Introduction
Salmonella is a gram-negative, facultative,
intracellular bacterium that frequently causes human and animal
diseases (1). According to the
WHO, >90 million people are infected by Salmonella
annually, 150,000 of whom will succumb to Salmonella
infection. The main clinical symptoms of Salmonella
infection, also called salmonellosis, are sepsis and
gastroenteritis (2). To date,
>2,600 subtypes of Salmonella have been discovered and
categorized (3). Salmonella
serotypes are typically associated with their host adaptation and
virulence capability, rendering serotyping to be a key tool for
Salmonella surveillance and outbreak investigations
(4). However, treatment of
invasive salmonellosis has been compromised due to the emergence of
Salmonella strains that are resistant to a variety of
first-line drugs such as ampicillin, chloramphenicol and
co-trimoxazole (5). Therefore, an
appropriate molecular typing method should be used in combination
with serotyping to investigate the epidemiology of
Salmonella.
Accurate typing and tracing are important for
microbial epidemiological investigation, food safety and public
health. Bacterial typing methods that are available include
phenotyping such as phage-typing, serotyping and ribotyping and
genotyping such as plasmid profile, pulsed-field gel
electrophoresis and multi-locus sequence typing (6). Among them, serotyping and multi-locus
sequence typing (MLST) are the most frequently used (7). Typing methods of Salmonella
can be divided into phenotypic typing based on their phenotypic
characteristics and molecular typing based on their gene expression
patterns (8). Serotyping using
standard agglutination methods has been the most common form of
typing since 1934(9). By contrast,
molecular typing techniques mainly include pulsed field gel
electrophoresis (PFGE) (10), MLST
and multi-locus variable-number tandem repeat analysis (8). MLST has become a particularly popular
molecular typing method due to its advantages: i) High resolution,
it is easy to discover differentiation of genetically related
bacterial isolates from nonambiguous sequencing data, which is
superior to serotyping and/or PFGE typing (11); and ii), reproducibility and
universality, MLST can be readily reproduced and does not require
access to specialized reagents or training (12). In particular, Salmonella
MLST is a molecular typing method that is based on the sequencing
of seven housekeeping genes of Salmonella, namely
chorismate synthase (aroC), β sliding clamp of DNA
polymerase III (dnaN), uroporphyrinogen-III
synthase (hemD), histidinol dehydrogenase
(hisD), phosphoribosylaminoimidazole carboxylase
catalytic subunit (purE), 2-oxoglutarate
dehydrogenase E1 component (sucA) and homoserine
dehydrogenase (thrA) (13). These genes are highly conserved and
only slowly accumulate site changes (14). Salmonella MLST is mainly
applied to understand the hereditary backgrounds, origins and
diversification of the various Salmonella sub-strains.
In the present study, the serotypes, MLST, drug
resistance, integrin class and distribution of 52 clinical isolates
of non-duplicated Salmonella which is not genome
contamination from children with diarrhea were analyzed.
Specifically, the possible correlation among the allelic profiles
of the housekeeping genes aroC, dnaN, hemD,
hisD, purE, sucA and thrA and the
Salmonella eBrust Groups (eBGs) and serotypes for potential
Salmonella serovar prediction were focused upon. In
addition, the association between drug resistance and integrin
distribution of the Salmonella strains was investigated.
In order to aid clinicians in improving the clinical
prevention and treatment of Salmonella infection, 1,725
diarrhea samples from children were collected to isolate the
Salmonella strain in the present study. The drug
sensitivities of those Salmonella strains were detected
focusing on 12 different antibiotics. Total Salmonella
genome DNA (gDNA) were extracted for MLST analysis of
characteristics of Salmonella. After sequence analysis,
eBURST analysis was applied for the various Salmonella
sequence types (STs). The class I, II, and III integron
distribution was also analyzed to map the integrated substructures
of the integrons.
Materials and methods
Strain conservation
The present study was approved by the Medical Ethics
Committee of Qingyuan People's Hospital, Qingyuan, China (approval
no. A0051). Written informed consent was obtained from each
participant's legal guardian. In total, 52 ‘non-duplicate’
(confirmed as not a biological contamination) Salmonella
strains were harvested from the feces of children (age, 3 months to
14 years) with diarrhea between September 2018 and April 2020 at
the Department of Clinical Laboratory, The Sixth Affiliated
Hospital of Guangzhou Medical University, Qingyuan People's
Hospital, Qingyuan, China. Patients aged ≤14 years who were
admitted to the Outpatient Department or hospitalized at Qingyuan
People's Hospital due to diarrhea were selected as candidates. A
total of 1,725 children (890 male and 835 female children, mean age
5.6±0.41, median age 5.1±0.69) were enrolled in our study, all of
whom exhibited one or more of the following clinical
manifestations: Diarrhea, fever and/or abdominal pain. The
exclusion criteria were: i) Children with diarrhea presenting with
watery stools, without red blood cells, white blood cells or pus
cells in routine stool test; ii) with low complete blood count; and
iii) with mild symptoms. The quality control strains used for
antibiotic susceptibility testing were Pseudomonas
aeruginosa (ATCC.27853), Staphylococcus aureus
(ATCC.25923) and E. coli (ATCC.25922) (all from American
Type Culture Collection) (15).
The positive integron reference strain was a clinical isolation
Klebsiella pneumoniae strain (1162281; GSK plc.) which
contained Class I, II, and III integrons (16).
Instruments and reagents
Instruments
BD Phoenix™ M50 automatic detector and bacterial
drug sensitivity analysis system (BD Biosciences). NMIC/ID-4
composite board for the drug sensitivity tests (BD Biosciences).
Thermal Cycler T100 (Bio-Rad Laboratories, Inc.). Gel Dox™ XR+ gel
imaging system (Bio-Rad Laboratories, Inc.).
Reagents. Triple sugar iron (TSI) agar medium
(Qingdao Hope Bio-Tcehnology Co.,Ltd). Bacterial group DNA
extraction kit (ab288102, Abcam), 2X Taq PCR MasterMix (KT121221,
Tiangen Biotech Co., Ltd.), Gel Red nucleic acid dye (SCT123,
Sigma-Aldrich; Merck KGaA), and Marker I, II and III DNA ladder
(MD101, MD102 and MD103; Tiangen Biotech Co., Ltd.). Primers
(Sangon Biotech Co., Ltd.).
Methods Sample collection and
bacterial culture
Fecal samples from children with diarrhea were
collected for Salmonella isolation and identification. A
small amount (10-50 mg) of feces from each child with diarrhea was
collected using sterile swabs, which were placed in 9 ml selenite
brilliant green sulfa enrichment broth (E-MA73; Eiken Chemical Co.,
Ltd.) and cultured at 36˚C for 18 h. After enrichment, 10 µl of the
samples was streaked onto a Salmonella chromogenic medium
agar plate (CM1007B; Thermo Fisher Scientific, Inc.), before
further culture at 36˚C for 18 h. Suspected colonies that are
purplish red or wine red with a diameter of 2-3 mm were selected
from the plate for biochemical identification.
Serotyping by slide agglutination. Salmonella
enterica isolates were cultured at 37˚C overnight. A drop of broth
was dropped on a glass slides to test somatic O antigen by slide
agglutination. Meanwhile, each Salmonella strain was grown
on Swarm agar plates at 37˚C overnight, and single colonies were
picked to test phases 1 and 2 of H antigens by slide agglutination.
Diagnostic sera for Salmonella antigens were purchased from
Tianrun Bio-Pharmaceutical Co. Ltd. and S&A Reagents Lab Ltd.
Serotyping and biotyping were performed according to the modified
Kauffmann-White scheme, which is a modification of the original
scheme from the 1930s (7).
Identification of Salmonella and drug sensitivity
detection. All Salmonella strains were inoculated onto
TSI agar plates and cultured for 12-18 h at 37˚C. Antibiotic
susceptibility test (AST) was performing using the BD Phoenix™ M100
Automated Microbiology System (BD Biosciences), a broth-based
microdilution method that utilizes a redox indicator
(colorimetric-oxidation-reduction) to enhance the detection of
bacterial growth. AST was performed on BD Phoenix™ NMIC-502 panels
(BD Biosciences). Samples were prepared and experimental conditions
were set following the manufacturer's protocols. Phoenix ID broth
(4 ml) was first inoculated with bacterial colonies from a pure
culture adjusted to a 0.5 McFarland standard using a CrystalSpec
nephelometer (BD Biosciences), before the suspension was poured
into the ID side of the Phoenix panel. Bacterial growth at 37˚C in
the panels was then monitored every 20 min to determine minimal
inhibitory concentration (MIC) of antibiotics. The interpretation
of MIC results were recorded as either susceptible (S),
intermediate (I) and resistant (R) according to the 2016 CLSl
M100-S26 susceptibility test guide (17). Drug resistance rate=(susceptible
strain number/total Salmonella strain number) x100%.
In total, 12 different antibiotics were used base on
the following antibiotic classification: i) Class A, including
carbenpenems imipenem and meropenem; ii) class C, such as
quinolones moxifloxacin; iii) class D, including cephalosporins
cefotaxime and cefepime; iv) class E, such as broad-spectrum
penicillins ampicillin, ampicillin/sulbactam, piperacillin and
piperacillin/tazobactam; v) class F, including β-lactam/β-lactamase
inhibitor combinations, including amoxicillin,
amoxicillin/clavulanate and aztreonam; and vi) class G, including
phenicols chloramphenicol.
Total bacterial DNA extraction. Salmonella
genomic DNA was extracted in accordance with the protocol of the
Bacterial group DNA extraction kit (ab288102; Abcam), transferred
to an aseptic 1.5 ml tube and stored at -80˚C.
Primer design. In total, seven housekeeping
genes of Salmonella, aroC, dnaN, hemD, hisD, purE,
sucA and thrA, were chosen for MLST analysis (18). The primer sequences were designed
according to the Salmonella MLST database (https://enterobase.readthedocs.io/en/latest/mlst/mlst-legacy-info-senterica.html)
and are listed in Table I. Primers
for the intI1, intI2, intI3 gene and variable
region were designed using primer3Web (version 4.1.0; http://primer3.ut.ee/) as described previously
(19) and are listed in Table II.
 | Table IPrimer sequences used for multi-locus
sequence typing. |
Table I
Primer sequences used for multi-locus
sequence typing.
| Gene | Oligonucleotide and
sequence (5'-3') | Length/bp |
|---|
| Chorismate
synthase | F:
CCTGGCACCTCGCGCTATAC | 826 |
| | R:
CCACACACGGATCGTGGCG | |
| β sliding clamp of
DNA polymerase III | F:
ATGAAATTTACCGTTGAACGTGA | 833 |
| | R:
AATTTCTCATTCGAGAGGATTGC | |
|
Uroporphyrinogen-III synthase | F:
GAAGCGTTAGTGAGCCGTCTGCG | 666 |
| | R:
ATCAGCGACCTTAATATCTTGCCA | |
| Histidinol
dehydrogenase | F:
GAAACGTTCCATTCCGCGCAGAC | 894 |
| | R:
CTGAACGGTCATCCGTTTCTG | |
|
Phosphoribosylaminoimidazole carboxylase
catalytic subunit | F:
ATGTCTTCCCGCAATAATCC | 510 |
| | R:
TCATAGCGTCCCCCGCGGATC | |
| 2-oxoglutarate
dehydrogenase E1 component | F:
AGCACCGAAGAGAAACGCTG | 643 |
| | R:
GGTTGTTGATAACGATACGTAC | |
| Homoserine
dehydrogenase | F:
GTCACGGTGATCGATCCGGT | 852 |
| | R:
CACGATATTGATATTAGCCCG | |
 | Table IIPrimer sequences of the three
integrase genes and variable areas. |
Table II
Primer sequences of the three
integrase genes and variable areas.
| Target gene | Oligonucleotide
(5'-3') | Length/bp |
|---|
| Int 1 | F:
GGTCAAGGATCTGGATTTCG | 493 bp |
| | R:
ACATGCGTGTAAATCATCGTC | |
| | | |
| Int 2 | F:
CACGGATATGCGACAAAAAGGT | 789 bp |
| | R:
GTAGCAAACGAGTGACGAAATG | |
| | | |
| Int 3 | F:
AGTGGGTGGCGAATGAGTG | 922 bp |
| | R:
TGTTCTTGTATCGGCAGGTG | |
| | | |
| Int-variable
area | 5'-CS:
GGCATCCAAGCAGCAAG | variable |
| | 3'-CS:
AAGCAGACTTGACCTGA | |
PCR for Salmonella MLST, integrase genes and
variable regions. The PCR mix for the seven housekeeping genes
was constructed as follows: 25 µl 2X Taq PCR MasterMix, 1 µl each
of the forward and reverse primers, 2 µl Salmonella gDNA
template and 22 µl ddH2O. The reaction conditions were
as follows: Initial denaturation at 94˚C for 5 min; followed by 32
cycles of denaturation at 95˚C for 30 secs, annealing at 60˚C for 1
min and extension at 72˚C for 1 min; and a final extension at 72˚C
for 10 min. The PCR products were visualized electrophoretically on
an agarose gel using Gel Red nucleic acid dye (Sigma-Aldrich; Merck
KGaA) and sent to Sangon Biotech Co., Ltd. for bidirectional Sanger
sequencing.
The PCR mix for the class I, II, and III integrons
comprised the following: 12.5 µl 2X Taq PCR MasterMix, 0.5 µl each
of the forward and reverse primer, 0.5 µl Salmonella gDNA
template and 11.5 µl ddH2O. The reaction conditions were
as follows: Initial denaturation at 95˚C for 5 min; followed by 26
cycles of denaturation at 94˚C for 30 sec, annealing at 62˚C for 30
sec and extension at 72˚C for 1 min; and a final extension at 72˚C
for 5 min. In total, 5 µl PCR products were loaded onto a 2%
agarose gel (60 min, 80 V) and visualized using Gel Red nucleic
acid dye and a gel imaging system (Bio-Rad Laboratories, Inc.).
The PCR mix for the variable regions of class I, II,
and III integrons comprised the following: 25 µl 2X Taq PCR
MasterMix, 0.5 µl each of the forward and reverse primer, 1 µl DNA
template and ddH2O to 50 µl. The reaction conditions
were as follows: Initial denaturation at 94˚C for 5 min; followed
by 35 cycles of denaturation at 94˚C for 30 sec, annealing at 58˚C
for 30 sec and extension at 72˚C for 2 min; and a final extension
at 72˚C for 7 min. PCR products (5 µl) were visualized
electrophoretically on a 1.2% agarose gel (60 min, 80 V) using Gel
Red nucleic acid dye and a gel imaging system (Bio-Rad
Laboratories, Inc.).
Sequence analysis of the variable
regions
The PCR products (displaying bright bands) were sent
to Sangon Biotech Co., Ltd. for sequencing. Corrected sequencing
data were obtained by removing failed signals with Chromas software
(version 2.6.6; http://technelysium.com.au/wp/chromas/). Subsequently,
sequencing data were compared and analyzed using BLAST (BLAST+
version 2.10.0; https://blast.ncbi.nlm.nih.gov/Blast.cgi). Results
with 100% coincidence were selected.
Genotyping analysis of Salmonella
MLST
The sequencing reads were subjected to the
Salmonella MLST database v1.1.3 (http://mlst.warwick.ac.uk/mlst/dbs/Senterica),
before the different allele values were obtained by allele/ST query
to identify the corresponding ST, yielding an allelic spectrum. The
ST classification data were then uploaded onto the Pub MLST Data
Analysis (http://pubmlst.org/salmonella/) tool to conduct eBURST
analysis to obtain the BURST group diagram (group definition:
Profiles match at n-2 loci to any other member, ‘n’ is the number
of loci in the scheme). The standard of clustering used is that if
four subunits in seven ST-labeled genes are the same, then they
would be considered to be in the same composite clone group
(20,21).
Statistical analysis
All statistical analyses were performed using SPSS
software version 16.0 (SPSS, Inc.). Drug resistance was analyzed
using the χ2 test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Serotyping
A total of 11 serotypes were obtained from the 52
Salmonella strains, including 33 strains of S.
Typhimurium, five strains of S. Enteritidis, four
strains of S. Stanley, three strains of S. Rigel and
one strain each of S. Paratyphi A, S. Derby, S. Dublin, S. San
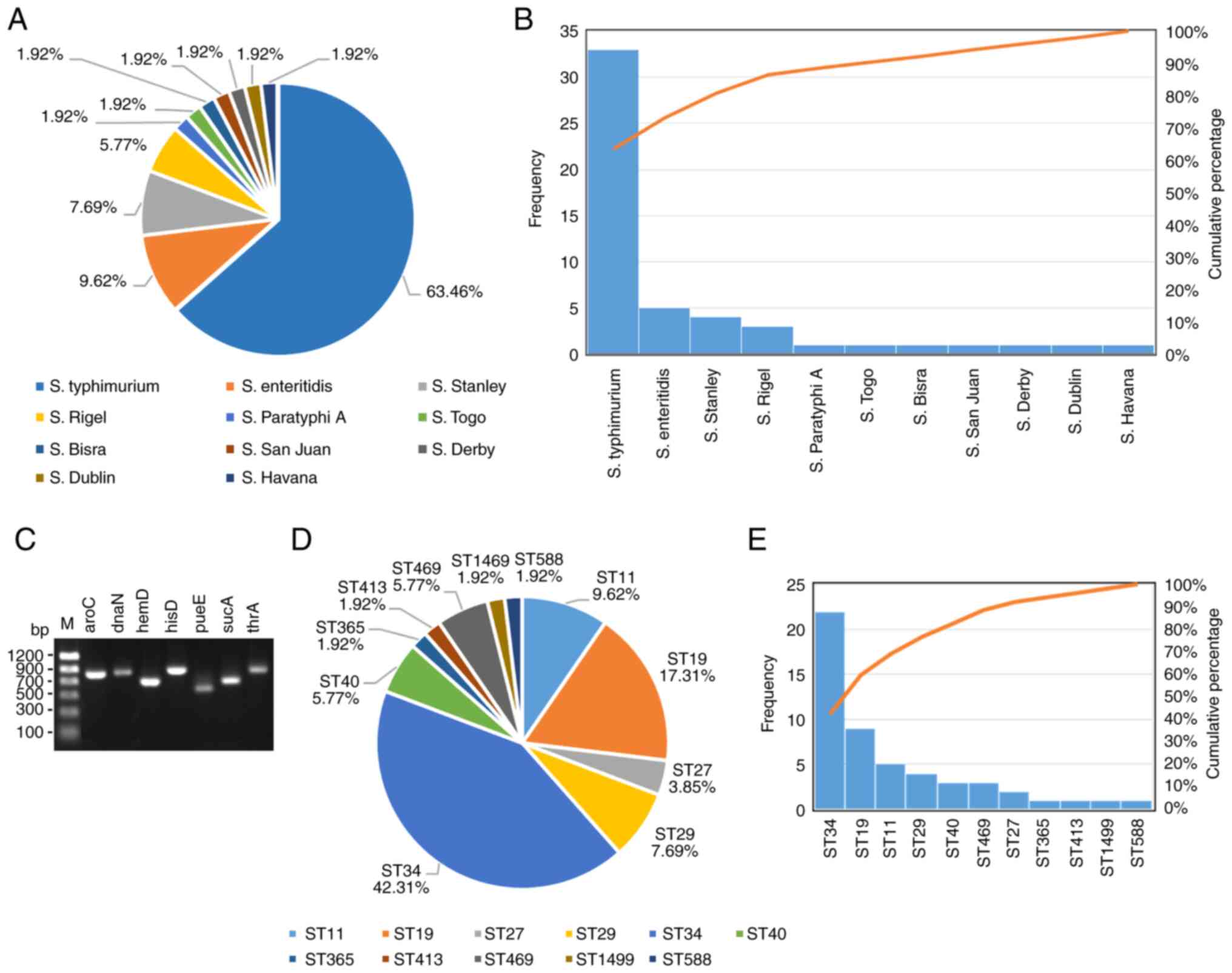
Juan, S. Togo, S. Bisra and S. Havana (Fig. 1A and B).
MLST results
To estimate the genetic correlations, MLST was
performed. In total, seven housekeeping gene loci, namely
aroC, dnaN, hemD, hisD, purE,
sucA and thrA, were chosen for MLST analysis of
Salmonella (Fig. 1C). A
total of 52 Salmonella strains were divided into 11 STs base
on MLST result (Table III),
including 22 strains with ST34 (42.31%), nine strains with ST19
(17.31%), five strains with ST11 (9.62%), four strains with ST29
(7.69%), three strains with ST40 (5.77%) and ST469 (5.77%), two
strains with ST27 (3.85%) and one strain with ST365 (1.92%), ST413
(1.92%), ST1499 (1.92%) and ST588 (1.92%) (Fig. 1D and E).
 | Table IIISalmonella multi-locus
sequence typing and distribution rate in 52 samples. |
Table III
Salmonella multi-locus
sequence typing and distribution rate in 52 samples.
| Chorismate
synthase | β sliding clamp of
DNA polymerase III |
Uroporphyrinogen-III synthase | Histidinol
dehydrogenase |
Phosphoribosylaminoimidazole carboxylase
catalytic subunit | 2-oxoglutarate
dehydrogenase E1 component | Homoserine
dehydrogenase | Sequence types | Distribution rate
% |
|---|
| 5 | 2 | 3 | 7 | 6 | 6 | 11 | 11 | 9.61 |
| 2 | 59 | 23 | 64 | 38 | 19 | 12 | 1499 | 1.92 |
| 10 | 7 | 12 | 9 | 5 | 9 | 2 | 19 | 17.31 |
| 5 | 14 | 79 | 9 | 6 | 12 | 17 | 27 | 3.85 |
| 186 | 35 | 78 | 75 | 39 | 182 | 97 | 588 | 1.92 |
| 16 | 16 | 20 | 18 | 5 | 12 | 18 | 29 | 7.69 |
| 10 | 19 | 12 | 9 | 5 | 9 | 2 | 34 | 42.31 |
| 130 | 97 | 25 | 125 | 84 | 9 | 101 | 365 | 1.92 |
| 19 | 20 | 3 | 20 | 5 | 22 | 22 | 40 | 5.77 |
| 15 | 70 | 93 | 78 | 113 | 6 | 68 | 413 | 1.92 |
| 92 | 107 | 79 | 156 | 64 | 151 | 87 | 469 | 5.77 |
Association between MLST and
serotyping
Among the 52 Salmonella samples, each
Salmonella serotype corresponded to ≥ one MLST type. The
most common serotype and ST type were S. Typhimurium (33/52)
and ST34 (22/52), respectively. These two types are the most
prevalent in the South of China. The three other predominant types
were S. Enteritideis, S. Stanley and S. Rigel,
accounting for ~80.77% in total. In the aforementioned serotypes,
four strains of ST11, nine strains of ST19, two strains of ST27,
four strains of ST29, 21 strains of ST34 and two strains of ST40
were identified, where ST34 was the most abundant ST within the
S. Typhimurium subtype (63.63%), whereas ST11 was the most
abundant ST of S. Enteritidis (80%; Table IV).
 | Table IVSalmonella ST type
corresponding to each serotype in 52 samples. |
Table IV
Salmonella ST type
corresponding to each serotype in 52 samples.
| Serotypes | ST types (number of
strains) |
|---|
|
Salmonella | ST19(9), ST29(2),
ST34(21), ST40(1) |
|
Typhimurium | |
|
Salmonella | ST11(4),
ST40(1) |
|
Enteritis | |
|
Salmonella | ST29(2),
ST27(2) |
| Stanley | |
|
Salmonella | ST469(3) |
| Rigel | |
|
Salmonella | ST365(1) |
| Paratyphi
A | |
|
Salmonella | ST34(1) |
| Togo | |
|
Salmonella | ST1499(1) |
| Bisra | |
|
Salmonella | ST413(1) |
| San
Juan | |
|
Salmonella | ST40(1) |
| Derby | |
|
Salmonella | ST11(1) |
| Dublin | |
|
Salmonella | ST588(1) |
| Havana | |
Analysis of the similarity,
variability and evolutionary relationships among different ST types
of Salmonella
eBURST is an algorithm that can identifies groups of
closely associated sequence types from MLST data (20). It was used to analyze the possible
similarity, variability and evolutionary relationships among
different ST types of Salmonella in the present study. The
genetic backgrounds were found to be diverse among the STs
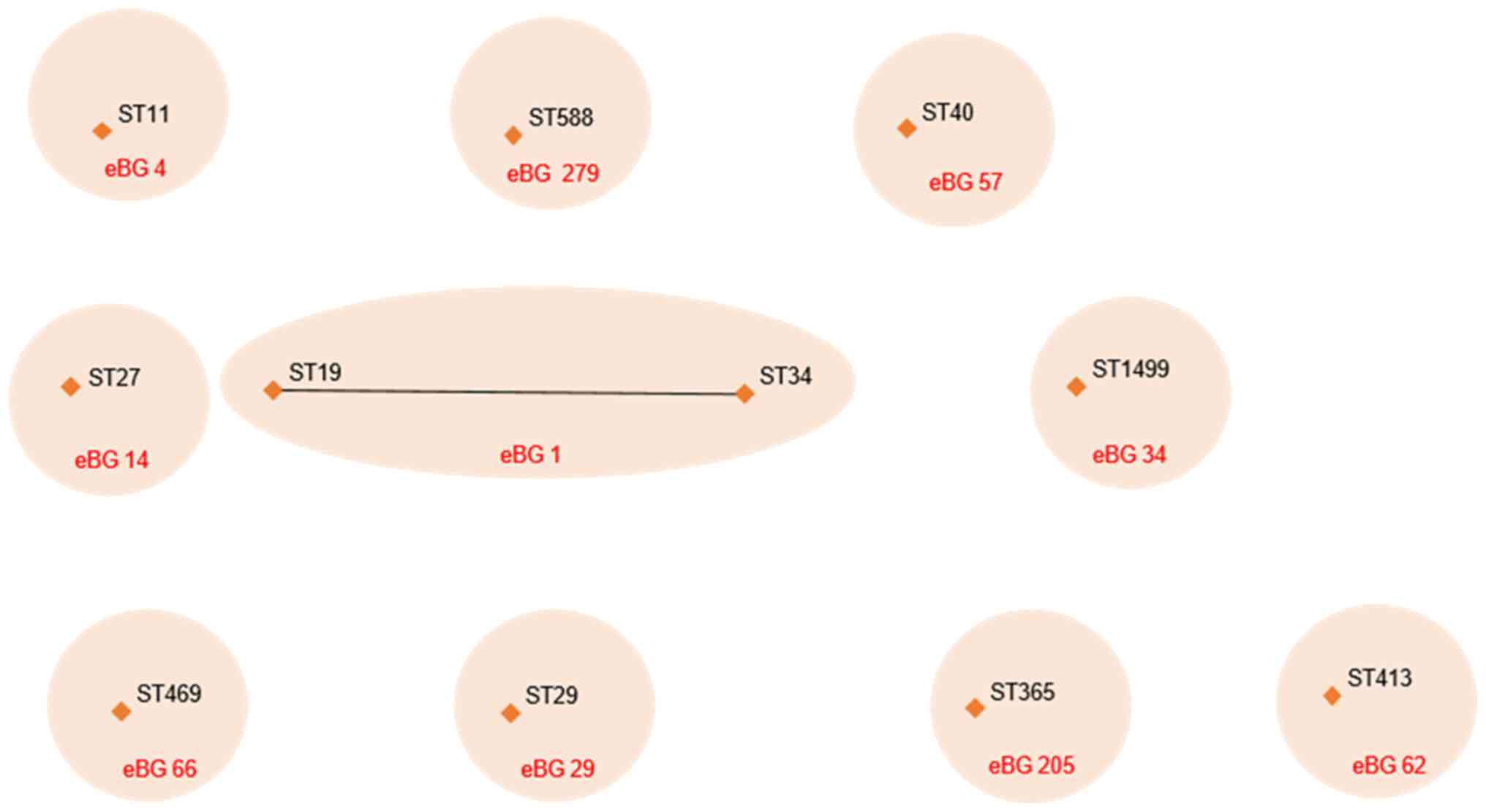
identified in the present study. In total, 11 STs belonged to 10
different eBURST groups (eBGs). Further analysis also indicated
that ST19 and ST34 are part of the same composite group (eBG1)
sharing a high genetic relationship (Fig. 2).
Association between class I integrons
and anti-bacterial resistance and anti-microbial resistance
profile
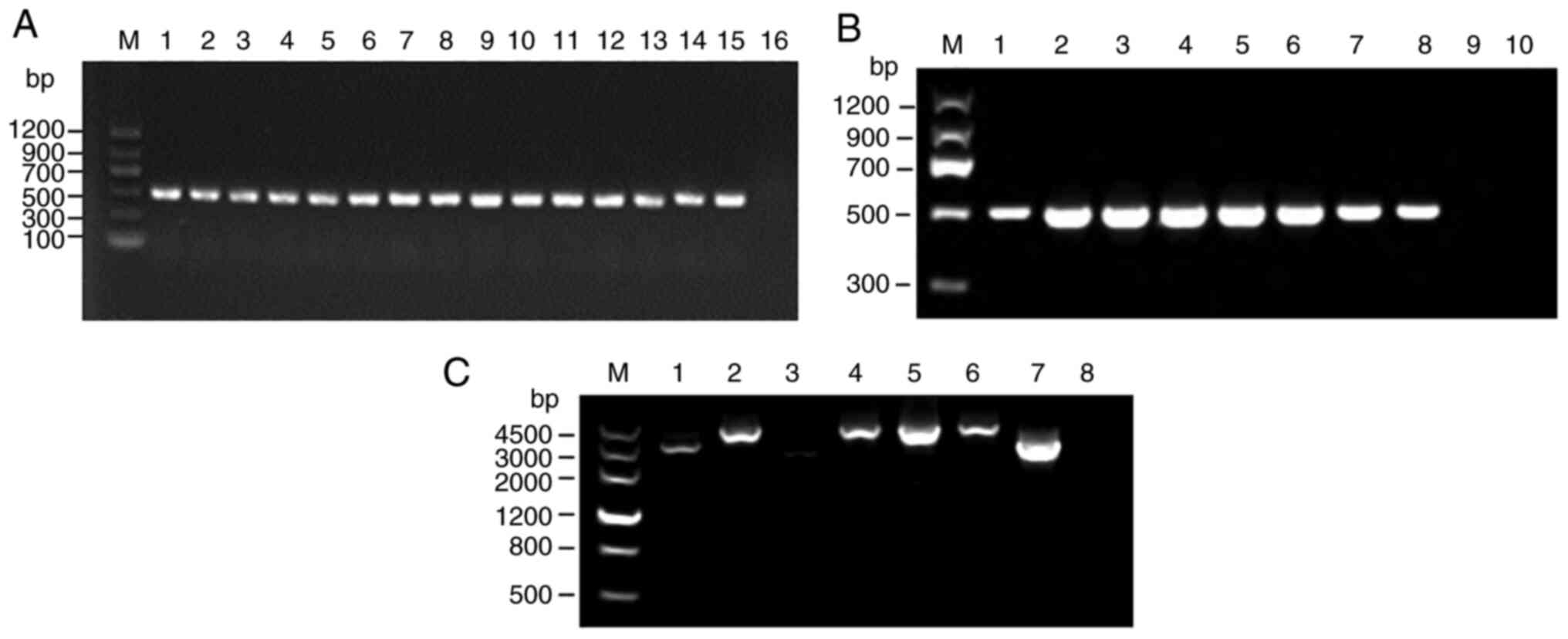
Among the 52 Salmonella strains, 20 harbored
class I integrase, where the detection rate was 38.46% (20/52).
However, none of the 52 strains harbored class II or III integrons
(Fig. 3A and B). The resistance rates of strains
harboring class I integrons toward ampicillin,
ampicillin/sulbactam, chloramphenicol and moxifloxacin were found
to be significantly higher compared with those of class I
intergroup-negative strains (P<0.05). Neither class I
integron-positive nor class I integron-negative strains were
resistant to amoxicillin/clavulanate, meropenem,
piperacillin/tazobactam or imipenem. The resistance rates of class
I integron-positive strains to aztreonam, piperacillin, cefepime
and cefotaxime were increased compared with those of class I
integron-negative strains, but no significance were found (Table V).
 | Table VDrug resistance phenotypes between 52
Salmonella profile I integron-positive and -negative
strains. |
Table V
Drug resistance phenotypes between 52
Salmonella profile I integron-positive and -negative
strains.
| | Profile Ⅰ integron
positive strains (n=20) | Profile Ⅰ integron
negative strains (n=32) | |
|---|
| Antibiotic | DRR (%) | DRR (%) | IR (%) | SR (%) | DRR (%) | IR (%) | SR (%) |
P-valuea |
|---|
|
Amoxicillin/Clavulanate | 0.0 | 0.0 | 13.3 | 86.7 | 0.0 | 0.0 | 100.0 | - |
| Ampicillin | 51.9 | 75.0 | 0.0 | 25.0 | 37.5 | 0.0 | 62.5 | <0.05 |
|
Ampicillin/Sulbactam | 19.2 | 40.0 | 35.0 | 25.0 | 6.25 | 28.1 | 65.6 | <0.05 |
| Aztreonam | 13.5 | 15.0 | 0.0 | 85.0 | 12.5 | 0.0 | 88.5 | >0.05 |
|
Chloramphenicol | 34.6 | 70.0 | 0.0 | 30.0 | 12.5 | 3.1 | 84.4 | <0.05 |
| Meropenem | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 100.0 | - |
| Moxifloxacin | 3.8 | 10.0 | 30.0 | 60.0 | 0.0 | 9.4 | 90.6 | <0.05 |
| Piperacillin | 48.1 | 65.0 | 10.0 | 25.0 | 37.5 | 3.1 | 59.4 | <0.05 |
|
Piperacillin/Tazobactam | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 100.0 | - |
| Cefepime | 14.0 | 20.0 | 0.0 | 80.0 | 10.7 | 0.0 | 89.3 | <0.05 |
| Cefotaxime | 17.3 | 25.0 | 0.0 | 75.0 | 12.5 | 0.0 | 87.5 | <0.05 |
| Imipenem | 0.0 | 0.0 | 5.0 | 95.0 | 0.0 | 12.5 | 87.5 | - |
Drug resistance gene cassette
distribution of class I integron-positive strains
A total of three class I integrin-positive strains
and one class I integron-negative strain (four strains in total)
were sent for genome sequencing (Fig.
3C), where 12 drug resistance gene cassettes were detected
(Table VI; Fig. 4). The actual drug resistance genes
of the successfully sequenced strains and the variable regions in
the gene cassette are summarized in Fig. 4. The integrated substructure is
presented in Fig. 4. The drug
resistance rates of Salmonella class I integrin-positive
strains against ampicillin, ampicillin/sulbactam, chloramphenicol,
moxifloxacin, piperacillin, cefepime and cefotaxime were
significantly higher than those of the negative strains
(P<0.05), indicating that the mechanism underlying the
acquisition of drug resistance in Salmonella was closely
related with the presence of class I integrons (Table V). However, the resistance rates of
class I integron-negative Salmonella strains to aztreonam,
moxifloxacin were similar to class I integrin-positive strains
(Table V), indicating that
multidrug resistance among Salmonella strains is not only
associated with class I integrons but also with other drug
resistance mechanisms which needs further investigation.
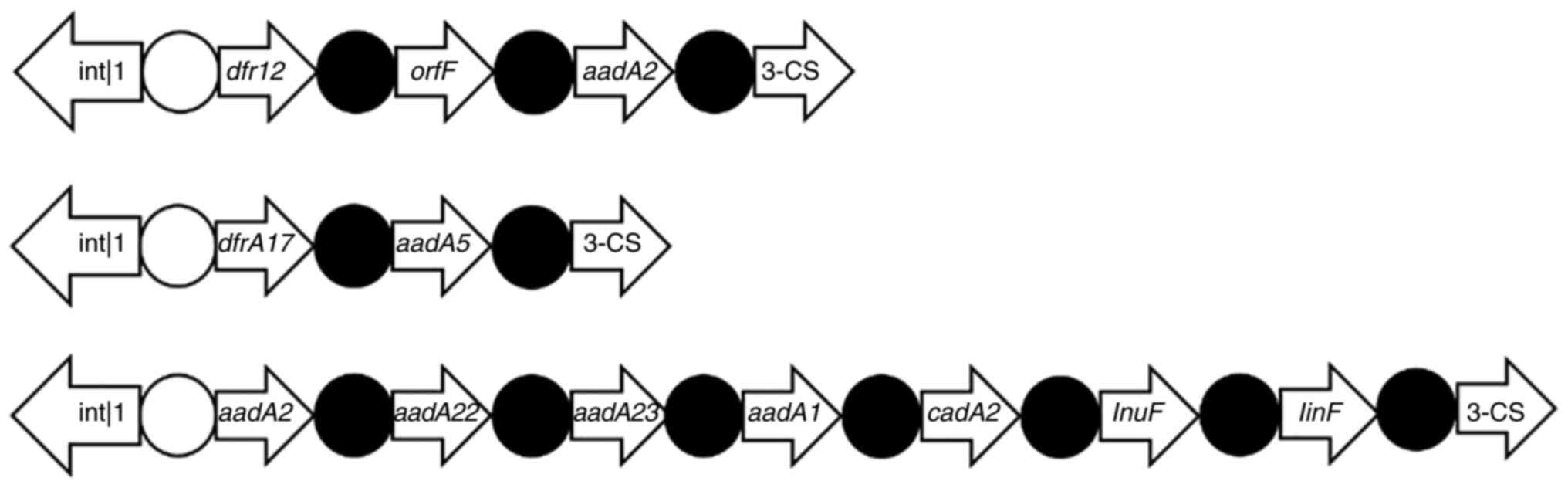 | Figure 4Integrated substructures of the
integrons in the present study. Among the 52 Salmonella
strains, 20 harbored class I integrases, where the detection rate
was 38.46% (20/52), but none of the 52 strains harbored class II or
class III integrons. White circle, integration site attI; black
circle, integration site attC. CS, conserved sequences; Dfr,
dihydrofolate reductase; Orf, open reading frame; aad,
aminoglycoside-adenylyltransferase; int, integrase; cadA2,
cadmium-translocating P-type ATPase 2; InuF, lincosamide. |
 | Table VIComparison between drug resistance
phenotypes and variable region gene boxes. |
Table VI
Comparison between drug resistance
phenotypes and variable region gene boxes.
| Sample number | Resistance
phenotypea | Size of the
variable area (bp) | Combination of the
variable region gene box |
|---|
| 19 | b, d, e, f | 2800 | dfrA12, orfF and
aadA2 |
| 35 | b, d, e, h | 2600 | dfrA17 and
aadA5 |
| 41 | b, d, e, g, h | 3000 | aadA2, aadA22,
aadA23 aadA1, cadA2 and lnuF, linF |
Discussion
Salmonella is a highly versatile pathogen
that can infect a wide range of hosts and cause different clinical
manifestations (22). To date,
>2,600 Salmonella serotypes have been reported worldwide,
of which 292 different serotypes belonging to 35 different somatic
(O) groups have been reported in China (23). Salmonella infections in
children with diarrhea in Guangdong have been reported to be mainly
caused by S. Typhimurium, S. Enteritis and S.
Stanley, which belong to five separate O groups (24). However, human infections caused by
non-typhoid Salmonella are becoming a global public
sanitation problem, leading to ~93.8 million cases of
gastroenteritis and 155,000 deaths every year (25).
In recent years, non-typhoid Salmonella is
emerging as one of the main pathogens in infants in China, causing
diarrhea, fever and abdominal pain (26-29).
Children have immature immune systems and weak gastrointestinal
systems that are particularly susceptible to Salmonella
(30). In the present study, all
Salmonella strains were collected from the fecal specimens
of children with diarrhea. S. Typhimurium was found
to be the most common serotype in the present study, with a
detection rate of 63.46% (33/52), followed by S.
Enteritis and S. Stanley, with detection rates of
9.62 and 7.69%, respectively.
Quinolones and third-generation cephalosporins are
currently the first-line therapeutic options for the treatment of
non-typhoid Salmonella infections (31). Due to the limitations of drug
administration to children, quinolones and aminoglycosides are
restricted in use (32).
Therefore, third-generation cephalosporin is the priority method
for the treatment of children with Salmonella infections
(33). In the present study, drug
sensitivity results showed that the resistance rate of non-typhoid
Salmonella strains was as high as 51.9% to ampicillin,
>48.1% to piperacillin and certainly >17.3% to cefotaxime.
These resistance rates are similar to those reported by a previous
study (34). The isolated
Salmonella strains had drug resistance to 3rd- or
4th-generation cephalosporins. However, amoxicillin/clavulanic
acid-, piperacillin/tazobactam-, imipenem- and meropenem-resistant
strains could not be detected in the present study. The treatment
of infectious diarrhea should be combined with drug sensitivity
testing to avoid the risk of multiple drug resistance strains
arising due to the overuse of clinical antibiotics.
Integrons are important genetic factors in the
capture, integration and expression of drug resistance genes and
are particularly abundant in Gram-negative bacteria (35,36).
Integrons can carry ≥ one drug resistance gene cassettes, which is
an important mechanism of the horizontal transmission of drug
resistance genes (37-39).
In the present study, class I, II and III integrons and variable
regions were sequenced and analyzed in the 52 non-typhoid
Salmonella strains. In total, 20 strains harbored class I
integrons with a positive rate of 38.46%, compared with the 57.0%
reported by a previous study (40). The prevalence of integrons found in
Salmonella varies from country to country and depends on the
origin of the isolates (41).
There have been several reports associating the prevalence of Class
I integrons in Salmonella isolates from different places in
China. Lu et al (42)
reported that 66.5% class I integrons in Salmonella enterica
serovar Indiana (87.2%) and Enteritidis (50.8%) were
isolated from chicken samples in Eastern China. In addition, class
I integrons were detected in 26.9% of the broiler chicken in
Shandong, China (43), whilst
34.7% class I integron-positive Salmonella were isolated in
duck farms and in a slaughterhouse in Shandong province, China
(44). However, another previous
study reported only 16.9% positivity in terms of class I integrons
in Salmonella isolated from farm animals in Shandong
province, China (45). Zhang et
al (46) also reported that
17.4% Salmonella isolated from healthy humans were positive
for class I integrons in Guangdong in China.
In the present study, 12 types of drug resistance
gene cassettes were detected, namely dfrA12, orfF,
aadA2, drfA17, aadA5, aadA2,
aadA22, aadA23, aadA1, cadA2,
InuF and linF. However, Class II and III integrons
could not be detected in the present study. The class I
integron-positive strain antibiotic resistance rate was found to be
significantly higher compared with that of the integron-negative
strains, except for sensitivity to amoxicillin/clavulanic acid,
piperacillin/tazobactam and imipenem, according to the drug
susceptibility analysis. These results suggest that integron
gene-positive strains are associated with significant multidrug
resistance, consistent with previous reports (47,48).
Class I integrons greatly increase the risk of horizontal drug
resistance gene transmission due to their mobile and integration
features (49). The cautious use
of antimicrobial agents is essential for preventing the emergence
and spread of drug-resistant strains of bacteria (50). The integron-positive and
antibiotic-resistant genes found in the Salmonella isolates
in the present study may contribute to the control and therapy of
Salmonella infection. However, further studies are necessary
to determine the significance of class I integron in the
distribution of antibiotic resistance.
MLST is a high-resolution typing technique first
proposed by Maiden et al (12) in 1998, which was developed based on
the technique of multi-site enzyme electrophoresis. Harbottle et
al (51) then used PFGE and
MLST to type 81 strains of Salmonella enteritis, indicating
that MLST was a suitable technique for the typing of the different
Salmonella serotypes. Although MLST has advantages that can
potentially replace and supplement serotyping, the principle of
MLST is different from that of serotype detection. The combination
of MLST and serotype detection can facilitate research on the
hereditary and evolutionary relationships of Salmonella. The
most common Salmonella sequence types were found to be ST19
and ST34 in the present study, where the corresponding serotype was
S. Typhimurium. This is consistent with findings from a
previous report, where the most prevalent Salmonella
subtypes found were ST34 and ST19 in the Guangdong province in
2007-2011(52). Observations from
the present study therefore provided important evidence and
confirmed further that these two types of Salmonella can
serve an important role in pediatric diarrhea in Guangdong, China.
Since the drug resistance characteristics of predominant
Salmonella ST and genetic drift between various
Salmonella ST could guide the clinical antibiotic treatment
of Salmonella infection and epidemics, the present study may
lay a foundation for a therapeutic strategy for pediatric diarrhea
caused by Salmonella infection in the future.
The characteristics of Salmonella infection
differ depending on the region. S. paratyphoid A
tended to dominate in Yunnan from 1995 to 2013, where the dominant
sequence types were ST85 and ST129(53). By contrast, the prevalent sequence
types were found to be ST11 and ST34 in Nanjing in
2014-2015(54). In the present
study, ST34 corresponded to S. Typhimurium and
S. Togo, while ST29 corresponded to S.
Stanley and S. Typhimurium. In addition,
although ST34 and ST29 were two different sequence types, they both
belong to the serotype group B (13), suggesting that these strains may
have been subjected to convergent evolution and are variations of
the same strain. According to eBURST cluster analysis, ST19 and
ST34 were highly associated, with only one pair of housekeeping
genes that were different. Among the housekeeping genes,
thrA comprised the largest number of alleles in the seven
groups of the ST types of 52 Salmonella strains. By
contrast, the least common was sucA, suggesting that
sucA was the most stable.
The present study suggests that multi-drug
resistance is closely associated with the presence of class I
integrons in isolated Salmonella strain. the isolated
Salmonella strains are particular resistant to ampicillin
(51.9%), chloramphenicol (34.6%) and piperacillin (48.1%). This is
consistent with results from a previous study conducted in
Guangdong, China (52). This
finding may facilitate the design of Salmonella antibiotics
for clinical practice. In particular, since class I integrons
appear to serve an important role in the acquisition of drug
resistance, they warrant immediate attention. Furthermore, the
present data show that MLST can be used for clinical
Salmonella genotyping. Due to the rapid emergence of
multi-drug resistance in Salmonella, further studies are
required to investigate the mechanism underlying bacterial drug
resistance, strictly monitor susceptible factors, control bacterial
drug resistance, strengthen disinfection and isolation methods in
clinical practice.
However, the present study remains associated with
a number of limitations. The number of cases examined in the
present study is small. In addition, the characterization of
multi-drug resistance was not performed, where the mechanism of
antibiotic resistance, virulence and transfer of resistance genes
were evaluated in the present study. The sensitivity and
specificity of the results of genotyping analysis of
Salmonella MLST were also not considered.
To conclude, Salmonella infection in
children with diarrhea was mainly caused by S.
Typhimurium, where the most prominent sequence types were
ST34 and ST19. The class I integrons found were closely associated
with Salmonella drug resistance. Deepening the research into
integrons is expected to serve an important role in understanding
the occurrence and transmission mechanism of Salmonella drug
resistance. Particular attention should be given to the 3rd- and
4th-generation cephalosporin resistance of Salmonella.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
Natural Science Foundation of China (grant no. 31770183) and the
Medical Science Technology Research Foundation of Guangdong (grant
no. A2015226), Guangdong Provincial Bureau of Traditional Chinese
Medicine (grant no. 20201407) and Qingyuan People's Hospital
Medical Scientific Research Fund Project (grant no. 20190209).
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
LX and QH performed the molecular genetic studies
and participated in the sequence alignment. YT and WW performed
species identification. LC and YL performed the antibiotics
susceptibility tests. YT and WW performed the PCR. CY and BF
conceived the study and participated in its design and
coordination. LX and BF confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical
Ethics Committee of Qingyuan People's Hospital (Qingyuan, China).
Written informed consent was obtained from each participant's legal
guardian.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
El-Tayeb MA, Ibrahim ASS, Al-Salamah AA,
Almaary KS and Elbadawi YB: Prevalence, serotyping and
antimicrobials resistance mechanism of Salmonella enterica isolated
from clinical and environmental samples in Saudi Arabia. Braz J
Microbiol. 48:499–508. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen HM, Wang Y, Su LH and Chiu CH:
Nontyphoid salmonella infection: Microbiology, clinical features,
and antimicrobial therapy. Pediatr Neonatol. 54:147–152.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jajere SM: A review of Salmonella enterica
with particular focus on the pathogenicity and virulence factors,
host specificity and antimicrobial resistance including multidrug
resistance. Vet World. 12:504–521. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jackson BR, Griffin PM, Cole D, Walsh KA
and Chai SJ: Outbreak-associated Salmonella enterica serotypes and
food Commodities, United States, 1998-2008. Emerg Infect Dis.
19:1239–1244. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Guo L and Zhao Y: Global spread and
molecular characterization of CTX-M-producing salmonella
typhimurium isolates. Antibiotics (Basel). 10(1417)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Darini AL, Magalhaes VD, Levy CL, Barth AL
and Coscina AL: Phenotyping and genotyping methods applied to
investigate the relatedness of Brazilian isolates of Enterobacter
cloacae. Braz J Med Biol Res. 32:1077–1081. 1999.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yan S, Zhang W, Li C, Liu X, Zhu L, Chen L
and Yang B: Serotyping, MLST, and Core Genome MLST Analysis of
salmonella enterica from different sources in China During
2004-2019. Front Microbiol. 12(688614)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ferrari RG, Panzenhagen PHN and
Conte-Junior CA: Phenotypic and genotypic eligible methods for
salmonella typhimurium source tracking. Front Microbiol.
8(2587)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Edwards PR and Kauffmann F: A
simplification of the Kauffmann-White schema. Am J Clin Pathol.
22:692–697. 1952.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ziebell K, Chui L, King R, Johnson S,
Boerlin P and Johnson RP: Subtyping of Canadian isolates of
salmonella enteritidis using multiple locus variable number tandem
repeat analysis (MLVA) alone and in combination with pulsed-field
gel electrophoresis (PFGE) and phage typing. J Microbiol Meth.
139:29–36. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kotetishvili M, Stine OC, Kreger A, Morris
JG Jr and Sulakvelidze A: Multilocus sequence typing for
characterization of clinical and environmental salmonella strains.
J Clin Microbiol. 40:1626–1635. 2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Maiden MC, Bygraves JA, Feil E, Morelli G,
Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, et al:
Multilocus sequence typing: A portable approach to the
identification of clones within populations of pathogenic
microorganisms. Proc Natl Acad Sci USA. 95:3140–3145.
1998.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Achtman M, Wain J, Weill FX, Nair S, Zhou
Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, et al:
Multilocus sequence typing as a replacement for serotyping in
Salmonella enterica. PLoS Pathog. 8(e1002776)2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kidgell C, Reichard U, Wain J, Linz B,
Torpdahl M, Dougan G and Achtman M: Salmonella typhi, the causative
agent of typhoid fever, is approximately 50,000 years old. Infect
Genet Evol. 2:39–45. 2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gugala N, Vu D, Parkins MD and Turner RJ:
Specificity in the susceptibilities of escherichia coli,
pseudomonas aeruginosa and staphylococcus aureus clinical isolates
to six metal antimicrobials. Antibiotics (Basel).
8(51)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Firoozeh F, Mahluji Z, Khorshidi A and
Zibaei M: Molecular characterization of class 1, 2 and 3 integrons
in clinical multi-drug resistant Klebsiella pneumoniae isolates.
Antimicrob Resist Infect Control. 8(59)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cui M, Zhang J, Gu Z, Li R, Chan EW, Yan
M, Wu C, Xu X and Chen S: Prevalence and molecular characterization
of mcr-1-positive Salmonella strains recovered from clinical
Specimens in China. Antimicrob Agents Chemother. 61:e02471–16.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bell RL, Gonzalez-Escalona N, Stones R and
Brown EW: Phylogenetic evaluation of the ‘Typhimurium’ complex of
Salmonella strains using a seven-gene multi-locus sequence
analysis. Infect Genet Evol. 11:83–91. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Machado E, Canton R, Baquero F, Galán JC,
Rollán A, Peixe L and Coque TM: Integron content of
extended-spectrum-beta-lactamase-producing Escherichia coli strains
over 12 years in a single hospital in Madrid, Spain. Antimicrob
Agents Chemother. 49:1823–1829. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Feil EJ, Li BC, Aanensen DM, Hanage WP and
Spratt BG: eBURST: Inferring patterns of evolutionary descent among
clusters of related bacterial genotypes from multilocus sequence
typing data. J Bacteriol. 186:1518–1530. 2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Spratt BG, Hanage WP, Li B, Aanensen DM
and Feil EJ: Displaying the relatedness among isolates of bacterial
species-the eBURST approach. FEMS Microbiol Lett. 241:129–134.
2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Uzzau S, Brown DJ, Wallis T, Rubino S,
Leori G, Bernard S, Casadesús J, Platt DJ and Olsen JE: Host
adapted serotypes of Salmonella enterica. Epidemiol Infect.
125:229–255. 2000.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kuang X, Hao H, Dai M, Wang Y, Ahmad I,
Liu Z and Zonghui Y: Serotypes and antimicrobial susceptibility of
Salmonella spp. isolated from farm animals in China. Front
Microbiol. 6(602)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li WW, Bai L, Zhang XL, Qiao X, Yang XR,
Chen YZ, Pei XY, Wu YN and Guo YC: Prevalence and antibiogram
distribution of Salmonella isolated from broiler production and
processing course in four provinces, China. Zhonghua Yu Fang Yi Xue
Za Zhi. 47:435–438. 2013.PubMed/NCBI(In Chinese).
|
|
25
|
Majowicz SE, Musto J, Scallan E, Angulo
FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A and Hoekstra RM:
International Collaboration on Enteric Disease ‘Burden of Illness’
Studies. The global burden of nontyphoidal Salmonella
gastroenteritis. Clin Infect Dis. 50:882–889. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu LJ, Luo Y, Shi GL and Li ZY:
Prevalence, clinical characteristics and changes of antibiotic
resistance in children with nontyphoidal Salmonella infections from
2009-2018 in Chongqing, China. Infect Drug Resist. 14:1403–1413.
2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Who PY, Yeung MPS, Nelson EAS and Goggins
WBI: Risk factors of non-typhoidal Salmonella gastroenteritis in
hospitalised young children: A case-control study. BMJ Paediatr
Open. 5(e000898)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ugboko HU, Nwinyi OC, Oranusi SU and
Oyewale JO: Childhood diarrhoeal diseases in developing countries.
Heliyon. 6(e03690)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gong B, Li H, Feng Y, Zeng S, Zhuo Z, Luo
J, Chen X and Li X: Prevalence, serotype distribution and
antimicrobial resistance of non-typhoidal Salmonella in
hospitalized patients in Conghua District of Guangzhou, China.
Front Cell Infect Microbiol. 12(805384)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Burns-Guydish SM, Olomu IN, Zhao H, Wong
RJ, Stevenson DK and Contag CH: Monitoring age-related
susceptibility of young mice to oral Salmonella enterica serovar
Typhimurium infection using an in vivo murine model. Pediatr Res.
58:153–158. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Angarone M and Snydman DR: AST ID
Community of Practice. Diagnosis and management of diarrhea in
solid-organ transplant recipients: Guidelines from the American
Society of Transplantation Infectious Diseases Community of
Practice. Clin Transplant. 33(e13550)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Patel K and Goldman JL: Safety concerns
surrounding quinolone use in children. J Clin Pharmacol.
56:1060–1075. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Al Kraiem AA, Yang G, Al Kraiem F and Chen
T: Challenges associated with ceftriaxone resistance in Salmonella.
Front Life Sci. 11:26–34. 2018.
|
|
34
|
Song Q, Xu Z, Gao H and Zhang D: Overview
of the development of quinolone resistance in Salmonella species in
China, 2005-2016. Infect Drug Resist. 11:267–274. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hall RM, Brookes DE and Stokes HW:
Site-specific insertion of genes into integrons: Role of the
59-base element and determination of the recombination cross-over
point. Mol Microbiol. 5:1941–1959. 1991.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Stokes HW and Hall RM: A novel family of
potentially mobile DNA elements encoding site-specific
gene-integration functions: Integrons. Mol Microbiol. 3:1669–1683.
1989.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cornaglia G, Giamarellou H and Rossolini
GM: Metallo-β-lactamases: A last frontier for β-lactams? Lancet
Infect Dis. 11:381–393. 2011.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fluit AC and Schmitz FJ: Resistance
integrons and super-integrons. Clin Microbiol Infect. 10:272–288.
2004.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu CC, Tang CY, Chang KC, Kuo HY and Liou
ML: A comparative study of class 1 integrons in Acinetobacter
baumannii. Gene. 544:75–82. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wu K, Wang F, Sun J, Wang Q, Chen Q, Yu S
and Rui Y: Class 1 integron gene cassettes in multidrug-resistant
Gram-negative bacteria in southern China. Int J Antimicrob Agents.
40:264–267. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Vo AT, van Duijkeren E, Gaastra W and
Fluit AC: Antimicrobial resistance, class 1 integrons, and genomic
island 1 in Salmonella isolates from Vietnam. PLoS One.
5(e9440)2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lu Y, Zhao H, Sun J, Liu Y, Zhou X, Beier
RC, Wu G and Hou X: Characterization of multidrug-resistant
Salmonella enterica serovars Indiana and Enteritidis from chickens
in Eastern China. PLoS One. 9(e96050)2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhao X, Hu M, Zhang Q, Zhao C, Zhang Y, Li
L, Qi J, Luo Y, Zhou D and Liu Y: Characterization of integrons and
antimicrobial resistance in Salmonella from broilers in Shandong,
China. Poult Sci. 99:7046–7054. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yang J, Ju Z, Yang Y, Zhao X, Jiang Z and
Sun S: Serotype, antimicrobial susceptibility and genotype profiles
of Salmonella isolated from duck farms and a slaughterhouse in
Shandong province, China. BMC Microbiol. 19(202)2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhao X, Yang J, Zhang B, Sun S and Chang
W: Characterization of integrons and resistance genes in Salmonella
isolates from farm animals in Shandong province, China. Front
Microbiol. 8(1300)2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang H, Shi L, Li L, Guo S, Zhang X,
Yamasaki S, Miyoshi S and Shinoda S: Identification and
characterization of class 1 integron resistance gene cassettes
among Salmonella strains isolated from healthy humans in China.
Microbiol Immunol. 48:639–645. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Bagheri-Nesami M, Rezai MS, Ahangarkani F,
Rafiei A, Nikkhah A, Eslami G, Shafahi K, Hajalibeig A and Khajavi
R: Multidrug and co-resistance patterns of non-fermenting
Gram-negative bacilli involved in ventilator-associated pneumonia
carrying class 1 integron in the North of Iran. Germs. 7:123–131.
2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gu B, Tong M and Zhao W, Liu G, Ning M,
Pan S and Zhao W: Prevalence and characterization of class I
integrons among Pseudomonas aeruginosa and Acinetobacter baumannii
isolates from patients in Nanjing, China. J Clin Microbiol.
45:241–243. 2007.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Deng Y, Bao XR, Ji L, Chen L, Liu J, Miao
J, Chen D, Bian H, Li Y and Yu G: Resistance integrons: Class 1, 2
and 3 integrons. Ann Clin Microbiol Antimicrob.
14(45)2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Prestinaci F, Pezzotti P and Pantosti A:
Antimicrobial resistance: A global multifaceted phenomenon. Pathog
Glob Health. 109:309–318. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Harbottle H, White DG, McDermott PF,
Walker RD and Zhao S: Comparison of multilocus sequence typing,
pulsed-field gel electrophoresis, and antimicrobial susceptibility
typing for characterization of Salmonella enterica serotype Newport
isolates. J Clin Microbiol. 44:2449–2457. 2006.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sun J, Ke B, Huang Y, He D, Li X, Liang Z
and Ke C: The molecular epidemiological characteristics and genetic
diversity of salmonella typhimurium in Guangdong, China, 2007-2011.
PLoS One. 9(e113145)2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Gu W, Yang Z, Chen Y, Yin J, Yang J, Li C,
Zhou Y, Yin J, Xu W, Zhao S, et al: Molecular characteristics of
Salmonella enterica Paratyphi A in Yunnan Province, southwest
China. Infect Genet Evol. 30:181–185. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Xu F, Wang T, Tan H, Li M, Jin Y and Guo
H: The multilocus sequence typing and drug resistance
characteristics of Salmonella in children with diarrhea. J Clin
Pediatr. 36:520–523. 2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|