TF expression by
immunofluorescent staining (scale bar, 100 µm). HNSCC, head and
neck squamous carcinoma; HR, hazard ratio (high vs. low
expression); FTH1, ferritin heavy chain 1; TF, transferrin; TFRC,
TF receptor; AUC, area under the ROC curve; ROC, receiver operating
characteristic. |
Based on the formal results, FTH1, TFRC and TF
expression was next validated using Oncomine and the Human Protein
Atlas database (Fig. 3C). The
brown color indicates positive IHC staining. FTH1 expression was
then observed in the digestive system heatmap, indicating that it
was mainly expressed in squamous epithelial cells and fibroblasts
(Fig. 2D). The above results
indicated that FTH1 is an independent prognostic factor for
HNSCC.
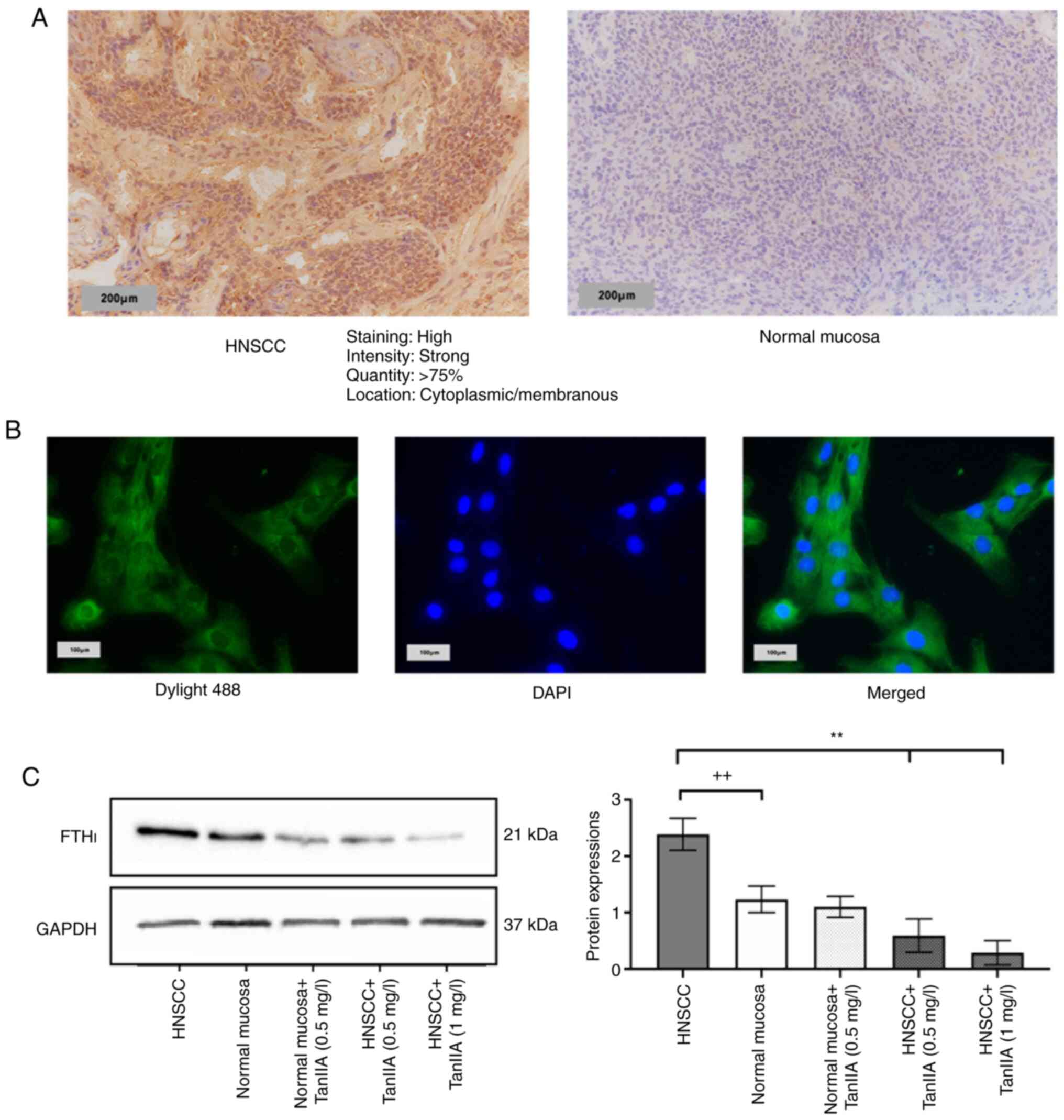
FTH1 protein expression
IHC observation and an IF assay were further
performed to determine the localization and FTH1 expression in
HNSCC cells. It is generally considered that FTH1 expression is
localized to the cytoplasm and membrane (18). The IHC staining result was fully
consistent with this (Fig. 4A). An
IF assay was then performed for further confirmation and the
results complied with IHC, i.e., fluorescence signals were present
in the cytoplasm, while no expression was observed in the nucleus
(Fig. 4B).
TanIIA downregulates FTH1 expression
in FaDu cells
FTH1 expression in FaDu cells was measured by
western blot analysis (Fig. 4C).
Normal mucosa cells were used as the control group. FTH1 in FaDu
cells was significantly overexpressed compared with the control
group (P<0.05). Different concentrations (0.25, 0.5 and 1 mg/l)
of TanIIA were adopted for FaDu cell treatment. The data obtained
confirmed that TanIIA at 0.5 mg/l effectively downregulated FTH1
expression in FaDu cells. Furthermore, it is noteworthy that the
P-value in the normal mucosa group indicated no significance
compared with that of the normal mucosa + TanIIA group. Whether
such a result indicated selectivity of TanIIA should be further
investigated in depth.
TanIIA effectively reduces FaDu cell
survival and invasion
A CCK-8 assay was adopted for detection of cell
survival (Fig. 5A-C). TanIIA
treatment was performed at concentrations of 0.25, 0.5 and 1.0
mg/l, and the survival rate was measured at 24, 48 and 72 h,
respectively, by determining the mean value. The proportion of live
cells in the treatment group (48 h: 70.3%; 72 h: 52.29%) decreased
significantly (P<0.05) as compared with the control group (48 h:
67.83%; 72 h: 54.35%). The effect of TanIIA exhibited a clear
dose-effect relationship (Fig.
5D). A Transwell assay was then performed to observe the
invasion of FaDu cells (Fig. 5E).
As indicated from the results, FaDu cells exhibited a stronger
migration and invasion ability than normal hypopharyngeal cells and
treatment with TanIIA significantly attenuated the invasion ability
of FaDu cells (P<0.05; Fig.
5F). Specifically, under treatment with TanIIA (0.5 mg/l)
(Fig. 5G), although the invasion
ability of HNSCC (mean=146) remained higher than that of the
control (mean=119), the difference was not statistically
significant (P=0.0557), thereby demonstrating that TanIIA treatment
effectively inhibited the invasion ability of FaDu cells.
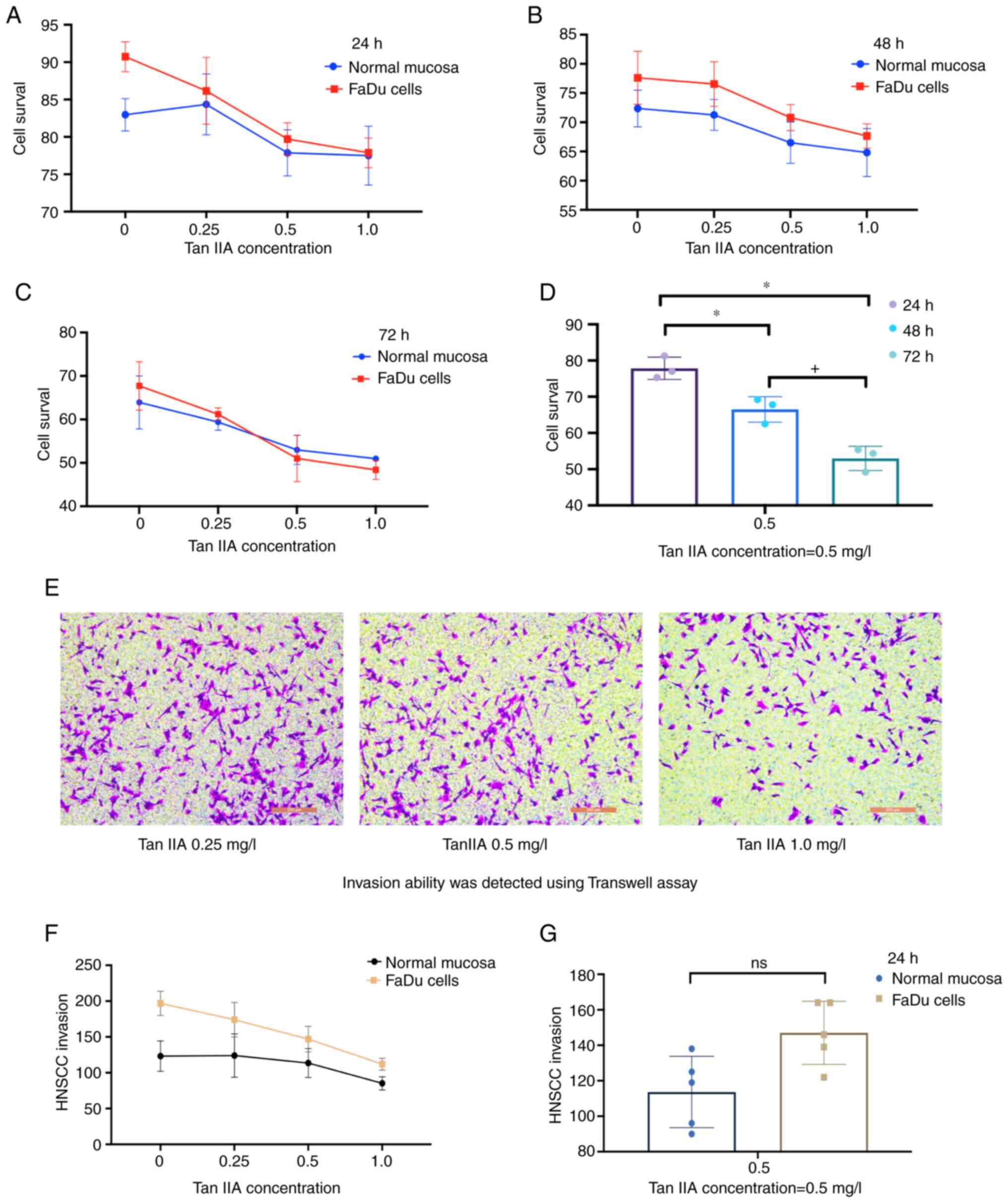 | Figure 5A Cell Counting Kit-8 assay was used
to detect the cell survival rate at (A) 24 h, (B) 48 h and (C) 72
h. The blue color represents the control group and the red color
represents the FaDu cell group. The survival rate of FaDu cells
after TanIIA treatment was lower than that of the control group
with a dose-dependent effect. (D) A comparison of each group at a
0.5 mg/l concentration of TanIIA was provided to assess whether the
effects were enhanced over time (*P<0.05, compared
with 24 h group; +P<0.05, compared with 48 h group).
(E) Invasion of FaDu cells in the presence of different
concentrations (0.25, 0.5 and 1 mg/l) of TanIIA measured by a
Transwell assay (scale bars, 200 µm). (F) Quantitative evaluation
of the Transwell assay results indicated that, compared with the
control group, FaDu cells exhibited increased invasion ability
in vitro (+P<0.05), which was attenuated by
TanIIA treatment (+P<0.05). (G) Compared with the
control group at 0.5 mg/l concentration TanIIA at 24 h. Statistical
analysis suggested (P=0.0557) no significant between TanIIA +FaDu
cells and Para cancer control, indicating that TanIIA effectively
reduced FaDu cells invasion. Tan IIA, Tanshinone IIA; ns, no
significance; HNSCC, head and neck squamous carcinoma. |
Discussion
The global annual occurrence of head and neck
cancers exceeds 0.5 million, out of which 90% are squamous cell
carcinoma, HNSCC (19). The
development of HNSCC refers to a multi-step process involving
multiple signal transduction pathways and complex cross-talk among
the pathways. The rapid advancement of microarray technology has
significantly boosted gene expression profiling, gene sequencing
and convenient diagnosis of diseases due to its high-throughput
characteristics and rapid detection ability. Numerous studies have
confirmed that ferroptosis has complex roles in tumor progression
(20) and regulate the tumor
microenvironment (21).
Increasing research studies have confirmed that
ferroptosis levels are closely related to HNSCC initiation,
progression, recurrence, metastasis and immune escape. Ferroptosis
(22) has been referred to as the
‘new land of cell death’. Distinct from apoptosis, necrosis and
autophagy, it refers to an iron-dependent, novel form of programmed
cell death with diverse and complex targets involved (e.g., PTGS2,
NOX1, FTH1, COX2, GPX4 and ACSl4). A tissue microarray study
indicated that patients with high expression of GPX4 had poor
survival and the expression of GPX4 has been negatively associated
with the immunogenic cell death-related protein calreticulin in an
HNSCC tissue array (23). An
increasing number of ferroptosis-related targets have been
identified as independent prognostic markers for HNSCC (24).
As a widely used TCM compound, TanIIA has already
been investigated as a primary or adjuvant therapy with beneficial
effects on HNSCC patient survival (10). The latest studies have confirmed
that TanIIA increased the level of lipid peroxides and decreased
glutathione levels in gastric cancer cells, both of which are
markers of ferroptosis (25). Guan
et al (8) proved that
TanIIA caused decreased intracellular glutathione levels and
cysteine levels and increased intracellular reactive oxygen species
(ROS) levels. Furthermore, p53 knockdown attenuated TanIIA-induced
lipid peroxidation and ferroptosis in a BGC-823 xenograft model.
The above studies revealed that TanIIA has significant impacts on
cancer cells, partially by inducing ferroptosis. Therefore, it is
reasonable to assume that TanIIA may regulate HNSCC progression
through ferroptosis-related marker genes.
In the present study, by listing all DEGs in HNSCC
and validated marker genes for ferroptosis, three possible targets
were screened out: FTH1, TFRC and TF. In a TGCA prognostic
analysis, FTH1 was indicated to be an independent prognostic factor
of HNSCC.
Iron is an essential element required by cells and
has been described as a key player in ferroptosis (26). Ferritin is a protein complex that
stores iron in a bioavailable, nontoxic form (27). FTH1 is the main subunit for
ferritin's function, which covers a ferroxidase active center
responsible for regulating the oxidation and integration of ferric
ions. Upstream of the transcription start site of ferritin heavy
chain contained an antioxidant response element, which could
protect cells from oxidative damage, thereby avoiding apoptosis by
responding to oxidative reactions (28). This feature makes FTH1 closely
related to numerous biological processes such as oxidative stress
(29), cell differentiation
(30) and neuronal functions
(31). Previous work has shown
that FTH1 expression is increased in several human cancers
including squamous cell carcinoma, the study by Yang et al
(32) indicated that long
non-protein-coding RNA FTH1 pseudogene 3 was significantly
up-regulated in Esophageal squamous cell carcinoma tissues and
cells and was critical to the course of tumor formation. Also, the
FTH1 expression level is associated with the survival rate of oral
squamous cell carcinoma (33).
Silencing of FTH1 or reducing its expression has been proved can
significantly inhibit carcinogenesis, mainly by promoting cell
death (34), inhibiting metastasis
(35), and attenuating tumor cell
invasion (36). Here we proved
that FTH1 is a validated ferroptosis marker gene that significantly
impacts HNSCC prognosis, which is mainly expressed in squamous
epithelial cells and fibroblasts in the human digestive system.
TanIIA is a pharmacologically active lipophilic
component of Salvia miltiorrhiza extract, which shares a
history of high repute in TCM (37). TanIIA has demonstrated potent tumor
inhibitory effects against various types of tumor cell (38); the underlying mechanism involves
regulation of the cell cycle (39), suppresses cell proliferation and
induces apoptosis (40), inhibits
tumor invasion and metastasis, inhibits angiogenesis (41) and reverses tumor multi-drug
resistance (42). Xu et al
(43) demonstrated that TanIIA
enhances the antitumor activity of chemotherapeutic drugs and
increases the sensitivity of FaDu cells to radiotherapy. Ding et
al (44) examined the
underlying mechanisms and indicated that TanIIA exerted a strong
radiosensitizing effect due to enhanced ROS generation and
autophagy. Here, CCK-8 and Transwell assays were used to determine
the effects of TanIIA on the cell survival and invasion ability in
an attempt to examine its pharmacological effects, indicating that
at certain concentrations, TanIIA significantly promoted FaDu cell
death and attenuated its invasion ability with a dose-effect
relationship.
It is generally accepted that FaDu cells exhibit the
biological characteristics of high proliferation and invasion
(45), which makes treatment of
HNSCC relatively difficult. In patients with early-stage HNSCC,
surgery and radiotherapy act as the mainstays of treatment, whereas
70-80% of patients are already locally advanced or advanced at the
initial presentation. For these patients, the 5-year survival rate
was only 40% or even less (46).
As reported by a previous study, the median survival time of
patients was only 10 months once HNSCC had recurred or metastasized
(47). Thus, adjuvant therapies
are critical to HNSCC treatment. To date, cisplatin remains the
treatment of choice for HNSCC, whereas its development of
resistance and toxicity cannot be ignored. Given all the above,
finding an effective drug with fewer and less severe side effects
is a promising solution. The present study proved that a certain
concentration of TanIIA (≥0.5 mg/l) was able to significantly
attenuate the protein expression of the prognostic factor FTH1.
In summary, the present study highlights a novel
perspective in the clinical treatment of HNSCC and provides a
theoretical basis for developing TanIIA for use in co-treatment for
HNSCC. By identifying the relationship between ferroptosis markers
and HNSCC, a ferroptosis coregulated gene was screened out, which
was able to significantly affect HNSCC prognosis. The present study
was the first, to the best of our knowledge, to indicate that
TanIIA treatment drastically inhibited FTH1 expression in FaDu
cells, significantly affected cell survival and suppressed the
invasive capacity of FaDu cells. However, the present study had
several limitations. FTH1 serves as a vital regulator of
ferroptosis, which is generally considered a suppressor of
ferroptosis (48), its specific
gene function still requires further study using the knockdown or
knockout method. The STRING data of the present study indicated
that FTH1 is associated with ferroptosis markers including GPX4,
SLC40A1, TFRC and NFE2L2, so the downstream signaling pathways are
worth further investigating. In addition, as TanIIA is a natural
antineoplastic drug, it is also important to perform clinical
research on whether dose escalation increases with long-term use
and if this would bring side effects.
Supplementary Material
Correlation analysis between verified
targets of ferroptosis.
Protein-protein interactions of
ferroptosis markers.
Venn diagrams of HNSCC and ferroptosis
markers.
Clinical characteristics of samples
from The Cancer Genome Atlas.
Acknowledgements
The authors would like to thank Dr Pin Dong
(Otolaryngology-Head and Neck Surgery, Shanghai General Hospital,
Shanghai, China) for reviewing this article for technical accuracy
and for providing valuable suggestions.
Funding
Funding: This study was supported by The National Natural
Science Foundation of China (grant no. 81800893).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WM and BW designed the experiments and drafted the
manuscript. WM and JD performed the experiments and analyzed and
interpreted the data. YL helped with data collection and analysis.
RH coordinated the research and participated in the experimental
design. All authors were involved in critically revising the
manuscript, and have read and approved the final manuscript. WM and
BW confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
This work was approved by the Ethics Committee of
Shanghai General Hospital (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Blanchard P, Landais C, Lacas B, Petit C,
Bourhis J and Pignon JP: SP-010: Update of the meta-analysis of
chemotherapy in head and neck cancer (MACH-NC). Radiother Oncol.
122(9)2017.
|
|
2
|
Ye J, Jiang X, Dong Z, Hu S and Xiao M:
Low-concentration PTX and RSL3 inhibits tumor cell growth
synergistically by inducing ferroptosis in mutant p53
hypopharyngeal squamous carcinoma. Cancer Manag Res. 11:9783–9792.
2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lee J, Ji HY, Kim MS and Roh JL:
Epigenetic reprogramming of epithelial-mesenchymal transition
promotes ferroptosis of head and neck cancer. Redox Biol.
37(101697)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Roh JL, Kim EH, Jang H and Shin D: Nrf2
inhibition reverses the resistance of cisplatin-resistant head and
neck cancer cells to artesunate-induced ferroptosis. Redox Biol.
11:254–262. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mou Y, Wang J, Wu J, He D, Zhang C, Duan C
and Li B: Ferroptosis, a new form of cell death: Opportunities and
challenges in cancer. J Hematol Oncol. 12(34)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kong N, Chen X, Feng J, Duan T, Liu S, Sun
X, Chen P, Pan T, Yan L, Jin T, et al: Baicalin induces ferroptosis
in bladder cancer cells by downregulating FTH1. Acta Pharm Sin B.
11:4045–4054. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhu S, Yu Q, Huo C, Li Y, He L, Ran B,
Chen J, Li Y and Liu W: Ferroptosis: A novel mechanism of
artemisinin and its derivatives in cancer therapy. Curr Med Chem.
28:329–345. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Guan Z, Chen J, Li X and Dong N:
Tanshinone IIA induces ferroptosis in gastric cancer cells through
p53-mediated SLC7A11 down-regulation. Biosci Rep.
40(BSR20201807)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Qian J, Cao Y, Zhang J, Li L, Wu J, Wei G,
Yu J and Huo J: Tanshinone IIA induces autophagy in colon cancer
cells through MEK/ERK/mTOR pathway. Transl Cancer Res. 9:6919–6928.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhao YX, Luo D, Zhang YH, Shen B, Wang BX
and Sun ZF: The effect of tanshinone ⅡA potentiates the effects of
cisplatin in Fadu cells in vitro through downregulation of
survivin. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi.
31:781–784. 2017.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
11
|
He L, Liu YY, Wang K, Li C, Zhang W, Li
ZZ, Huang XZ and Xiong Y: Tanshinone IIA protects human coronary
artery endothelial cells from ferroptosis by activating the NRF2
pathway. Biochem Biophys Res Commun. 575:1–7. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhou N and Bao J: FerrDb: A manually
curated resource for regulators and markers of ferroptosis and
ferroptosis-disease associations. Database (Oxford).
2020(baaa021)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261.
2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Obuchowski NA and Bullen JA: Receiver
operating characteristic (ROC) curves: Review of methods with
applications in diagnostic medicine. Phys Med Biol.
63(07TR01)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based human protein atlas. Nat
Biotechnol. 28:1248–1250. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xu LP: Studies on the inhibition of AT#9
on three tumour cells proliferation by cell counting-kit 8. Journal
of North Pharmacy. 9:43–44. 2012.(In Chinese).
|
|
18
|
Di Sanzo M, Quaresima B, Biamonte F,
Palmieri C and Faniello MC: FTH1 pseudogenes in cancer and cell
metabolism. Cells. 9(2554)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu C, Guo T, Xu G, Sakai A, Ren S,
Fukusumi T, Ando M, Sadat S, Saito Y, Khan Z, et al:
Characterization of alternative splicing events in HPV-negative
head and neck squamous cell carcinoma identifies an oncogenic DOCK5
variant. Clin Cancer Res. 24:5123–5132. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hassannia B, Vandenabeele P and
Vanden*Berghe T: Targeting ferroptosis to iron out cancer. Cancer
Cell. 35:830–849. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang K, Ping L, Du T, Liang G, Huang Y,
Li Z, Deng R and Tang J: A ferroptosis-related lncRNAs signature
predicts prognosis and immune microenvironment for breast cancer.
Front Mol Biosci. 8(678877)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jiang X, Stockwell BR and Conrad M:
Ferroptosis: Mechanisms, biology and role in disease. Nat Rev Mol
Cell Biol. 22:266–282. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhao YY, Lian JX, Lan Z, Zou KL, Wang WM
and Yu GT: Ferroptosis promotes anti-tumor immune response by
inducing immunogenic exposure in HNSCC. Oral Dis: Nov 12, 2021
(Epub ahead of print).
|
|
24
|
Tang Y, Li C, Zhang YJ and Wu ZH:
Ferroptosis-related long non-coding RNA signature predicts the
prognosis of head and neck squamous cell carcinoma. Int J Biol Sci.
17:702–711. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ni H, Ruan G, Sun C, Yang X, Miao Z, Li J,
Chen Y, Qin H, Liu Y, Zhang L, et al: Tanshinone IIA inhibits
gastric cancer cell stemness through inducing ferroptosis. Environ
Toxicol. 37:192–200. 2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jiang Y, Mao C, Yang R, Yan B, Shi Y, Liu
X, Lai W, Liu Y, Wang X, Xiao D, et al: EGLN1/c-Myc induced
lymphoid-specific helicase inhibits ferroptosis through lipid
metabolic gene expression changes. Theranostics. 7:3293–3305.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Plays M, Müller S and Rodriguez R:
Chemistry and biology of ferritin. Metallomics.
13(mfab021)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Scaramuzzino L, Lucchino V, Scalise S,
Conte ML, Zannino C, Sacco A, Biamonte F, Parrotta EI, Costanzo FS
and Cuda G: Dissecting the molecular response of human Escs to
iron-mediated oxidative stress by genetic silencing of FTH1 gene.
Res Sq. 1:1–28. 2021.
|
|
29
|
Lee JH, Jang H, Cho EJ and Youn HD:
Ferritin binds and activates p53 under oxidative stress. Biochem
Biophys Res Commun. 389:399–404. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Di Sanzo M, Aversa I, Santamaria G,
Gagliardi M, Panebianco M, Biamonte F, Zolea F, Faniello MC, Cuda G
and Costanzo F: FTH1P3, a novel H-ferritin pseudogene
transcriptionally active, is ubiquitously expressed and regulated
during cell differentiation. PLoS One. 11(e0151359)2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mu T, Qin Y, Liu B, He X, Liao Y, Sun J,
Qiu J, Li X, Zhong Y and Cai J: In vitro neural differentiation of
bone marrow mesenchymal stem cells carrying the FTH1 reporter gene
and detection with MRI. Biomed Res Int.
2018(1978602)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang L, Sun K, Chu J, Qu Y, Zhao X, Yin H,
Ming L, Wan J and He F: Long non-coding RNA FTH1P3 regulated
metastasis and invasion of esophageal squamous cell carcinoma
through SP1/NF-kB pathway. Biomed Pharmacother. 106:1570–1577.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang CZ: Long non-coding RNA FTH1P3
facilitates oral squamous cell carcinoma progression by acting as a
molecular sponge of miR-224-5p to modulate fizzled 5 expression.
Gene. 607:47–55. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Muhammad JS, Bajbouj K, Shafarin J and
Hamad M: Estrogen-induced epigenetic silencing of FTH1 and TFRC
genes reduces liver cancer cell growth and survival. Epigenetics.
15:1302–1318. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li J, Lama R, Galster SL, Inigo JR, Wu J,
Chandra D, Chemler SR and Wang X: Small-molecule MMRi62 induces
ferroptosis and inhibits metastasis in pancreatic cancer via
degradation of ferritin heavy chain and mutant p53. Mol Cancer
Ther. 21:535–455. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Huang HX, Yang G, Yang Y, Yan J, Tang XY
and Pan Q: TFAP2A is a novel regulator that modulates ferroptosis
in gallbladder carcinoma cells via the Nrf2 signalling axis. Eur
Rev Med Pharmacol Sci. 24:4745–4755. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ansari MA, Khan FB, Safdari HA, Almatroudi
A, Alzohairy MA, Safdari M, Amirizadeh M, Rehman S, Equbal MJ and
Hoque M: Prospective therapeutic potential of Tanshinone IIA: An
updated overview. Pharmacol Res. 164(105364)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang Y, Jiang P, Ye M, Kim SH, Jiang C
and Lü J: Tanshinones: Sources, pharmacokinetics and anti-cancer
activities. Int J Mol Sci. 13:13621–13666. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Che XH, Park EJ, Zhao YZ, Kim WH and Sohn
DH: Tanshinone II A induces apoptosis and S phase cell cycle arrest
in activated rat hepatic stellate cells. Basic Clin Pharmacol
Toxicol. 106:30–37. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Won SH, Lee HJ, Jeong SJ, Lee HJ, Lee EO,
Jung DB, Shin JM, Kwon TR, Yun SM, Lee MH, et al: Tanshinone IIA
induces mitochondria dependent apoptosis in prostate cancer cells
in association with an inhibition of phosphoinositide 3-kinase/AKT
pathway. Biol Pharm Bull. 33:1828–1834. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sui H, Zhao J, Zhou L, Wen H, Deng W, Li
C, Ji Q, Liu X, Feng Y, Chai N, et al: Tanshinone IIA inhibits
β-catenin/VEGF-mediated angiogenesis by targeting TGF-β1 in
normoxic and HIF-1α in hypoxic microenvironments in human
colorectal cancer. Cancer Lett. 403:86–97. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fan Q, Fan GJ, Yang PM and Zhao JY: Effect
of tanshinone microemulsion on reversing MDR in human tumor cells.
Zhongguo Zhong Yao Za Zhi. 29:1079–1081. 2004.PubMed/NCBI(In Chinese).
|
|
43
|
Xu H, Hao YL, Xu LN, Chen L and Xu FW:
Tanshinone sensitized the antitumor effects of irradiation on
laryngeal cancer via JNK pathway. Cancer Med. 7:5187–5193.
2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ding L, Wang S, Qu X and Wang J:
Tanshinone IIA sensitizes oral squamous cell carcinoma to radiation
due to an enhanced autophagy. Environ Toxicol Pharmacol.
46:264–269. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Johnson DE, Burtness B, Leemans CR, Lui
VWY, Bauman JE and Grandis JR: Head and neck squamous cell
carcinoma. Nat Rev Dis Primers. 6(92)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ortiz-Cuaran S, Bouaoud J, Karabajakian A,
Fayette J and Saintigny P: Precision medicine approaches to
overcome resistance to therapy in head and neck cancers. Front
Oncol. 11(614332)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Gougis P, Moreau Bachelard C, Kamal M, Gan
HK, Borcoman E, Torossian N, Bièche I and Le*Tourneau C: Clinical
development of molecular targeted therapy in head and neck squamous
cell carcinoma. JNCI Cancer Spectr. 3(pkz055)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li X, Si W, Li Z, Tian Y, Liu X, Ye S,
Huang Z, Ji Y, Zhao C, Hao X, et al: miR-335 promotes ferroptosis
by targeting ferritin heavy chain 1 in in vivo and in
vitro models of Parkinson's disease. Int J Mol Med.
47(61)2021.PubMed/NCBI View Article : Google Scholar
|