Introduction
Clear cell renal cell carcinoma (ccRCC) is one of
the commonest tumors of the urinary system. Each year 202,000 cases
of ccRCC are diagnosed and there are 102,000 ccRCC-related deaths
worldwide (1). In 2014 alone,
there were ~66,800 newly confirmed cases and 13,860 ccRCC-related
deaths in China (2). Although
ultrasound and computed tomography have been widely used in
clinical practice, ~1/3 of patients with ccRCC have already
developed local or distant metastasis at the time of initial
diagnosis and these patients have a poor prognosis (3). Therefore, it is important to explore
and investigate novel diagnostic and therapeutic approaches for the
clinical treatment of patients with ccRCC.
A-kinase interaction protein 1 (AKIP1) was initially
reported as novel human breast cancer-associated gene 3, which
encodes a selectively splicing proline-rich protein (4). Increasing evidence suggests that
AKIP1 dysregulation is associated with physiological and
pathological changes in numerous human malignancies and may be
involved in tumor metastasis. It has previously been reported that
in esophageal squamous cell carcinoma (ESCC) cell lines and
clinical samples AKIP1 is upregulated which is associated with a
poor prognosis in patients with ESCC (5). In addition, another study
demonstrated that AKIP1 is a downstream target of NK2 homeobox 8
and a transcription factor of vascular endothelial growth factor
(VEGF)C, which can induce lymphangiogenesis and lymphatic
metastasis of ESCC (6).
Furthermore, AKIP1 has been considered to be a potential metastatic
agent in numerous types of human cancers (7-9).
However, the clinical significance of its molecular function and
the underlying mechanisms that promote distant metastasis are not
well understood, especially in ccRCC. The latest clinical data
suggests that AKIP1 expression is increased in ccRCC tissues and
that AKIP1 expression levels are positively associated with TNM
stage. These data also indicate that higher AKIP1 expression is
associated with a worse prognosis (10). However, whether AKIP1 serves a role
in promoting tumor processes, such as proliferation, invasion and
migration in ccRCC cells, remains to be elucidated and its
potential underlying mechanism remains unknown.
A previous study reported that AKIP1 binds to
Rac1(11), which is involved in
promoting the formation of cancer stemness in ccRCC (12). Rac1 inhibition can reduce the
malignant progression of pituitary tumor-transforming
gene-1-positive ccRCC (13), which
indicates that Rac1 is involved in promoting ccRCC progression.
Furthermore, a previous study demonstrated that Rac1 activates the
ERK signaling pathway to promote colon cancer invasion and
migration (14). In addition,
Kyoto Encyclopedia of Genes and Genomes analysis indicated that ERK
can activate the downstream cellular (c)-Myc signaling pathway.
The aim of the present study was to investigate
whether AKIP1 can affect the proliferation, invasion, migration and
angiogenesis of ccRCC cells via its interaction with Rac1.
Furthermore, the influence of AKIP1 and Rac1 on the expression of
the downstream ERK/c-Myc signaling pathway was explored.
Materials and methods
Cell culture
Normal renal epithelial cell lines (HKC-5 and HK-2),
human umbilical vein endothelial cells (HUVECs) and ccRCC cell
lines (NRCC, Caki-1 and A498) were purchased from BioVector NTCC,
Inc. HKC-5 and HK-2 cells were cultured in minimum essential medium
(MEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin. ccRCC cells were cultured in DMEM/F12
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS and 1%
penicillin-streptomycin. HUVECs were cultured in endothelial cell
basal medium (EBM; Cambrex Bio Science Rockland Ltd.) containing
endothelial cell growth medium supplements (EGM; Cambrex Bio
Science Rockland Ltd.), 10% FBS, 100 IU/ml penicillin, and 100
mg/ml streptomycin. All cells were cultured at 37˚C in a humidified
atmosphere of 95% air and 5% CO2. The approval of Ethics
Committee of Beijing Ditan Hospital was received for the use of
human primary cell lines.
Cell transfection
The small interfering (si)RNA-negative control (NC),
siRNA-AKIP1-1/2, empty vector (Ov-NC) and Ov-Rac1 constructs were
purchased from Guangzhou Ribo Biotechnology Co., Ltd. (https://www.ribobio.com/). ccRCC cells were seeded
into 6-well plates at a density of 1x105 cells/well in
complete medium and were cultured at 37˚C. When cells reached 70%
confluency they were transfected with siRNA-NC (siB06525141922-1-5;
60 nM), siRNA-AKIP1-1 (siG1281084018-1-5; 60 nM), siRNA-AKIP1-2
(siG1281084034-1-5; 60 nM), Ov-NC and Ov-Rac1 at 37˚C using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 24 h. The cells were collected for further
experimentation following 48 h of transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from ccRCC cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and
complementary DNA was synthesized from RNA using the PrimeScript RT
reagent kit (Takara Bio, Inc.). The following temperature protocol
was used for reverse transcription: 37˚C for 50 min followed by
70˚C for 15 min. Subsequently, qPCR was performed using the Kapa
SYBR® FAST qPCR Master Mix (Takara Bio, Inc.) and was
analyzed using the MyGo PCR detection system (IT-IS Life Science
Ltd.). The thermocycling conditions were: 10 min initial
denaturation at 94˚C, 15 sec denaturation at 94˚C and 30 sec of
annealing at 55˚C (40 cycles) and final extension for 1 min at
72˚C. The mRNA expression levels of AKIP1, VEGFA, VEGFR2 and Rac1
were quantified using the 2-∆∆Cq method (15) and were normalized to the internal
reference gene GAPDH. The primer sequences used in the present
study are as follows: AKIP1 forward,
5'-AGAACATCTCTAAGGACCTCTACAT-3' and reverse,
5'-TCCAGAATCAACTGCTACCACAT-3'; VEGFA forward,
5'-GGGCAGAATCATCACGAAGT-3' and reverse, 5'-TGGTGATGTTGGACTCCTCA-3';
VEGFR2 forward, 5'-CTCTTGGCCGTGGTGCCTTTG-3' and reverse,
5'-GTGTGTTGCTCCTTCTTTCAAC-3'; Rac1 forward,
5'-AAAATGTCCGTGCAAAGTGGT-3' and reverse,
5'-CTCGATCGTGTCTTTATCATCCC-3'; GAPDH forward,
5'-AATGGACAACTGGTCGTGGAC-3' and reverse,
5'-CCCTCCAGGGGATCTGTTTG-3'.
Western blotting
Total protein in ccRCC cells was extracted using
RIPA buffer (Beyotime Institute of Biotechnology) and the protein
concentration was determined using a BCA Protein Assay Kit (Thermo
Fisher Scientific, Inc.). Total protein (40 µg total protein/lane)
was separated using 10% SDS-PAGE. Separated protein was transferred
to a PVDF membrane (MilliporeSigma). Subsequently membranes were
blocked with 5% non-fat milk for 1 h at room temperature and were
then incubated with primary antibodies overnight at 4˚C. The
primary antibodies used were as follows: Anti-AKIP1 (1:1,000; cat.
no. ab135996; Abcam), anti-Ki67 (1:5,000; cat. no. ab92742; Abcam),
anti-proliferating cell nuclear antigen (PCNA; 1:5,000; ab92552;
Abcam), anti-MMP2 (1:5,000; ab92536; Abcam), anti-MMP9 (1:5,000;
ab76003; Abcam), anti-VEGFA (1:1,000; ab214424; Abcam), anti-VEGFR2
(1:5,000; ab134191; Abcam), anti-Rac1 (1:1,000; ab155938; Abcam),
anti-p-ERK (1:1,000; ab201015; Abcam), anti-ERK (1:10,000;
ab184699; Abcam), anti-c-Myc (1:1,000; ab32072; Abcam) and
anti-GAPDH (1:2,500; ab9485; Abcam). Membranes were then washed in
TBS with 0.1% Tween-20 three times. Following the primary
incubation membranes were incubated with the HRP-conjugated
secondary antibody (1:2,000; cat. no. ab6721; Abcam) at room
temperature for 1 h. The blots were visualized using ECL reagent
(Pierce; Thermo Fisher Scientific, Inc.) and the band intensity was
semi-quantified using ImageJ 1.51 software (National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
Following transfection, ccRCC cells were seeded into
96-well plates at a density of 2x103 cells/well and were
incubated for 24, 48 and 72 h at 37˚C. A total of 10 µl CCK-8
solution (Dojindo Molecular Technologies, Inc.) was added to each
well for 2 h at 37˚C. The optical density at 450 nm was assessed
using a microplate reader.
5-Ethynyl-2'-deoxyuridine (EdU)
staining assay
Following transfection, ccRCC cells were seeded into
a culture flask and incubated with diluted EdU solvent for 24 h at
room temperature. Subsequently, the cells were fixed with 4%
paraformaldehyde for 10 min and stained with Alexa Fluor 594 (cat.
no. ab150084; 1:200; Abcam) overnight at room temperature. Cell
nuclei were counterstained with DAPI for 15 min at room
temperature. Cell proliferation was observed using a fluorescence
microscope (Eclipse 80i; Nikon Corporation).
Transwell assay
Following transfection, ccRCC cells were seeded at a
density of 4x104 cells into the upper Transwell chamber
that was pre-coated with Matrigel (BD Biosciences) for 1 h at 37˚C.
In the lower chamber 1 ml DMEM/F12 medium was added. Subsequently
the cells were incubated at 37˚C for 24 h. After removing the cells
on the upper surface of the chamber the remaining cells were fixed
in 4% paraformaldehyde for 10 min at room temperature. The cells
were then stained with 0.1% crystal violet (Sangon Biotech Co.,
Ltd.) for 1 h at room temperature. The number of cells was
quantified using a light microscope (Olympus Corporation) and
invasive ability was assessed using ImageJ 1.51 software.
Wound healing assay
Following transfection, ccRCC cells were seeded into
6-well plates and incubated until >90% confluency. A sterile 200
µl pipette tip was used to create a 0.5-1 cm horizontal line. After
washing with PBS the scratched cells were cultured in serum-free
medium for 24 h. The width of the scratch was imaged at 0 and 24 h
using a light microscope (Olympus Corporation) and the migratory
ability of the cells was quantified using ImageJ 1.51 software
(National Institutes of Health).
HUVEC tube-formation assay
The transfected ccRCC cells were cultured in MEM
until they reached 70% confluency. Following washing with PBS,
ccRCC cells were cultured in serum-free MEM for 48 h at 37˚C. The
culture media (CM) was collected following centrifugation at 500 x
g at room temperature for 10 min. Early passage (<Phase 4)
HUVECs were seeded at 3x104 cells/well on
Matrigel-coated 48-well plates. HUVECs were then incubated with the
CM for 6 h at 37˚C. HUVEC tube formation was observed and imaged
using a phase-contrast inverted microscope.
Co-immunoprecipitation (co-IP)
Following transfection, ccRCC cells were lysed using
RIPA lysis buffer (Beyotime Institute of Biotechnology) containing
1% PMSF and 1% protease inhibitor, centrifuged at 24,148.8 x g for
10 min at 4˚C. A part of the cell lysate was isolated as input and
250 µl of lysates were incubated with 1 µg anti-AKIP1 (cat. no.
LS-C309384; LSBio) or anti-Rac1 (cat. no. sc-514583; Santa Cruz
Biotechnology, Inc.) and homologous IgG antibodies (cat. no.
ab172730; Abcam and cat. no. sc-2025; Santa Cruz Biotechnology,
Inc.) at 4˚C overnight. Subsequently, 25 µl protein A/G beads (Cell
Signaling Technology, Inc.) were added to the lysis solution and
the solution was incubated for 1 h at 4˚C. Then, the supernatant
was centrifuged for 3 min at 1,509.3 x g at 4˚C. The beads were
extracted and were incubated with protein loading buffer at 100˚C
for 15 min to isolate the proteins, which were subsequently
detected via western blotting.
Statistical analysis
Data are presented as the mean ± SD. All statistical
analysis was performed using GraphPad Prism 8.0 software (GraphPad
Software, Inc.). One-way ANOVA followed by Tukey's post hoc test
was used to perform statistical analysis among more than two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
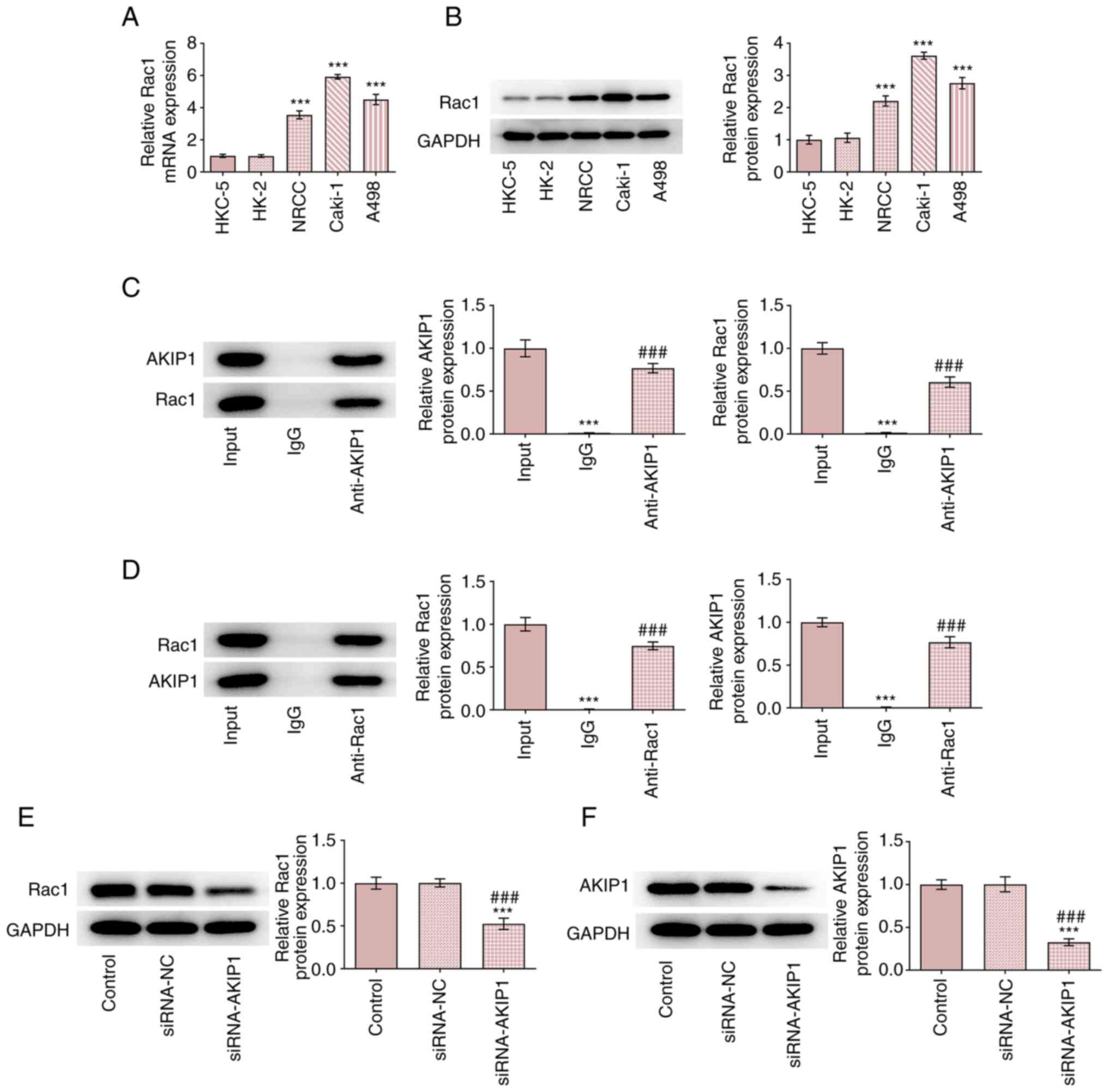
AKIP1 expression levels increase in
ccRCC cells
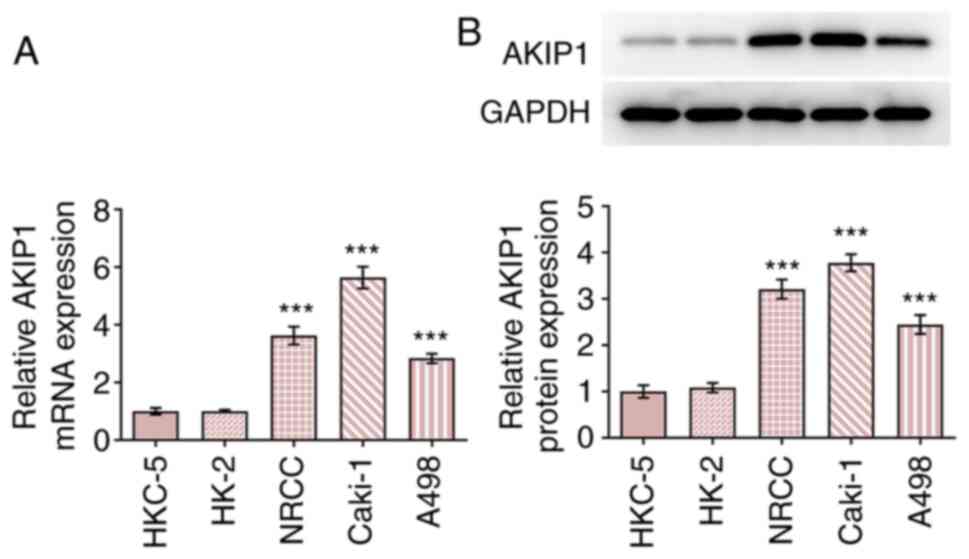
The results demonstrated that AKIP1 mRNA and protein
expression levels were increased in ccRCC cells compared with
normal renal epithelial cells. Furthermore, AKIP1 mRNA and protein
expression levels in Caki-1 cells were the highest among all ccRCC
cell lines tested (Fig. 1).
AKIP1 knockdown inhibits ccRCC cell
proliferation
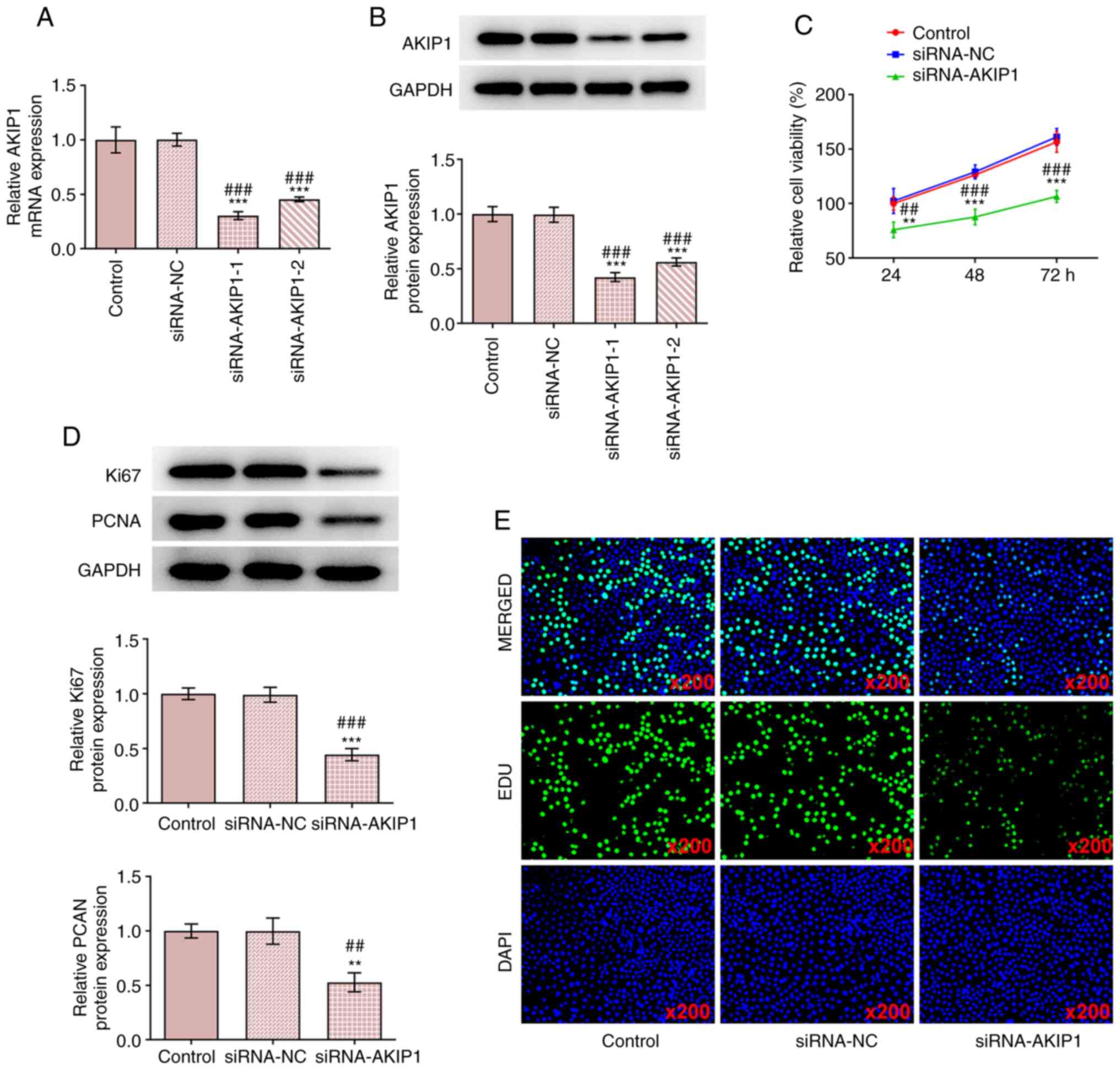
ccRCC cells were transfected with siRNA-NC or
siRNA-AKIP1-1/2 and AKIP1 mRNA, and protein expression levels were
determined. The results demonstrated that AKIP1 mRNA and protein
expression levels were markedly decreased in the siRNA-AKIP1-1/2
groups compared with siRNA-NC group. The transfection efficiency in
the siRNA-AKIP1-1 group was greater than that of the siRNA-AKIP1-2
group and therefore the siRNA-AKIP1-1 group was selected for use in
the subsequent experiments (Fig.
2A and B). Following
siRNA-AKIP1 transfection, the viability of Caki-1 cells (Fig. 2C) was decreased and the protein
expression levels of proliferation markers including Ki67 and PCNA
were also downregulated, which implied the inhibition of cell
proliferation (Fig. 2D). The
proliferation of Caki-1 cells was also suppressed by AKIP1
knockdown (Fig. 2E).
AKIP1 knockdown inhibits the invasion
and migration of ccRCC cells
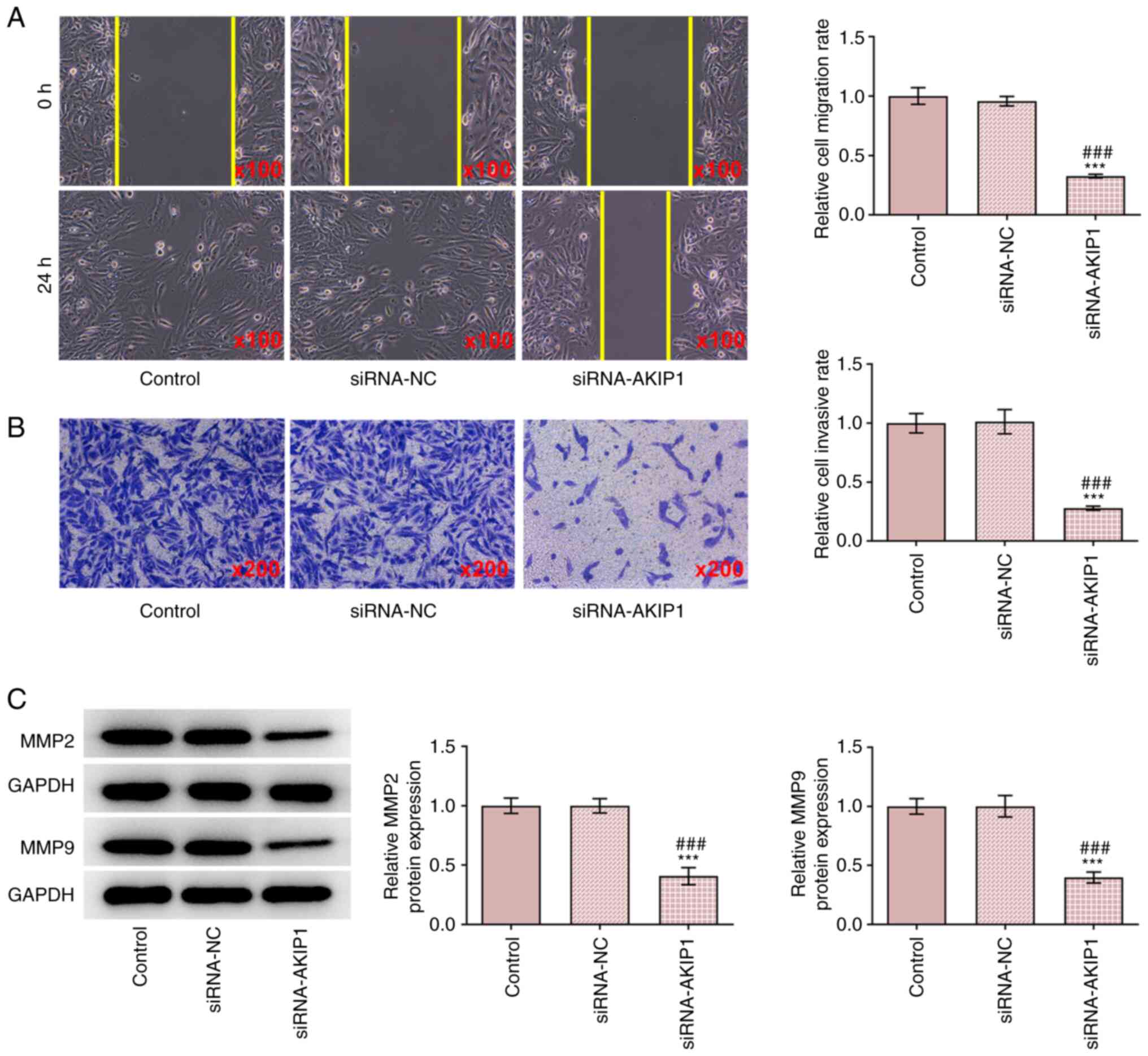
The results demonstrated that the invasion and
migration of Caki-1 cells were inhibited in the siRNA-AKIP1 group
compared with the control group, whereas no change was observed
between the siRNA-NC and control groups (Fig. 3A and B). The protein expression levels of
metastasis-associated proteins including MMP2 and MMP9 were
downregulated by AKIP1 knockdown, suggesting that AKIP1 deficiency
hampered the metastasis in ccRCC cells and they were not affected
in the siRNA-NC group (Fig.
3C).
AKIP1 knockdown inhibits angiogenesis
in ccRCC cells
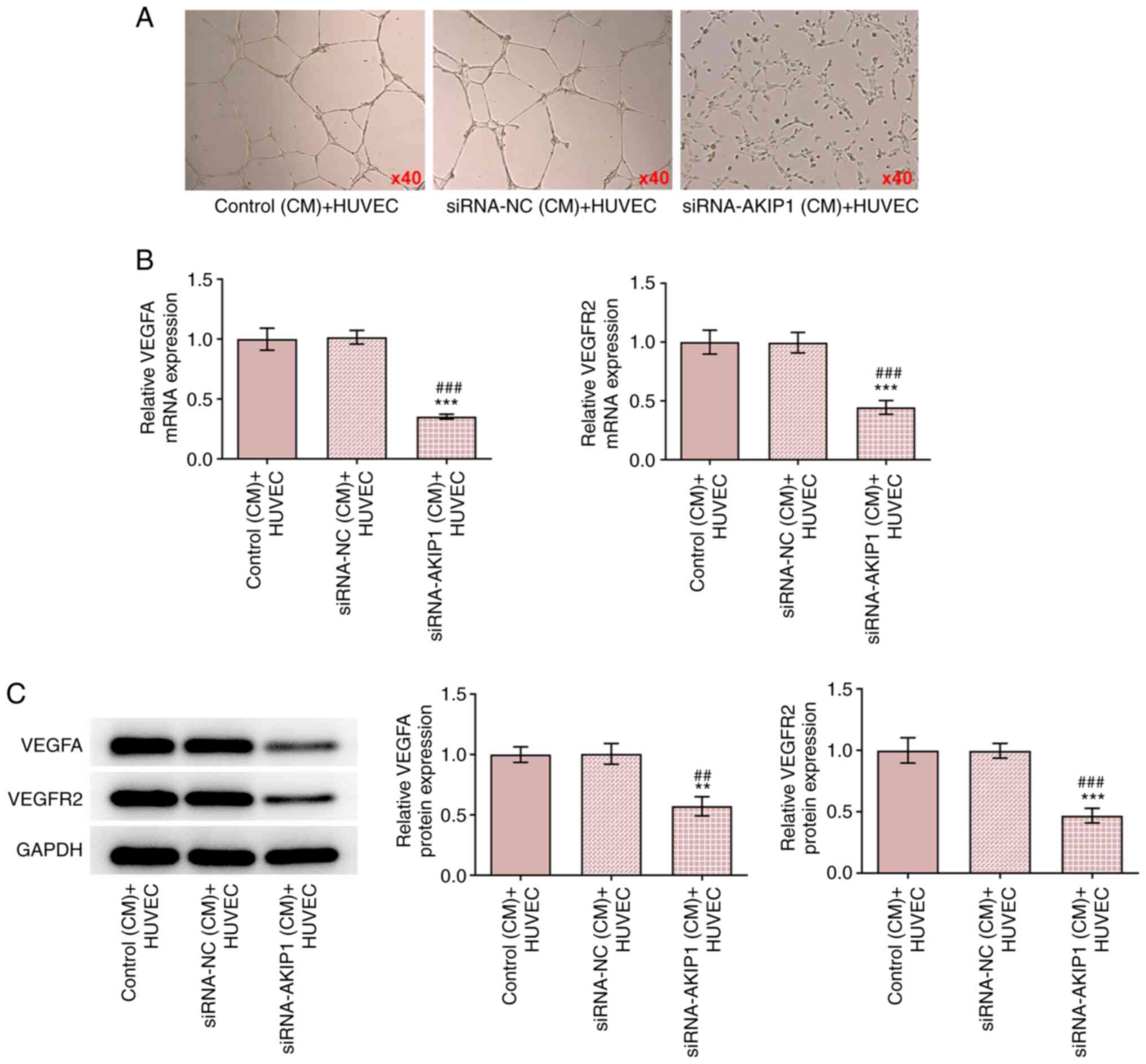
The CM of siRNA-NC-transfected Caki-1 cells had no
effect on HUVEC tube-formation, whereas tube formation was
suppressed by AKIP1 knockdown (Fig.
4A). In addition, the mRNA and protein expression levels of
VEGFA and VEGFR2 in HUVECs were decreased in the CM of
siRNA-AKIP1-transfected Caki-1 cells (Fig. 4B and C).
AKIP1 potentially binds to Rac1 in
ccRCC cells and AKIP1 downregulation inhibits Rac1 expression
The results demonstrated that Rac1 mRNA and protein
expression levels were increased in ccRCC cells and that the
highest expression levels of Rac1 were demonstrated in Caki-1 cells
(Fig. 5A and B). co-IP experiments demonstrated that
Rac1 co-precipitated with AKIP1 when an anti-AKIP1 antibody was
used and AKIP1 co-precipitated with Rac1when an anti-Rac1 antibody
was used (Fig. 5C and D). Following Caki-1 cell transfection
with siRNA-NC and siRNA-AKIP1 the results demonstrated that there
was no obvious change in expression levels of Rac1 and AKIP1 in the
siRNA-NC group, whereas expression of Rac1 and AKIP1 decreased in
the siRNA-AKIP1 group compared with the control (Fig. 5E and F).
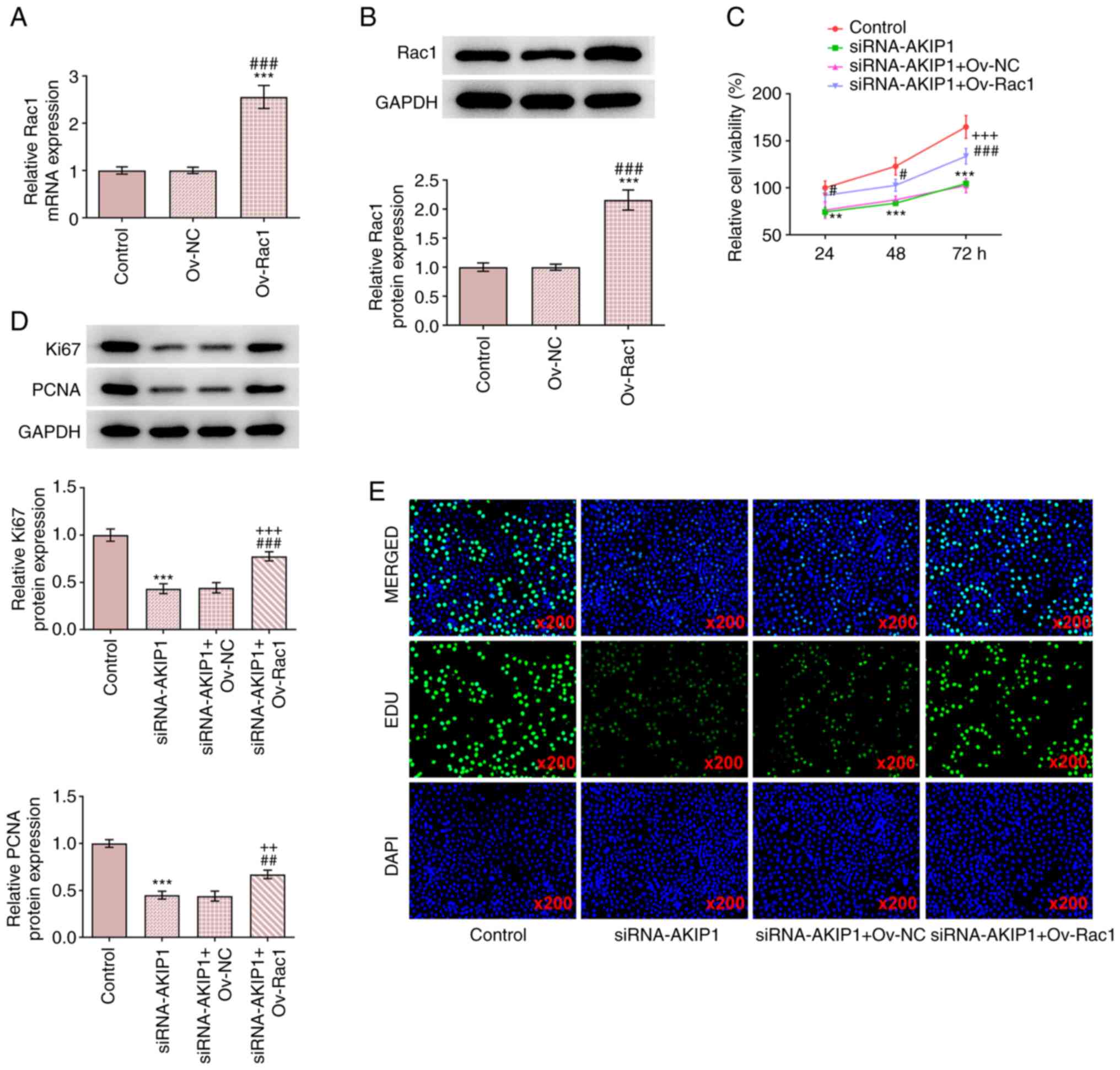
Rac1 overexpression reverses the
effects of AKIP1 silencing on the proliferation of ccRCC cells
The results demonstrated that Ov-Rac1-transfected
Caki-1 cells exhibited upregulated Rac1 mRNA and protein expression
levels (Fig. 6A and B). Rac1 overexpression also improved the
viability of Caki-1 cells and promoted the protein expression of
Ki67 and PCNA compared with the siRNA-AKIP1 group (Fig. 6C and D). In addition, EdU staining indicated
that Rac1 overexpression reversed the effect of AKIP1 knockdown and
enhanced the proliferation of Caki-1 cells (Fig. 6E).
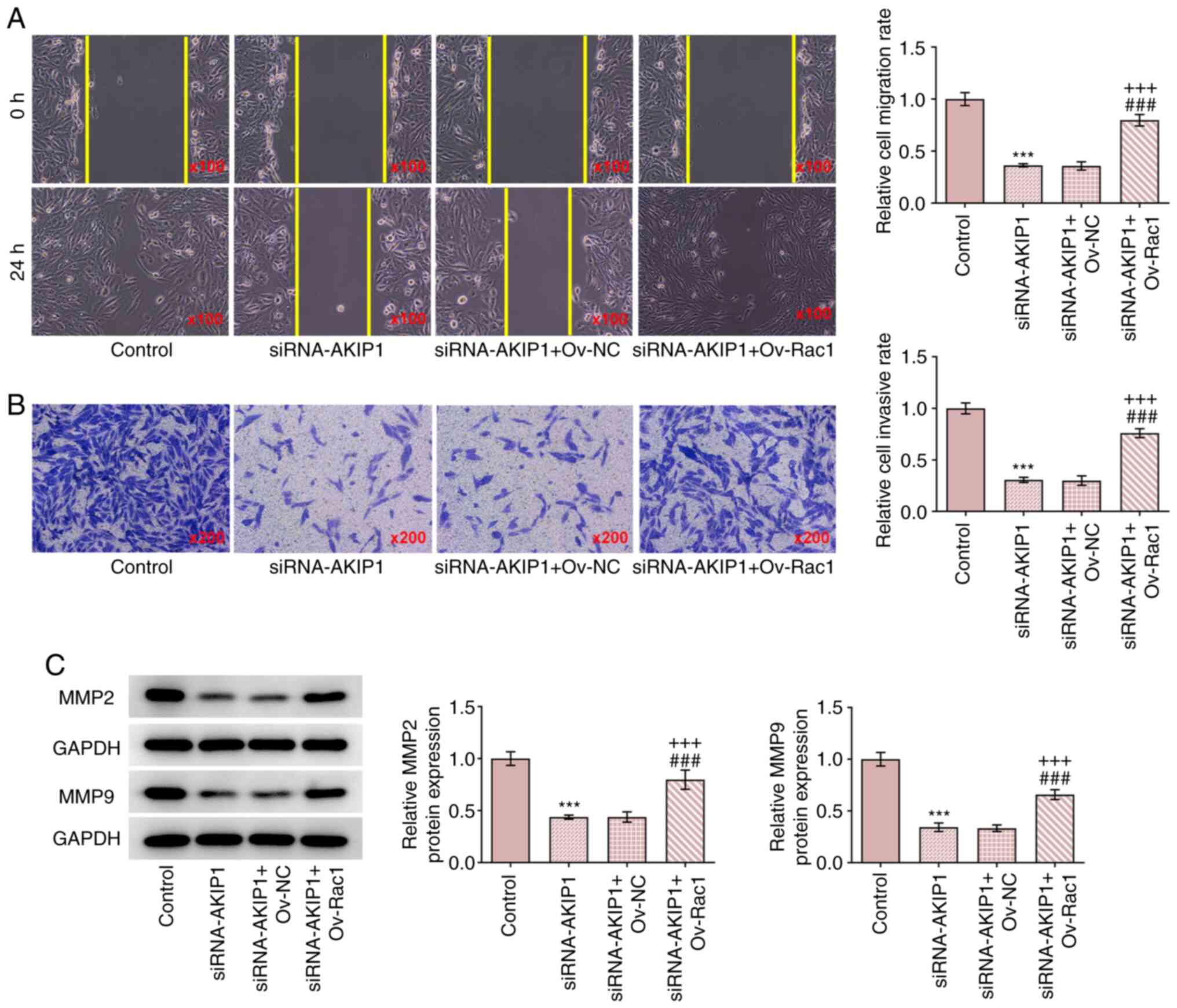
Rac1 overexpression reverses the
impacts of AKIP1 silencing on the invasion and migration of ccRCC
cells
The results demonstrated that Rac1 overexpression
increased the invasion and migration of Caki-1 cells transfected
with siRNA-AKIP1 (Fig. 7A and
B). In addition, Rac1
overexpression upregulated the protein expression levels of MMP2
and MMP9 in Caki-1 cells transfected with siRNA-AKIP1 (Fig. 7C). These results indicated that
Rac1 overexpression may be able to reverse the effect of AKIP1
knockdown on the invasion and migration of Caki-1 cells.
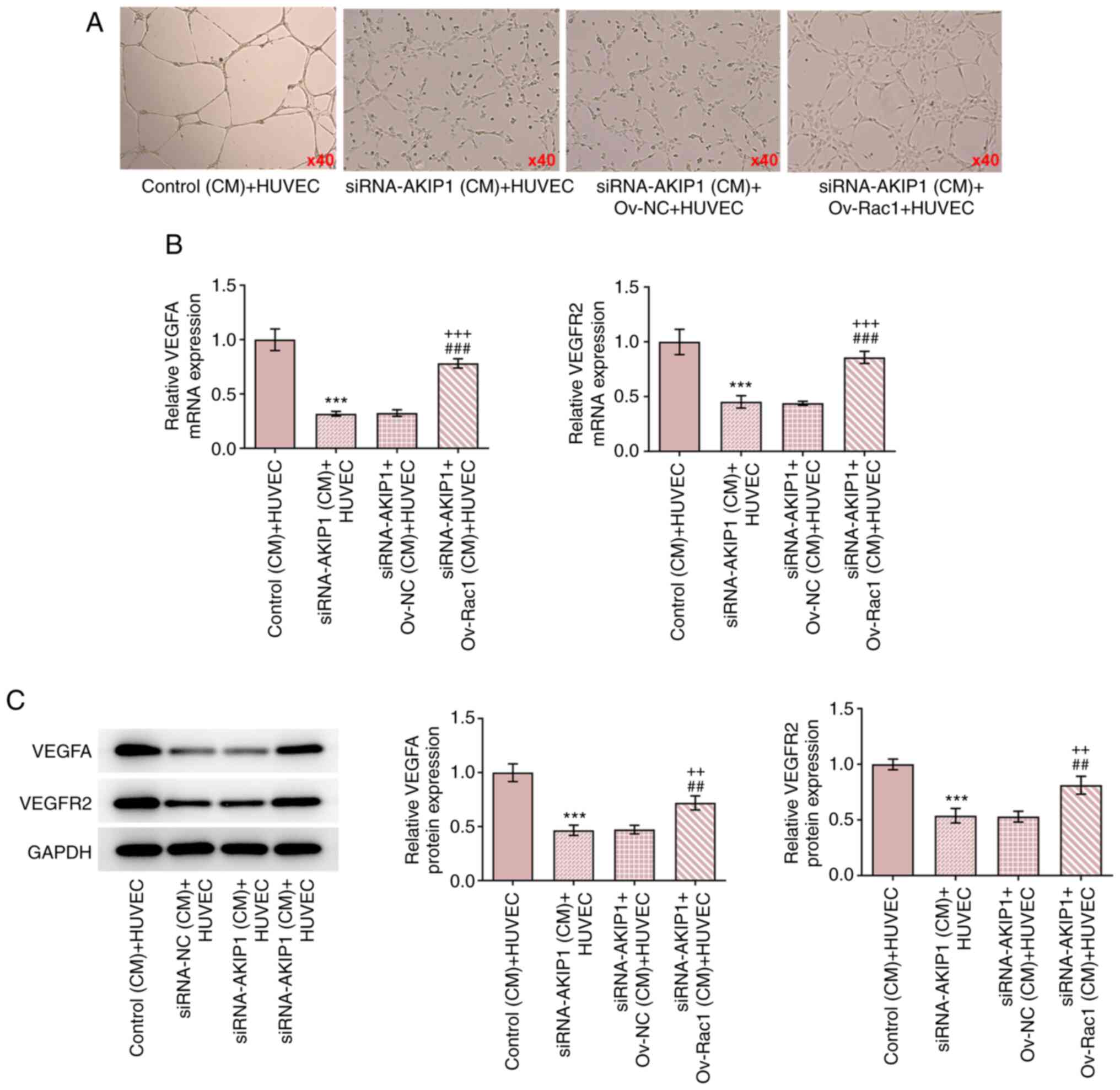
Rac1 overexpression reverses the
impacts of AKIP1 knockdown on the angiogenesis in ccRCC cells
The results demonstrated that HUVEC tube-formation
was increased in the CM of Caki-1 cells transfected with
siRNA-AKIP1 and Ov-Rac1, compared with the CM of Caki-1 cells
transfected with siRNA-AKIP1 alone (Fig. 8A). The mRNA and protein expression
levels of VEGFA and VEGFR2 were upregulated in HUVECs cultured in
the CM of Caki-1 cells transfected with siRNA-AKIP1 and Ov-Rac1,
compared with the CM of Caki-1 cells transfected with siRNA-AKIP1
alone (Fig. 8B and C).
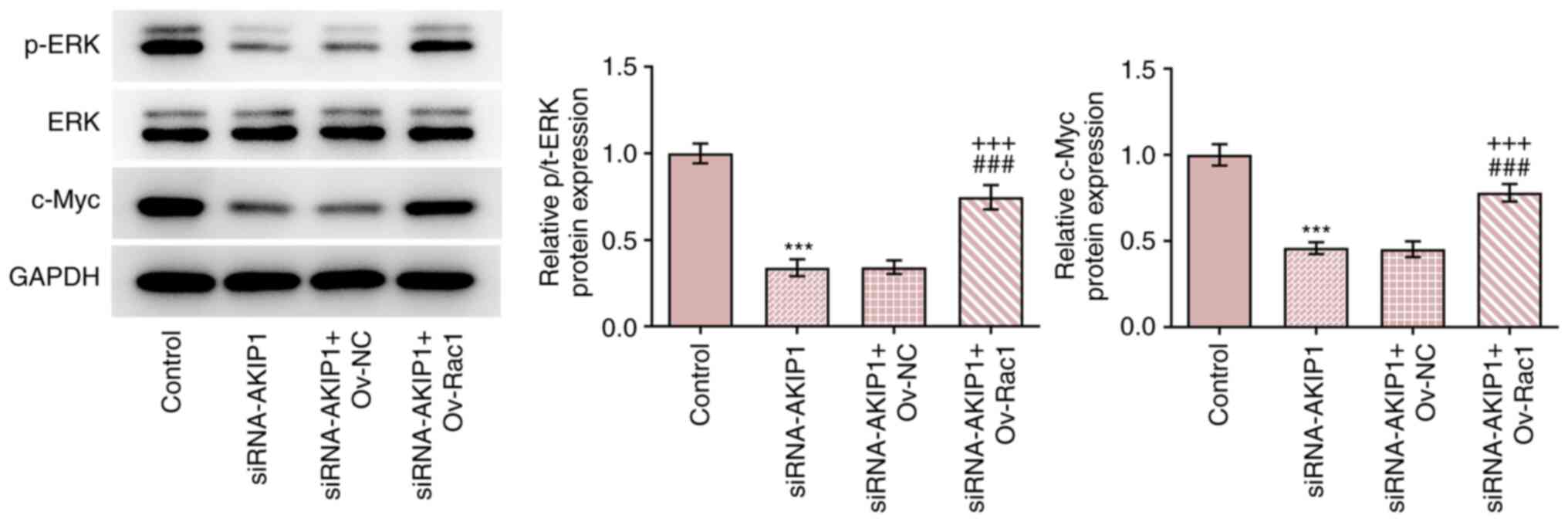
Rac1 elevation countervails the
impacts of AKIP1 depletion on the expression of ERK/c-Myc signaling
pathway-associated proteins in ccRCC cells
The results demonstrated that AKIP1 knockdown
inhibited the expression of phosphorylated (p)/total (t)-ERK and
c-Myc in Caki-1 cells. Furthermore, Rac1 overexpression increased
the expression levels of p/t-ERK and c-Myc in Caki-1 cells
transfected with siRNA-AKIP1 (Fig.
9).
Discussion
AKIP1 was first identified in breast and prostate
cancer cell lines and serves an important role in normal
physiological processes (4).
Recently, AKIP1 has been reported to act as an oncogene in common
malignant tumors, including lung, liver and breast cancer,
participating in the physiological and pathological changes of
malignant tumors and promoting malignant biological behaviors, such
as tumor proliferation, invasion and metastasis (6,9,16).
AKIP1 is highly expressed in cervical cancer cells and the
inhibition of AKIP1 expression in cervical cancer cells suppresses
cell proliferation, tumor growth and angiogenesis in BALB/c nude
mice (17). AKIP1 overexpression
is also involved in cell proliferation, clonal formation and
angiogenesis (17). A previous
study reported that AKIP1 expression is upregulated in liver cancer
tissues and is associated with early recurrence and a poor
prognosis in patients. In vitro, AKIP1 promotes tumor cell
invasion and colony growth and in vivo it promotes
intrahepatic and lung metastasis (18). Although AKIP1 expression was
previously demonstrated to be increased in ccRCC tissues, the
regulating role of AKIP1 in the proliferation, invasion, migration
and angiogenesis of ccRCC cells remains unknown. The results of the
present study demonstrated that AKIP1 expression levels were
upregulated in ccRCC cells consistent with previous studies. To
assess the biological function of AKIP1 in ccRCC cells, the
expression levels of AKIP1 in Caki-1 cells were knocked down using
siRNA-AKIP1. The results demonstrated that AKIP1 knockdown
potentially inhibited the proliferation, invasion, migration and
angiogenesis of Caki-1 cells.
A previous study reported that AKIP1 can bind to
Rac1(11). The present study
demonstrated an interaction between AKIP1 and Rac1, and that AKIP1
knockdown downregulated Rac1 expression levels. Rac1, as a member
of the Rho family, serves a role in cell proliferation, cell
survival, cytoskeleton remodeling and gene transcription (19). Rac1 is expressed at low levels or
not at all in normal human tissues, but it is increased in
pancreatic (20), prostate
(21), breast (22) and colorectal cancer (23), and other malignant tumor tissues or
cells. In addition, this high expression of Rac1 is closely related
to the degree of differentiation of tumor cells, the pathological
classification, TNM staging and other phenotypes. A previous study
using renal cancer tissues also demonstrated that Rac1 activation
leads to renal cancer infiltration (24). The present study demonstrated that
Rac1 expression levels were also upregulated in ccRCC cells, which
was consistent with Rac1 expression levels in other types of
cancer. These results indicated that Rac1 overexpression could
potentially reverse the downregulation of AKIP1 to promote the
proliferation, invasion, migration and angiogenesis of Caki-1
cells.
Rac1 activates the ERK signaling pathway in
hepatocellular carcinoma and colon cancer to regulate the
proliferation, invasion and migration of cancer cells (25,26).
Zhao et al (27) reported
that the slit guidance ligand 2/roundabout guidance receptor 1 axis
promotes the malignant progression of osteosarcoma cells via
activation of the proto-oncogene tyrosine protein kinase
Src/ERK/c-Myc/6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
2 signaling pathway. ERK1/2 promotes aerobic glycolysis of
hepatocellular carcinoma cells via the regulation of
differentiation inhibitor 1 expression and via the formation of an
intracellular message transfer chain with c-Myc (28). The ERK/c-Myc signaling pathway is
also suppressed to reduce the proliferation of human epithelial
ovarian cancer cells (29). In the
present study it was demonstrated that AKIP1 knockdown suppressed
the expression levels of p-ERK and c-Myc by downregulating Rac1
expression. However, Rac1 overexpression reversed this inhibitory
effect, which potentially activated the ERK/c-Myc signaling
pathway.
In conclusion, AKIP1 knockdown potentially
suppressed the cell proliferation, invasion, migration and
angiogenesis of ccRCC cells and may have suppressed the ERK/c-Myc
signaling pathway by binding to Rac1. However, this phenomenon was
potentially reversed by Rac1 overexpression. Therefore, the results
of the present study provided a theoretical basis for the treatment
of ccRCC and showed an interaction between AKIP1 and Rac1, while
the functional relationship between these two proteins will be
explored in the future.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by Beijing Medical Award
Foundation (grant no. YXJL-2021-0800-0410).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ, HZ, ZH and QL contributed to the study design.
XW, XL, PY and SJ performed the experiments and analyzed the data.
YZ and HZ contributed to the experiments and writing and performed
the secondary data analyses and revising the manuscript for
intellectual and scientific content. ZH and XW contributed to the
conception of the study. YZ and HZ confirm the authenticity of all
the raw data. All authors read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wu Y, Wang YQ, Weng WW, Zhang QY, Yang XQ,
Gan HL, Yang YS, Zhang PP, Sun MH, Xu MD and Wang CF: A
serum-circulating long noncoding RNA signature can discriminate
between patients with clear cell renal cell carcinoma and healthy
controls. Oncogenesis. 5(e192)2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kitching R, Li H, Wong MJ, Kanaganayakam
S, Kahn H and Seth A: Characterization of a novel human breast
cancer associated gene (BCA3) encoding an alternatively spliced
proline-rich protein. Biochim Biophys Acta. 1625:116–121.
2003.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lin C, Song L, Liu A, Gong H, Lin X, Wu J,
Li M and Li J: Overexpression of AKIP1 promotes angiogenesis and
lymphangiogenesis in human esophageal squamous cell carcinoma.
Oncogene. 34:384–393. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mo D, Li X, Li C, Liang J, Zeng T, Su N,
Jiang Q and Huang J: Overexpression of AKIP1 predicts poor
prognosis of patients with breast carcinoma and promotes cancer
metastasis through Akt/GSK-3β/Snail pathway. Am J Transl Res.
8:4951–4959. 2016.PubMed/NCBI
|
|
7
|
Jiang W, Yang W, Yuan L and Liu F:
Upregulation of AKIP1 contributes to metastasis and progression and
predicts poor prognosis of patients with colorectal cancer. Onco
Targets Ther. 11:6795–6801. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sun Y, Shi G, Ma C, Jiao J, Liu Y, Gao Q,
Zhang X and Feng Q: Upregulation of a kinase interacting protein 1
in tongue squamous cell carcinoma correlates with lymph node
metastasis and poor overall survival. Medicine (Baltimore).
100(e25278)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Guo X, Zhao L, Cheng D, Mu Q, Kuang H and
Feng K: AKIP1 promoted epithelial-mesenchymal transition of
non-small-cell lung cancer via transactivating ZEB1. Am J Cancer
Res. 7:2234–2244. 2017.PubMed/NCBI
|
|
10
|
Peng H, Zhang R and Zhang H: A-kinase
interacting protein 1 high expression correlates with advanced
tumor stage and poor overall survival in surgical patients with
clear cell renal cell carcinoma. Medicine (Baltimore).
99(e20742)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yao C, Yu KP, Philbrick W, Sun BH, Simpson
C, Zhang C and Insogna K: Breast cancer-associated gene 3 interacts
with Rac1 and augments NF-κB signaling in vitro, but has no effect
on RANKL-induced bone resorption in vivo. Int J Mol Med.
40:1067–1077. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen L, Zhang D, Ding T, Liu F, Xu X, Tian
Y, Xiao J and Shen H: LncRNA NR2F2-AS1 Upregulates Rac1 to increase
cancer stemness in clear cell renal cell carcinoma. Cancer Biother
Radiopharm. 35:301–306. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hsieh YY, Liu TP and Yang PM: In silico
repurposing the Rac1 inhibitor NSC23766 for treating PTTG1-high
expressing clear cell renal carcinoma. Pathol Res Pract.
215(152373)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang T, Wang Z, Liu Y, Huo Y, Liu H, Xu
C, Mao R, Zhu Y, Liu L, Wei D, et al: Plastin 1 drives metastasis
of colorectal cancer through the IQGAP1/Rac1/ERK pathway. Cancer
Sci. 111:2861–2871. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ma D, Li M, Su J and Zhang S: BCA3
contributes to the malignant progression of hepatocellular
carcinoma through AKT activation and NF-κB translocation. Exp Cell
Res. 362:142–151. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang W, Wu Q, Wang C, Yang L, Liu P and
Ma C: AKIP1 promotes angiogenesis and tumor growth by upregulating
CXC-chemokines in cervical cancer cells. Mol Cell Biochem.
448:311–320. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cui Y, Wu X, Lin C, Zhang X, Ye L, Ren L,
Chen M, Yang M, Li Y, Li M, et al: AKIP1 promotes early recurrence
of hepatocellular carcinoma through activating the
Wnt/β-catenin/CBP signaling pathway. Oncogene. 38:5516–5529.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bosco EE, Mulloy JC and Zheng Y: Rac1
GTPase: A ‘Rac’ of all trades. Cell Mol Life Sci. 66:370–374.
2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang S, Shi W, Hu W, Ma D, Yan D, Yu K,
Zhang G, Cao Y, Wu J, Jiang C and Wang Z: DEP Domain-Containing
Protein 1B (DEPDC1B) promotes migration and invasion in pancreatic
cancer through the Rac1/PAK1-LIMK1-Cofilin1 signaling pathway. Onco
Targets Ther. 13:1481–1496. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kato T, Kawai K, Egami Y, Kakehi Y and
Araki N: Rac1-dependent lamellipodial motility in prostate cancer
PC-3 cells revealed by optogenetic control of Rac1 activity. PLoS
One. 9(e97749)2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tian Y, Xu L, He Y, Xu X, Li K, Ma Y, Gao
Y, Wei D and Wei L: Knockdown of RAC1 and VASP gene expression
inhibits breast cancer cell migration. Oncol Lett. 16:2151–2160.
2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Xie N, Meng Q, Zhang Y, Luo Z, Xue F, Liu
S, Li Y and Huang Y: MicroRNA-142-3p suppresses cell proliferation,
invasion and epithelial-to-mesenchymal transition via RAC1-ERK1/2
signaling in colorectal cancer. Mol Med Rep. 24(568)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wacker I and Behrens J: Activin B
antagonizes RhoA signaling to stimulate mesenchymal morphology and
invasiveness of clear cell renal cell carcinomas. PLoS One.
9(e111276)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang LL, Luo J, He ZH, Liu YQ, Li HG, Xie
D and Cai MY: STEAP3 promotes cancer cell proliferation by
facilitating nuclear trafficking of EGFR to enhance
RAC1-ERK-STAT3 signaling in hepatocellular
carcinoma. Cell Death. Dis. 12(1052)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jiang QH, Wang AX and Chen Y: Radixin
enhances colon cancer cell invasion by increasing MMP-7 production
via Rac1-ERK pathway. ScientificWorldJournal.
2014(340271)2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao SJ, Shen YF, Li Q, He YJ, Zhang YK,
Hu LP, Jiang YQ, Xu NW, Wang YJ, Li J, et al: SLIT2/ROBO1 axis
contributes to the Warburg effect in osteosarcoma through
activation of SRC/ERK/c-MYC/PFKFB2 pathway. Cell Death Dis.
9(390)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sharma BK, Kolhe R, Black SM, Keller JR,
Mivechi NF and Satyanarayana A: Inhibitor of differentiation 1
transcription factor promotes metabolic reprogramming in
hepatocellular carcinoma cells. FASEB J. 30:262–275.
2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Dai L, Wang W, Liu Y, Song K and Di W:
Inhibition of sphingosine kinase 2 down-regulates ERK/c-Myc pathway
and reduces cell proliferation in human epithelial ovarian cancer.
Ann Transl Med. 9(645)2021.PubMed/NCBI View Article : Google Scholar
|