Introduction
Hepatocellular carcinoma (HCC) is a common type of
cancer in clinical practice, which poses a great threat to human
health (1). According to global
cancer data, it is estimated that in 2020 there were 905,677 new
diagnoses of liver cancer and 830,180 deaths from liver cancer
(2). Rapid invasion and metastasis
are some of the common characteristics of HCC that make it
difficult to treat (3). In
addition, in most cases, patients are diagnosed at later stages of
tumor progression owing to the absence of specific indicators at
the early diagnostic stage (4).
Hence, it seems imperative to elucidate the molecular mechanisms
underlying HCC progression, as well as to analyze the roles of less
investigated factors that are involved in this process.
It has been reported that the cytoskeleton serves
essential roles in malignant tumor invasion and metastasis
(5). As the main component of the
cytoskeleton, microtubule kinesin can act in intracellular
transport, cell division and bipolar spindle formation (6). In addition, recent research has
confirmed that microtubule kinesin is closely associated with tumor
progression and development (7).
The kinesin-13 family includes kinesin family member 2 (KIF2)A,
KIF2B and KIF2C. Through depolymerization of tubulin, kinesin-13
can participate in spindle assembly, regulate dynamic changes in
the cytoskeleton and, thus, promote cell proliferation and
migration (8,9). Recent studies have suggested that
KIF2A is a crucial factor in the invasion and metastasis of various
tumors, such as gastric cancer, papillary thyroid carcinoma and
non-small-cell lung cancer (10-12).
Additionally, it has been verified that KIF2A is upregulated in HCC
tissues and is associated with biomarkers for tumor metastasis and
shorter relapse-free survival times (13,14).
However, the potential impact of KIF2A on the malignant progression
of HCC has not been fully understood.
Notch is a transmembrane receptor that is part of
the evolutionarily highly conserved Notch signaling cascade; it
serves a pivotal role in regulating a number of fundamental
cellular processes (15). The
Notch receptor family contains four homologous proteins known as
Notch1-4(16). Previous studies
have reported that aberrant activation of Notch signaling has been
causally linked to HCC progression (17-19).
The members of the Notch receptor family can participate in the
development of HCC by regulating tumor microenvironment,
tumorigenesis, angiogenesis, invasion, metastasis and
epithelial-mesenchymal transition (17-19).
The present study was conducted to identify the
biological functions of KIF2A in HCC and to investigate the
molecular mechanisms underlying the involvement of KIF2A in the
malignant progression of HCC.
Materials and methods
Cell culture
HHL-5 normal human hepatocytes, and Li-7 and Huh7
HCC cell lines were obtained from the Cell Bank of the Chinese
Academy of Sciences. The MHCC97 HCC cell line was obtained from
Procell Life Science & Technology Co., Ltd.. The immortalized
hybrid HUVEC/EAhy926 cells were obtained from American Type Culture
Collection. All of the cells were cultured in Dulbecco's Modified
Eagle's Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin
at 37˚C in a 5% CO2 incubator.
Cell transfection
Small interfering RNA (siRNA) targeting KIF2A
(siRNA-KIF2A, cat. no. A10001; siRNA-KIF2A-1,
5'-AAACAAAGACAGCAGUUAUAU-3'; siRNA-KIF2A-2,
5'-AAACAAAGAGAAUGUAAUAAA-3') and the scrambled negative control
(siRNA-NC, 5'-UUCUCCGAACGUGUCACGU-3') were constructed by Shanghai
GenePharma Co., Ltd. The Notch1 overexpression plasmid (Ov-Notch1)
was established by inserting the Notch1 gene into the pcDNA3.1
vector (Shanghai GenePharma Co., Ltd.), whereas an empty vector
served as the NC (Ov-NC). These vectors were transfected into Huh7
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) strictly as instructed by the
manufacturer's guidelines. Briefly, siRNAs (3 µl) or pcDNA3.1
vectors (4 µg) and Lipofectamine 2000 reagent (10 µl) were added to
Opti-MEM (250 µl; Gibco; Thermo Fisher Scientific, Inc.) and
incubated for 5 min at room temperature. Subsequently, diluted
siRNAs or pcDNA3.1 vectors were mixed with diluted Lipofectamine
2000 and then incubated for 20 min at room temperature. HCC cells
were then re-plated in serum-free DMEM, the transfection mixtures
were separately added to the cells when the cell confluence reached
80-85%, and the cells were cultured for 4 h at 37˚C. Finally, the
medium was replaced with complete DMEM and cells were cultured at
37˚C for 48 h before further experiments.
Bioinformatics analysis
Cancer Cell Line Encyclopedia (CCLE) is a
compilation of gene expression, chromosomal copy number and
massively parallel sequencing data from ~1,000 human cancer cell
lines (20). KIF2A mRNA expression
in liver cancer cell lines was analyzed using the Broad Institute
Cancer Cell Line Encyclopedia database (https://portals.broadinstitute.org/ccle). Raw
sequencing data used for CCLE database analysis are available
through the Sequence Read Archive under accession number
PRJNA523380. The interaction between KIF2A and Notch1 was
predicated by BioGRID database (https://thebiogrid.org/).
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was used to measure cell viability.
The transfected or untransfected Huh7 cells were incubated in
96-well plates at a density of 5x103 cells/well for 24,
48 or 72 h at 37˚C. Subsequently, 10 µl CCK-8 reagent (Beyotime
Institute of Biotechnology) was added into each well and incubated
at 37˚C for an additional 4 h. The optical density was measured at
450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
5-ethynyl-2'-deoxyuridine (EdU)
staining
Cell proliferation was evaluated using the EdU kit
(Beijing Solarbio Science & Technology Co., Ltd.). EdU reagent
(10 µmol/l) was added to the Huh7 cells at a density of
5x103 cells/well in 96-well plates according to the
instructions of the EdU fluorescence staining cell proliferation
kit and was then incubated at 37˚C for 2 h. Subsequently, cells
were fixed in 4% paraformaldehyde for 15 min at room temperature
and then permeabilized in 0.3% Triton X-100 for 10 min at room
temperature. The cells were subsequently washed with PBS, incubated
with the click reaction solution in the dark for 30 min at room
temperature and stained with DAPI (Beyotime Institute of
Biotechnology) in the dark for 10 min at room temperature. The
EdU-positive cells in five randomly selected fields were observed
using a fluorescence microscope (magnification, x200; Olympus
Corporation) and were quantified using ImageJ 1.51 software
(National Institutes of Health).
Cell migration assay
Cell migratory ability was assessed by wound healing
assay. Briefly, Huh7 cells were seeded on 6-well plates and grown
to 90% confluence. A wound was created by scratching the monolayer
of cells with a 200-µl pipette tip, and the detached cells were
washed twice with PBS. Subsequently, cells were cultured in fresh
serum-free DMEM for 24 h. Images of the wounds were captured at 0
and 24 h under a light microscope (magnification, x100; Leica
Microsystems GmbH). The distance of cell migration was quantified
using the following equation: Migration (%)=[(0 h average scratch
distance-24 h average scratch distance)/0 h average scratch
distance] x100.
Cell invasion assay
Cell invasive ability was assessed by Transwell
invasion assay using Transwell chambers (Corning, Inc.). Huh7 cells
were suspended in fresh serum-free DMEM. Subsequently, a total of
5x104 cells/ml were seeded into the upper chamber of
Transwell plates precoated with Matrigel (BD Biosciences) at 37˚C
for 30 min, and 600 µl DMEM containing 10% FBS was applied as a
chemoattractant in the lower chamber. After 24 h of
incubation at 37˚C, non-invasive cells were gently removed using
cotton swabs. The invasive cells in the lower chamber were fixed
with 4% paraformaldehyde at room temperature for 15 min and stained
with 0.1% crystal violet (Beijing Solarbio Science & Technology
Co., Ltd.) at room temperature for 10 min. Finally, images of the
stained cells were captured and counted in five randomly selected
fields under a light microscope (magnification, x200; Leica
Microsystems GmbH).
Tube formation assay
Briefly, the conditioned media (CM) of untransfected
Huh7 cells, Huh7 cells transfected with siRNA-NC, Huh7 cells
transfected with siRNA-KIF2A-1, Huh7 cells co-transfected with
siRNA-KIF2A-1 + Ov-NC and Huh7 cells co-transfected with
siRNA-KIF2A-1 + Ov-Notch1 were collected ~24 h post-incubation at
37˚C. HUVECs at a density of 2x104 cells/well were
seeded on 96-well plates precoated with Matrigel (BD Biosciences)
at 37˚C for 30 min and then incubated with 250 µl CM at 37˚C for 24
h. Tube formation was observed under a light microscope
(magnification, x40; Leica Microsystems GmbH).
Western blotting
Total proteins were extracted from HHL-5, Li-7, Huh7
and MHCC97 cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology) and BCA Protein Assay Kit (Beyotime Institute of
Biotechnology) was used to determine protein concentrations. Equal
amounts of protein samples (30 µg) were separated by SDS-PAGE on
5-10% gels and then transferred onto PVDF membranes. Non-specific
binding was blocked with 5% non-fat milk for 1.5 h at room
temperature. Subsequently, the membranes were incubated with
primary antibodies against KIF2A (1:1,000; cat. no. ab197988;
Abcam), Ki67 (1:1,000; cat. no. ab16667; Abcam), proliferating cell
nuclear antigen (PCNA; 1:1,000; cat. no. ab92552; Abcam), MMP2
(1:5,000; cat. no. ab92536; Abcam), MMP7 (1:1,000; cat. no.
ab207299; Abcam), MMP9 (1:10,000; cat. no. ab76003; Abcam),
vascular endothelial growth factor A (VEGFA; 1:1,000; cat. no.
ab46154; Abcam), VEGF receptor 1 (VEGFR1; 1:1,000; cat. no.
ab32152; Abcam), VEGFR2 (1:1,000; cat. no. ab134191; Abcam), Notch1
(1:1,000; cat. no. ab52627; Abcam) and GAPDH (1:2,500; cat. no.
ab9485; Abcam) overnight at 4˚C. Following incubation with primary
antibodies, the membranes were incubated with an HRP-conjugated
goat anti-rabbit secondary antibody (1:50,000; cat. no. ab205718;
Abcam) for 1 h at room temperature. Protein bands were developed
with BeyoECL Plus (Beyotime Institute of Biotechnology). Protein
expression was semi-quantified using ImageJ v1.6 (National
Institutes of Health) with GAPDH as the internal reference.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from HHL-5, Li-7, Huh7 and
MHCC97 cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). A total of 1 µg RNA was reverse
transcribed to cDNA using a PrimeScript 1st strand cDNA Synthesis
Kit (Takara Bio, Inc.) according to the manufacturer's protocol.
Subsequently, qPCR analysis was carried out on an ABI 7500 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR
Premix Ex Taq kit (Takara Bio, Inc.). The qPCR thermocycling
conditions were as follows: Initial denaturation at 95˚C for 10
min; followed by 40 cycles of 95˚C for 15 sec and 64˚C for 30 sec.
The following primer sequences were used for qPCR: KIF2A forward,
5'-CTGCTGCTCCAGATGAGGTG-3' and reverse,
5'-TGCTGGTATACTGTGAACTCGT-3'; Notch1 forward,
5'-GAGGCGTGGCAGACTATGC-3' and reverse, 5'-CTTGTACTCCGTCAGCGTGA-3';
GAPDH forward, 5'-CAGGAGGCATTGCTGATGAT-3' and reverse,
5'-GAAGGCTGGGGCTCATTT-3'. Relative gene expression levels were
calculated using the 2-∆∆Cq method (21) with GAPDH as the internal reference
gene.
Co-immunoprecipitation (Co-IP)
Co-IP was used to analyze the interaction between
KIF2A and Notch1. Briefly, 4x107 Huh7 cells were lysed
using IP lysis buffer (Beyotime Institute of Biotechnology).
Subsequently, anti-KIF2A (5 µg/mg lysate; cat. no. A300-914A;
Invitrogen; Thermo Fisher Scientific, Inc.), anti-Notch1 (5 µg/mg
lysate; cat. no. A301-894A; Invitrogen; Thermo Fisher Scientific,
Inc.) or 1 µg control IgG (cat. no. ab172730; Abcam) were added
into 250 µl cell lysates and incubated overnight at 4˚C.
Subsequently, cell lysates were cultivated with 25 µl protein A/G
agarose beads (Santa Cruz Biotechnology, Inc.) for 2 h at 4˚C. The
solution was centrifuged at 2,500 x g for 4 min at 4˚C. The
precipitated sample was washed and analysis of the immunocomplexes
was carried out through western blot analysis.
Statistical analysis
Data analysis was performed using GraphPad Prism 6
(GraphPad Software, Inc.). All experiments were repeated three
times. Differences among multiple groups were analyzed using
one-way analysis of variance followed by Tukey's post hoc test.
Experimental data are expressed as the mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference.
Results
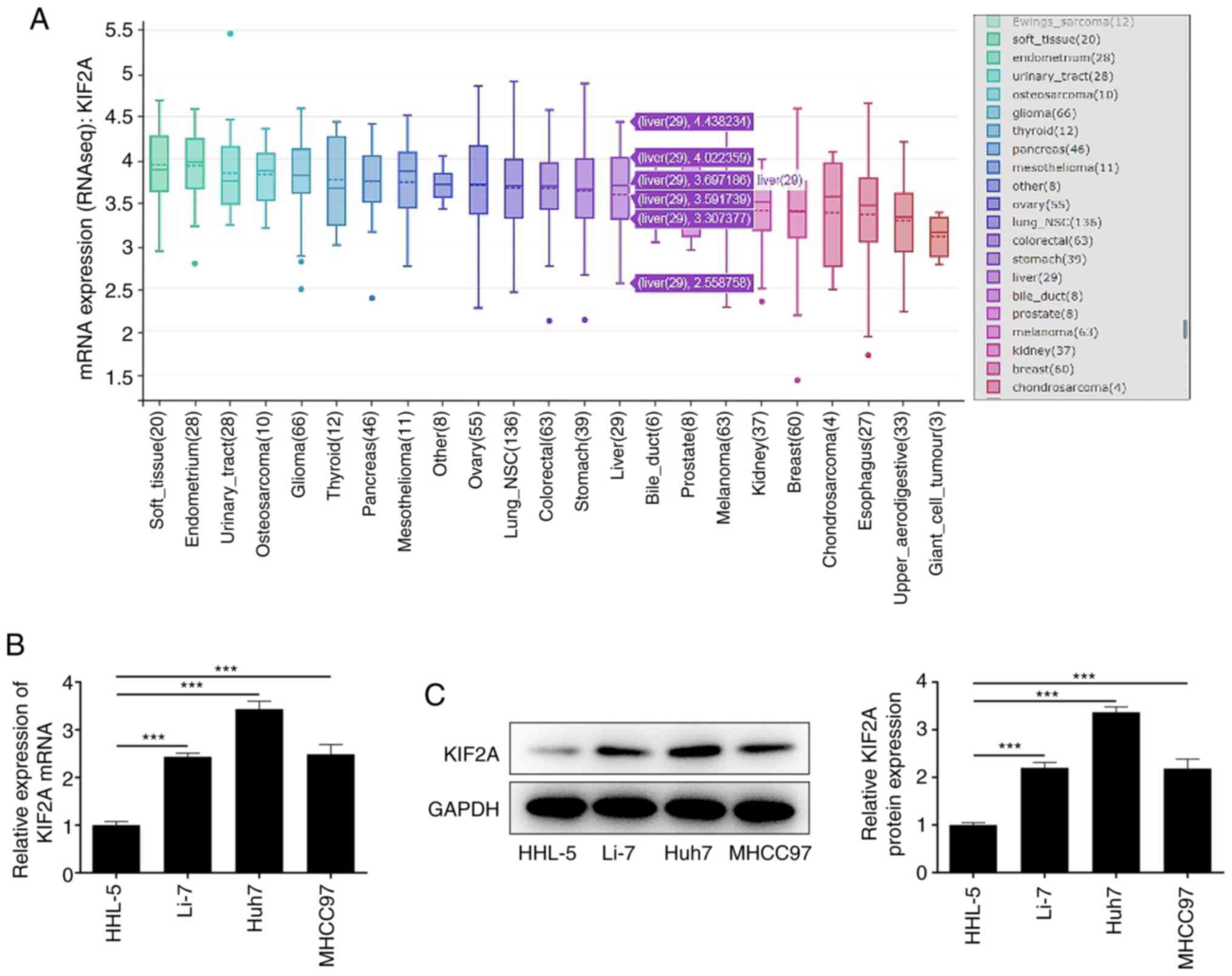
KIF2A is upregulated in HCC cells
The Broad Institute Cancer Cell Line Encyclopedia
database indicated that KIF2A was highly expressed in HCC cells
(Fig. 1A). In addition,
differences in expression levels of KIF2A between HHL-5 human
hepatocytes and HCC cell lines Li-7, Huh7, MHCC97 were examined by
RT-qPCR and western blot analysis. In contrast to those in HHL-5
cells, KIF2A mRNA (Fig. 1B) and
protein expression levels (Fig.
1C) were significantly higher in HCC cells, especially in Huh7
cells. Therefore, Huh7 cells were selected for the follow-up
experiments.
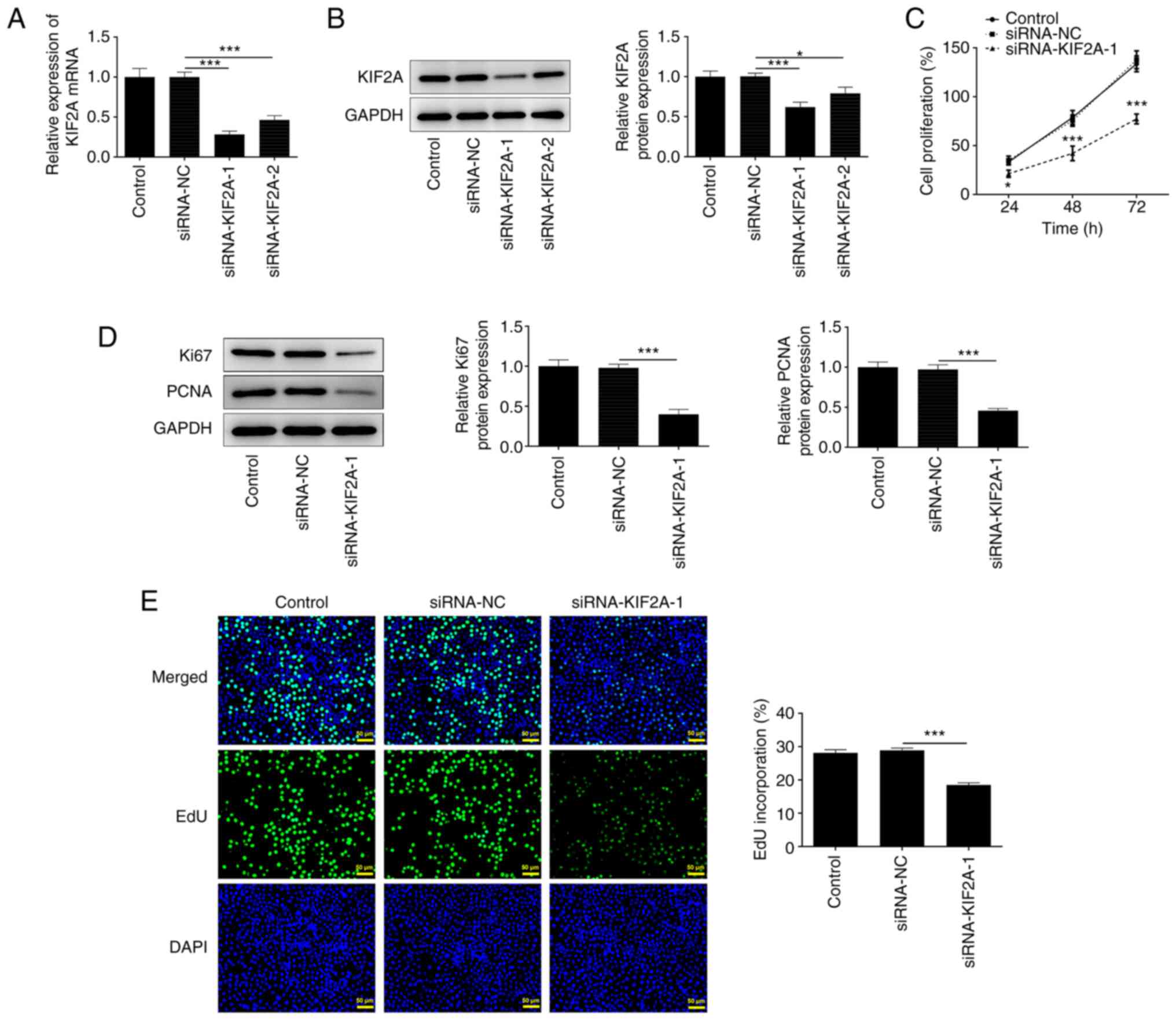
Downregulation of KIF2A suppresses
Huh7 HCC cell proliferation
To examine the impact of KIF2A on HCC progression,
Huh7 cells were transfected with siRNA-KIF2A-1/2 or siRNA-NC.
Transfection efficiency was determined by RT-qPCR (Fig. 2A) and western blot analysis
(Fig. 2B), both of which showed
that KIF2A expression was downregulated following transfection.
siRNA-KIF2A-1 with optimized transfection efficiency was selected
for the functional experiments. Results of the CCK-8 assay
indicated that transfection with siRNA-KIF2A-1 suppressed HCC cell
proliferation (Fig. 2C).
Furthermore, the reduced protein expression levels of Ki67 and PCNA
(Fig. 2D), as well as the
reduction in EdU-positive stained cells (Fig. 2E), in the siRNA-KIF2A-1 group
suggested that KIF2A knockdown could suppress HCC cell
proliferation.
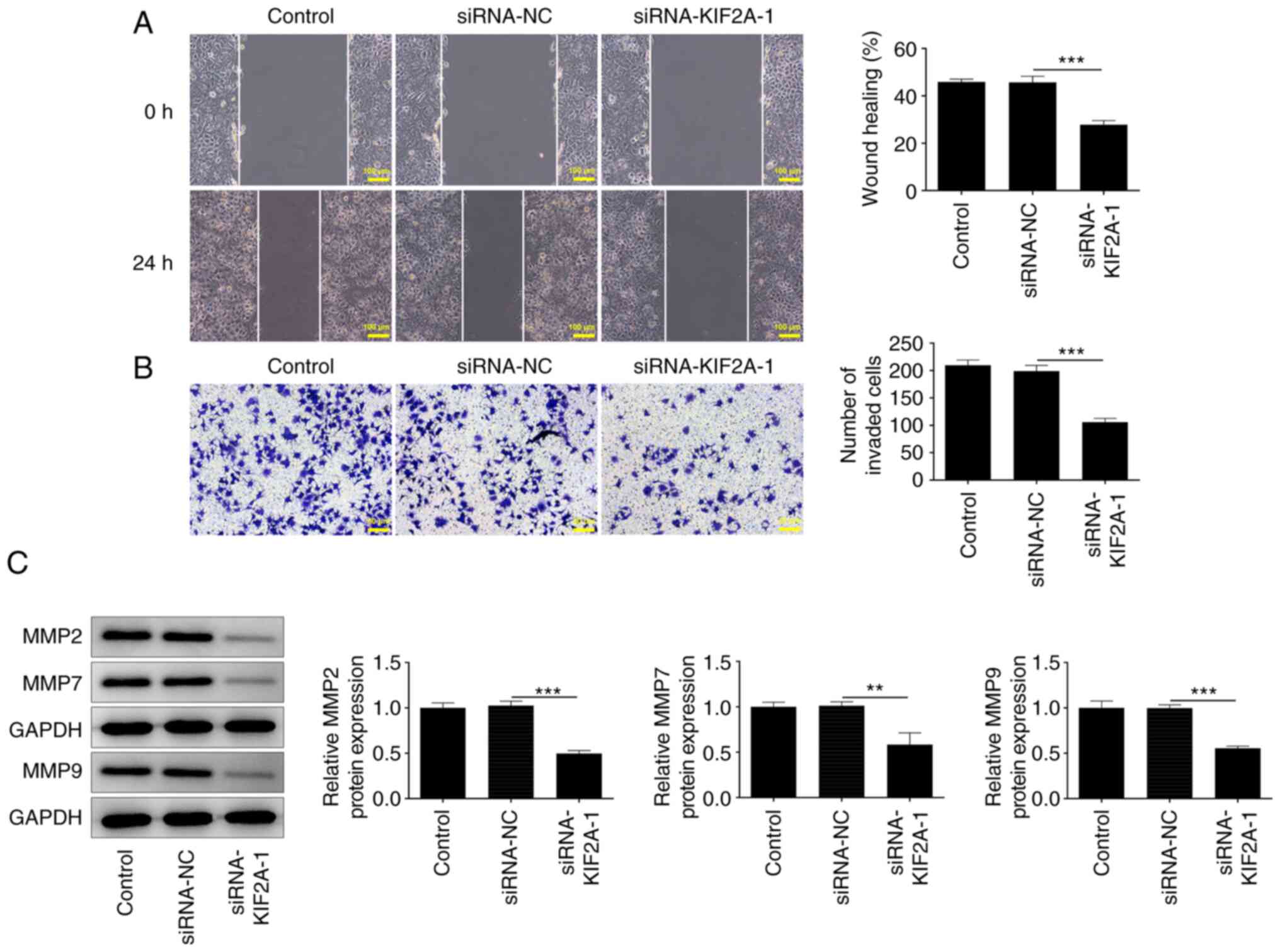
Downregulation of KIF2A inhibits Huh7
HCC cell migration and invasion
Wound healing and Transwell assays were performed to
investigate whether KIF2A was functionally involved in HCC cell
migration and invasion. It was observed that silencing of KIF2A
strongly inhibited migration and invasion in HCC cells (Fig. 3A and B). The decreased expression levels of
MMP2, MMP7 and MMP9 also demonstrated that KIF2A knockdown
suppressed HCC cell migration and invasion in vitro
(Fig. 3C).
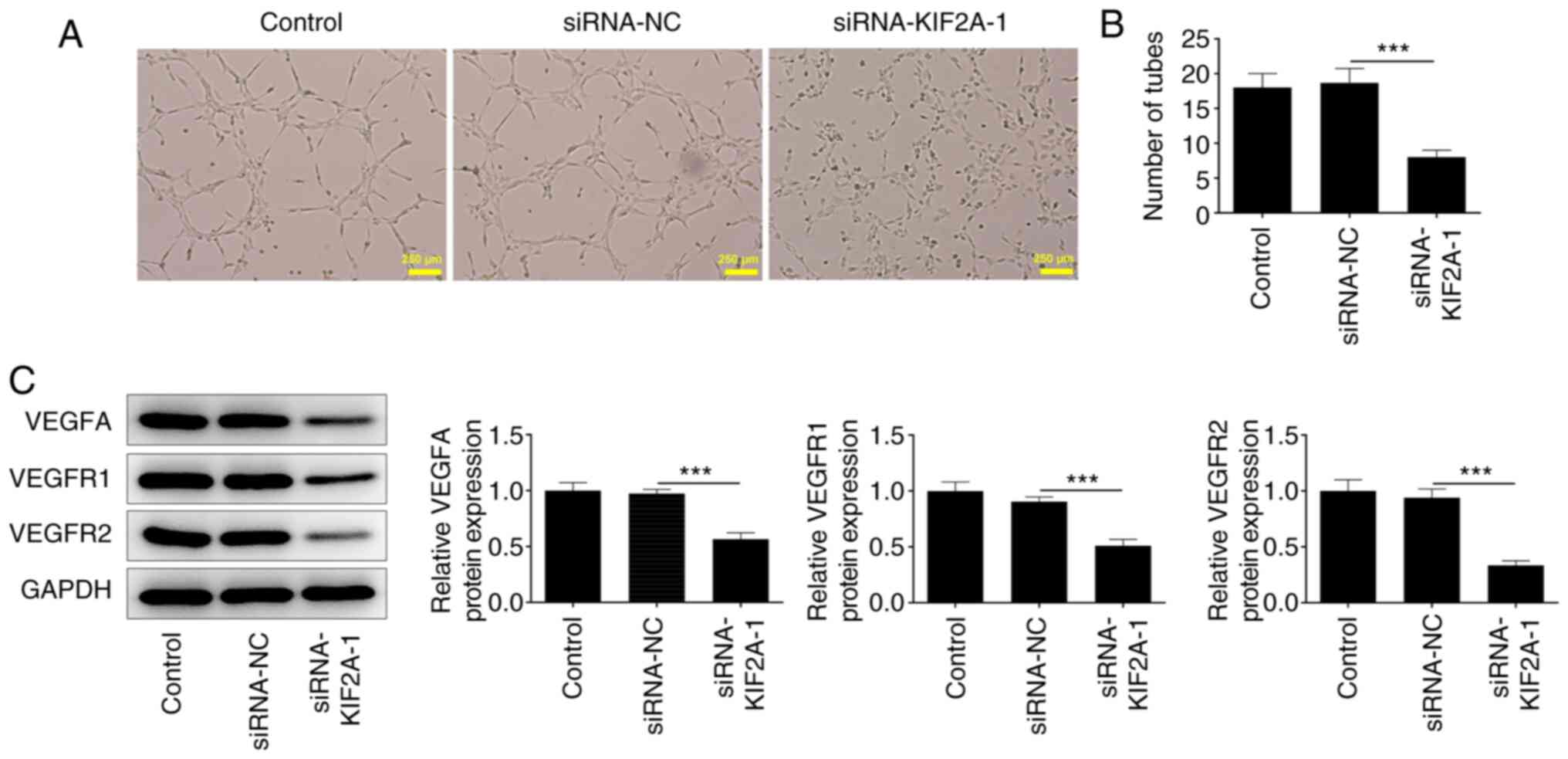
Downregulation of KIF2A induces weaker
angiogenesis in vitro
It is well known that tumor growth and metastasis
need angiogenesis for nutritional provision (22). A tube formation assay using HUVECs
suggested that KIF2A knockdown may suppress angiogenesis (Fig. 4A and B). Additionally, the decreased expression
levels of VEGFA, VEGFR1 and VEGFR2 indicated that silencing of
KIF2A may be causally associated with weaker angiogenesis in
vitro (Fig. 4C).
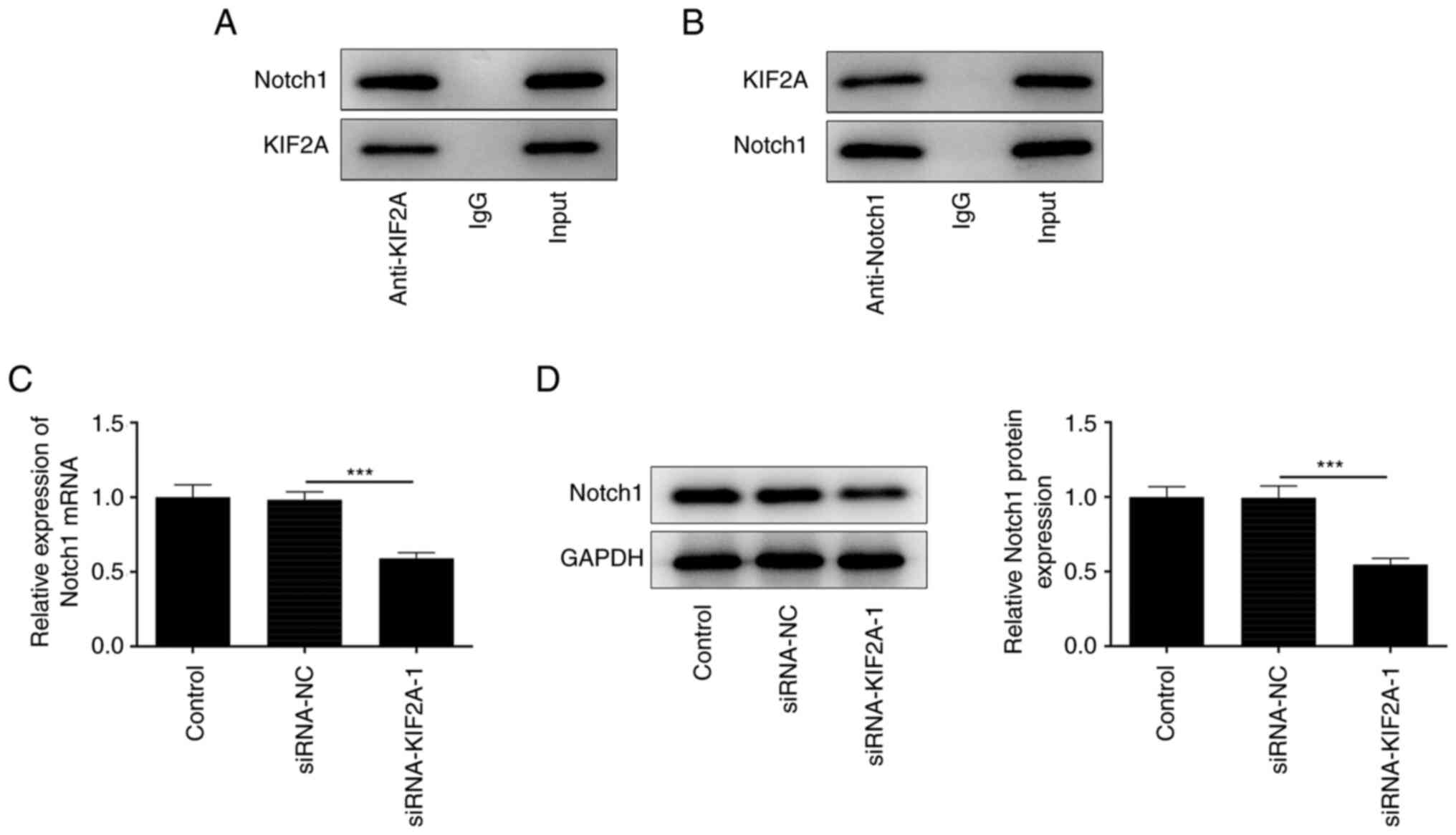
KIF2A interacts with Notch1
To further explore the molecular mechanisms
underlying the participation of KIF2A in HCC progression, the
possible interaction between KIF2A and Notch1 was predicted by
BioGRID database and verified by a Co-IP assay. Notch1 protein was
present in the anti-KIF2A group (Fig.
5A) and KIF2A protein was detected in the anti-Notch1 group
(Fig. 5B). Co-IP assay results
suggested that KIF2A may interact with and bind to Notch1. In
addition, silencing of KIF2A downregulated Notch1 expression in
Huh7 HCC cells (Fig. 5C and
D), which suggested a positive
association between KIF2A and Notch1 expression.
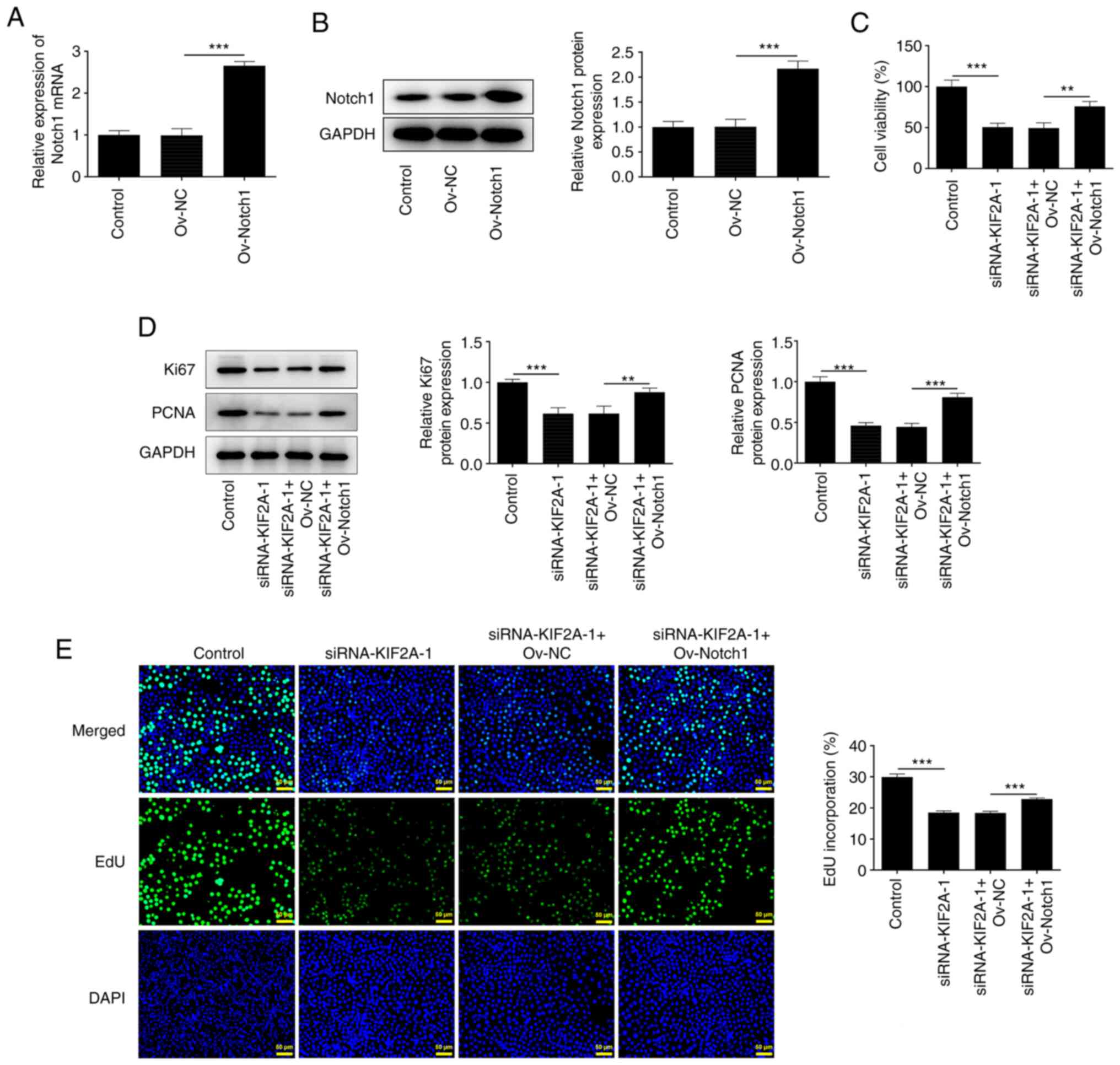
Downregulation of KIF2A suppresses
Huh7 HCC cell proliferation by suppressing Notch1
The Ov-Notch1 vector was transfected into Huh7 cells
to upregulate Notch1 expression and the transfection efficacy was
assessed by RT-qPCR and western blot analysis. The mRNA and protein
expression levels of Notch1 were significantly increased following
transfection with Ov-Notch1 compared with the Ov-NC-transfected
group (Fig. 6A and B). Results of CCK-8 analysis showed that
KIF2A knockdown suppressed HCC cell viability, which was partially
reversed upon Notch1 overexpression (Fig. 6C). In addition, increases in Ki67
and PCNA protein expression levels also indicated that upregulation
of Notch1 reduced the suppressive effect of KIF2A knockdown on HCC
cell proliferation (Fig. 6D).
Furthermore, the increased number of EdU-positive cells
demonstrated that the suppressed proliferation caused by KIF2A
silencing was partially reversed by Notch1 overexpression (Fig. 6E). These results suggested that
KIF2A knockdown may suppress the proliferative capability of HCC
cells by downregulating Notch1 expression.
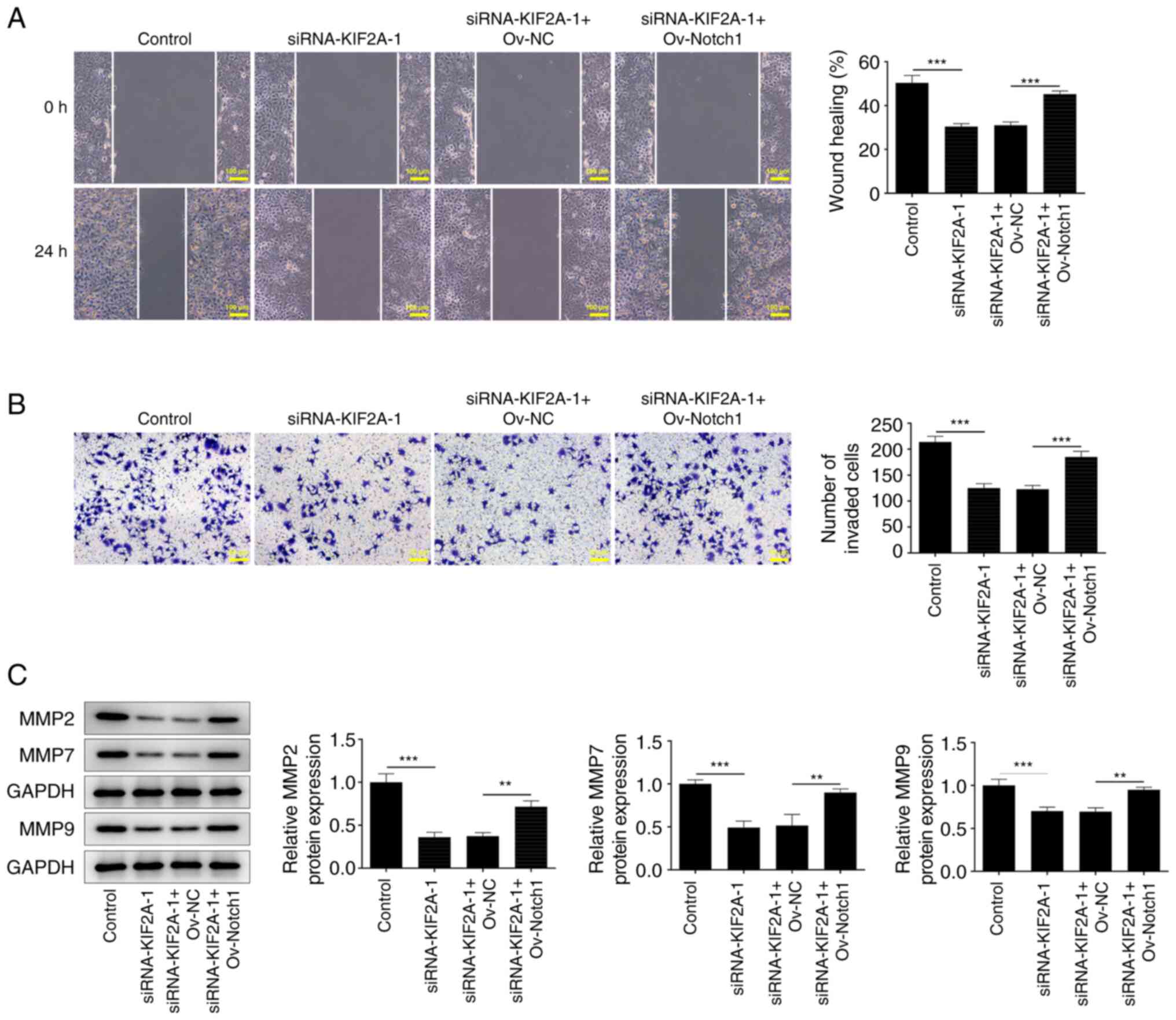
Downregulation of KIF2A inhibits Huh7
HCC cell migration and invasion by suppressing Notch1
The results of the wound healing and Transwell
assays indicated that the suppressive effects of KIF2A knockdown on
HCC cell migration and invasion, respectively, were reversed upon
upregulation of Notch1 (Fig. 7A
and B). Furthermore, the increase
in MMP2, MMP7 and MMP9 expression levels also suggested that the
KIF2A knockdown-induced suppression of HCC cell migration and
invasion was reversed by Notch1 overexpression (Fig. 7C). Collectively, these results
indicated that KIF2A knockdown may suppress the migration and
invasion of HCC cells by downregulating Notch1.
Downregulation of KIF2A induces weaker
in vitro angiogenesis by suppressing Notch1
It was observed that silencing of KIF2A lowered the
tube formation ability of HUVECs, whereas upregulation of Notch1
reversed this phenomenon (Fig. 8A
and B). In addition, the increased
expression levels of VEGFA, VEGFR1 and VEGFR2 caused by Notch1
overexpression suggested that the suppressive effects of KIF2A
knockdown on in vitro angiogenesis of Huh7 HCC cells were
partially reversed by upregulation of Notch1 (Fig. 8C and D). Overall, KIF2A knockdown may induce
weaker angiogenesis in vitro by downregulating Notch1
expression.
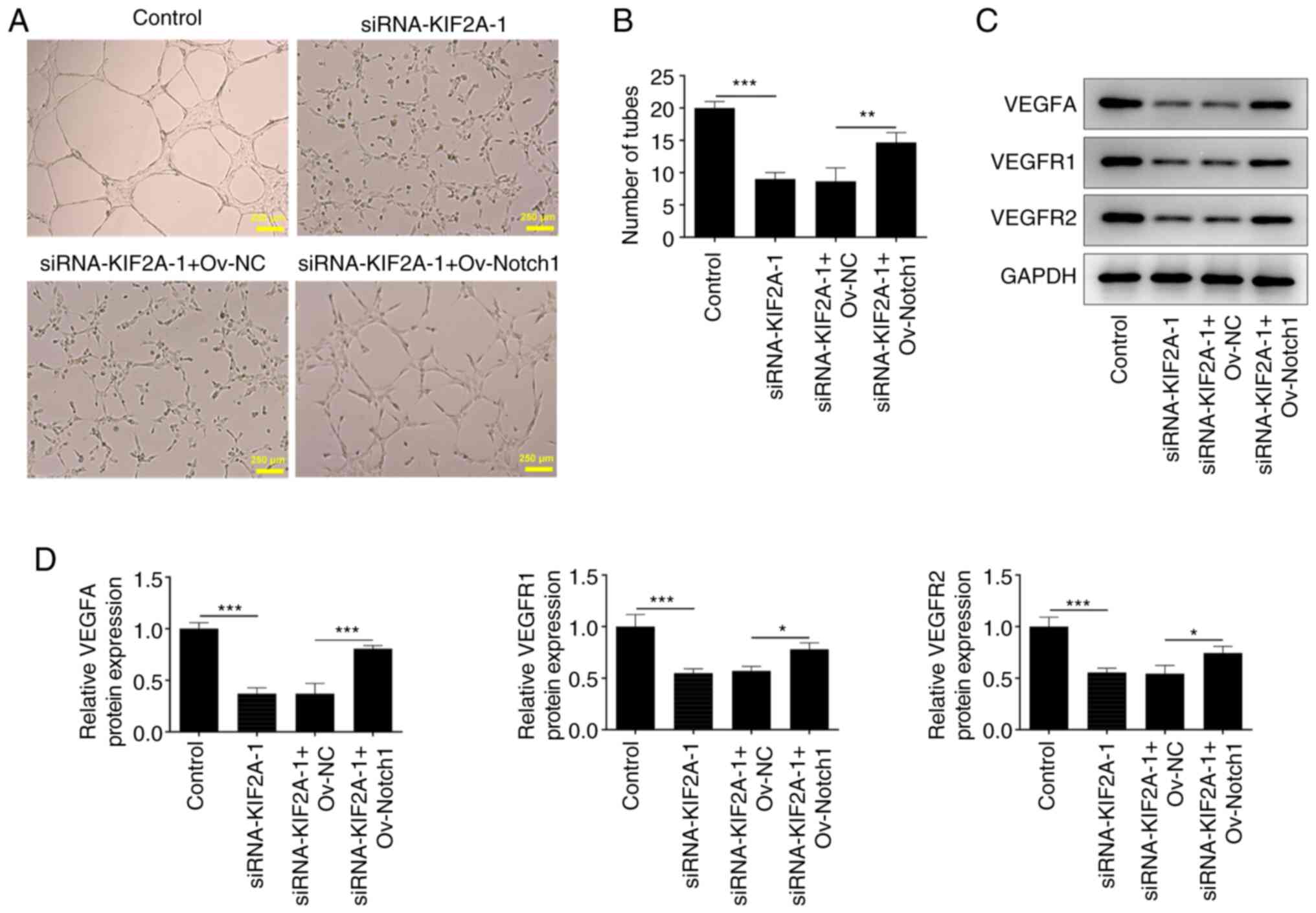 | Figure 8Downregulation of KIF2A induces
impaired angiogenesis in vitro by Notch suppression. The CM
of Huh7 HCC cells transfected with siRNA-KIF2A-1 or co-transfected
with siRNA-KIF2A-1 and Ov-Notch1 was collected and HUVECs were
subsequently incubated with the CM at 37˚C for 24 h. (A and B)
In vitro angiogenesis of HUVECs was evaluated by tube
formation assay. (C and D) VEGFA, VEGFR1 and VEGFR2 protein
expression levels in HUVECs were detected by western blot analysis.
Data are presented as the mean ± SD of three independent
experiments. *P<0.05, **P<0.01 and
***P<0.001. CM, conditioned media; HCC,
hepatocellular carcinoma; KIF2A, kinesin family member 2A; NC,
negative control; Ov, overexpression; siRNA, small interfering RNA;
VEGF, vascular endothelial growth factor; VEGFR, vascular
endothelial growth factor receptor. |
Discussion
HCC is a highly malignant tumor with a poor
prognosis (1). The metastatic
capacity of HCC is a key factor that affects recurrence and
prognosis after surgical resection (23). A recent clinical trial verified
that KIF2A is closely correlated with tumor size and clinical stage
of patients with tumors (24).
Additionally, Chen et al (14) demonstrated that KIF2A was
upregulated in HCC tissues, and was positively associated with
biomarkers for cell invasion and migration, such as MMP2, MMP7 and
MMP9. Furthermore, it has been reported that KIF2A is related to
neoplastic pathological grading and tumor-node-metastasis staging
in HCC (13). Hence, the present
study was designed to systematically determine the biological role
of KIF2A in HCC and to improve our understanding of the molecular
mechanism underlying the involvement of KIF2A in the malignant
progression of HCC.
A number of studies have revealed that KIF2A serves
a vital role in the development of several malignancies. For
example, it has been reported that silencing of KIF2A could
markedly block the proliferation, migration and invasion of
osteosarcoma cells (24).
Furthermore, Zhang et al (25) discovered that downregulation of
KIF2A could promote apoptosis, as well as inhibit proliferation,
migration and invasion of gastric cancer cells. In the present
study, it was observed that KIF2A expression was aberrantly
increased in HCC cells. KIF2A knockdown suppressed Huh7 HCC cell
proliferation, migration and invasion, and impaired angiogenesis
in vitro.
A number of studies have also demonstrated that
abnormal activation of the Notch1 signaling pathway contributes to
the development of various malignant tumors, and this has emerged
as a common topic in oncology research (17,18).
Notch1 has been identified to be highly expressed in HCC tissues
and cell lines, and to be positively associated with advanced tumor
progression and poorer prognosis of patients (17). Lu et al (26) reported that inactivation of the
Notch1 signaling pathway could suppress the metastasis of HCC
cells. Liu et al (27)
reported that downregulation of Notch1 could inhibit invasion and
angiogenesis of human breast cancer cells by inhibition of the
NF-κB signaling. In the present study, it was demonstrated that
KIF2A protein interacted with Notch1 protein. Furthermore, KIF2A
silencing decreased the mRNA and protein expression levels of
Notch1, indicating that there was a positive association between
KIF2A and Notch1 expression. The suppressive effects of KIF2A
knockdown on HCC cell proliferation, migration, invasion and in
vitro angiogenesis were partially reversed by Notch1
overexpression.
In conclusion, downregulation of KIF2A suppressed
HCC cell proliferation, migration and invasion, and arrested in
vitro angiogenesis by suppressing Notch1. Thus the present
results suggested that KIF2A may contribute to the malignant
progression of HCC via activation of the Notch1 signaling pathway.
These findings suggested that KIF2A may be an important target for
HCC, providing a promising approach for the treatment of HCC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The Zhejiang
Provincial Medical and Health Science and Technology Planning
Project of China (grant no. 2022KY705).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QW, XR, YC and BZ searched the literature and
designed the study. QW, XR, YC, YJ, XZ, CL and BZ participated in
the experimental process, performed data analysis and wrote the
manuscript. QW, XR, YC and BZ critically revised the manuscript. QW
and BZ confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing
interests.
References
|
1
|
Sivasudhan E, Blake N, Lu ZL, Meng J and
Rong R: Dynamics of m6A RNA methylome on the hallmarks of
hepatocellular carcinoma. Front Cell Dev Biol.
9(642443)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Xu G, Zhang P, Liang H, Xu Y, Shen J, Wang
W, Li M, Huang J, Ni C, Zhang X, et al: Circular RNA
hsa_circ_0003288 induces EMT and invasion by regulating
hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma.
Cancer Cell Int. 21(212)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yuan P, Mu J, Wang Z, Ma S, Da X, Song J,
Zhang H, Yang L, Li J and Yang J: Down-regulation of SLC25A20
promotes hepatocellular carcinoma growth and metastasis through
suppression of fatty-acid oxidation. Cell Death Dis.
12(361)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Moodley S, Lian EY, Crupi MJF, Hyndman BD
and Mulligan LM: RET isoform-specific interaction with scaffold
protein Ezrin promotes cell migration and chemotaxis in lung
adenocarcinoma. Lung Cancer. 142:123–131. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Maia AF, Tanenbaum ME, Galli M, Lelieveld
D, Egan DA, Gassmann R, Sunkel CE, van den Heuvel S and Medema RH:
Genome-wide RNAi screen for synthetic lethal interactions with the
C. elegans kinesin-5 homolog BMK-1. Sci Data.
2(150020)2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang W, Zhang R, Wang X, Wang N, Zhao J,
Wei Z, Xiang F and Wang C: Suppression of KIF3A inhibits triple
negative breast cancer growth and metastasis by repressing Rb-E2F
signaling and epithelial-mesenchymal transition. Cancer Sci.
111:1422–1434. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Welburn JP and Cheeseman IM: The
microtubule-binding protein Cep170 promotes the targeting of the
kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol Biol
Cell. 23:4786–4795. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang X, Ma C, Wang Q, Liu J, Tian M, Yuan
Y, Li X and Qu X: Role of KIF2A in the progression and metastasis
of human glioma. Mol Med Rep. 13:1781–1787. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang Z, Liu X, Liu X and Niu D: Long
non-coding RNA BLACAT1 promotes the tumorigenesis of gastric cancer
by sponging microRNA-149-5p and targeting KIF2A. Cancer Manag Res.
12:6629–6640. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang YF, Li MY, Tang YF, Jia M, Liu Z and
Li HQ: Circular RNA circEIF3I promotes papillary thyroid carcinoma
progression through competitively binding to miR-149 and
upregulating KIF2A expression. Am J Cancer Res. 10:1130–1139.
2020.PubMed/NCBI
|
|
12
|
Zhu Y, Ma C, Lv A and Kou C: Circular RNA
circ_0010235 sponges miR-338-3p to play oncogenic role in
proliferation, migration and invasion of non-small-cell lung cancer
cells through modulating KIF2A. Ann Med. 53:693–706.
2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu W, Xu C, Meng Q and Kang P: The
clinical value of kinesin superfamily protein 2A in hepatocellular
carcinoma. Clin Res Hepatol Gastroenterol.
45(101527)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen J, Li S, Zhou S, Cao S, Lou Y, Shen
H, Yin J and Li G: Kinesin superfamily protein expression and its
association with progression and prognosis in hepatocellular
carcinoma. J Cancer Res Ther. 13:651–659. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Capaccione KM and Pine SR: The notch
signaling pathway as a mediator of tumor survival. Carcinogenesis.
34:1420–1430. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bazzoni R and Bentivegna A: Role of notch
signaling pathway in glioblastoma pathogenesis. Cancers (Basel).
11(292)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang L, Chen J, Yong J, Qiao L, Xu L and
Liu C: An essential role of RNF187 in Notch1 mediated metastasis of
hepatocellular carcinoma. J Exp Clin Cancer Res.
38(384)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jue C, Lin C, Zhisheng Z, Yayun Q, Feng J,
Min Z, Haibo W, Youyang S, Hisamitsu T, Shintaro I, et al: Notch1
promotes vasculogenic mimicry in hepatocellular carcinoma by
inducing EMT signaling. Oncotarget. 8:2501–2513. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang Q, Li J, Zheng J and Wei A: The
carcinogenic role of the notch signaling pathway in the development
of hepatocellular carcinoma. J Cancer. 10:1570–1579.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yehya AHS, Asif M, Petersen SH,
Subramaniam AV, Kono K, Majid AMSA and Oon CE: Angiogenesis:
Managing the culprits behind tumorigenesis and metastasis. Medicina
(Kaunas). 54(8)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang YC, Xu Z, Zhang TF and Wang YL:
Circulating microRNAs as diagnostic and prognostic tools for
hepatocellular carcinoma. World J Gastroenterol. 21:9853–9862.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang ZX, Ren SC, Chang ZS and Ren J:
Identification of kinesin family member 2A (KIF2A) as a promising
therapeutic target for osteosarcoma. Biomed Res Int.
2020(7102757)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang X, Wang Y, Liu X, Zhao A, Yang Z,
Kong F, Sun L, Yu Y and Jiang L: KIF2A promotes the progression via
AKT signaling pathway and is upregulated by transcription factor
ETV4 in human gastric cancer. Biomed Pharmacother.
125(109840)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lu L, Liu S, Dong Q and Xin Y: Salidroside
suppresses the metastasis of hepatocellular carcinoma cells by
inhibiting the activation of the Notch1 signaling pathway. Mol Med
Rep. 19:4964–4972. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu Y, Su C, Shan Y, Yang S and Ma G:
Targeting Notch1 inhibits invasion and angiogenesis of human breast
cancer cells via inhibition nuclear Factor-κB signaling. Am J
Transl Res. 8:2681–2692. 2016.PubMed/NCBI
|