Introduction
Clear cell renal cell carcinoma (ccRCC) is a primary
subtype of RCC. Approximately 80% of patients with RCC are
diagnosed with ccRCC, with males exhibiting a higher incidence of
ccRCC than females (1-3).
Early detection of ccRCC is difficult due to its insidious
occurrence and lack of reliable biomarkers; thus, the majority of
patients are diagnosed during the late stages of the disease
(4,5). In addition, ccRCC is insensitive to
traditional chemoradiotherapy (6-8).
Moreover, even following surgical treatment, there are high rates
of metastasis and recurrence (9-11).
The outcome of surgical treatment is closely related to the
clinical stage, with survival rates of 81% for stage I, 74% for
stage II, 53% for stage III and 8% for stage IV (12). Therefore, identifying biomarkers
for ccRCC is key for early diagnosis and improving prognosis.
Krüppel-like factors (KLFs) belong to a family of
transcription factors that have a Cys2-His2 zinc-finger domain at
the C-terminal region, which binds to GC and CGGA-boxes of DNA. At
the N-terminal region, KLFs have a transcription regulatory motif
that binds to transcription-activation or repressive factors
(13-15).
The first KLF (KLF1) was identified in mammalian red blood cells in
1993(16). Following
identification of KLF1 and KLF17, a number of KLF genes were found
in the human genome (17). These
KLF proteins are expressed in a variety of human tissue and exhibit
diverse functions in physiological processes such as maintenance of
internal environmental homeostasis, immune response, inflammation,
neurogenesis and organ development (18-25).
Genome-wide knockout of KLF family members leads to developmental
abnormality and mortality. For example, klf15-/-
mice have enlarged hearts (26),
klf6-/- or klf1-/- mice have
abnormal hematopoietic system (27,28).
klf2-/- or klf5-/- mice die in
utero during embryonic life (29,30).
Previous studies have revealed that KLFs participate in
proliferation, apoptosis, epithelial-mesenchymal transition (EMT),
angiogenesis and other malignant biological behavior of cancer
(14,31-33).
KLF4 inhibits EMT and proliferation of endometrial cancer cells,
whilst KLF8 promotes EMT and proliferation of bladder cancer cells
(34,35); these findings suggest that the KLF
gene family may serve a critical role in cancer genesis and
prognosis.
The present study systematically analyzed expression
and clinical application of KLF genes in ccRCC. KLF gene expression
levels and their association with clinical prognosis of ccRCC were
analyzed using public databases and findings were validated using
in vitro cellular assays.
Materials and methods
The cancer genome atlas (TCGA)
database
The clinical data and gene expression of patients
with ccRCC were obtained from TCGA database (portal.gdc.cancer.gov; data collected June 2021).
Using the archive of the TCGA Kidney Renal Clear Cell Carcinoma
(KIRC) project (portal.gdc.cancer.gov; data collected June 2021),
transcriptome maps of KLFs were extracted to obtain gene
transcription information. Demographic and clinical
characteristics, including age, tumor grade and stage at diagnosis
(TNM classification), were obtained from the electronic
records.
DNA methylation and genetic
alteration
CBioPortal Cancer Genomics (cbioportal.org; data collected June 2021) is an open
resource web platform that integrates multiple cancer genome
databases, such as TCGA and the International Cancer Genome
Consortium. Using cBioPortal, KLF2 and KLF11 mutations were
analyzed in TCGA ccRCC cohort (portal.gdc.cancer.gov; data collected June 2021). This
tool provides real-time access and visualization of DNA methylation
profiles and gene expression from TCGA. The methylation level of
promoter region of KLF2 was retrieved from MethHC (bioinfo-zs.com/smartapp/). Significant
methylation sites were selected as candidate sites according to
P<0.05 based on results from univariate Cox regression analysis.
The correlation between two factors was evaluated by the Pearson's
correlation test.
Biological pathway enrichment
analysis
Gene Ontology and Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis was conducted using Metascape
3.5 (metascape.org). Gene set variation analysis
(GSVA) was also used to assess the variations in pathway enrichment
among patients with high and low risk score) via the ‘GSVA’ R
package. A total of 50 hallmark gene sets were obtained from the
Molecular Signatures Database (MSigDB; gsea-msigdb.org/gsea/msigdb/). Analysis of the
intersection of low level of KLF2 and KLF11 activated genes using
Venn diagram.
Cell culture
The human ccRCC cell lines 769-P, 786-O and OR-SC-2
and HK-2, 293(T) (normal renal tubular epithelial cell line) were
obtained from the Affiliated Hospital of Yangzhou University
(Suzhou, China). 293(T) cells were cultured in DMEM (Biological
Industries) and other cells were cultured in RPMI 1640 medium
(Biological Industries), supplemented with 10% fetal bovine serum
(VivaCell Biosciences, Ltd.) and 1% penicillin/streptomycin
solution (cat. no. C0222; Beyotime Institute of Biotechnology). The
cell culture was maintained at 37˚C and 5% CO2. Cells
were treated with 5-aza-2'-deoxycytidine 5 µM (5-AZA-CdR; cat. no.
HY-A0004; MedChemExpress) for 24 h in a cell incubator at 37˚C with
5% CO2.
Lentivirus transduction and cell
transfection
For KLF2 overexpression plasmid (human cDNA was
cloned and inserted into a pCDH-CMV-MCS-EF1-copGFP-T2A-Puro) and
2nd packaging plasmids psPAX2, pMD2.G from GENERAL BIOL. The psPAX2
(12 µg) packaging and pMD2.G (9 µg) envelope plasmids were
co-transfected with pCDH-KLF2 (4 µg). The plasmid ratio was
pCDH-KLF2: psPAX2: pMD2.G=4:3:1. Subsequently, the overexpression
plasmid of KLF2 and control plasmid were transfected into 293(T)
cells using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) in a 100-mm dish (5x106 cells)
and maintained in an incubator at 37˚C with 5% CO2. The
medium was replaced with fresh medium after 6 h. Following
incubation for 48 h, the viral supernatant was harvested at 4˚C for
5 min at 2,000 g, immediate added to 786-O and 769-P cells and
incubated with 8 µg/ml polybrene (cat. no. C0351; Beyotime
Institute of Biotechnology). The multiplicity of infection (MOI)
infected cells was 10. Stably transfected cells were selected with
2.5 µg/ml puromycin (cat. no. ST551; Beyotime Institute of
Biotechnology) for three days following 48 h viral infection. Cells
were cultured in RPMI 1640 medium containing 1%
penicillin/streptomycin solution, while 2.5 µg/ml puromycin was
added for maintenance.
Reverse transcription-quantitative
(RT-q) PCR
According to the manufacturer's protocol, total cell
RNA in six-well cell culture plates was isolated using
TRIzol® reagent (cat. no. 10296028; Invitrogen; Thermo
Fisher Scientific, Inc.). Each well contained ~2x105
cells. Subsequently, total RNA (1 µg) was reverse transcribed into
cDNA using NovoScript® Plus All-in-one 1st Stand cDNA
Synthesis SuperMix (gRNA Purge; cat. no. E047-01A; Suzhou
Novoprotein Technology, Ltd.) according to the manufacturer's
protocol. NovoStart® SYBR Green Color qPCR SuperMix kit
(cat. no. E168-01B; Suzhou Novoprotein Technology, Ltd.) was used
for amplification. Thermocycling conditions were as follows: 95˚C
for 30 sec, followed by 40 cycles of 95˚C for 5 sec and 60˚C for 30
sec. The expression of genes was normalized to that of GAPDH, which
was used as an endogenous control. The 2-∆∆Cq method
(36) was performed to analyze
mRNA expression levels. The primers are shown in Table I.
 | Table IPrimers for reverse-transcription
quantitative PCR assay. |
Table I
Primers for reverse-transcription
quantitative PCR assay.
| Gene | Primer sequence
(5'-3') |
|---|
| KLF2-Forward |
CACCAAGAGTTCGCATCTGA |
| KLF2-Reverse |
CGTGTGCTTTCGGTAGTGG |
| GAPDH-Forward |
GGGGTCATTGATGGCAACAATA |
| GAPDH-Reverse |
ATGGGGAAGGTGAAGGTCG |
Western blotting
Cells were washed three times with cold PBS. Total
protein was extracted using RIPA lysis buffer (cat. no. P00013B;
Beyotime Institute of Biotechnology) and the protein concentration
was measured by BCA (cat. no. P0009; Beyotime Institute of
Biotechnology). Then, the protein was separated by 10% SDS-PAGE (20
µg per well and transferred onto a PVDF membrane. Subsequently, the
membrane was blocked with 5% milk at room temperature for 1 h and
incubated with primary antibodies against KLF2 (1:1,000; cat. no.
340341; Zen-Bio, Inc.), GAPDH (1:1,000; cat. no. R24404; Zen-Bio,
Inc.), E-cadherin (1:1,000; cat. no. 340341; Zen-Bio, Inc.),
N-cadherin (1:1,000; cat. no. 380671; Zen-Bio, Inc.) and vimentin
(1:1,000; cat. no. 380771; Zen-Bio, Inc.) at 4˚C overnight. The
membrane was washed and incubated with horseradish
peroxidase-labelled secondary antibody (cat. no. #S001; 1:5,000;
Affinity Biosciences, Ltd.) for 2 h at room temperature. The
membrane was washed three times with PBS-0.1% Tween-20 for 5 min
each. A chemiluminescent substrate kit (cat. no. BL520A; Biosharp
Life Sciences) and iBright CL1000 imaging system (Invitrogen;
Thermo Fisher Scientific, Inc.) were used to detect the proteins.
Each set of experiments was repeated at least three times, with
GAPDH as the internal reference control, and the relative amounts
of protein bands were analyzed using ImageJ software (Version 1.48;
National Institutes of Health).
Wound healing assay
To analyze cell migration, normal and KLF2
overexpressing 786-O and 769-P cells were seeded in a 6-well plate
at a concentration of 1x105 cells/well with 80%
confluency. Using the tip of a 10-µl sterile pipette, a wound was
scratched throughout the center of the well. Next, wells were
gently rinsed twice with PBS to remove isolated cells and residual
serum. Subsequently, all wells were refilled with fresh serum-free
RPMI-1640 (Biological Industries) and cells were cultured for 24
and 48 h at 37˚C and 5% CO2. Photographs of cell
migration were taken at 24 and 48 h after injury by using an
inverted fluorescence microscope (MF53-N; Guangzhou Micro-short
Technology Co., Ltd.) with bright field at x40 magnification. The
wound area was calculated using ImageJ 1.48v (National Institutes
of Health) and migration rates were calculated from area ratios.
Mobility was calculated as follows: Mobility rate=(area of the
starting scratch-area of the current scratch)/area of the starting
scratch x100, and the result was used to determine the migration
capacity of the cells.
Tissue microarray (TMA) and
immunohistochemistry (IHC)
A total of 20 tissue samples from patients with
ccRCC were obtained from The Second Affiliated Hospital, School of
Medicine, The Chinese University of Hong Kong, and a waiver of
informed consent was granted by the hospital ethics committee.
Tissue was fixed in 4% paraformaldehyde for 2-3 days, dehydrated,
embedded in paraffin and cut into 4 µm thick sections. TMAs were
dewaxed and rehydrated. Sections were placed sequentially in xylene
I for 15 min-xylene II for 15 min-xylene III for 15 min-anhydrous
ethanol I for 5 min-anhydrous ethanol II for 5 min-85% alcohol for
5 min-75% alcohol for 5 min-distilled water wash. Antigen repair
was performed using EDTA (pH 9.0) by heating at 100˚C in a
microwave oven for 8 min to boiling, ceasing for 8 min and then for
7 min. 3% H2O2 to block endogenous peroxidase
activity then incubating with 2% BSA (cat. no. G5001; Wuhan
Servicebio Technology Go, Ltd.) for 30 min at room temperature.
Finally, anti-KLF2 (1:100; cat. no. #DF13602; Affinity Biosciences,
Ltd.) was incubated overnight at 4˚C. The sections were incubated
for 50 min at room temperature with horseradish
peroxidase-conjugated secondary antibody (1:200; cat. no. GB23303;
Wuhan Servicebio Technology Co., Ltd.). Tissue specimens were
subsequently stained with 1X 3,3'-diaminobenzidine (cat. no.
ZLI-9018; Beijing, ZSGB-BIO Technology Co, Ltd.) for 2 min under
the inverted fluorescence microscope (Zesiss Aixo Vert. A1; Carl
Zeiss AG) with bright field, then washed with PBS. Images obtained
under a Nikon E100 microscope NIKON DS-U3 system scan. CaseViewer
2.4 (3DHistech, Ltd.) software was observed and intercepted at x10
and x40 magnification, respectively.
As previously described, H-score was determined
based on the number and staining intensity of positive cells
(37,38). Briefly, H-score was calculated as
follows: (% weak intensity cells x 1) + (% moderate intensity cells
x 2) + (% strong intensity cells x 3). The image scanning software
AIpathwell (v1.0, Wuhan Servicebio Technology Co, Ltd.) analyses
and calculates the H-score according. The intensity of positive
cells was graded as 0 (negative, unstained), 1 (weak), 2 (moderate)
and 3 (strong).
Statistical analysis
The gene expression values were converted into a
non-overlapping number of exon fragments per kilobase to attain the
normalized expression of KLFs. Survival analysis was evaluated
using the R package ‘survival’ (Version:3.2-11, cran.r-project.org/web/packages/survival/index.html).
The survival rates of patients in different test groups were
analyzed using Kaplan Meier (K-M) curve. The final prognostic K-M
plots were constructed using a log-rank P-value, 95% confidence
interval (CI) and hazard ratio (HR). For recurrence-free survival
with HR and 95% CI, univariate and multivariate Cox proportional
risk regression analysis was performed for KLFs and clinical
characteristics. The risk score formula was as follows: Risk
score=0.025587 x age + [0.406461 x (Grade 3 + Grade 4)] + [1.088888
x (Stage III + Stage IV)] + [-0.2134 x KLF2] + [-0.197943 x KLF11].
The K-M curve showed two sets of survival states; receiver
operating characteristics (ROC) curve assesses the predictive value
of the prediction. P<0.05 was considered to indicate a
statistically significant difference. The statistical analysis of
in vitro expression results was performed using paired
Student's t-test and one-way ANOVA followed by post hoc Tukey's
test with GraphPad Prism 8.0.2 (GraphPad Software, Inc.). Data are
presented as mean ± SD from at least three independent
experiments.
Results
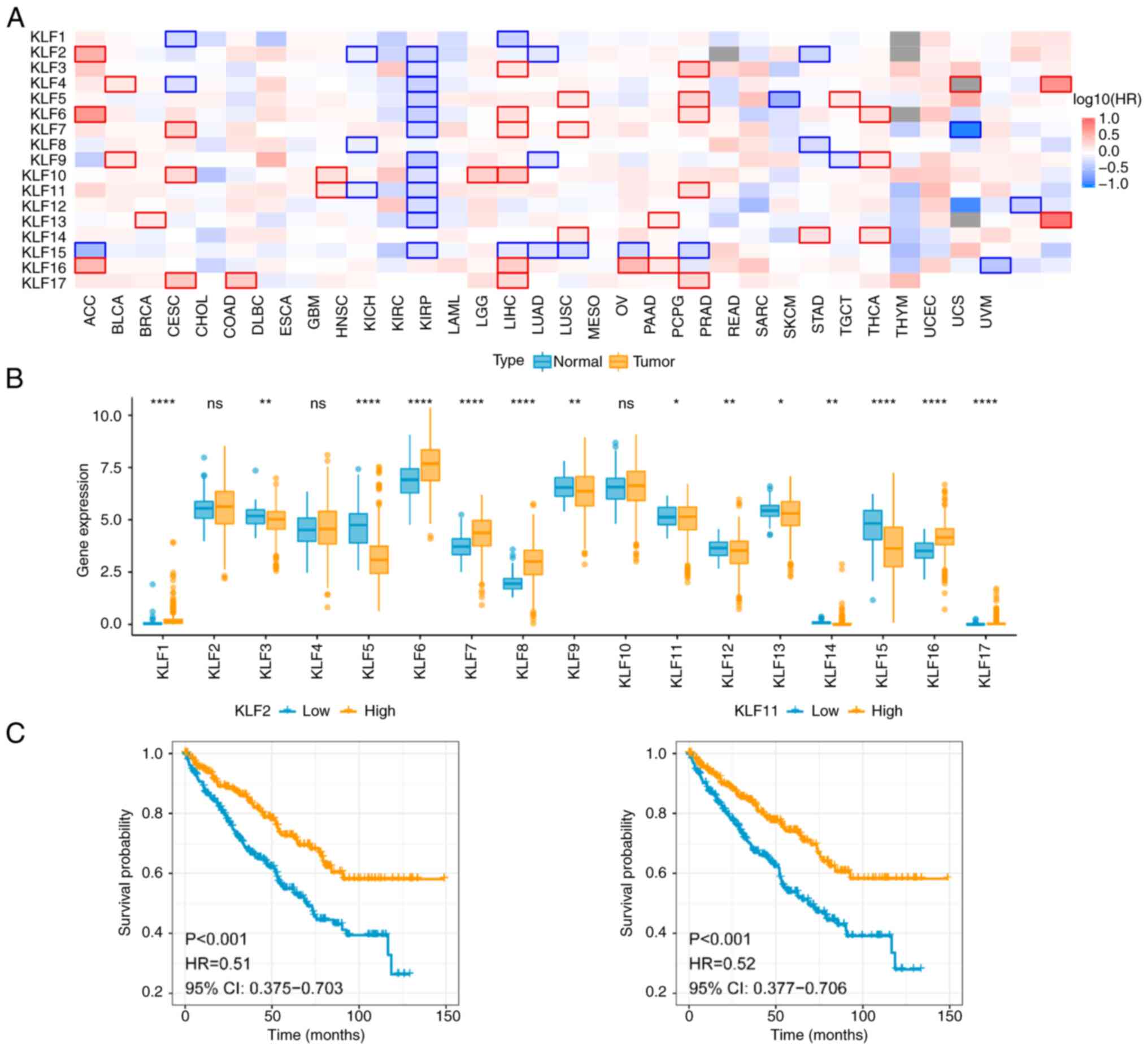
Exploration of KLFs and ccRCC
prognosis
To investigate the association between KLFs and
clinical prognosis of ccRCC, KLF mRNA expression in TCGA database
was analyzed. KLFs were downregulated in ccRCC tissue (Fig. 1A). By contrast, data from TCGA
database indicated that KLFs were expressed in numerous types of
human cancer, such as adrenocortical carcinoma and uterine corpus
endometrial carcinoma. These findings suggested that KLFs may serve
an inhibitory role in progression of urological tumor. The mRNA
levels of 17 KLFs in ccRCC and normal tissue were examined in the
TCGA-KIRC database. The mRNA expression profiles of KLFs indicated
that the majority of KLFs were downregulated in ccRCC compared with
in normal tissue, including KLF3, KLF5, KLF9, KLF11, KLF12, KLF13,
KLF14 and KLF15 (Fig. 1B). By
contrast, KLF1, KLF6, KLF7, KLF8, KLF16 and KLF17 were highly
expressed in tumor tissue. KLF2, KLF4 and KLF10 showed no
significant difference between normal and tumor tissue.
Collectively these data suggest a role for KLFs in ccRCC
progression.
Univariate and multivariate Cox
analysis of KLFs and clinical prognosis
To evaluate the prognostic value of the expression
of KLFs, univariate Cox regression analysis was performed (Table II). Age, grade, stage and KLF2,
KLF3, KLF4, KLF5, KLF6, KLF7, KLF8, KLF9, KLF10, KLF11, KLF12,
KLF13 and KLF15 expression were positive associated with ccRCC
prognosis. Multivariate Cox proportional hazard regression
analysis, based on the sixteen positive factors, revealed that age,
grade, stage, KLF2 and KLF11 were independent factors for ccRCC
prognosis (Table II). K-M
survival curves of KLF2 and KLF11 (Fig. 1C) showed that patients with ccRCC
who had low KLF2 and KLF11 expression had a lower survival
probability. It suggests that the expression of KLF2 and KLF11 were
associated with poor prognosis of ccRCC.
 | Table IIUnivariate and multivariate Cox risk
ratio analysis of KLF gene expression and overall survival in
patients with kidney renal clear cell carcinoma with The Cancer
Genome Atlas data. |
Table II
Univariate and multivariate Cox risk
ratio analysis of KLF gene expression and overall survival in
patients with kidney renal clear cell carcinoma with The Cancer
Genome Atlas data.
| | Univariate Cox | Multivariate
Cox |
|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age | 1.027 | 1.015-1.040 |
2.42x10-5 | 1.023 | 1.009-1.037 | 0.001326 |
| Sex | 0.959 | 0.702-1.310 |
7.91x10-1 | - | - | - |
| Grade | 2.638 | 1.872-3.718 |
3.04x10-8 | 1.485 | 1.013-2.176 | 0.042861 |
| Stage | 3.733 | 2.715-5.132 |
5.09x10-16 | 2.874 | 2.033-4.063 |
0.000229x10-5 |
| KLF2 | 0.703 | 0.615-0.804 |
2.39x10-7 | 0.769 | 0.633-0.934 | 0.008103 |
| KLF3 | 0.638 | 0.522-0.780 |
1.11x10-5 | 1.405 | 0.820-2.407 | 0.216050 |
| KLF4 | 0.728 | 0.641-0.826 |
8.71x10-7 | 0.992 | 0.787-1.251 | 0.947531 |
| KLF5 | 0.747 | 0.641-0.870 |
1.86x10-4 | 0.854 | 0.713-1.024 | 0.088489 |
| KLF6 | 0.681 | 0.594-0.782 |
4.45x10-8 | 0.889 | 0.678-1.165 | 0.393358 |
| KLF7 | 0.692 | 0.590-0.813 |
6.99x10-6 | 1.381 | 0.904-2.110 | 0.135895 |
| KLF8 | 0.843 | 0.714-0.996 |
4.48x10-2 | 1.096 | 0.858-1.400 | 0.462576 |
| KLF9 | 0.639 | 0.555-0.737 |
6.53x10-10 | 0.926 | 0.646-1.328 | 0.676057 |
| KLF10 | 0.756 | 0.663-0.863 |
3.24x10-5 | 1.272 | 0.960-1.685 | 0.094032 |
| KLF11 | 0.666 | 0.572-0.775 |
1.51x10-7 | 0.594 | 0.366-0.964 | 0.035164 |
| KLF12 | 0.622 | 0.521-0.742 |
1.44x10-7 | 0.895 | 0.546-1.467 | 0.660072 |
| KLF13 | 0.654 | 0.556-0.768 |
2.58x10-7 | 0.917 | 0.645-1.303 | 0.629127 |
| KLF15 | 0.751 | 0.664-0.849 |
4.73x10-6 | 0.864 | 0.729-1.023 | 0.090373 |
| KLF16 | 1.056 | 0.848-1.314 |
6.28x10-1 | - | - | - |
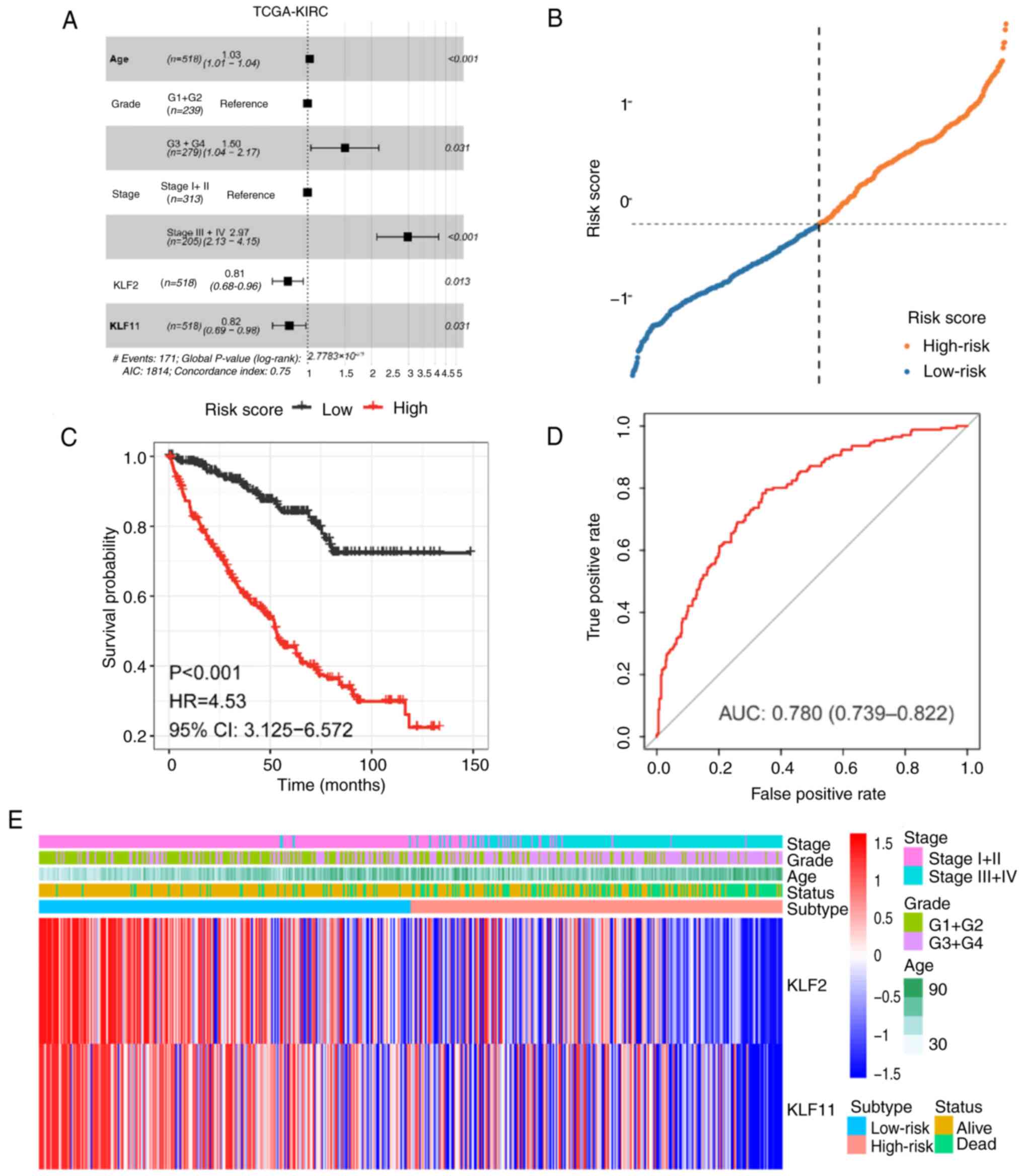
Prognostic models to assess the impact
of KLFs and clinical characteristics on the prognosis of ccRCC
KLF2 and KLF11, which were screened from the
multiple Cox regression analysis, were used to construct a
predictive model. To investigate their effect on ccRCC prognosis,
risk score was calculated as follows (Fig. 2A; Table III). Next, based on the median
risk score, patients with ccRCC were divided into low- and
high-risk groups (Fig. 2B). Heat
map analysis was performed for the gene expression levels in the
high and low-risk groups (Fig.
2E). K-M curve showed that patients in the high-risk group had
worse survival than patients in the low-risk group (Fig. 2C). In addition, the area under the
ROC curve of risk score model was 0.780, indicating that it had an
average diagnostic performance, as previously described (39) (Fig.
2D). These findings suggested that expression levels of KLF2
and KLF11 may be an independent predictor of ccRCC prognosis,
according to the results of multivariate Cox regression analysis
and prognostic predictive model. KLF2 and KLF11 were independent
prognostic factors in ccRCC.
 | Table IIIMultivariate Cox regression
analysis. |
Table III
Multivariate Cox regression
analysis.
| Factor | coef | exp(coef) | se(coef) | Z-score | P-value |
|---|
| Age | 0.025587 | 1.025917 | 0.006851 | 3.735 | 0.000188 |
| Grade 3 + 4 | 0.406461 | 1.501494 | 0.188450 | 2.157 | 0.031016 |
| Stage III + IV | 1.088888 | 2.970967 | 0.170346 | 6.392 |
0.000164x10-6 |
| KLF2 | -0.213400 | 0.807830 | 0.085902 | -2.484 | 0.012982 |
| KLF11 | -0.197940 | 0.820422 | 0.091603 | -2.161 | 0.030709 |
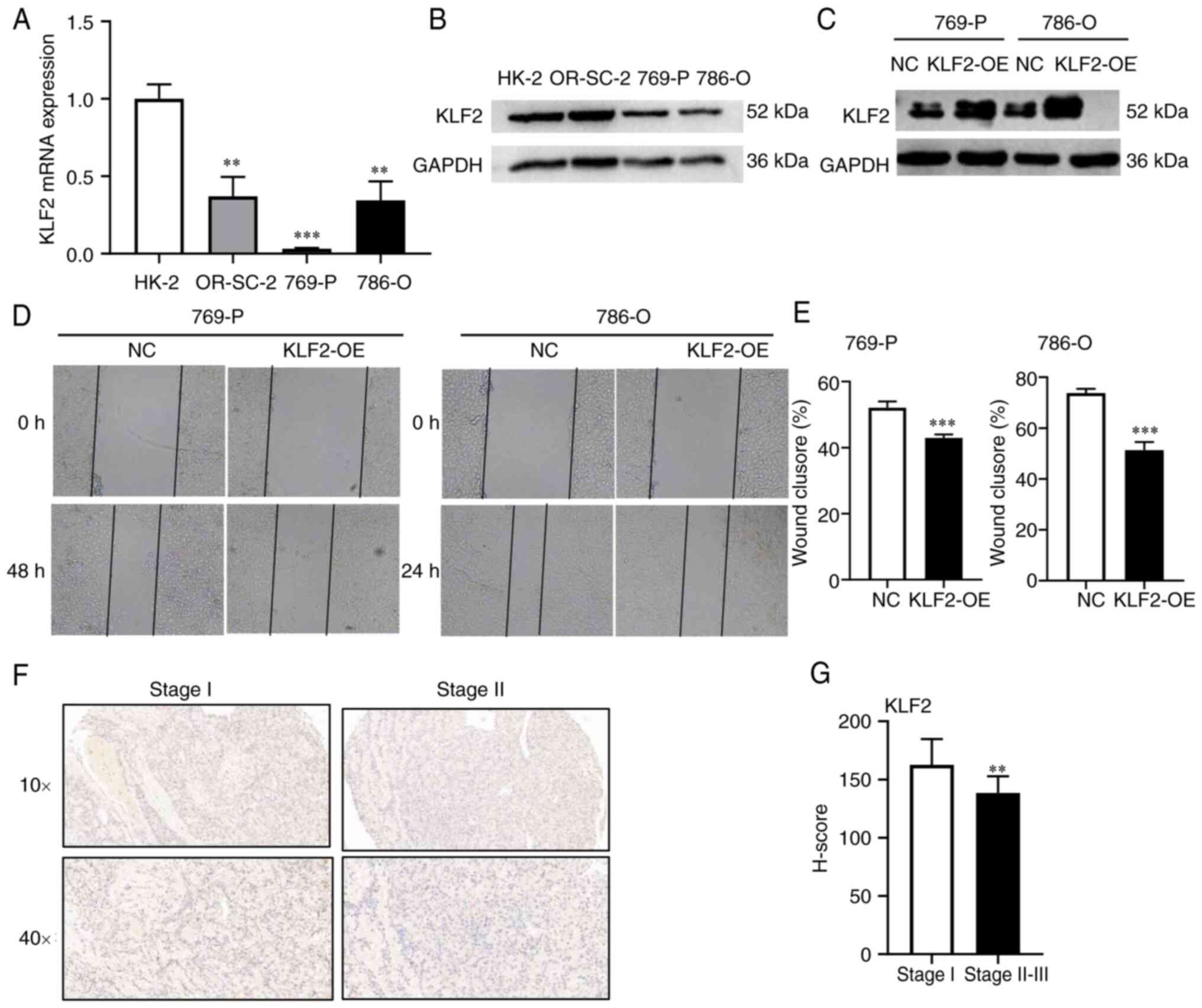
KLF2 overexpression inhibits cell
migration of ccRCC
To validate the effect of KLFs on ccRCC cell lines,
the expression of KLF2 and KLF11 in different ccRCC cell lines was
examined using RT-qPCR and western blotting. KLF2 had lower
expression in 786-O and 769-P cells than in HK-2 cells (Fig. 3A and B). Since KLF11 exhibit low expression in
ccRCC (40), KLF2 was selected for
further validation. To confirm the role of KLF2 in ccRCC migration,
KLF2 plasmid was constructed and transfected into 769-P and 786-O
cells for KLF2 overexpression (Fig.
3C). In the wound healing assay, the scratch width in the
PCDH-CMV-KLF2 group was notably wider than in the control (Fig. 3D and E). These findings suggested that
increased expression of KLF2 impaired the cell migration. To
validate the function of KLF2 in ccRCC, KLF2 expression was
detected using a commercially available ccRCC TMA. IHC was
performed on ccRCC clinical samples (Table IV) and the results indicated that
the KLF2 protein level was significantly lower in stage II-III
tissue compared with stage I tissue (Fig. 3F and G). These findings were consistent with
the aforementioned results of bioinformatics analysis. KLF2 was
lowly expressed in cells and tissues, which overexpression of kLF2
inhibited cell migration.
 | Table IVClinical characteristics of 20
patients with clear cell renal cell carcinoma. |
Table IV
Clinical characteristics of 20
patients with clear cell renal cell carcinoma.
| Characteristic | n (%) |
|---|
| Sex | |
|
Male | 15(75) |
|
Female | 5(25) |
| Age, years | |
|
≤55 | 8(40) |
|
>55 | 12(60) |
| Tumor grade | |
|
I | 4(20) |
|
II +
III | 16(80) |
| TNM stage | |
|
T1 + T2 | 20(100) |
|
T3 + T4 | 0 (0) |
| Pt stage | |
|
T1 + T2 | 20(100) |
|
T3 + T4 | 0 (0) |
| pM stage | |
|
M0 | 20(100) |
|
M1 | 0 (0) |
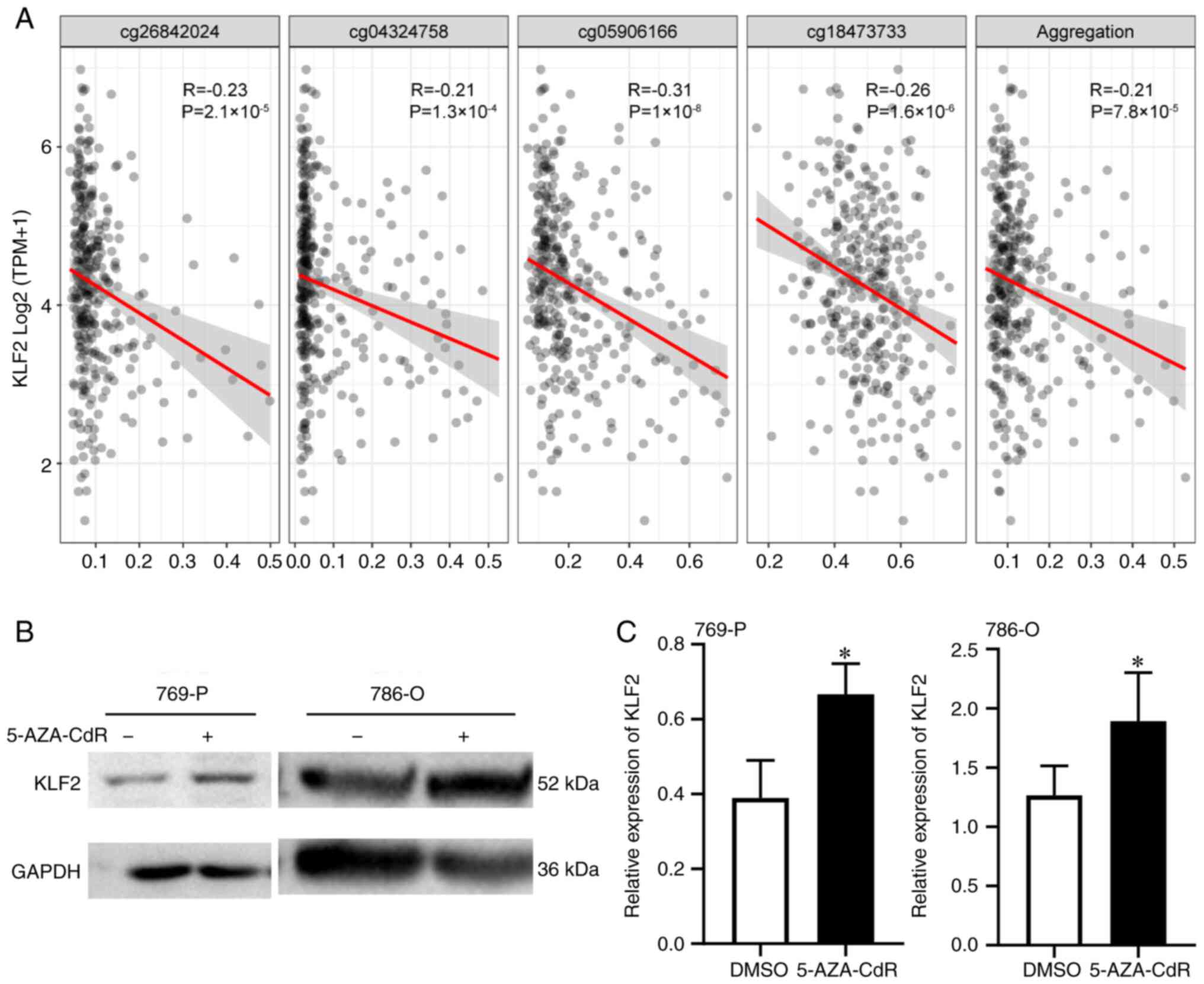
KLF2 promoter methylation
alterations
Examination of TCGA database using cBioPortal
revealed that no genetic variations (data not shown) or methylation
affected survival rate of patients with ccRCC. KLF2 exhibited 10
methylation sites (Table V).
Univariate Cox analysis identified 8 candidate methylation sites
for KLF2 (CG03725130, CG03725130, CG05906166, CG10819847,
CG15496085, CG18473733, CG25266327 and CG26842024). Correlation
analysis found significant differences between candidate
methylation sites and expression of the KLF2 gene was significantly
associated with candidate methylation sites (Fig. 4A). This implied that differences in
KLF2 gene expression were associated with methylation. Based on
this analysis, it was concluded that low expression of KLF2 was
associated with poor survival and methylation. Therefore, KLF2
expression levels were detected after treatment of 769-P and 786-O
cells with 5-AZA-CdR (a DNA methyltransferase inhibitor). The
results showed that KLF2 protein expression increased after
treatment (Fig. 4B and C). Overall, KLF2 expression levels are
associated with poor prognosis, and DNA methylation may be
involved.
 | Table VUnivariate analysis of KLF2
methylation sites. |
Table V
Univariate analysis of KLF2
methylation sites.
| CpG | HR | 95% CI | P-value |
|---|
|
KLF2-Body-Island-cg02668248 | 1.409 | 0.873-2.275 | 0.160 |
|
KLF2-TSS200-Island-cg03725130 | 0.332 | 0.190-0.578 | <0.001 |
|
KLF2-Body-Island-cg04324758 | 3.252 | 1.740-6.079 | <0.001 |
|
KLF2-Body-Island-cg05906166 | 8.040 | 3.263-19.808 | <0.001 |
|
KLF2-TSS1500-Island-cg10819847 | 0.416 | 0.233-0.744 | 0.003 |
|
KLF2-TSS1500-Island-cg15496085 | 2.130 | 1.213-3.740 | 0.008 |
|
KLF2-Body-Island-cg18473733 | 1.687 | 1.004-2.836 | 0.048 |
|
KLF2-TSS1500-N_Shore-cg22247553 | 0.765 | 0.474-1.234 | 0.273 |
|
KLF2-5'UTR-Island-cg25266327 | 0.488 | 0.298-0.798 | 0.004 |
|
KLF2-Body-Island-cg26842024 | 2.179 | 1.438-3.304 | <0.001 |
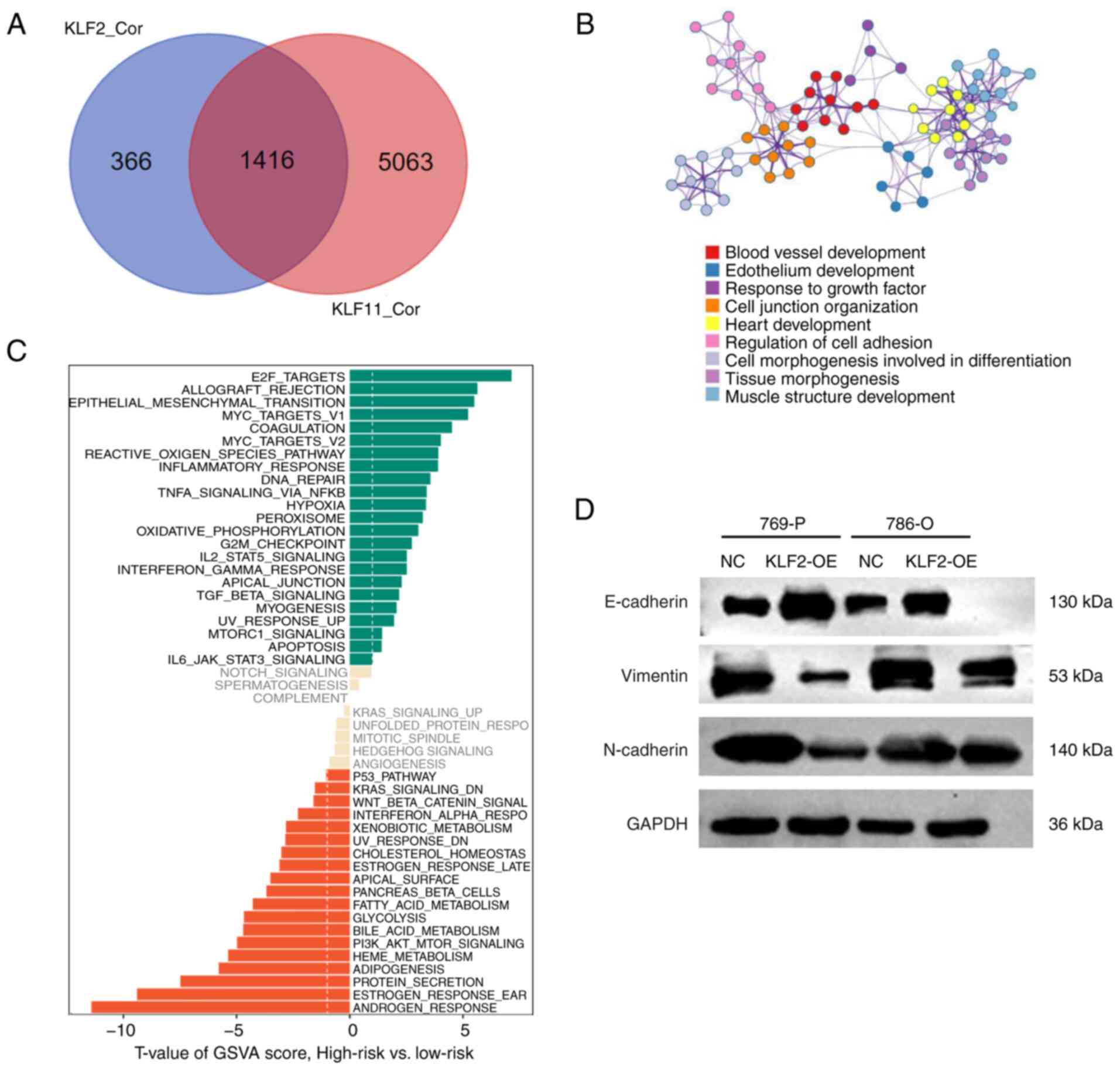
Identify different pathway enrichment
between high and low risk groups
GSVA enrichment analysis was used to determine the
underlying mechanisms in the high- and low-risk groups. This
analysis revealed that the high-risk group was primarily enriched
in ‘E2F targets’, ‘allograft rejection’ and ‘EMT’ (Fig. 5C). The Venn diagram identified
co-negative regulation of KLF2 and KLF11 for 1,416 co-regulated
genes, which means these genes had highly correlated expression
with both KLF2 and KLF11 (Fig.
5A). GO analysis of co-regulated genes demonstrated enrichment
in ‘regulation of cell adhesion’ and ‘cell junction organization’
(Fig. 5B). Western blot analysis
detected changes in expression of proteins involved in the EMT
process. The level of E-cadherin was significantly downregulated,
whereas the levels of N-cadherin and vimentin were increased
following overexpression of KLF2 in 76P-P and 786-O cells (Fig. 5D). These findings suggested that
the expression levels of KLF2 associated with EMT proteins.
Discussion
KLF family members are zinc-finger proteins that
bind to DNA transcription regions, thus serving a vital role in
transcriptional regulation (41).
The involvement of KLFs in tumor progression has been widely
reported (42-44).
In the present study, gene expression levels and clinical factors
were integrated to assess the prognostic value of KLFs. A
prognostic model consisting of KLF2, KLF11, age, stage and grade
was constructed from a TCGA cohort. The results showed that KLF2
was an independent prognostic factor for ccRCC. KLF2 acts as a
nuclear transcription factor in pathophysiological processes,
including immune inflammation (45,46),
angiogenesis (47) and
osteoclastogenesis (48) and is
lowly expressed in numerous types of cancer. For example,
overexpression of KLF2 inhibits cell proliferation, migration and
metastasis in pancreatic ductal adenocarcinoma (49). Xue et al (50) demonstrated that KLF2 was
downregulated in clinical tissue samples and cell lines of prostate
cancer and observed that migration and invasion of prostate cancer
cells were inhibited following over-expression of KLF2. Xu et
al (51) showed that KLF2
affects proliferation and apoptosis of gastric cancer cells by
regulating transcription and expression of cyclin-dependent kinase
genes. These aforementioned studies indicated that KLF2 serves a
key role in cancer development. In addition, previous studies have
shown that KLF2 functions as a vascular protective factor in
nephropathy (52) and ccRCC
resistance (53). Several studies
(53-55)
have reported the role of KLF2 in ccRCC. Lu et al (54) showed that KLF2 suppresses cell
migration and invasion via ferroptosis in metastatic ccRCC.
However, a recent study showed that, compared with paraneoplastic
tissue, KLF2 is highly expressed in non-metastatic ccRCC (55). Thus, KLF2 serves different roles in
different progressive stages of ccRCC. Similar studies on KLF2 have
been conducted in hepatocellular carcinoma (56,57).
However, only a few studies have reported the function of KLF2 in
ccRCC (53-55).
Therefore, the role of KLF2 in ccRCC requires further
investigation.
The present study found that KLF2 was significantly
under expressed in ccRCC; this was correlated with a poor
prognosis. In vitro experiments confirmed that
overexpression of KLF2 inhibited migration in the 786-O and 769-P
cell lines. The present results were consistent a previous study
(54). However, the mechanism of
KLF2 in ccRCC needs to be further investigated. The present study
highlighted methylation as an important mechanism for regulating
KLF2 expression. Treatment with 5-AZA-CdR, a DNA methyltransferase
inhibitor commonly used to deoxygenate DNA, restored KLF2
expression in ccRCC cells. Gene methylation is associated with
tumor genesis and KLF2 methylation has been demonstrated in
non-small cell lung cancer (58).
However, to the best of our knowledge, the present study is first
to report methylation of KLF2 in ccRCC. Whether changes in KLF2
methylation occur in ccRCC tissue remains to be determined in
future studies. In addition, the present study performed simple
signaling pathways assessment, which showed that KLF2 and KLF11 may
decrease ‘cell adhesion’ and ‘cell junction organization’. GSVA
analysis revealed that ‘EMT’ was enriched in the high-risk group.
Cell adhesion and migration are preconditions for cancer metastasis
and 90% of patients with cancer to disease due to cancer-associated
metastasis (59). In line with
this, the present study found that, following overexpression of
KLF2 in 769-P cells and 786-O cells, expression of EMT-associated
proteins was significantly inhibited. These findings suggested that
KLF2 may be a potential biomarker for predicting ccRCC development
and progression. However, further studies on the potential
prognostic value of KLF2 in ccRCC are necessary.
In conclusion, the present study analyzed expression
of KLF2 in multiple types of cancer and highlighted its role in the
prognosis of ccRCC. A risk model of KLFs was constructed and KLF2
expression appeared to have a favorable prognostic value. These
results indicated that the expression of KLF2 may predict the
prognosis of patients with ccRCC. Furthermore, novel ccRCC
prognostic indicators may improve early diagnosis and access to
therapy and increase patient survival. Future studies should
evaluate the synergistic effect of KLF genes for immunotherapy.
Further studies may provide comprehensive insight into the
potential association between KLFs and ccRCC prognosis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
Natural Science Founding of Anhui Provincial (grant no.
1808085MH247), Key Projects of Natural Science Research Projects in
Anhui Universities (grant no. KJ2021A0202) and General Project of
Natural Science Research Project of Anhui University (grant no.
YJS20210280).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FT, JZ, GP and JD contributed to the conception,
design and interpretation of the study. FH and YR performed the
experiments and wrote the manuscript. ZW, HZ, YL and MW were
responsible for the tissue preservation collection, statistical
analysis and bioinformatics analysis in preparation of figures and
tables. FH and YR confirmed the authenticity of all raw data. All
the authors participated in revision of the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of The Second Affiliated Hospital, School of Medicine,
The Chinese University of Hong Kong and a waiver of informed
consent was granted (approval no. 2022004).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mehdi A and Riazalhosseini Y: Epigenome
aberrations: Emerging driving factors of the clear cell renal cell
carcinoma. Int J Mol Sci. 18(1774)2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Linehan WM and Ricketts CJ: The cancer
genome atlas of renal cell carcinoma: Findings and clinical
implications. Nat Rev Urol. 16:539–552. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lucarelli G, Loizzo D, Franzin R,
Battaglia S, Ferro M, Cantiello F, Castellano G, Bettocchi C,
Ditonno P and Batting M: Metabolomic insights into
pathophysiological mechanisms and biomarker discovery in clear cell
renal cell carcinoma. Expert Rev Mol Diagn. 19:397–407.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Allen A, Gau D, Francoeur P, Sturm J, Wand
Y, Martin R, Maranchie J, Duensing A, Kaczorowski A, Duensing S, et
al: Actin-binding protein profilin1 promotes aggressiveness of
clear-cell renal cell carcinoma cells. J Bio Chem. 295:15636–15649.
2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang Q, Zhang H, Chen Q, Wan Z, Gao X and
Qian W: Identification of METTL14 in kidney renal clear cell
carcinoma using bioinformatics analysis. Dis Markers.
2019(5648783)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Boustany J, Abdessater M, Hachem CE,
Khoury ZE, Khoury WE and Khoury RE: Recurrent metastatic clear cell
renal carcinoma with sarcomatoid dedifferentiation treated with
surgery and Cabozantinib. Oncotarget. 11:1922–1928. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cakici MC, Kır G, Akalın MK and Yıldırım
A: Clear cell renal cell carcinoma with osseous metaplasia: Two
extremely rare cases and review of the literature. Arch Esp Uro.
73:651–654. 2020.PubMed/NCBI(In English, Spanish).
|
|
9
|
Zhao J and Eyzaguirre E: Clear cell
papillary renal cell carcinoma. Arch Pathol Lab Med. 143:1154–1158.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Aeppli S, Eboulet EI, Eisen T, Escudier B,
Fischer S, Larkin J, Gruenwald V, McDermott D, Oldenburg J, Omlin
A, et al: Impact of COVID-19 pandemic on treatment patterns in
metastatic clear cell renal cell carcinoma. ESMO Open. 5 (Suppl
3)(e000852)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bersanelli M, Brunelli M, Gnetti L,
Maestroni U and Buti S: Pazopanib as a possible option for the
treatment of metastatic non-clear cell renal carcinoma patients: A
systematic review. Ther Adv Med Oncol.
12(1758835920915303)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Rajandram R, Perumal K and Yap NY:
Prognostic biomarkers in renal cell carcinoma: Is there a
relationship with obesity? Transl Androl Urol. 8 (Suppl
2):S138–S146. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Oates AC, Pratt SJ, Vail B, Yan YI, Ho RK,
Johnson SL, Postlethwait JH and Zon LI: The zebrafish klf gene
family. Blood. 98:1792–1801. 2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Moore DL, Blackmore MG, Hu Y, Kaestner KH,
Bixby JL, Lemmon VP and Goldberg JL: KLF family members regulate
intrinsic axon regeneration ability. Science. 326:298–301.
2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chen Z, Lei T, Chen X, Zhang J, Yu A, Long
Q, Long H, Jin D, Gan L and Yang Z: Porcine KLF gene family:
Structure, mapping, and phylogenetic analysis. Genomics.
95:111–119. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Miller IJ and Bieker JJ: A novel,
erythroid cell-specific murine transcription factor that binds to
the CACCC element and is related to the Krüppel family of nuclear
proteins. Mol Cell Biol. 13:2776–2786. 1993.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ali A, Zhang P, Liangfang Y, Wenshe S,
Wang H, Lin X, Dai Y, Feng XH, Moses R, Wang D, et al: KLF17
empowers TGF-β/Smad signaling by targeting Smad3-dependent pathway
to suppress tumor growth and metastasis during cancer progression.
Cell Death Dis. 6(e1681)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Reidling JC and Said HM: Regulation of the
human biotin transporter hSMVT promoter by KLF-4 and AP-2:
Confirmation of promoter activity in vivo. Am J Physiol Cell
physiol. 292:C1305–C1312. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Saifudeen Z, Dipp S, Fan H and El-Dahr SS:
Combinatorial control of the bradykinin B2 receptor promoter by
p53, CREB, KLF-4, and CBP: Implications for terminal nephron
differentiation. Am J Physiol Renal Physiol. 288:F899–F909.
2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Seiler K, Soroush Noghabi M, Karjalainen
K, Hummel M, Melchers F and Tsuneto M: Induced pluripotent stem
cells expressing elevated levels of sox-2, oct-4, and klf-4 are
severely reduced in their differentiation from mesodermal to
hematopoietic progenitor cells. Stem Cells Dev. 20:1131–1142.
2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shao M, Ge GZ, Liu WJ, Xiao J, Xia HJ, Fan
Y, Zhao F, He BL and Chen C: Characterization and phylogenetic
analysis of Krüppel-like transcription factor (KLF) gene family in
tree shrews (Tupaia belangeri chinensis). Oncotarget.
8:16325–16339. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shimeld SM: C2H2 zinc finger genes of the
Gli, Zic, KLF, SP, Wilms' tumour, Huckebein, Snail, Ovo, Spalt,
Odd, Blimp-1, Fez and related gene families from Branchiostoma
floridae. Dev Genes Evol. 218:639–649. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sun Y, Li Y, Wang M, Yue M, Bai L, Bian J,
Hao W, Sun J, Zhang S and Liu H: Increased AT2R
expression is induced by AT1R autoantibody via two axes,
Klf-5/IRF-1 and circErbB4/miR-29a-5p, to promote VSMC migration.
Cell Death Dis. 11(432)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sunadome K, Yamamoto T, Ebisuya M, Kondoh
K, Sehara-Fujisawa A and Nishida E: ERK5 regulates muscle cell
fusion through Klf transcription factors. Dev Cell. 20:192–205.
2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Suske G, Bruford E and Philipsen S:
Mammalian SP/KLF transcription factors: Bring in the family.
Genomics. 85:551–556. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fisch S, Gray S, Heymans S, Halder SM,
Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, et al:
Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy.
Pro Natl Acad Sci USA. 104:7074–7079. 2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Matsumoto N, Kubo A, Liu H, Akita K, Laub
F, Ramirez F, Keller G and Friedman SL: Developmental regulation of
yolk sac hematopoiesis by Kruppel-like factor 6. Blood.
107:1357–1365. 2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Nuez B, Michalovich D, Bygrave A,
Ploemacher R and Grosveld F: Defective haematopoiesis in fetal
liver resulting from inactivation of the EKLF gene. Nature.
375:316–318. 1995.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wani MA, Means RT and Lingrel JB: Loss of
LKLF function results in embryonic lethality in mice. Transgenic
Res. 7:229–238. 1998.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shindo T, Manabe I, Fukushima Y, Tobe K,
Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami
H, et al: Krüppel-like zinc-finger transcription factor KLF5/BTEB2
is a target for angiotensin II signaling and an essential regulator
of cardiovascular remodeling. Nat Med. 8:856–863. 2002.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Han M, Wang Y, Guo G, Li L, Dou D, Ge X,
Lv P, Wang F and Gu Y: microRNA-30d mediated breast cancer
invasion, migration, and EMT by targeting KLF11 and activating
STAT3 pathway. J Cell Biochem. 119:8138–8145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang X, Li X, Huang C, Li L, Qu H, Yu X,
Ni H and Cui Q: Kruppel-like factor 4 (KLF-4) inhibits the
epithelial-to-mesenchymal transition and proliferation of human
endometrial carcinoma cells. Gynecol Endocrinol. 32:772–776.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Meng J, Lu X, Zhou Y, Zhang M, Gao L, Gao
S, Yan F and Liang C: Characterization of the prognostic values and
response to immunotherapy/chemotherapy of Krüppel-like factors in
prostate cancer. J Cell Mol Med. 24:5797–5810. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wen Y, Lu X, Ren J, Privratsky JR, Yang B,
Rudemiller NP, Zhang J, Griffiths R, Jain MK, Nedospasov SA, et al:
KLF4 in macrophages attenuates TNFα-mediated kidney injury and
fibrosis. J Am Soc Nephrol. 30:1925–1938. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Liang K, Liu T, Chu N, Kang J, Zhang R, Yu
Y and Li D and Li D: KLF8 is required for bladder cancer cell
proliferation and migration. Biotechnol Appl Biochem. 62:628–633.
2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Maclean A, Bunni E, Makrydima S,
Withington A, Kamal AM, Valentijn AJ and Hapangama DK: Fallopian
tube epithelial cells express androgen receptor and have a distinct
hormonal responsiveness when compared with endometrial epithelium.
Hum Reprod. 35:2097–2106. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Dogan S, Vasudevaraja V, Xu B, Serrano J,
Ptashkin RN, Jung HJ, Chiang S, Jungbluth AA, Cohen MA, Ganly I, et
al: DNA methylation-based classification of sinonasal
undifferentiated carcinoma. Mod Pathol. 32:1447–1459.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yu J, Mao W, Sun S, Hu Q, Wang C, Xu Z,
Liu R, Chen S, Xu B and Chen M: Identification of an m6A-related
lncRNA signature for predicting the prognosis in patients with
kidney renal clear cell carcinoma. Front Oncol.
11(663263)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xi Z, Zhang R, Zhang F, Ma S and Feng T:
KLF11 expression predicts poor prognosis in glioma patients. Int J
Gen Med. 14:2923–2929. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Dai X, Ren T, Zhang Y and Nan N:
Methylation multiplicity and its clinical values in cancer. Expert
Rev Mol Med. 23(e2)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Luo Y and Chen C: The roles and regulation
of the KLF5 transcription factor in cancers. Cancer Sci.
112:2097–2117. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Marrero-Rodríguez D, la Cruz HA,
Taniguchi-Ponciano K, Gomez-Virgilio L, Huerta-Padilla V,
Ponce-Navarrete G, Andonegui-Elguera S, Jimenez-Vega F,
Romero-Morelos P, Rodriguez-Esquivel M, et al: Krüppel like factors
family expression in cervical cancer cells. Arch Med Res.
48:314–322. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and krüppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Turpaev KT: Transcription factor KLF2 and
its role in the regulation of inflammatory processes. Biochemistry
(Mosc). 85:54–67. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jha P and Das H: KLF2 in regulation of
NF-κB-mediated immune cell function and inflammation. Int J Mol
Sci. 18(2383)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Novodvorsky P and Chico TJ: The role of
the transcription factor KLF2 in vascular development and disease.
Prog Mol Biol Transl Sci. 124:155–188. 2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Rolph D and Das H: Transcriptional
regulation of osteoclastogenesis: The emerging role of KLF2. Front
Immunol. 11(937)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zhang D, Dai Y, Cai Y, Suo T and Liu H,
Wang Y, Cheng Z and Liu H: KLF2 is downregulated in pancreatic
ductal adenocarcinoma and inhibits the growth and migration of
cancer cells. Tumor Biol. 37:3425–3431. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xue P, Yan M, Wang K, Gu J, Zhong B and Tu
C: Up-regulation of LINC00665 facilitates the malignant progression
of prostate cancer by epigenetically silencing KLF2 through EZH2
and LSD1. Front Oncol. 11(639060)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen
WM, Huang MD and Shu YQ: SP1-induced upregulation of the long
noncoding RNA TINCR regulates cell proliferation and apoptosis by
affecting KLF2 mRNA stability in gastric cancer. Oncogene.
34:5648–5661. 2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Rane MJ, Zhao Y and Cai L: Krüppel-like
factors (KLFs) in renal physiology and disease. EBioMedicine.
40:743–750. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Holland WS, Tepper CG, Pietri JE, Chinn
DC, Gandara DR, Mack PC and Lara PN Jr: Evaluating rational
non-cross-resistant combination therapy in advanced clear cell
renal cell carcinoma: Combined mTOR and AKT inhibitor therapy.
Cancer Chemother Pharmacol. 69:185–194. 2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lu Y, Qin H, Jiang B, Lu W, Hao J, Cao W,
Du L, Chen W, Zhao X and Guo H: KLF2 inhibits cancer cell migration
and invasion by regulating ferroptosis through GPX4 in clear cell
renal cell carcinoma. Cancer Lett. 522:1–13. 2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Li M, Zhang M, Chen M, Xiao J, Mu X, Peng
J and Fan J: KLF2-induced circZKSCAN1 potentiates the tumorigenic
properties of clear cell renal cell carcinoma by targeting the
miR-1294/PIM1 axis. Cell Cycle. 1–15. 2022.PubMed/NCBI View Article : Google Scholar : (Epub ahead of
print).
|
|
56
|
Zou K, Lu X, Ye K, Wang C, You T and Chen
J: Krüppel-like factor 2 promotes cell proliferation in
hepatocellular carcinoma through up-regulation of c-myc. Cancer
Biol Ther. 17:20–26. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jin L, He Y, Tang S and Huang S: LncRNA
GHET1 predicts poor prognosis in hepatocellular carcinoma and
promotes cell proliferation by silencing KLF2. J Cell Physiol.
233:4726–4734. 2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Jiang W, Xu X, Deng S, Luo J, Xu H, Wang
C, Sun T, Lei G, Zhang F, Yang C, et al: Methylation of
kruppel-like factor 2 (KLF2) associates with its expression and
non-small cell lung cancer progression. Am J Transl Res.
9:2024–2037. 2017.PubMed/NCBI
|
|
59
|
Zhang Y and Weinberg RA:
Epithelial-to-mesenchymal transition in cancer: Complexity and
opportunities. Front Med. 12:361–373. 2018.PubMed/NCBI View Article : Google Scholar
|