Introduction
Lung transplants are one of the most important
methods of saving the lives of patients with end-stage lung disease
(1). However, the shortage of
donor lung organs limits the number of lung transplants performed
(2). To reduce the gap between the
number of lung donors and patients who require a transplant,
several research centers have taken a series of measures, such as
expanding donor standards and implementing extracorporeal lung
perfusion after cardiac death (3).
At present, the majority of donor lungs come from patients who have
been declared brain-dead (4).
Compared with other donor organs from who have been declared
brain-dead, the utilization rate of lung donors is not high and
there are regional differences in different countries (5-62%)
(5-7).
And the incidence rate of complications following lung
transplantation, such as graft rejection, after brain death is
higher compared with heart transplantation (33 vs. 17%) (8-10).
Therefore, to improve the quality of donor lungs and reduce the
number of complications after transplantation, there is a need to
improve lung injury after brain death (BD).
A previous study has suggested that lung endoplasmic
reticulum stress is involved in lung apoptosis during BD in rats
(11). Endoplasmic reticulum
stress is characterized by the accumulation of unfolded or
misfolded proteins in the endoplasmic reticulum that can cause an
unfolded protein response (UPR) and endoplasmic
reticulum-associated degradation (ERAD) (12). This can escalate to involve the
ubiquitin proteasome system (UPS), degrading ubiquitinated
misfolded and unfolded proteins (13). Therefore UPR, ERAD and UPS interact
in a coordinated manner to maintain the intracellular protein
balance (14). The most common
proteasome in UPS, the 26S proteasome, is composed of the 20S
proteasome and 19S regulatory particles, both of which participate
in degrading ubiquitin-tagged proteins (15). The 20S proteasome has α and β
subunits, and the 20S proteasome β subunits display chymotrypsin
(β5)-, trypsin (β2)- and caspase (β1)-like activity (16). 20S proteasome dysfunction is
involved in the pathophysiology of various acute and chronic lung
diseases (17,18). Proteasome inhibitors can restore
protein homeostasis, reduce oxidative stress and apoptosis and
improve organ ischemia-reperfusion injury (19). Previous investigation has revealed
their potential role in protection of kidney transplantation
(20). However, information on the
effect of the proteasome inhibitor on lung injury in brain-dead
donors is sparse.
The aim of the current study was to explore whether
the 20S proteasome was involved in lung injury after BD in rats, as
well as the effect and mechanism of the proteasome inhibitor MG132
on this lung injury, to provide a potential new method to improve
the lung donor function after BD.
Materials and methods
Experimental animals and groups
A total of 60 adult male Sprague Dawley rats, 2-4
months old, weighing 200-250 g, were housed at 22˚C, with a 12-h
light/dark cycle, and with relative humidity maintained at 40-60%
and with free access to food and water in the Henan Provincial
Experimental Animal Center, Zhengzhou, China. The experimental
protocols complied with the Guide for the Care and Use of
Laboratory Animals of the National Institutes of Health (21).
Induction of BD
Animals were anesthetized with 1% pentobarbital
sodium (60 mg/kg) by intraperitoneal injection (ip), and their
arterial pressure was monitored via the femoral artery. Venous
access was established through the femoral vein and the urine
volume was monitored by cystostomy. The rats were intubated by
tracheotomy mechanically, with an oxygen fraction of 100% (Harvard
Apparatus). Next, a 2-mm hole was drilled with a marathon-3 dental
grinder (Saeyang Co., Ltd.) through the skull, 4-mm lateral to the
sagittal suture. Next, a 3F Fogarty Catheter (Edwards Lifesciences)
was placed and inflated with 20 µl fluid every 5 min until BD was
achieved (11). During the entire
experimental procedure, SpO2 remained >90% and the
SBP remained ≥90 mmHg.
BD was confirmed by induction of a deep coma,
spontaneous respiratory arrest, mydriasis, absence of brainstem
reflexes and an amplitude of the electroencephalogram <0.02
v.
Animal groups
Animals were randomly assigned into the following
groups: i) Sham operation group (sham, n=6); ii) BD groups (the BD
subgroups were divided into BD 0.5, 1, 2, 4 and 6 h according to
the time of specimen collection; n=6 per group); iii) MG132 groups,
in which MG132 (cat. no. m1902; Abmole Bioscience Inc.) was
dissolved in DMSO, diluted to 10 mmol/l with PBS and was
administered at a dose of 10 mg/kg (ip) ~30 min before BD induction
(22) (the MG132 subgroups were
divided into BD 2 h + MG132 and BD 6 h + MG132; n=6 per group); and
iv) control groups in which an equivalent volume of DMSO to that in
the MG132 group 30 min before BD induction (the control subgroups
were divided into BD 2 h + control and BD 6 h + control; n=6 per
group).
Specimen collection
The blood was drawn from the femoral artery to
measure partial artery pressure of oxygen (PaO2). Rats
were sacrificed by exsanguination of the abdominal aorta until
cardiac arrest was achieved. The right lower lung was fixed with 4%
paraformaldehyde at room temperature for 7 days, the right middle
lung after removal was quickly placed in liquid nitrogen and then
transferred to be frozen in a -80˚C refrigerator for the next
experiments within 3 months and the weight of the left lung was
measured (wet weight and dry weight after 1 week in a 60˚C oven).
Bronchoalveolar lavage fluid (BALF) was obtained by lavage of the
left main bronchus with 2 ml normal saline repeated three times
after clipping the right main bronchus according to previous
research methods (11). After
mixing, 10 µl of BALF was collected into the cell counter (Jiangsu
Jimbio Technology Co., Ltd.) to measure the cell concentration in
BALF, and these data are displayed below. The BALF was centrifuged
at 13,800 x g/min for 10 min at 4˚C, the cell sediment was
resuspended with 500 µl normal saline to make cell smears to
observe the cell morphology by using hematoxylin and eosin
(H&E) staining as described below, and the supernatant was
obtained to measure the protein concentration using a bicinchoninic
acid assay.
H&E staining
The right lower lobe was fixed in 4%
paraformaldehyde for 7 days at room temperature, embedded in
paraffin, sectioned at 4-µm and stained with H&E staining
(11). The paraffin sections were
heated at 60˚C oven for 1 h, dewaxed twice in xylene solutions for
10 min each and rehydrated in descending alcohol series. The
paraffin slides were stained with hematoxylin for 5 min at room
temperature and differentiated with 0.1% hydrochloric acid ethanol
for 1 min, followed with eosin for 5 min at room temperature,
dehydrated in ascending alcohol series, followed by being dewaxed
twice in xylene solutions for 10 min each. The cell smears were
fixed with 95% alcohol for 10 min, and the following steps of
staining were the same as those for paraffin sections. The slides
were captured using a conventional light microscope (Axiolab 5;
Zeiss GmbH) at a magnification of x200. A semiquantitative
severity-based scoring system was used as previously described in
the reference (23): i) Intra- and
extra-alveolar hemorrhage; ii) intra-alveolar edema; iii)
inflammatory infiltration of the inter-alveolar septa and airspace;
iv) over-inflation; and v) erythrocyte accumulation below the
pleura. Variables i) -iv) were graded as: 0=Negative, 1=slight,
2=moderate, 3=high and 4=severe. Variable v) was scored as 0=absent
or 1=present. Lungs were scored by two blinded investigators,
across 10 random, non-coincidental fields per section and then the
mean values were analyzed.
Immunohistochemistry staining
The paraffin sections were heated at 60˚C oven for 1
h, dewaxed twice in xylene solutions for 15 min each and rehydrated
in descending alcohol series. Antigen retrieval was performed with
sodium citrate buffer (cat. no. C1031; Beijing Solarbio Science
& Technology Co., Ltd. China) at 100˚C for 10 min. The slides
were immersed in 3% H2O2 at room temperature
for 30 min to inhibit endogenous peroxidase activity. The slides
were then blocked with 10% normal goat serum (cat. no. WGAR1009-5;
Wuhan Servicebio Technology Co., Ltd.) at room temperature for 1 h.
The slides were incubated with the primary antibody at 4˚C
overnight followed with secondary antibody at room temperature for
1 h. The primary antibody used was a 20S proteasome β1 antibody
(1:100 diluted in 1% BSA; cat. no. sc-374405; Santa Cruz
Biotechnology, Inc.). The secondary antibody used was
Biotin-conjugated Affinipure goat anti-mouse IgG (1:100; cat. no.
SA00004-1; ProteinTech Group, Inc.). DAB was added for color
development for 10 min and then counterstained with hematoxylin for
5 min at room temperature. Single sections from five rats per a
group were evaluated. The slides were captured using a Nikon
ECLIPSE Ni-E400 fluorescence microscope (Nikon Corporation).
Semi-quantitative image analysis was performed by using the
open-source software Image J 1.53e (National Institutes of Health)
plugin the IHC profiler (24). The
staining score was scored as 4 (high positive), 3 (positive), 2
(low positive) and 1 (negative); the staining positive number score
was defined in at least five areas (x400 magnification per section)
in a blinded manner and scored as 1 (<10%), 2 (10-49%), 3
(50-74%) and 4 (75-100%). The final score was defined as staining
number score multiplied by staining color score as described before
(25).
Immunofluorescence staining
Immunofluorescence staining was performed as
described previously (26). The
paraffin sections were heated at 60˚C oven for 1 h, washed twice in
xylene solutions for 15 min each, rehydrated in descending alcohol
series, blocked with sodium citrate buffer (cat. no. C1031; Beijing
Solarbio Science & Technology Co., Ltd. China) at 100˚C for 10
min, incubated with primary antibodies for 6 h at 4˚C, followed by
secondary antibodies for 6 h at 4˚C in an opaque wet box and
stained with DAPI for 10 min at room temperature in the dark. The
primary antibodies used were 20S proteasome β1 (1:100 diluted with
1% BSA; cat. no. sc-374405, Santa Cruz Biotechnology, Inc.) and
inducible nitric oxide synthase (iNOS; 1:100; cat. no. GB11119;
Wuhan Servicebio Technology Co., Ltd.), myeloperoxidase (MPO;
1:100; cat. no. GB11224; Wuhan Servicebio Technology Co., Ltd.) and
CD31 (1:100; cat. no. GB113151; Wuhan Servicebio Technology Co.,
Ltd.). The secondary antibodies used were Cy3-conjugated Affinipure
goat anti-rabbit IgG (1:100; cat. no. SA00009-2; ProteinTech Group,
Inc.) or Coralite488-conjugated goat anti-mouse IgG (1:100; cat.
no. SA00013-1; ProteinTech Group, Inc.). An Olympus fluorescence
microscope (Olympus Corporation) was used to obtain images at
excitation/emission wavelengths of 547/570 nm (Cy3, red), 494/520
nm (Coralite488, green), and 360/460 nm (DAPI, blue) (original
magnification x400).
Reverse transcription-quantitative PCR
(RT-qPCR)
The mRNA levels of Psmb1 were measured using
RT-qPCR. Total RNA was extracted from lung tissues using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
and reverse transcription was performed as described previously
(27). Synthesis of cDNA and
sample preparation were performed according to the manufacturer's
instructions for the PrimeScript RT reagent kit (Takara Bio, Inc.)
and SYBR Premix Ex Taq kit (Takara Bio, Inc.), respectively. qPCR
was performed using a QuantStudio 5 Real-Time PCR System (Thermo
Fisher Scientific, Inc.). The thermocycling conditions were 95˚C
for 30 sec, 95˚C for 5 sec and 60˚C for 35 sec (40 cycles). The
method of quantification (2-ΔΔCq method) was performed
as described previously (28). The
primer sequences (Invitrogen; Thermo Fisher Scientific, Inc.) used
in this experiment are provided in Table SI.
Cell hypoxia/reoxygenation (H/R)
Rat alveolar macrophages (NR8383 cell line; The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences)
have been demonstrated to closely mimic the important biological
characteristics of normal alveolar macrophages previously, and have
been used instead of primary alveolar macrophages (29,30).
The cells were cultured in a semi-suspension with Ham's F-12
Nutrient Mixture (cat. no. GNM21700; Genom) supplemented with 20%
FBS (cat. no. WGG8001-100; Wuhan Servicebio Technology Co., Ltd.)
at 37˚C, in a humidified incubator supplied with 20% O2
and 5% CO2. NR8383 cells were serum starved for 6 h to
ensure synchronization of the cell cycle and were pretreated with
10 µM MG132 (MG132 group) or an equivalent volume of DMSO vehicle
(control group) for 1 h. The cells were then cultured in an
incubator supplied with 1% oxygen for 2 h or 6 h to mimic hypoxia
(30). Cells were then treated as
follows: Half of the medium was absorbed, and the same amount of
40% FBS and medium along with the same drug concentration was added
and reoxygenated in the normoxic incubator for 2 h.
Apoptosis analysis of NR8383 cells
using flow cytometry
A total of 1x106 cells/ml NR8383 cells
were prepared according to the instructions of the Annexin
V-FITC/PI Apoptosis Detection kit (cat. no. ca1020; Beijing
Solarbio Science & Technology Co., Ltd.). Briefly, NR8383 cells
were harvested and resuspended with 100 µl binding buffer. Cells
were incubated with 5 µl Annexin V-FITC at room temperature for 5
min in the dark, then 5 µl PI and 400 µl of PBS were added.
Apoptosis was detected using a flow cytometry (BD FACSCantoII; BD
Biosciences) and analyzed using BD FACSDiva Software v8.0.1 (BD
Biosciences).
Western blotting
The proteins were extracted from lung tissue or
cells and lysed by using RIPA buffer (high; cat. no. R0010; Beijing
Solarbio Science & Technology Co., Ltd.) on ice for >30 min.
Protein levels were quantified using BCA reagent. Samples were
loaded 10 µl per lane and electrophoresed by 10% SDS-PAGE and
transferred to polyvinylidene difluoride membranes. The membranes
were blocked in 5% non-fat milk for 2 h at room temperature. The
membranes were incubated with primary antibodies at 4˚C overnight.
The primary antibodies used were: 20S proteasome β1 (1:500; cat.
no. sc-374405; Santa Cruz Biotechnology, Inc.), iNOS (1:1,000; cat.
no. AF0199; Affinity Biosciences), cleaved caspase 3 (1:1,000; cat.
no. 9661; Cell Signaling Technology, Inc.), p-JNK (1:1,000; cat.
no. ET1609-42; HUABIO, Inc.), total JNK Antibody (1:500; cat. no.
ET1601-28; HUABIO, Inc.) and GAPDH (1:5,000; cat. no. 60004-1-lg;
ProteinTech Group, Inc.). The secondary antibodies used were
horseradish peroxide-conjugated goat anti-mouse IgG (1:5,000; cat.
no. SA00001-1; ProteinTech Group, Inc.) and goat anti-rabbit IgG
(1:5,000; cat. no. SA00001-2; ProteinTech Group, Inc.). The bands
were visualized using the Chemiluminescent Substrate kit (cat. no.
PE0010; Beijing Solarbio Science & Technology Co., Ltd.)
combined with a Bio-Rad exposure system (Bio-Rad Laboratories,
Inc.) and analyzed using ImageJ 1.53e.
Statistical analysis
All experiments were repeated three times.
Statistical analysis was performed using GraphPad Prism version 5.0
(GraphPad Software, Inc.). The ordinal data (hispathological injury
scores, IHC scores) are presented as median and range, and
differences between sham group and BD groups were analyzed using
the Kruskal-Wallis test followed by Dunn post hoc tests. Other data
are presented as the mean ± SD, and differences in characters
[PaO2/fractional concentration of inspired oxygen
(FiO2), Wet/Dry weight ratio of left lung, BALF cells
count, BALF protein content, relative protein level, relative
expression of Psmb1) between sham group and BD groups were analyzed
using a one-way ANOVA followed by Bonferroni's test in the post-hoc
comparison. Differences in other characters between control groups
and MG132 groups were analyzed using a one-way ANOVA followed by
unpaired Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Lung injury gradually increases in
rats following BD
To evaluate the lung injury after BD in rats, the
oxygenation index (PaO2/FiO2), the wet/dry
weight ratio, H&E staining, and histological lung injury scores
were used to evaluate the standard of lung injury in lung tissue at
different time points after BD. The PaO2/FiO2
of the BD groups were significantly lower compared with that of the
sham group (Fig. 1A). The wet/dry
weight ratios of the left lung in the BD groups were significantly
higher compared with that of the sham group (Fig. 1B). Compared with the sham group,
pulmonary alveolar wall thickening, pulmonary hemorrhage, and
neutrophil infiltration were observed in the BD rats (Fig. 1C); the histological lung injury
scores increased over time and were significantly higher compared
with that in the sham group (Fig.
1D). Based on the microscopic images, a notable increase in the
proportion of cells in the BALF was observed (Fig. 1E). The cell count and the protein
content in the BALF was higher in the experimental group compared
with that in the sham group (Fig.
1F and G). These results
indicated that lung injury gradually increased in rats after
BD.
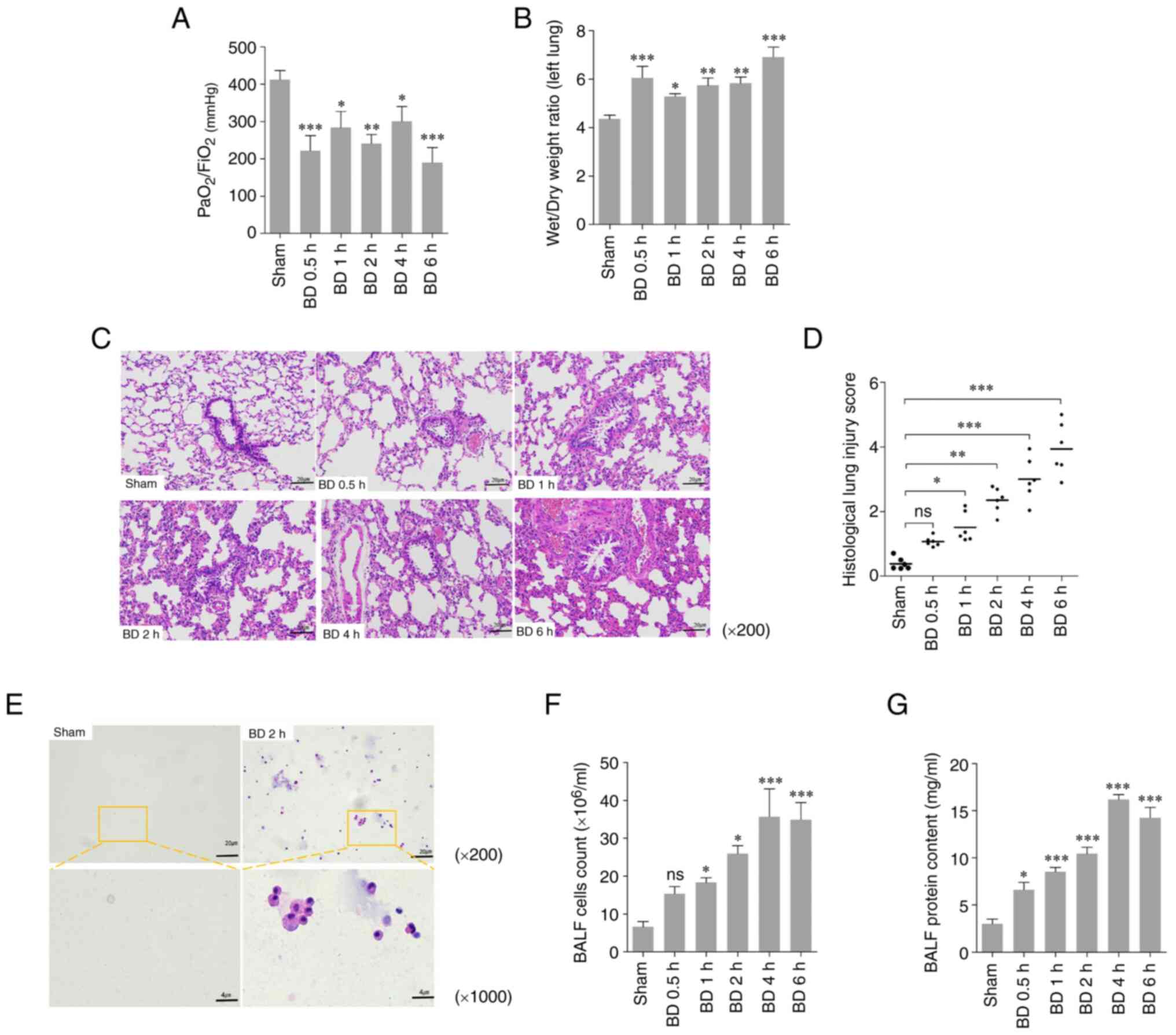 | Figure 1Lung injury after BD in rats. (A)
PaO2/FiO2 (mmHg) ratio in the rats amongst
the different groups. (B) Wet/dry weight ratio of the left lung in
the rats amongst the different groups. (C) Histopathological
sections of H&E staining (original magnification x200) and (D)
the histological lung injury score evaluated through H&E
staining in the rats amongst the different groups as median and
range. (E) Cells in the BALF in the rats amongst the different
groups (original magnification: Top row, x,200; bottom row,
x1,000). (F) Cell count and (G) the protein content in BALF amongst
the different groups. *P<0.05, **P<0.01
and ***P<0.001 vs. sham. ns/no significance; BD,
brain death; PaO2, partial artery pressure of oxygen;
FiO2, fractional concentration of inspired oxygen;
H&E, hematoxylin and eosin; BALF, bronchoalveolar lavage
fluid. |
Expression of the 20S proteasome β1 is
increased in the lung tissues of BD rats
The 20S proteasome β1 is a subunit of the 20S
proteasome with caspase-like activity that is inhibited by
MG132(31). Immunohistochemical
semi-quantitative analysis, western blotting and RT-qPCR were used
to detect the expression of the 20S proteasome β1. As presented in
Fig. 2, induction of BD
significantly increased the expression of the 20S proteasome β1 in
the lung tissues compared with the sham group (Fig. 2A, B, F and
G).
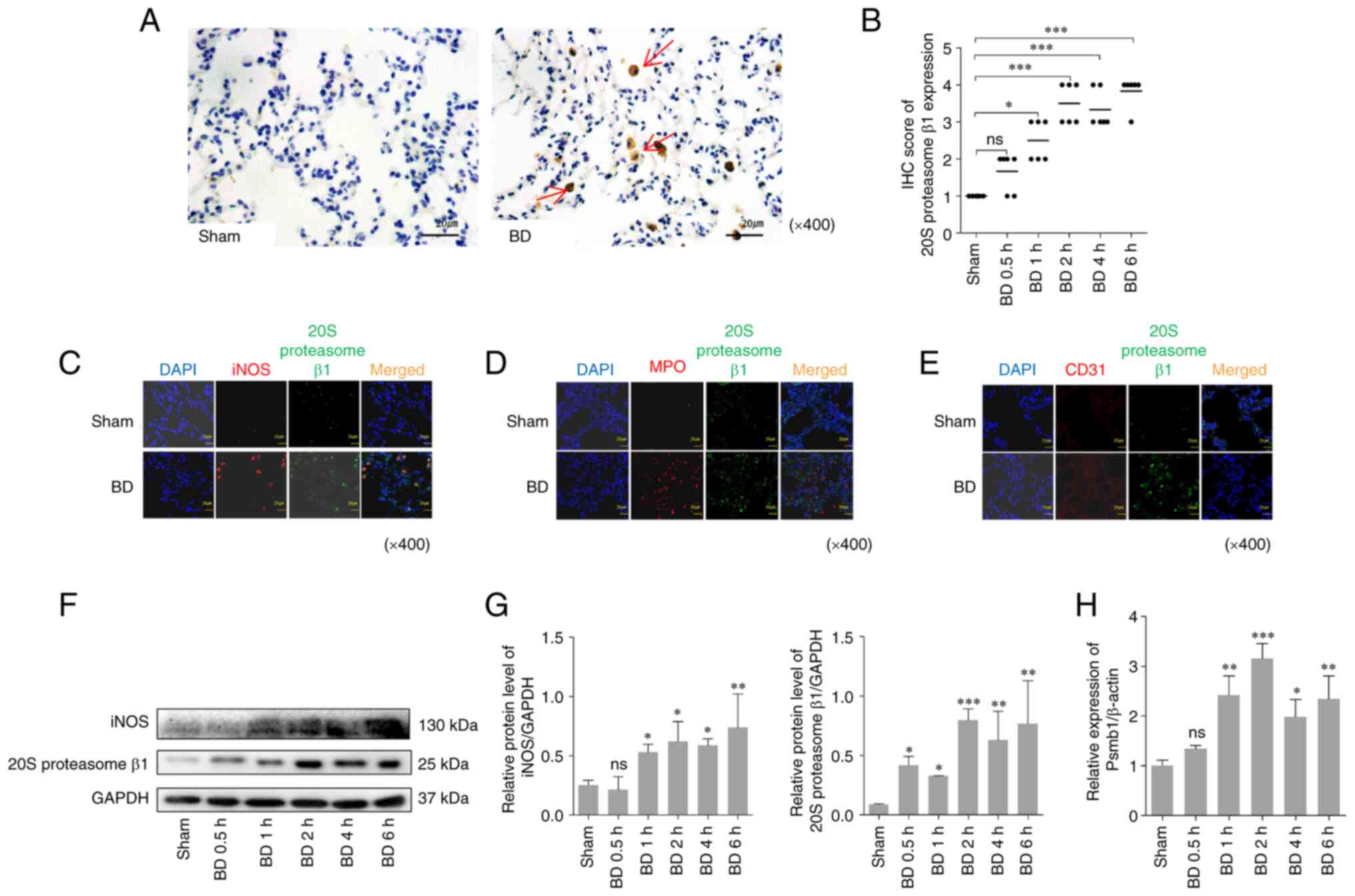 | Figure 2Expression of the 20S proteasome β1
in lung tissues after BD in rats. (A) IHC staining of the 20S
proteasome β1 in lung tissues 2 h after BD and in the sham group
(original magnification x400). The arrows show the 20S proteasome
β1 positive cells. (B) Semi-quantitative evaluation of 20S
proteasome IHC staining in lung tissues amongst the different
groups. (C-E) Immunofluorescence expression of 20S proteasome β1
and (C) iNOS, (D) MPO and (E) CD31 in lung tissues 2 h after BD and
in the sham group (original magnification x400). Green, 20S
proteasome β1; red, iNOS/MPO/CD31; orange, merged expression
showing colocalization. (F) Western blotting and (G) quantification
of the protein expression levels of the 20S proteasome β1 and iNOS
from extracts of the lung tissue amongst the different groups. (H)
Relative Psmb1 levels in lung tissues after BD detected by RT-qPCR.
*P<0.05, **P<0.01,
***P<0.001 vs. sham. ns/no significance; BD, brain
death; IHC, immunohistochemistry; iNOS, inducible nitric oxide
synthase; MPO, myeloperoxidase. |
Notably, the 20S proteasome β1 positive cells
exhibited an alveolar macrophage-like morphology. To confirm the
upregulation of the 20S proteasome β1 in alveolar cells from the
lung tissues following BD, immunofluorescence staining was used to
detect the co-localization of 20S proteasome β1 and iNOS/MPO/CD31
in lung tissue sections of rats after BD (MPO is considered as one
of markers of neutrophil activation, CD31 is one of the markers of
endothelial cell and iNOS is considered one of the markers of
macrophages). The results indicated the presence and the
differences in the co-localization of the 20S proteasome β1 and
iNOS/MPO/CD31 in the lung tissues, and 20S proteasome β1 mostly
colocalized with iNOS, but not with MPO and CD31 (Fig. 2C-E) to rule out the increased high
expression of 20S proteasome β1 on neutrophils/endothelial cells.
Compared with the sham group, induction of BD significantly
increased the protein expression of the 20S proteasome β1 and iNOS
(Fig. 2F and G) and also increased the relative Psmb1
levels (Fig. 2H).
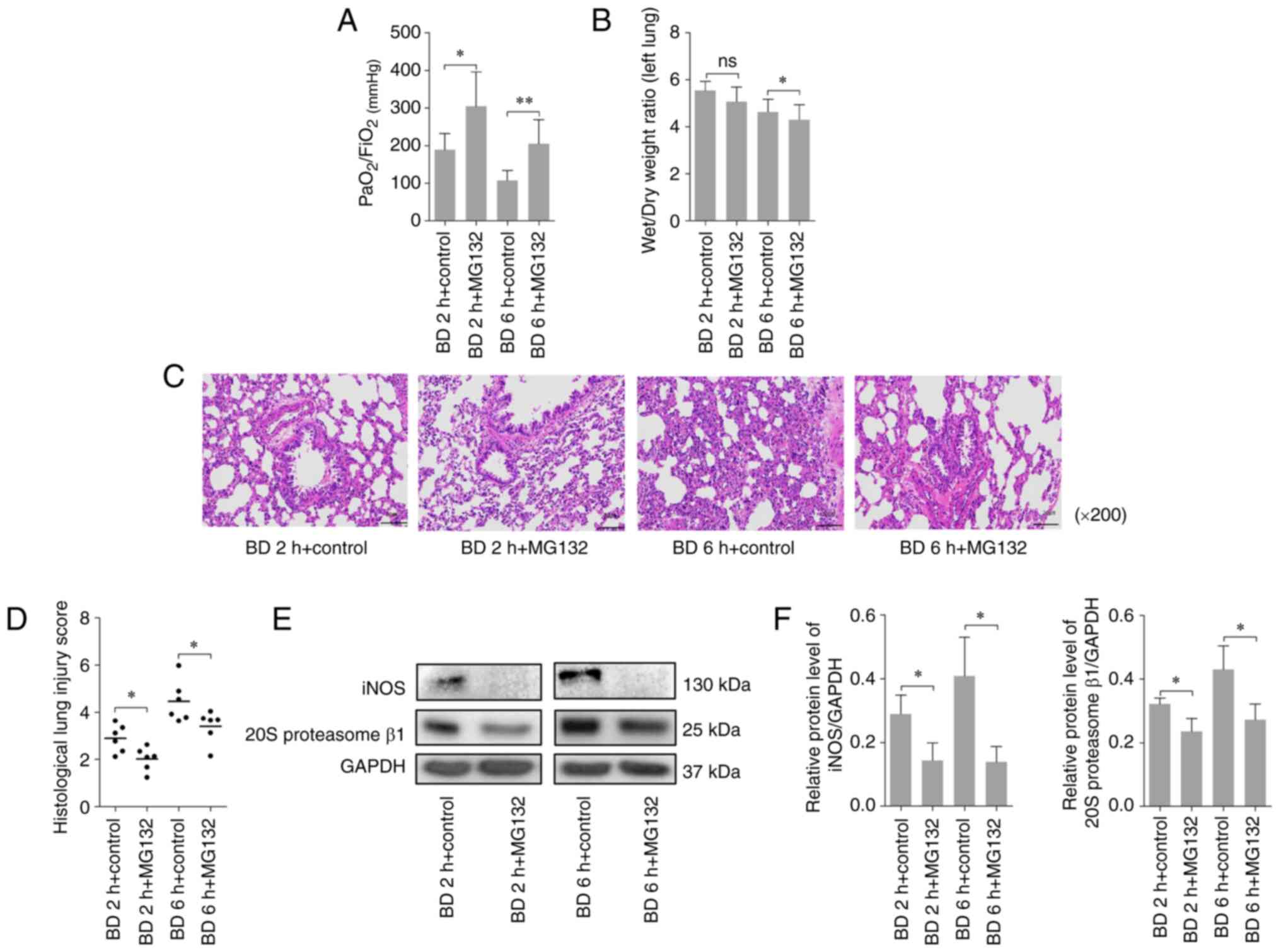
Inhibition of the proteasome by MG132
reduces lung injury following BD
To explore the effect of proteasomal inhibition on
lung injury following BD in rats, a proteasome inhibitor, MG132,
was administered before induction of BD and the specimens were
collected for analysis at different time points following BD.
Compared with the control group, MG132 treatment was revealed to
significantly increase the PaO2/FiO2 after BD
(Fig. 3A) and significantly reduce
the left lung wet/dry weight ratio 6 h after BD (Fig. 3B). However, MG132 treatment also
alleviated the thickening of the alveolar wall, pulmonary
hemorrhage, and neutrophil infiltration (Fig. 3C), whilst also significantly
decreasing the histological lung injury scores compared with the
control group (Fig. 3D). As
presented in Fig. 3E and F, MG132 treatment significantly reduced
the protein expression levels of the 20S proteasome β1 and iNOS in
lung tissues following BD at 2 and 6 h. These results suggested
that inhibition of the proteasome by MG132 decreased lung injury
after BD.
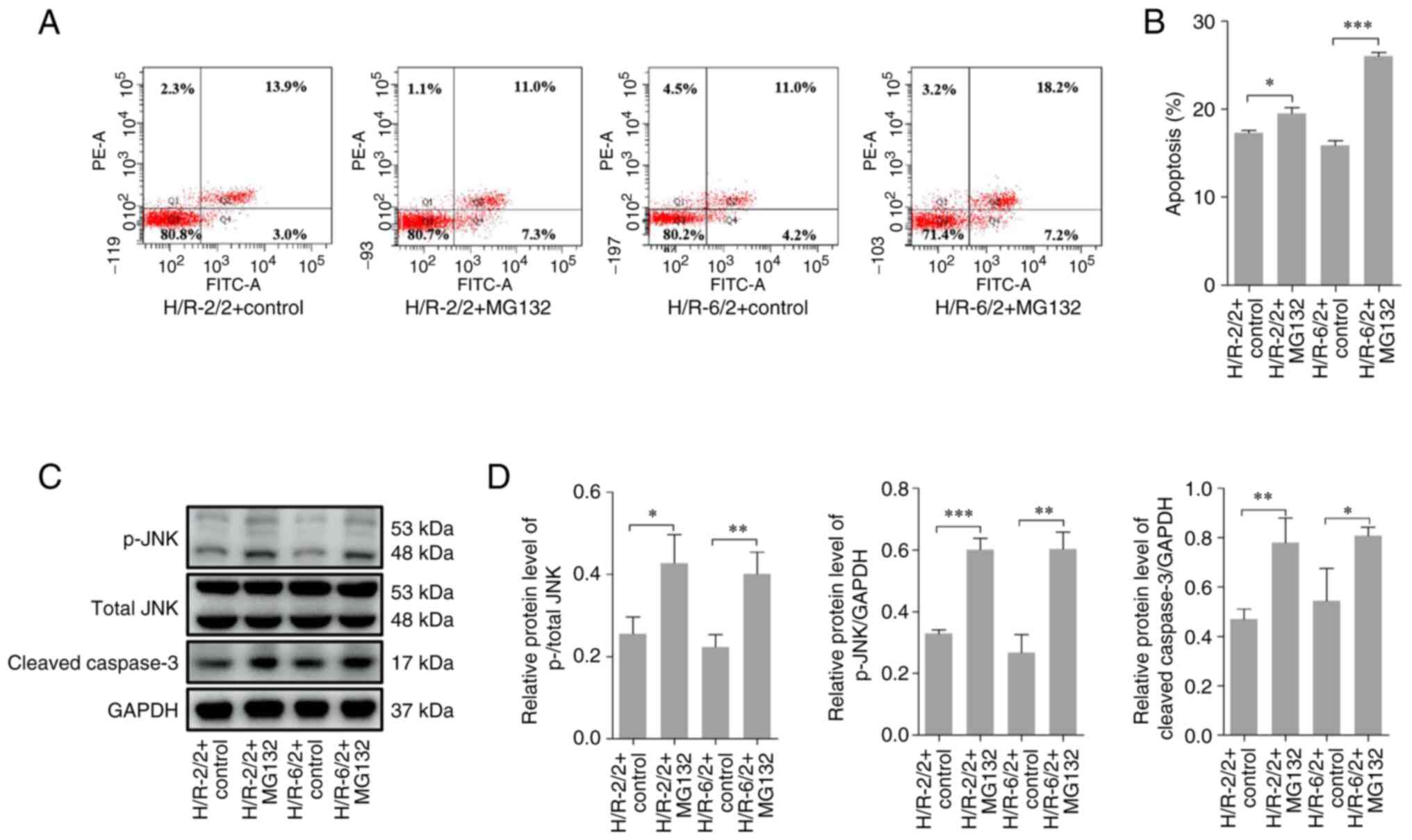
MG132 increases apoptosis following
H/R in cultured rat alveolar macrophages
Previous studies suggest that in the acute stage of
infection or BD, alveolar macrophages are rapidly recruited and
they secrete a large quantity of harmful substances, which in turn
attack the lung tissues and induce lung tissue injury (32,33).
As the upregulation of 20S proteasome β1 in alveolar macrophages
was confirmed, whether MG132 treatment protected lung tissues via
induction of apoptosis of alveolar macrophages was next assessed.
To test this hypothesis, NR83883 cells (rat alveolar macrophage
cells) were used as an in vitro model. Flow cytometry and
western blotting were used to detect the apoptosis of NR8383 cells
after hypoxia treatment for 2 or 6 h, followed by re-oxygenation
for 2 h. Compared with the control group, MG132 treatment
significantly increased the apoptotic rate of NR8383 cells
(Fig. 4A and B) after 2 h of hypoxia and 2 h of
re-oxygenation (H/R-2/2), and after 6 h of hypoxia and 2 h of
re-oxygenation (H/R-6/2). In addition, MG132 administration
significantly increased the protein expression levels of p-JNK and
cleaved-caspase 3 in NR8383 cells after H/R-2/2 and H/R-6/2
compared with the control group (Fig.
4C and D). These results
highlighted that inhibition of proteasomal activity using MG132
increased cell apoptosis after H/R in cultured rat alveolar
macrophages.
Discussion
BD can lead to a series of pathophysiological
changes, including hemodynamic, metabolomic, inflammatory and
neuroendocrinal abnormalities (34). All of these can contribute to lung
injury. The donor lung after BD is associated with a high incidence
rate of complications following lung transplantation (8-10).
Thus, there is a need to study the mechanism of lung injury after
BD and to develop effective therapeutics to decrease lung
injury.
In the present study, a decrease in the arterial
oxygenation index and an increase in the left lung wet/dry weight
ratio and the aggravation of lung pathological injury were observed
after BD in rats, in accordance with published results describing
the lung after BD (35,36).
The UPS is an important pathway for maintaining
protein homeostasis in eukaryotic cells (37). The 20S proteasome β1 subunit is a
component of the 26S proteasome (15,16).
The biological activity of the 20S proteasome can be detected in
the alveolar space of patients with acute lung injury (17). In addition, the protein
concentration of the 20S proteasome is also significantly increased
in the alveolar space of patients with acute lung injury and in the
circulation of patients with sepsis (38). In agreement with the previous data,
in the present study the expression of the 20S proteasome increased
over time in the lung tissues of rats after BD compared with the
sham group samples. These results suggested that the 20S proteasome
was involved in lung injury after BD.
The present results demonstrated that MG132, as a
proteasome inhibitor, improved the arterial blood oxygenation
index, alleviated lung pathological manifestations and reduced the
left lung wet/dry weight ratio in rats after BD, suggesting that
MG132 could decrease lung injury after BD in rats. In addition,
MG132 was revealed to downregulate the protein expression levels of
the 20S proteasome β1 subunit at the same time. It has previously
been demonstrated that MG132 can decrease the levels of
inflammatory cytokines in BALF in a model of sepsis-induced acute
lung injury, suggesting that MG132 crosses the blood-air barrier
and gains access to lumen facing cells such as the alveolar
macrophages (39,40). Therefore, it was speculated that
that MG132 may play a role in lung protection by inhibiting the
expression of the proteasome.
The current study revealed that the 20S proteasome
β1 and iNOS were colocalized in lung tissues after BD using an
immunofluorescence assay and the protein expression levels were
also significantly augmented in lung tissues following BD. Previous
studies have detected iNOS expression in lung tissues after BD and
reveal that in response to inflammatory stimuli, activated alveolar
macrophages express high levels of iNOS and produce large amounts
of NO, eventually resulting in tissue damage (41,42).
Alveolar macrophages in the lungs express iNOS and are important
sources of endogenous pulmonary NO production in inflammatory
states, such as septic ALI (43-45).
MG132 treatment can inhibit the protein expression
of iNOS through inhibition of the JNK/c-Myc signaling pathway and
plays a protective role in sepsis-induced ALI (46). The current study demonstrated that
MG132 treatment could inhibit the protein expression of 20S
proteasome β1 and iNOS in lung tissues following BD in rats,
indicating that the effect of MG132 treatment may be associated
with alveolar macrophages. The NR8383 cell line has been
demonstrated to closely mimic important biological characteristics
of normal alveolar macrophages (47); thus, it was selected in the current
study as a model cell hypoxia reoxygenation model for a cell
experiment, which revealed that MG132 could promote the cell death
of rat alveolar macrophages after H/R injury.
In addition, MG132 acts as a proteasome inhibitor,
primarily affecting the caspase-related pathways (30). The JNK signal transduction pathway
is an important member of the MARK pathway, which mediates
intracellular signal transduction and induces apoptosis by
activating the caspase family protein kinase (48). Previous studies have indicated that
MG132 can promote the activity of JNKs49,50). The current study
revealed that MG132 upregulated p-JNK protein expression and
activated caspase 3, which promoted NR8383 apoptosis after H/R.
Therefore, it is speculated that MG132 may promote the apoptosis of
rat alveolar macrophages by mediating the JNK-caspase pathway, thus
protecting the lungs after BD.
In conclusion, the 20S proteasome was demonstrated
to be involved in lung injury after BD in rats, and MG132 could
effectively reduce lung injury. This may be associated with the
ability of MG132 to inhibit the expression of the proteasome in
lung tissues and promote the apoptosis of alveolar macrophages. As
such, this drug should be further explored regarding its potential
to protect potential donor lungs following BD.
Supplementary Material
Primer sequences for reverse
transcription-quantitative PCR
Acknowledgements
We would like to thank Dr H.W. Tang (Henan Key
Laboratory of Digestive Organ Transplantation, First Affiliated
Hospital of Zhengzhou University, Zhengzhou, China) for their
technical assistance.
Funding
Funding: This study was supported by the National Natural
Science Foundation of China (grant no. 81971881).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HS, DY, and ZH performed the experiments. YL and HS
performed the literature search and analyzed the data. WG and SZ
interpreted the data. SZ, WG and HS designed the study. HS, DY and
YL prepared and wrote the study. HS and SZ confirm the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The protocol used in the present study was approved
by the Institutional Animal Care and Use Committee of Zhengzhou
University (approval no. 2019-ky-019).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nathan SD: The future of lung
transplantation. Chest. 147:309–316. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Young KA and Dilling DF: The Future of
Lung Transplantation. Chest. 155:465–473. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jin Z, Hana Z, Alam A, Rajalingam S,
Abayalingam M, Wang Z and Ma D: Review 1: Lung transplant-from
donor selection to graft preparation. J Anesth. 34:561–574.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Van Raemdonck D, Keshavjee S, Levvey B,
Cherikh WS, Snell G, Erasmus M, Simon A, Glanville AR, Clark S,
D'Ovidio F, et al: Donation after circulatory death in lung
transplantation-five-year follow-up from ISHLT Registry. J Heart
Lung Transplant. 38:1235–1245. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kute V, Ramesh V, Shroff S, Guleria S and
Prakash J: Deceased-Donor organ transplantation in India: Current
status, challenges, and solutions. Exp Clin Transplant. 18 (Suppl
2):S31–S42. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yeo HJ, Yoon SH, Lee SE, Jeon D, Kim YS,
Cho WH and Kim DH: Current status and future of lung donation in
Korea. J Korean Med Sci. 32:1953–1958. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Paraskeva MA, Levin KC, Westall GP and
Snell GI: Lung transplantation in Australia, 1986-2018: More than
30 years in the making. Med J Aust. 208:445–450. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ware LB, Wang Y, Fang X, Warnock M, Sakuma
T, Hall TS and Matthay M: Assessment of lungs rejected for
transplantation and implications for donor selection. Lancet.
360:619–620. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zweers N, Petersen AH, van der Hoeven JA,
de Haan A, Ploeg RJ, de Leij LF and Prop J: Donor brain death
aggravates chronic rejection after lung transplantation in rats.
Transplantation. 78:1251–1258. 2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Takahashi T, Terada Y, Pasque MK, Itoh A,
Nava RG, Puri V, Kreisel D, Patterson AG and Hachem RR: Comparison
of outcomes in lung and heart transplant recipients from the same
multiorgan donor. Clin Transplant. 34(e13768)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Tang H, Zhang J, Cao S, Yan B, Fang H,
Zhang H, Guo W and Zhang S: Inhibition of endoplasmic reticulum
stress alleviates lung injury induced by brain death. Inflammation.
40:1664–1671. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hwang J and Qi L: Quality control in the
endoplasmic reticulum: Crosstalk between ERAD and UPR pathways.
Trends Biochem Sci. 43:593–605. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xia SW, Wang ZM, Sun SM, Su Y, Li ZH, Shao
JJ, Tan SZ, Chen AP, Wang SJ, Zhang ZL, et al: Endoplasmic
reticulum stress and protein degradation in chronic liver disease.
Pharmacol Res. 161(105218)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cybulsky AV: The intersecting roles of
endoplasmic reticulum stress, ubiquitin-proteasome system, and
autophagy in the pathogenesis of proteinuric kidney disease. Kidney
Int. 84:25–33. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kopp F, Hendil KB, Dahlmann B, Kristensen
P, Sobek A and Uerkvitz W: Subunit arrangement in the human 20S
proteasome. Proc Natl Acad Sci USA. 94:2939–2944. 1997.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Everly JJ, Walsh RC, Alloway RR and Woodle
ES: Proteasome inhibition for antibody-mediated rejection. Curr
Opin Organ Transplant. 14:662–666. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kerrin A, Weldon S, Chung AH, Craig T,
Simpson AJ, O'Kane CM, McAuley DF and Taggart CC: Proteolytic
cleavage of elafin by 20S proteasome may contribute to inflammation
in acute lung injury. Thorax. 68:315–321. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Semren N, Welk V, Korfei M, Keller IE,
Fernandez IE, Adler H, Günther A, Eickelberg O and Meiners S:
Regulation of 26S proteasome activity in pulmonary fibrosis. Am J
Respir Crit Care Med. 192:1089–1101. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kukan M: Emerging roles of proteasomes in
ischemia-reperfusion injury of organs. J Physiol Pharmacol. 55 (1
Pt 1):3–15. 2004.PubMed/NCBI
|
|
20
|
Lo S, MacMillan-Crow LA and Parajuli N:
Renal cold storage followed by transplantation impairs proteasome
function and mitochondrial protein homeostasis. Am J Physiol Renal
Physiol. 316:F42–F53. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
National Research Council (US), Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals. 8th
edition. National Academies Press (US),Washington, DC, 2011.
|
|
22
|
Chen X, Li SL, Wu T and Liu JD: Proteasome
inhibitor ameliorates severe acute pancreatitis and associated lung
injury of rats. World J Gastroenterol. 14:3249–3253.
2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
van Zanden JE, Rebolledo RA, Hoeksma D,
Bubberman JM, Burgerhof JG, Breedijk A, Yard BA, Erasmus ME,
Leuvenink HGD and Hottenrott MC: Rat donor lung quality
deteriorates more after fast than slow brain death induction. PLoS
One. 15(e242827)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Varghese F, Bukhari AB, Malhotra R and De
A: IHC Profiler: An open source plugin for the quantitative
evaluation and automated scoring of immunohistochemistry images of
human tissue samples. PLoS One. 9(e96801)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Guo Z, Zhang X, Zhu H, Zhong N, Luo X,
Zhang Y, Tu F, Zhong J, Wang X, He J and Huang L: TELO2 induced
progression of colorectal cancer by binding with RICTOR through
mTORC2. Oncol Rep. 45:523–534. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lee HY and Oh SH: Autophagy-mediated
cytoplasmic accumulation of p53 leads to apoptosis through DRAM-BAX
in cadmium-exposed human proximal tubular cells. Biochem Biophys
Res Commun. 534:128–133. 2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chen S, Fang H, Li J, Shi J, Zhang J, Wen
P, Wang Z, Yang H, Cao S, Zhang H, et al: Microarray analysis for
expression profiles of lncRNAs and circRNAs in rat liver after
brain-dead donor liver transplantation. Biomed Res Int.
2019(5604843)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hino M, Oda M, Yoshida A, Nakata K, Kohchi
C, Nishizawa T, Inagawa H, Hori H, Makino K, Terada H and Soma G:
Establishment of an in vitro model using NR8383 cells and
Mycobacterium bovis Calmette-Guerin that mimics a chronic infection
of Mycobacterium tuberculosis. In Vivo. 19:821–830. 2005.PubMed/NCBI
|
|
30
|
Fan T, Huang Z, Wang W, Zhang B, Xu Y, Mao
Z, Chen L, Hu H and Geng Q: Proteasome inhibition promotes
autophagy and protects from endoplasmic reticulum stress in rat
alveolar macrophages exposed to hypoxia-reoxygenation injury. J
Cell Physiol. 233:6748–6758. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kisselev AF: Site-Specific proteasome
inhibitors. Biomolecules. 12(54)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sutherland AJ, Ware RS, Winterford C and
Fraser JF: The endothelin axis and gelatinase activity in alveolar
macrophages after brain-stem death injury: A pilot study. J Heart
Lung Transplant. 26:1040–1047. 2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Brieland JK, Kunkel RG and Fantone JC:
Pulmonary alveolar macrophage function during acute inflammatory
lung injury. Am Rev Respir Dis. 135:1300–1306. 1987.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Avlonitis VS, Fisher AJ, Kirby JA and Dark
JH: Pulmonary transplantation: The role of brain death in donor
lung injury. Transplantation. 75:1928–1933. 2003.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sammani S, Park KS, Zaidi SR, Mathew B,
Wang T, Huang Y, Zhou T, Lussier YA, Husain AN, Moreno-Vinasco L,
et al: A sphingosine 1-phosphate 1 receptor agonist modulates brain
death-induced neurogenic pulmonary injury. Am J Respir Cell Mol
Biol. 45:1022–1027. 2011.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wauters S, Somers J, De Vleeschauwer S,
Verbeken E, Verleden GM, van Loon J and Van Raemdonck DE:
Evaluating lung injury at increasing time intervals in a murine
brain death model. J Surg Res. 183:419–426. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Goldberg AL: Protein degradation and
protection against misfolded or damaged proteins. Nature.
426:895–899. 2003.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Roth GA, Moser B, Krenn C, Roth-Walter F,
Hetz H, Richter S, Brunner M, Jensen-Jarolim E, Wolner E,
Hoetzenecker K, et al: Heightened levels of circulating 20S
proteasome in critically ill patients. Eur J Clin Invest.
35:399–403. 2005.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wu B, Miao X, Ye J and Pu X: The
protective effects of protease inhibitor MG-132 on sepsis-induced
acute lung rats and its possible mechanisms. Med Sci Monit.
25:5690–5699. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Caldeira MV, Salazar IL, Curcio M,
Canzoniero LM and Duarte CB: Role of the ubiquitin-proteasome
system in brain ischemia: Friend or foe? Prog Neurobiol. 112:50–69.
2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Vieira RF, Breithaupt-Faloppa AC,
Matsubara BC, Rodrigues G, Sanches MP, Armstrong-Jr R, Ferreira SG,
Correia CJ, Moreira LFP and Sannomiya P: 17β-Estradiol protects
against lung injuries after brain death in male rats. J Heart Lung
Transplant. 37:1381–1387. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Forstermann U and Sessa WC: Nitric oxide
synthases: Regulation and function. Eur Heart. J 33:829–837,
837a-837d. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kobzik L, Bredt DS, Lowenstein CJ, Drazen
J, Gaston B, Sugarbaker D and Stamler JS: Nitric oxide synthase in
human and rat lung: Immunocytochemical and histochemical
localization. Am J Respir Cell Mol Biol. 9:371–377. 1993.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Farley KS, Wang LF, Razavi HM, Law C,
Rohan M, McCormack DG and Mehta S: Effects of macrophage inducible
nitric oxide synthase in murine septic lung injury. Am J Physiol
Lung Cell Mol Physiol. 290:L1164–L1172. 2006.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fujii Y, Goldberg P and Hussain SN:
Contribution of macrophages to pulmonary nitric oxide production in
septic shock. Am J Respir Crit Care Med. 157 (5 Pt 1):1645–1651.
1998.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang Y, Huang T, Jiang L, Gao J, Yu D, Ge
Y and Lin S: MCP-induced protein 1 attenuates sepsis-induced acute
lung injury by modulating macrophage polarization via the JNK/c-Myc
pathway. Int Immunopharmacol. 75(105741)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Helmke RJ, German VF and Mangos JA: A
continuous alveolar macrophage cell line: Comparisons with freshly
derived alveolar macrophages. In Vitro Cell Dev Biol. 25:44–48.
1989.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bogoyevitch MA and Kobe B: Uses for JNK:
The many and varied substrates of the c-Jun N-terminal kinases.
Microbiol Mol Biol Rev. 70:1061–1095. 2006.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wu HM, Wen HC and Lin WW: Proteasome
inhibitors stimulate interleukin-8 expression via Ras and apoptosis
signal-regulating kinase-dependent extracellular signal-related
kinase and c-Jun N-terminal kinase activation. Am J Respir Cell Mol
Biol. 27:234–243. 2002.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Tarjanyi O, Haerer J, Vecsernyes M, Berta
G, Stayer-Harci A, Balogh B, Farkas K, Boldizsár F, Szeberényi J
and Sétáló G Jr: Prolonged treatment with the proteasome inhibitor
MG-132 induces apoptosis in PC12 rat pheochromocytoma cells. Sci
Rep. 12(5808)2022.PubMed/NCBI View Article : Google Scholar
|