Introduction
Osteosarcoma, the most common bone malignancy in
children and adolescents (1),
often manifests as serious pain in the bones and joints, as well as
in the local mass. It is a highly malignant tumor with a poor
prognosis (1). The treatment for
high-grade osteosarcoma includes preoperative chemotherapy followed
by surgical resection and postoperative chemotherapy (2,3).
Although multimodal chemotherapy increases beneficial outcomes in
patients with osteosarcoma, metastasis and recurrence contribute to
poor prognosis in patients with advanced osteosarcoma (4,5).
Thus, identifying effective prognostic markers in patients with
osteosarcoma and clarifying the underlying molecular mechanisms is
crucial to guide the clinical diagnosis of osteosarcoma.
Proline- and serine-rich 2 (PROSER2) is encoded by
the 47th open reading frame on human chromosome 10 (6,7). A
genome-wide associated study analysis involving 3,230 patients with
pediatric fractures identified one locus on chromosome 10
(rs112635931), near PROSER2 antisense RNA 1 and PROSER2, which may
be the candidate gene associated with pediatric fractures (8). PROSER2 accurately predicts survival
in patients with breast cancer (9). Additionally, transcriptome analysis
identified an optimal multivariable Cox regression model including
five predictors (CD180, Myc, PROSER2, dynein axonemal intermediate
chain 1 and fetal and adult testis expressed 1) in 82 osteosarcoma
samples (10). However, the
molecular mechanism of PROSER2 in regulating proliferation and
metastasis of human osteosarcoma has remained unclear.
The Wnt signaling pathway is involved in cell
proliferation, differentiation and embryonic development (11-13).
Abnormal activation of this pathway has recently been reported to
be involved in the pathogenesis, invasion and migration of various
benign and malignant tumors (14,15).
It includes two major pathways by activating distinct Wnt
receptors: β-catenin-dependent and -independent (canonical and
non-canonical, respectively) Wnt signaling pathways (16). The Wnt/Ca2+ cascade is a
branch of non-canonical Wnt signaling activated in the presence of
Wnt ligands, including Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7a and
Wnt11(17). Wnt ligands attract
and activate Dishevelled by binding to Frizzled family proteins and
further activating phospholipase C. This catalyzes the conversion
of phosphatidylinositol 4,5-bisphosphate to 1,4,5-triphosphate
inositol and diacylglycerol, leading to release of intracellular
calcium (18). Increased
intracellular Ca2+ activates calcineurin (CaN),
Ca2+/calmodulin-dependent kinase II and protein kinase
C, which stimulate the activation of calcium-related pathway
transcription factor and nuclear factor of activated T-cells (NFAT)
and regulate transcription and expression of downstream target
proteins (19). It has been
reported that the NFATc family is a class of
Ca2+/CaN-dependent transcription factors, including
NFATc1, NFATc2, NFATc3, NFATc4 and NFAT5, which are widely
distributed in various human tissue (19). CaN is key for skeletal muscle
differentiation and regeneration and conversion of type II skeletal
muscle fibers to type I. The NFAT family regulates expression of
type I and II skeletal muscle fiber genes. Specifically, NFATc1
activates the promoter of type I skeletal muscle fiber gene and
upregulates its expression, but inhibits the promoter of the type
II skeletal muscle fiber gene (20). CaN/NFATc1 pathway is involved in
bone resorption and reconstruction, as well as changes in bone
microenvironment, which is associated with occurrence of bone
cancer (20). Thus, the present
study investigated changes in the NFATc1-regulated pathway in
osteosarcoma cells.
To the best of our knowledge, the mediation of the
canonical Wnt/Ca2+ pathway by PROSER2 to regulate
progression and metastasis of osteosarcoma has not been reported.
Thus, in the present study, the prognostic value of PROSER2 in
patients with osteosarcoma and the molecular mechanism of PROSER2
in the progression and metastasis of osteosarcoma was investigated.
It is key to find new prognostic biomarkers, which is helpful to
evaluate the clinical diagnosis and treatment effects in patients
with osteosarcoma.
Materials and methods
Cell lines, reagents and clinical
specimens
Human fetal osteoblasts (hFOB1.19, cat. no.
CL-0353), MG63 (Cat. No. CL-0157) and U2OS (cat. no. CL-0236, all
Procell Life Science &Technology Co., Ltd.) cells were cultured
in Dulbecco's modified Eagle's medium (cat. no. 11965092; Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(cat. no. SH30071.03; HyClone; Cytiva) and 1X
penicillin-streptomycin solution (cat. no. P1400, Beijing Solarbio
Science & Technology Co., Ltd.) at 37˚C in a humidified
atmosphere containing 5% CO2. The complete medium for
hFOB1.19 cells was supplemented with 0.3 mg/ml geneticin G418 (cat.
no. A1720; Sigma-Aldrich; Merck KGaA). Lipofectamine™ 2000 (cat.
no. 11668-027) was purchased from Invitrogen (Thermo Fisher
Scientific, Inc.). MTT was obtained from Sigma-Aldrich (Merck
KGaA). The NFAT inhibitor VIVIT (cat. no. HY-P1026) was obtained
from MedChemExpress. Clinical specimens collected from the
Orthopedics Department at Chengdu Fifth People's Hospital (Chengdu,
China) between February 2021 and January 2022 comprised four pairs
of osteosarcoma specimens and paired paratumor (distance, <3 cm)
tissue. The total number of patients was 4 including 1 male and 3
females. The age range of patients was 12.5-17.6 years old. The
patients did not receive chemotherapy, radiotherapy or other
clinical therapy. The specimens were obtained following routine
surgery. The present study was conducted in accordance with the
Declaration of Helsinki (2013) and approved by the Ethics Committee
of Chengdu Fifth People's Hospital (approval no. 2020A-00173;
Chengdu, China). All patients or their parent/legal guardian
provided written informed consent for participation.
Analysis of PROSER2 expression and
clinicopathological indicators
Patients with osteosarcoma in TCGA database
(ocg.cancer.gov/programs/target) were divided into two
groups according to the median value of PROSER2 expression,
including 50 cases in the low and 51 cases in the high PROSER2
expression group. R software (R version 4.2.1) was used to analyze
the correlation between PROSER2 expression and clinicopathological
parameters of patients with osteosarcoma using the Wilcoxon
rank-sum (continuous variables) or Pearson's χ2 test
(rank variables). Hazard ratio (HR) was calculated using univariate
and multivariate analyses.
Survival analysis of patients with
osteosarcoma
The survival probability of patients with
osteosarcoma in low and high PROSER2 groups was analyzed using the
Xiantao platform (https://xiantao.love). The survival probability of
subgroup analysis was also determined by sex, metastasis, tumor
region and primary site progression. HR and its corresponding
log-rank P-value (P<0.05) were calculated for overall survival
(OS).
Gene Ontology (GO)/Kyoto Encyclopedia
of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis
(GSEA)
Xiantao database (xiantao.love/products) is
primarily used for gene expression, co-expression, enrichment and
interaction network analysis and contains the RNA sequencing and
microarray data from TCGA database and part of the Gene Expression
Omnibus database. Numerous researchers have published articles
using the Xiantao database and similar procedures to the present
study (21-24).
GO, KEGG and GSEA analyses were performed on PROSER2 differentially
expressed genes (DEGs) using Xiantao platform. Firstly, single gene
difference analysis was performed using the DESeq2 package (version
1.26.0) (25). GO and KEGG
analysis criteria were absolute value log2 fold change
>1 and adjusted P-value (p.adj)<0.01. GSEA was conducted
according to ID and log2 fold change values of the DEGs.
Gene set permutations were performed 1,000 times for each analysis
and the screening criteria were false discovery rate (FDR)<0.25
and p.adj<0.05.
PROSER2 short hairpin (sh)RNA
lentiviral particle transduction
PROSER2 human shRNA (locus ID 254427; cat. no.
TL306312V) and control lentiviral particles (cat. no. TR30021V)
were purchased from OriGene Technologies, Inc. Human shRNA
lentiviral particles contained four unique 29-mer target-specific
shRNA with lentiviral titer >1x107 TU/ml.
Multiplicity of infection used to infect osteosarcoma cells was 30.
The transduction of PROSER2 and control shRNA lentiviral particles
(generation system used was 3rd) was performed according to the
manufacturer's protocol (cat. no. TL306312V, OriGene Technologies).
Briefly, 5x104 MG63 or U2OS cells were plated in 24-well
plates and incubated for 18 h at 37˚C with 5% CO2.
Appropriate amounts of lentiviral particles were supplemented with
8 µg/ml Polybrene® (cat. no. sc-134220, Santa Cruz
Biotechnology, Inc.) to a total volume of 500 µl. After 24 h at
37˚C, the culture medium was removed and 1 ml fresh complete DMEM
without Polybrene was added. Stable clones expressing PROSER2 or
control shRNA were screened by splitting cells at a 1:10 ratio in
complete DMEM containing 10 µg/ml puromycin (Santa Cruz
Biotechnology, Inc.). Finally, fresh puromycin-containing medium
was replaced every 5 days until resistant colonies were identified
for stable PROSER2 or control shRNA expression. The concentration
of puromycin used for selection and maintenance is 10 and 2 µg/ml.
The screening time continued for 1 month.
Wound healing assay
The stable cells infected with PROSER2 and control
shRNA lentivirus were cultured on 6-well plates without serum
(3x105 cells/well). Following adherence, cells at 100%
confluence were scratched horizontally across the wall with a
disposable pipette tip. The dislodged cells were washed with PBS
buffer three times and discarded. Then, cells were cultured with
serum-free DMEM for 24 and 48 h. The cells were photographed using
a microscope(Olympus CX43, 40X magnification).
Migration and invasion assay
Migration and invasion assays were performed using
Boyden chambers with Transwell membrane filter inserts (cat. no.
3422; Corning Costar, Inc.). Briefly, 3x104 cells/well
in serum-free DMEM were seeded into the upper chambers of a
24-well. DMEM containing 10% FBS were added into the lower
chambers. Transwell chamber (pore size, 8 µm) for the migration
assay and incubated at 37˚C for 24 and 48 h. MG63 cells on the
lower surface of the filter were fixed with 10% formalin solution
for 30 min and stained with 0.1% crystal violet for 30 min at room
temperature. The number of migratory cells from five randomly
selected fields of view in a single chamber of three samples was
counted under a light microscope (mean ± SEM). For the invasion
assay, Matrigel (cat. no. 354248; BD Biosciences) was applied at
37˚C for 5 h to coat the upper chamber, and the other steps were
the same as that of the migration assay.
Nuclear and cytoplasmic protein
extraction
PROSER2 and control shRNA lentiviruses infected MG63
cells were treated with 10 µM VIVIT at 37˚C for 24 h. Then, cells
were collected and nuclear and cytoplasmic proteins were extracted
using the Nuclear and Cytoplasmic Protein Extraction kit (cat. no.
P0027; Beyotime Institute of Biotechnology). This kit uses
cytoplasmic protein extraction reagents A and B to fully expand
cells under low osmotic pressure conditions, then destroy the cell
membrane, release cytoplasmic proteins and then obtain cell nucleus
pellets by centrifugation at 12,000 g 10 min, at 4˚C. Finally,
nuclear protein was extracted using high-salt nuclear protein
extraction reagent (cat. no. P0027; Beyotime Institute of
Biotechnology).
Western blot analysis
The primary antibodies were as follows: Anti-Wnt5a
(cat. no. ab235966; Abcam), NFATC1 polyclonal (A01; cat. no.
H00004772-A01; Abnova), anti-cyclooxygenase-2 (COX2; cat. no.
ab198646; Abcam), anti-MMP2 (EPR17003-25; cat. no. ab181286;
Abcam), monoclonal anti-MMP9 (cat. no. SAB5300247; Sigma-Aldrich;
Merck KGaA) and β-actin (SP124; cat. no. ab115777; Abcam). Cell
lysate was prepared using Cell lysis buffer for Western and IP
(cat. no. P0013; Beyotime Institute of Biotechnology). Protein
determination method is BCA assay and 20 µg of protein loaded per
lane. Total protein was separated by 10% SDS-PAGE and transferred
onto nitrocellulose membranes at 300 mA for 2 h. The membranes were
blocked with 5% bovine serum albumin (Thermo Fisher Scientific,
Inc.) for 1 h at room temperature and incubated with primary
antibody at 1:1,000 dilution at 4˚C overnight. The membranes were
washed three times for 5 min with 1X Tris-buffered saline + 0.05%
Tween 20. The membrane was incubated with secondary antibody at a
dilution of 1:10,000 for 1 h and washed three times as
aforementioned. Goat anti-rabbit IgG H&L (cat. No. ab6721;
Abcam) HRP, 1:10,000 dilution; incubated at room temperature for 1
h. Bands were visualized using enhanced chemiluminescent kit
(Pierce, Thermo Fisher Scientific, Inc.). ImageJ software (version
1.8.0; National Institutes of Health) was used to analyze the
densitometry of the bands.
Apoptosis assay
MG63 cells were infected with PROSER2 and control
shRNA lentiviruses and stable clones with PROSER2 shRNA and control
shRNA were selected (2x105 cells/plate). Each group was
treated with 3 µg/ml cyclosporine A (CsA, MedChemExpress LLC) for
24 h at 37˚C. The cells were centrifuged twice at 400 g for 10 min
at 4˚C. The cell apoptosis rate (early + late apoptosis) was
determined by FACS using BD LSRFortessa X-20 flow cytometer and BD
FACSDiva™ software version 6.0 (BD Biosciences) with Annexin V-FITC
dual staining kit (cat. no. C1062S-1; Beyotime Institute of
Biotechnology) according to the manufacturer's instructions.
Confocal microscopy imaging
Intracellular Ca2+ was measured by
incubation with Ca2+-selective fluorescent indicator
Fura-2 AM (cat. no. ab120873; Abcam) with a purity of >99%.
Briefly, human osteosarcoma MG63 cells were infected with PROSER2
shRNA and control shRNA lentiviral particles for 48 h. The cells in
each group were loaded with 2 µM Fura-2 AM in DMSO/HBSS for 30 min
at room temperature in the dark. The cells were washed with
prewarmed HBSS and incubated for 30 min at 37˚C in the dark.
Finally, the cells were washed again with pre-warmed HBSS and live
cells were imaged using a confocal microscope (excitation laser,
405 nm; emission gate center, 519 nm; magnification, x200).
Statistical analysis
The data from MTT, migration and FACS assays were
analyzed using SPSS software version 24 (IBM Corp.). The results
are presented as the mean ± standard deviation. One-way ANOVA
followed by Tukey's post hoc test or two-way ANOVA, followed by
Bonferroni's post hoc test was used for comparisons between >2
groups. Matched data, such as tumor and adjacent normal tissue,
were compared using a paired t test (parametric data). All
experiments were repeated twice with technical duplicates.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Correlation of PROSER2 expression and
clinicopathological parameters
The data of patients with osteosarcoma was analyzed
(Table I). Briefly, according to
the PROSER2 expression level, 101 patients with osteosarcoma were
divided into high and low PROSER2 groups using Wilcoxon rank-sum
test (continuous variables). The overall survival rate was
significantly different between the two groups. Univariate and
multivariate analyses were performed, and HRs were calculated
(Table II). The results showed
that expression of PROSER2 and metastasis were independent
prognostic factors for patients with osteosarcoma.
 | Table IBaseline data in patients with
osteosarcoma. |
Table I
Baseline data in patients with
osteosarcoma.
| Characteristic | Low expression of
PROSER2 (n=50.00) | High expression of
PROSER2 (n=51.00) | P-value |
|---|
| Age, years (%) | | | 0.597 |
|
<18 | 37.00 (36.60) | 41.00 (40.60) | |
|
≥18 | 13.00 (12.90) | 10.00 (9.90) | |
| Sex, n (%) | | | 0.626 |
|
Female | 22.00 (21.80) | 19.00 (18.80) | |
|
Male | 28.00 (27.70) | 32.00 (31.70) | |
| Race, n (%) | | | Not analyzed |
|
Native
American or Indigenous Alaskan | 0.00 (0.00) | 1.00 (1.30) | |
|
Asian | 4.00 (5.30) | 3.00 (3.90) | |
|
Black or
African American | 3.00 (3.90) | 7.00 (9.20) | |
|
Native
Hawaiian or Pacific Islander | 0.00 (0.00) | 0.00 (0.00) | |
|
White | 35.00 (46.10) | 23.00 (30.30) | |
| Metastasis, n
(%) | | | 0.074 |
|
No | 42.00 (41.60) | 34.00 (33.70) | |
|
Yes | 8.00 (7.90) | 17.00 (16.80) | |
| Primary site
progression, n (%) | | | >0.999 |
|
No | 14.00 (27.50) | 18.00 (35.30) | |
|
Yes | 8.00 (15.70) | 11.00 (21.60) | |
| Overall survival
event, n (%) | | | <0.001 |
|
Alive | 38.00 (38.40) | 20.00 (20.20) | |
|
Dead | 12.00 (12.10) | 29.00 (29.30) | |
| Median age, years
(IQR) | 15.35 (12.78,
18.06) | 15.06 (12.23,
17.56) | 0.632 |
 | Table IIUnivariate and multivariate analysis
of patient characteristics. |
Table II
Univariate and multivariate analysis
of patient characteristics.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Characteristic | Total, n | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age, years | 99 | | | | |
|
<18 | 76 | Reference | | | |
|
≥18 | 23 | 0.732
(0.325-1.653) | 0.454 | | |
| Metastasis | 99 | | | | |
|
No | 75 | Reference | | | |
|
Yes | 24 | 3.679
(1.964-6.892) |
<0.001a | 4.102
(2.162-7.783) |
<0.001a |
| Race | 74 | | | | |
|
White | 57 | Reference | | | |
|
Black or
African | 17 | 1.246
(0.494-3.142) | 0.641 | | |
|
American and
Asian | | | | | |
| Primary site
progression | 50 | | | | |
|
No | 31 | Reference | | | |
|
Yes | 19 | 1.769
(0.864-3.626) | 0.119 | | |
| PROSER2
expression | 99 | | | | |
|
Low | 50 | Reference | | | |
|
High | 49 | 3.662
(1.860-7.208) |
<0.001a | 4.016
(2.020-7.984) |
<0.001a |
Prognostic value of PROSER2 in
patients with osteosarcoma
The association between PROSER2 expression and
survival in patients with osteosarcoma was investigated using
Kaplan-Meier plotter in the Xiantao database. High expression of
PROSER2 was significantly associated with poorer OS (HR=3.66; 95%
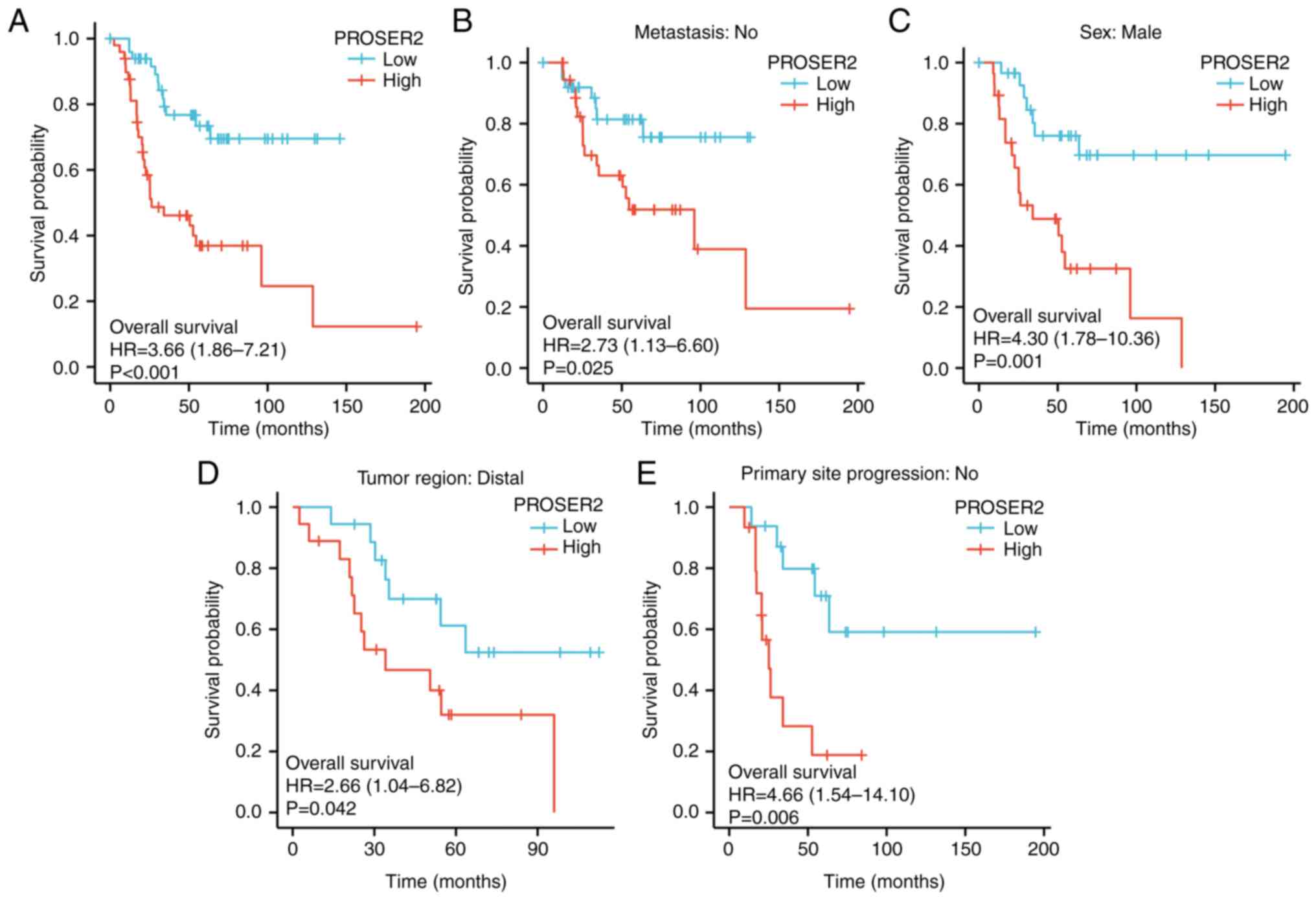
CI, 1.86-7.21; Fig. 1).
Significant effects in subgroup analysis were exhibited only in
patients with osteosarcoma without metastasis, with distal tumor
regions, no primary site progression and in males with
osteosarcoma. The HR and 95% CIs were 2.73 (1.13-6.60), 2.66
(1.04-6.82), 4.66 (1.54-14.10) and 4.30 (1.78-10.36). All data
demonstrated that high PROSER2 expression was associated with poor
survival probability in patients with osteosarcoma. Collectively,
these results suggest that PROSER2 was a valuable prognostic
predictor in patients with osteosarcoma.
Expression of PROSER2 is upregulated
in clinical specimens and osteosarcoma cell lines
Expression of PROSER2 was detected in four pairs of
osteosarcoma clinical specimens, including tumor and paired
paratumor tissue. Expression of PROSER2 was significantly higher in
osteosarcoma than in paratumor tissue (Fig. 2A). Expression of PROSER2 in human
osteoblast and osteosarcoma cell lines was detected by western blot
analysis. Expression of PROSER2 was significantly increased in MG63
and U2OS cells compared with that in hFOB1.19 cells (Fig. 2B). These data suggested that
PROSER2 expression was higher in osteosarcoma cancer cells.
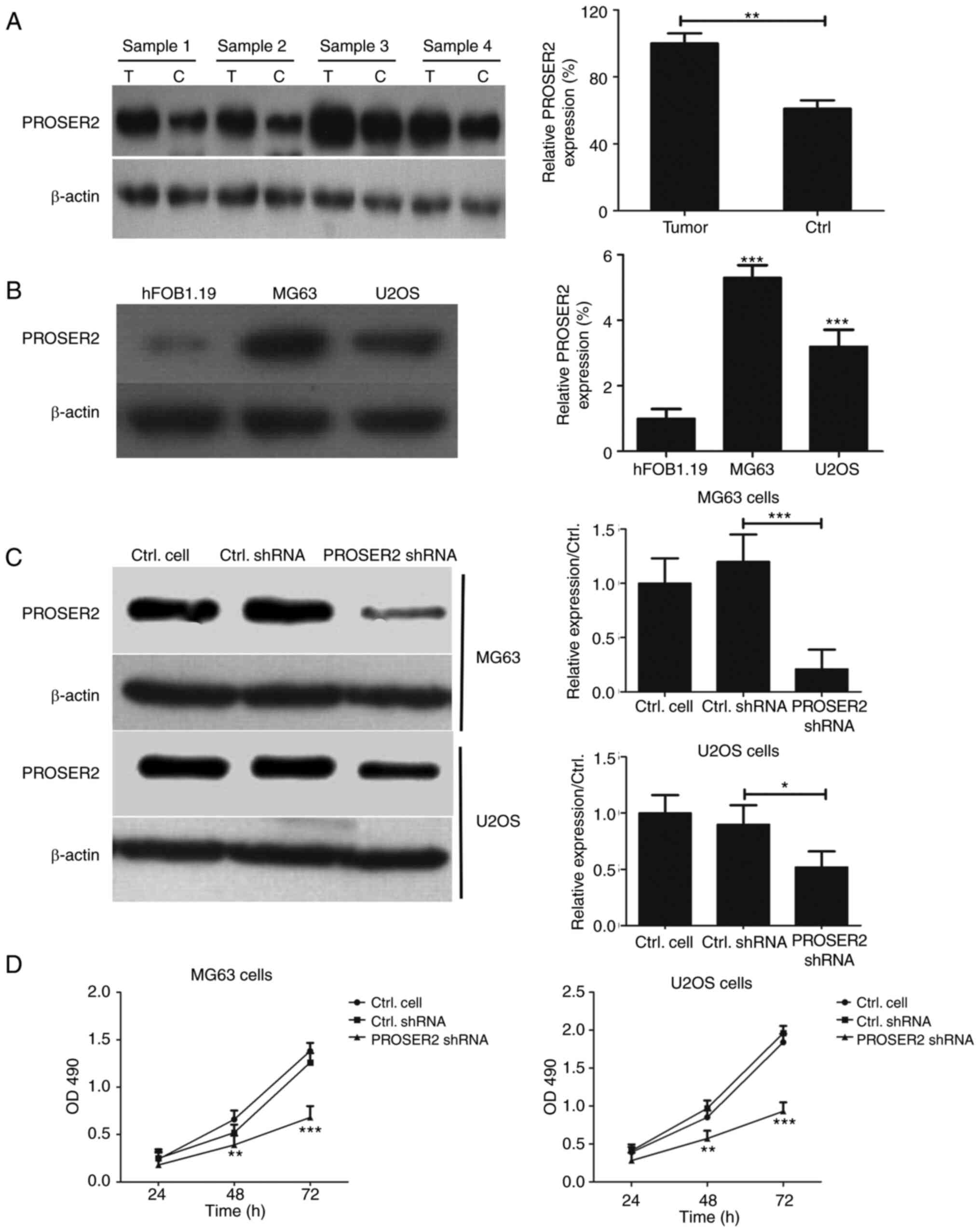 | Figure 2PROSER2 knockdown inhibits viability
of osteosarcoma cells. (A) Expression of PROSER2 was determined by
western blot analysis in four pairs of clinical specimens.
**P<0.01. (B) PROSER2 levels were determined by
western blot analysis in hFOB1.19, MG63 and U2OS cells.
***P<0.001 vs. hFOB1.19. (C) MG63 and U2OS cells were
infected with PROSER2 and control shRNA lentivirus for 24 h and
PROSER2 expression was determined by western blotting analysis.
*P<0.05, ***P<0.001. (D) Viability of
MG63 and U2OS cells infected with PROSER2 or Ctrl shRNA lentivirus
and cultured for 24, 48 and 72 h was assessed by MTT assay.
**P<0.01, ***P<0.001 vs. Ctrl shRNA.
PROSPER2, proline- and serine-rich 2; Ctrl, control; T, tumor
tissue; C, paratumor tissue; OD, optical density; sh, short
hairpin. |
PROSER2 knockdown inhibits viability
of MG63 cells and U2OS cells
Next, PROSER2 and control shRNA lentiviruses were
used to infect osteosarcoma cells and western blotting was
performed to test expression of PROSER2 in MG63 and U2OS cells.
Expression of PROSER2 was significantly suppressed in PROSER2 shRNA
lentivirus-infected MG63 and U2OS cells compared with control shRNA
lentivirus-infected MG63 and U2OS cells (Fig. 2C).
To investigate whether interference with PROSER2
affects osteosarcoma cell proliferation, MTT assay was performed to
test the viability of PROSER2 and control shRNA lentivirus-infected
MG63 and U2OS cells. PROSER2 knockdown inhibited the viability of
MG63 and U2OS cells compared with control shRNA lentivirus at 48
and 72 h (Fig. 2D).
PROSER2 knockdown inhibits cell
proliferation, migration and invasion of MG63 cells
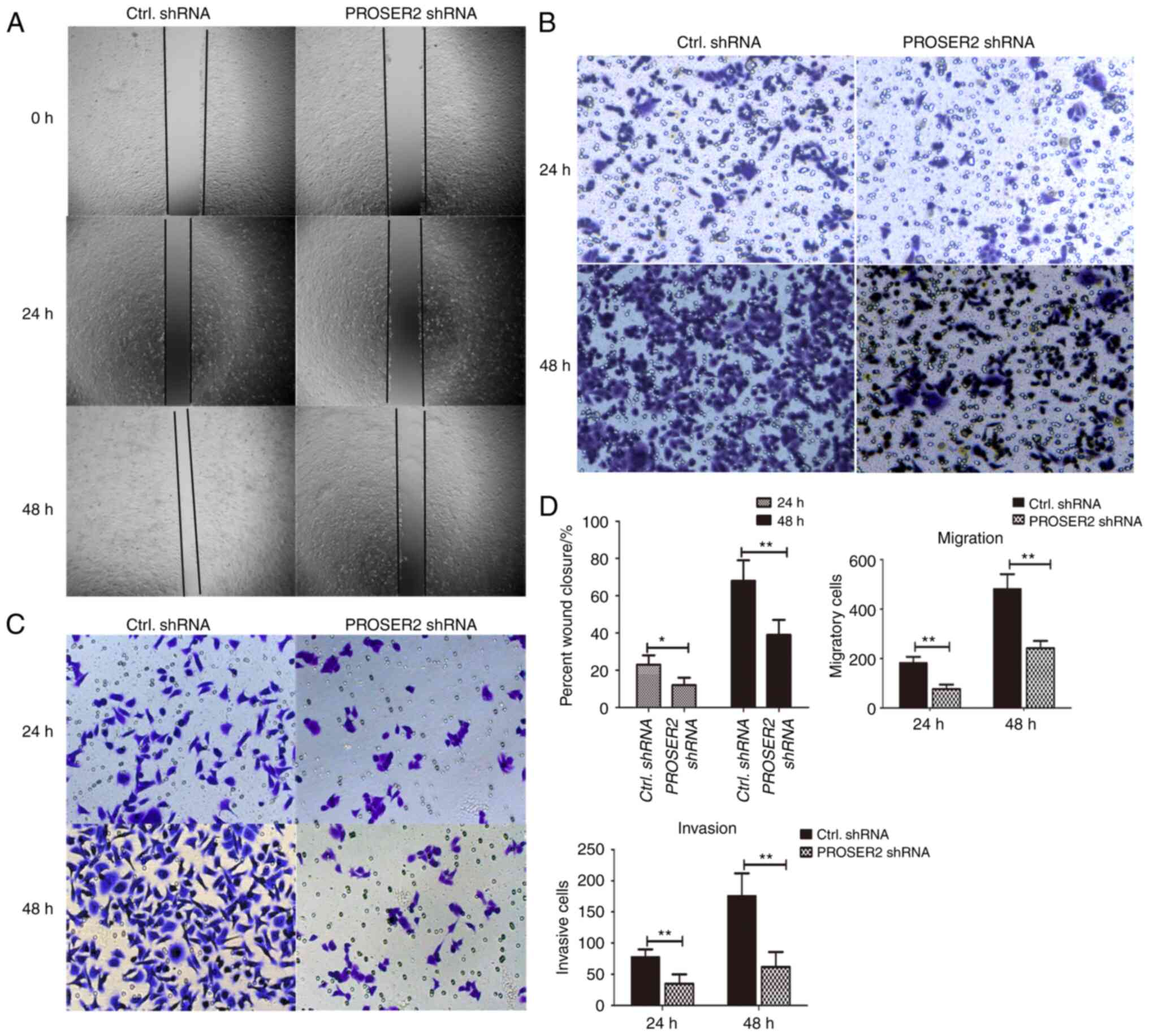
To identify whether PROSER2 regulates proliferation,
migration and invasion of osteosarcoma cells, wound closure, cell
migration and invasion assays were performed. MG63 cells were
infected with PROSER2 and control shRNA lentiviruses for 24 and 48
h (Fig. 3A and C). Wound closure was significantly lower
in PROSER2 shRNA lentivirus-infected MG63 cells than in control
shRNA-infected MG63 cells at both timepoints (Fig. 3D). The migratory ability of PROSER2
shRNA lentivirus-infected MG63 cells was assessed using Transwell
assay. The migratory ability of PROSER2 shRNA lentivirus-infected
MG63 cells was significantly decreased compared with that of
control shRNA lentivirus-infected cells at 24 and 48 h (Fig. 3B and D) suggesting the invasive ability of MG63
cells were inhibited after PROSER2 knockdown. These data revealed
that PROSER2 knockdown inhibited proliferation, migration and
invasion of MG63 cells.
GSEA and GO/KEGG analysis
GSEA and GO/KEGG analyses were performed using the
Xiantao platform. Firstly, patients with osteosarcoma were divided
into high and low PROSER2 expression groups. DEGs were screened
with the threshold values of |log2(FC)|>1 and
p.adj<0.01, from which 2,348 coding genes were selected for
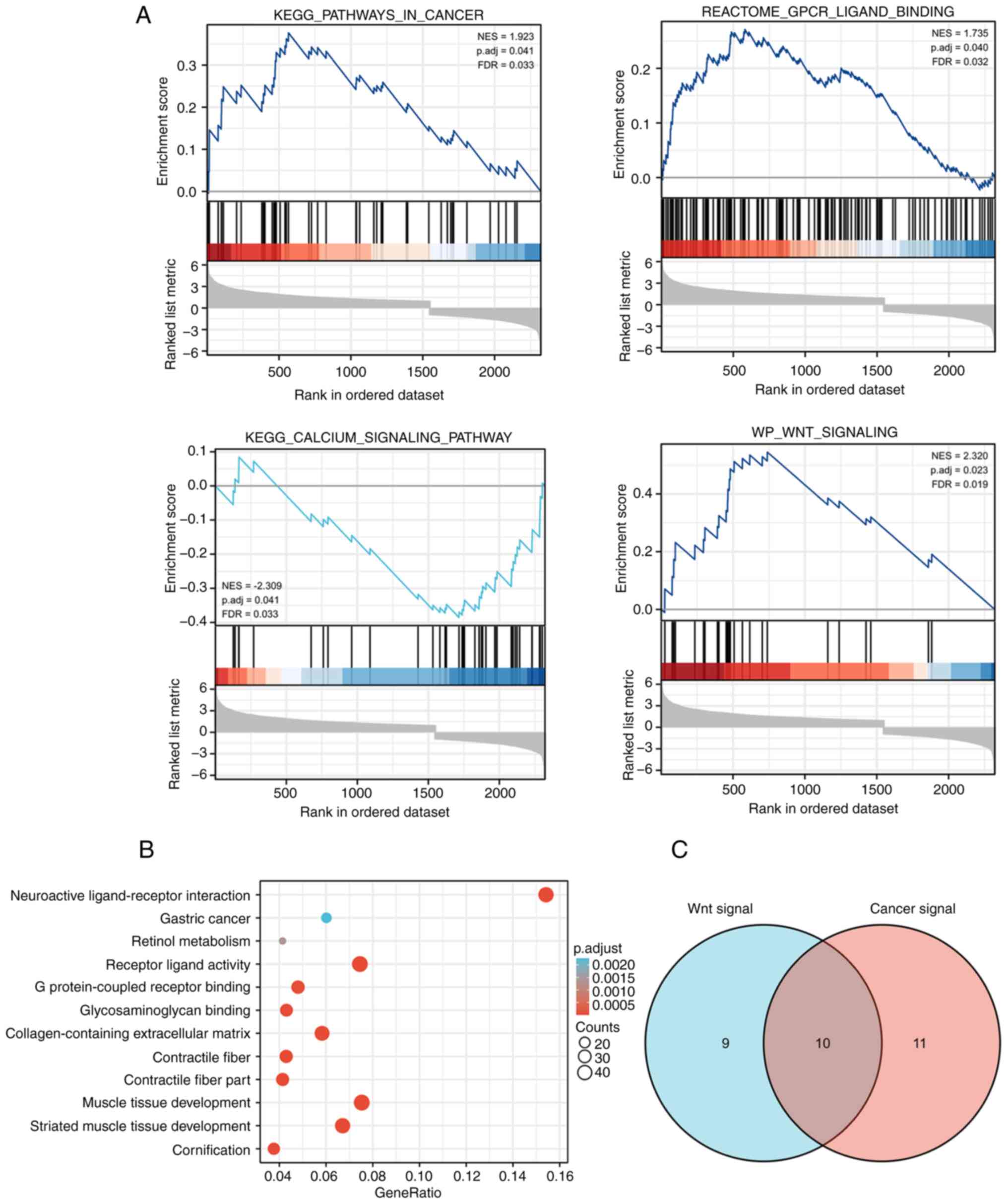
GSEA. A total of 105 signaling pathways were significantly enriched
at FDR<0.05 and p.adj<0.05. The differentially enriched terms
included ‘KEGG pathways in cancer’, ‘GPCR ligand binding’, ‘calcium
signaling pathway’ and ‘Wnt signaling pathway’ (Fig. 4A; Table SI). KEGG analysis results
(Fig. 4B; Table SII) revealed that PROSER2 DEGs
were primarily involved in pathways such as ‘neuroactive
ligand-receptor interactions’, ‘gastric cancer’ and ‘retinol
metabolism’. GO analysis revealed that PROSER2 DEGs were primarily
involved in ‘muscle tissue development’, ‘regulation of ion
transmembrane transport’, ‘cellular calcium ion homeostasis’ and
‘calcium ion transport’ in the biological process group (Table SII). In the molecular function
group, the DEGs were primarily enriched in ‘receptor-ligand
activity’, ‘DNA-binding transcription activator activity’, ‘ion
channel activity’ and ‘G protein-coupled receptor binding’. In the
cellular component group, DEGs were primarily enriched in
‘collagen-containing extracellular matrix’, ‘transmembrane
transporter complex’, ‘ion channel complex’ and ‘transporter
complex’.
To determine the molecular mechanisms of
PROSER2-regulated proliferation, migration and invasion of MG63
cells, a Venn diagram was constructed based on ‘Wnt signaling
pathway’ and ‘pathways in cancer’ terms and ten overlapping genes
were screened: Frizzled 10(FZD10), WNT10B, WNT1, WNT4, WNT3A,
WNT11, WNT2B, FZD9, WNT2 and WNT6 (Fig. 4C). Among these, WNT4, WNT11 and
WNT6 are key molecules regulating the Wnt/Ca2+ signaling
pathway in cancer progression (17).
PROSER2 is involved in the
Wnt/Ca2+ signaling pathway in MG63 cells
As aforementioned, calcium and Wnt signaling
pathways were significantly enriched based on the GSEA. Therefore,
whether PROSER2 regulated the proliferation, migration and invasion
of osteosarcoma cells via the Wnt/Ca2+ signaling pathway
was evaluated. Expression of Wnt5a, COX2, MMP-2 and MMP-9 was
significantly decreased in PROSER2 shRNA lentivirus-infected MG63
compared with control shRNA lentivirus-infected MG63 cells
(Fig. 5A and B). Moreover, to test whether nuclear
translocation of NFATc1 was changed in PROSER2 shRNA-transfected
MG63 cells, nuclear NFATc1 expression was examined by western blot
analysis. Expression of nuclear NFATc1 was significantly decreased
in PROSER2 shRNA lentivirus-infected MG63 cells compared with that
in control cells (Fig. 5C and
D). CaN promotes nuclear
translocation of NFATc1 via dephosphorylation and activates
downstream target genes (19).
Therefore, expression of the catalytic subunit of CaN (CNA) was
measured in the cytoplasm of PROSER2 shRNA lentivirus-infected MG63
cells using western blotting. The expression of CNA was
significantly decreased in PROSER2 shRNA lentivirus-infected MG63
cells compared with that in control cells (Fig. 5C and D). Moreover, the intracellular
Ca2+ concentration in PROSER2 shRNA and control shRNA
lentivirus-infected osteosarcoma cells was assessed using the
Ca2+ sensing probe, Fura-2 AM. Ca2+
concentration was notably decreased in PROSER2 shRNA
lentivirus-infected osteosarcoma cells compared with that in
control shRNA lentivirus-infected osteosarcoma cells (Fig. 5E). These data revealed that PROSER2
regulated migration and invasion of osteosarcoma cells, potentially
via the Wnt/Ca2+/NFATc1 signaling pathway.
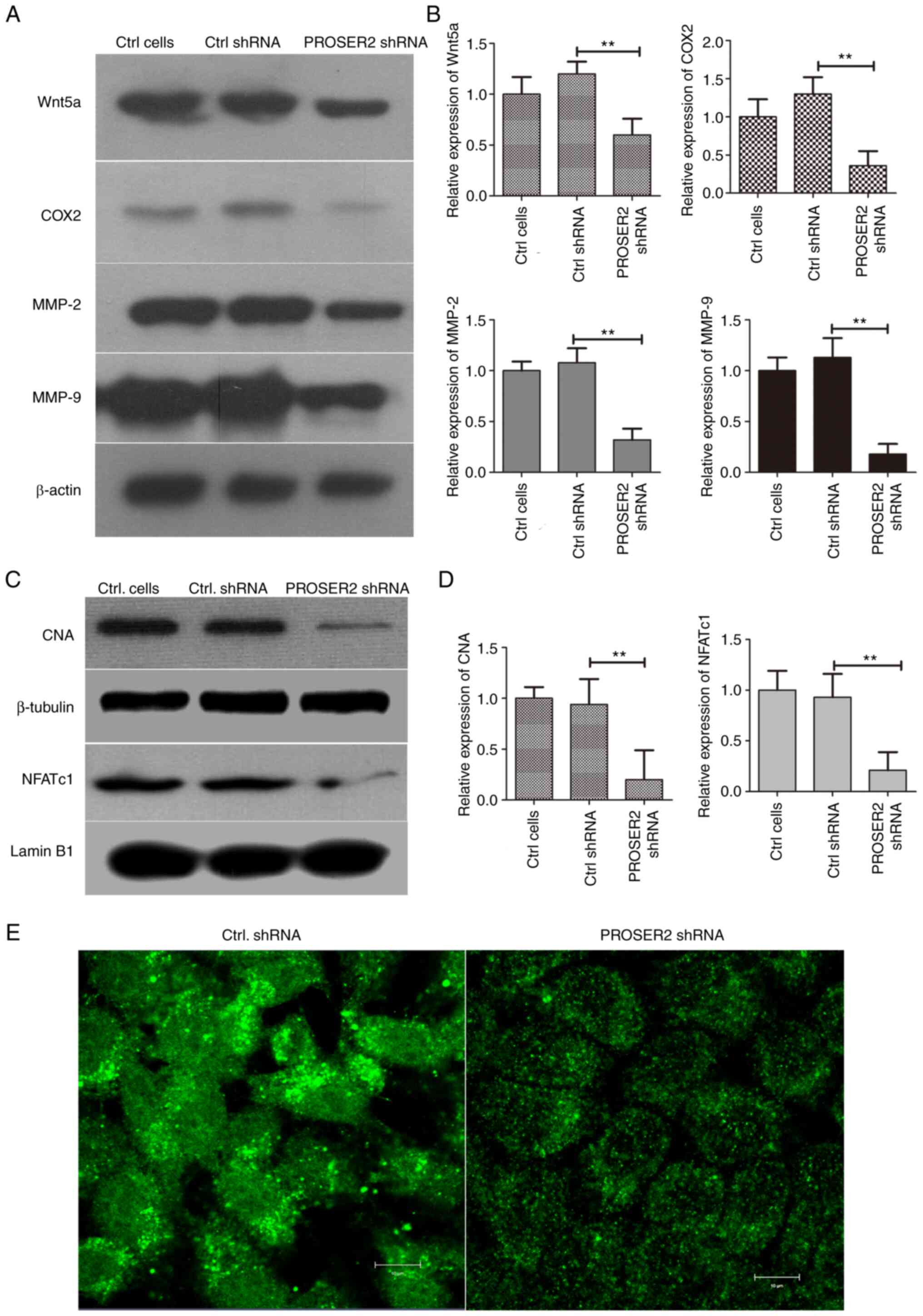 | Figure 5PROSER2 is involved in the
Wnt/Ca2+ signaling pathway in MG63 cells. (A) MG63 cells
were infected with PROSER2 and Ctrl shRNA lentivirus for 24 h. The
expression of Wnt5a, COX2, MMP-2 and MMP-9 was detected by western
blot analysis in PROSER2 and Ctrl shRNA lentivirus-infected MG63
cells. (B) Histograms of relative expression of Wnt5a, COX2, MMP-2
and MMP-9. (C) Nuclear expression of NFATc1 and cytoplasmic CNA
were detected by western blotting analysis in PROSER2 and Ctrl
shRNA lentivirus-infected MG63 cells. (D) Histograms of relative
expression of NFATc1 and CNA. **P<0.01. (E) Confocal
microscopy imaging. Intracellular Ca2+ was measured by
incubation with the Ca2+ selective fluorescent
indicator, Fura-2 AM. Scale bar, 10 µM. PROSPER2, proline- and
serine-rich 2; NFATc1, nuclear factor of activated T-cells 1; CNA,
catalytic subunit of calcineurin; Ctrl, control; sh, short hairpin;
COX, cyclooxygenase; MMP, matrix metalloproteinase. |
Interference with PROSER2 promotes
apoptosis of MG63 cells via suppression of nuclear localization of
NFATc1 mediated by VIVIT and CsA
Whether PROSER2 affected the nuclear translocation
of NFATc1 was investigated. VIVIT is a specific inhibitor of NFAT
and is a cell-permeable peptide that selectively inhibits
CaN-mediated dephosphorylation of NFAT (19). PROSER2 and control shRNA lentivirus
MG63 cells were treated with 10 µM VIVIT at 37˚C for 24 h. Nuclear
expression of NFATc1, as well as total Wnt5a, COX2, MMP-9 and MMP-2
expression levels, were detected by western blot analysis. Nuclear
NFATc1 expression, as well as total COX2, MMP-9 and MMP-2
expression were significantly decreased in VIVIT-treated PROSER2
shRNA lentivirus-infected MG63 cells compared with
non-VIVIT-treated cells (Fig. 6).
However, expression of Wnt5a was not significantly changed in
VIVIT-treated and non-VIVIT-treated groups, suggesting that
interference with PROSER2 inhibited the expression of Wnt5a but
VIVIT treatment did not affect the expression of Wnt5a in MG63
cells. These results revealed that interference with PROSER2
promoted inhibition of NFATc1 by VIVIT in MG63 cells (Fig. 6).
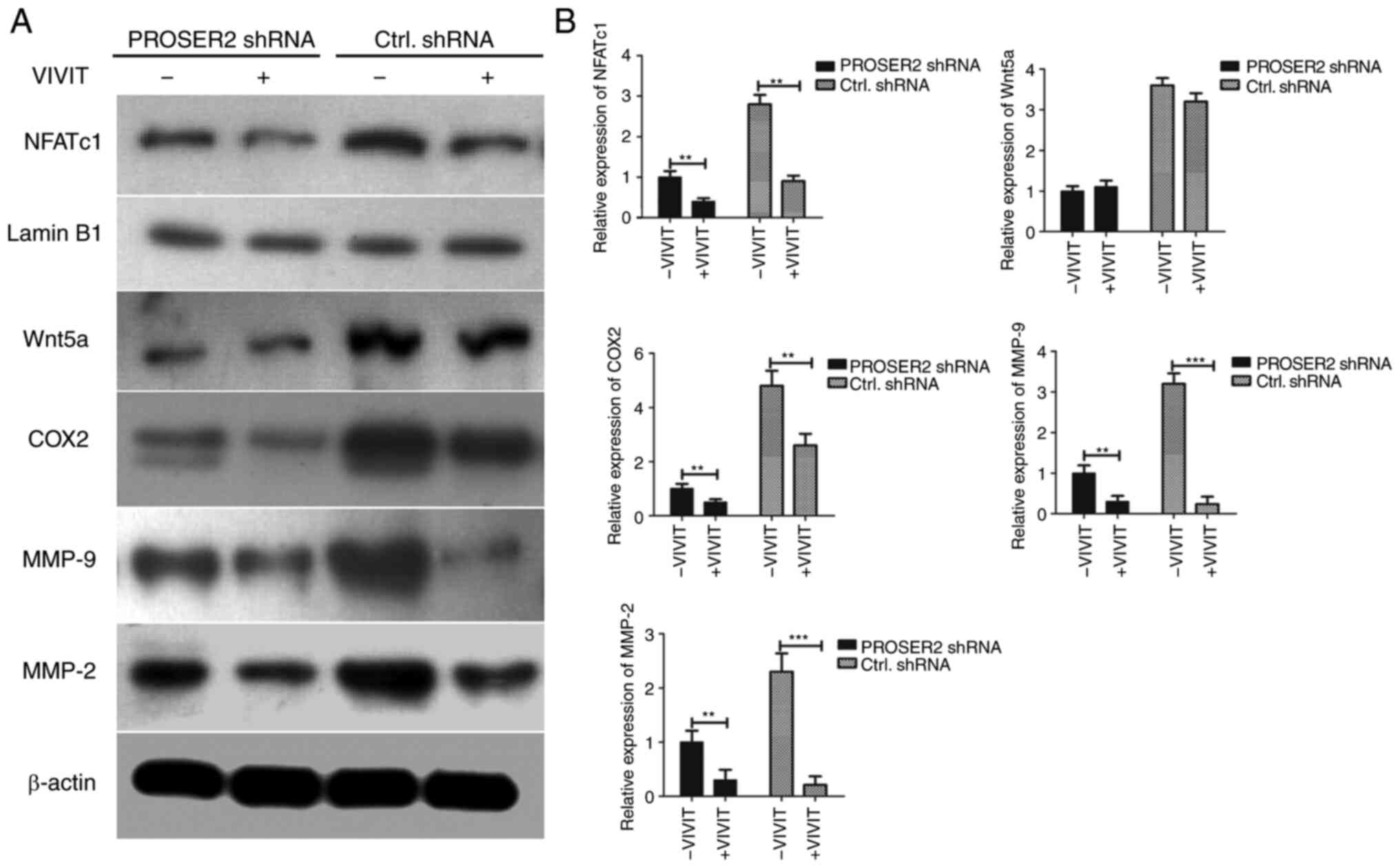 | Figure 6Interference with PROSER2 promotes
VIVIT-induced inhibition of NFATc1 in MG63 cells via suppressing
nuclear localization of NFATc1. (A) MG63 cells were transfected
with PROSER2 or Ctrl shRNA lentivirus and treated with 10 µM VIVIT
for 24 h. The nuclear expression of NFATc1 and lamin B1 was
detected by western blotting. Lamin B1 was used as a nuclear
internal reference protein. The expression of Wnt5a, COX2, MMP-2
and MMP-9 were determined by western blotting analysis. (B)
Histograms of relative expression of NFATc1, Wnt5a, COX2, MMP-2 and
MMP-9. **P<0.01 and ***P<0.001.
PROSPER2, proline- and serine-rich 2; NFATc1, nuclear factor of
activated T-cells 1; Ctrl, control; sh, short hairpin; COX,
cyclooxygenase; MMP, matrix metalloproteinase. |
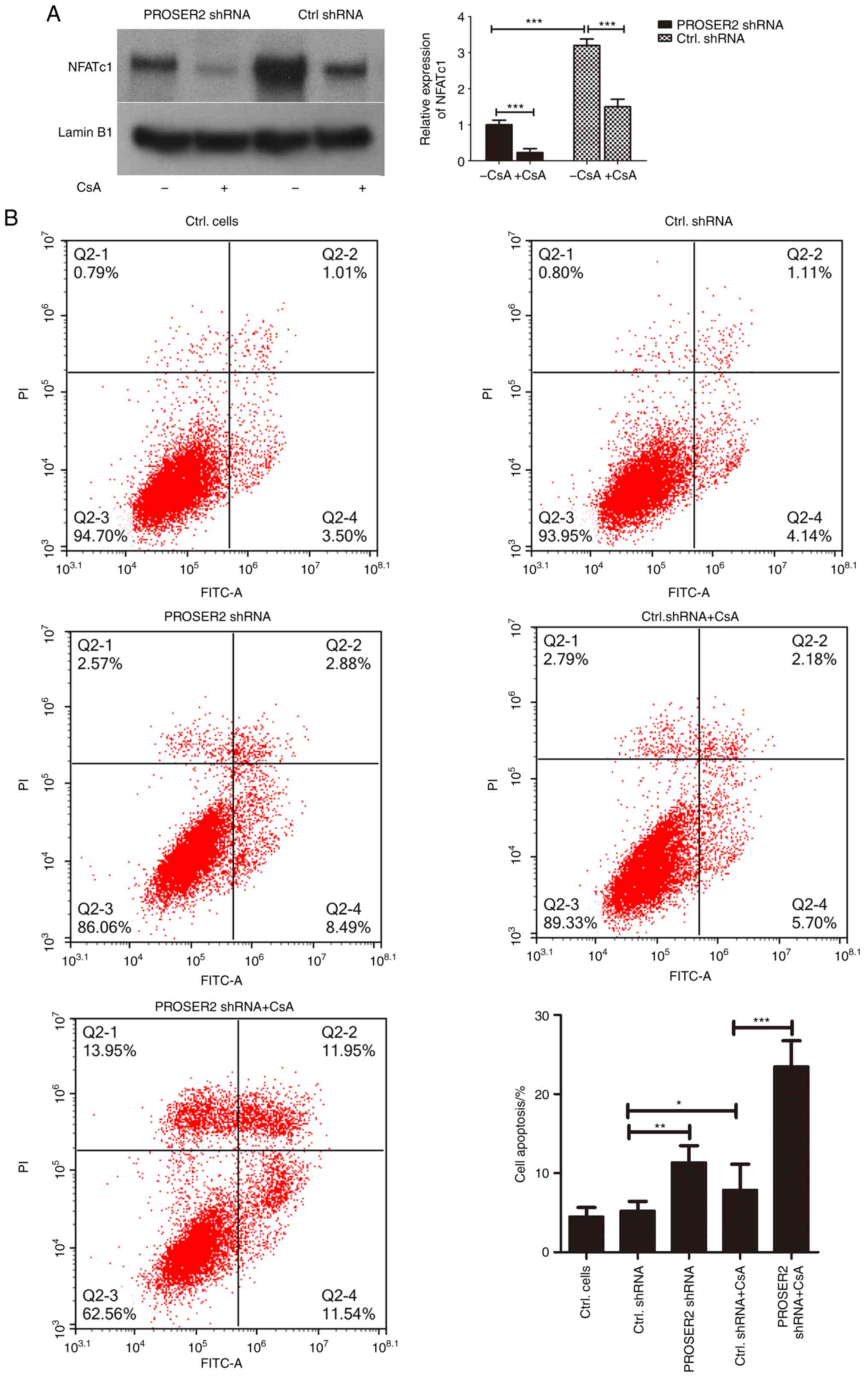
PROSER2 knockdown with lentivirus significantly
inhibited nuclear translocation of NFATc1 in MG63 cells (Fig. 7A). CsA was used to inhibit NFATc1
translocation to the nucleus of MG63 cells. Interference with
PROSER2 promoted inhibition of NFATc1 by CsA in MG63 cells
(Fig. 7A). The apoptosis rate of
PROSER2 shRNA lentivirus-infected osteosarcoma cells was examined
by FACS assay. Apoptosis in MG63 cells infected with PROSER2 shRNA
lentivirus for 48 h was significantly upregulated compared with
cells infected with control shRNA lentivirus (Fig. 7B). Moreover, CsA promoted apoptosis
of MG63 cells and interference with PROSER2 by lentivirus promoted
CsA-mediated apoptosis of MG63 cells (Fig. 7B).
Discussion
Osteosarcoma predominantly occurs in children and
young adults prone to malignant transformation and metastasis
(26). The present study of the
pathogenesis of osteosarcoma may provide an effective reference for
the clinical treatment of patients with osteosarcoma. Here,
Therapeutically Applicable Research to Generate Effective
Treatments (ocg.cancer.gov/programs/target) transcripts per
million reads (TPM) format data in mRNA-sequencing data files from
the Osteosarcoma project in TCGA database was analyzed and revealed
that PROSER2 was an independent prognostic biomarker for patients
with osteosarcoma. High expression of PROSER2 was associated with
poor prognosis in patients. Next, expression of PROSER2 in four
pairs of clinical specimens from patients with osteosarcoma was
detected. Western blot analysis revealed that PROSER2 levels were
significantly higher in osteosarcoma tissue than in paired normal
control tissue. Moreover, PROSER2 levels were significantly higher
in the osteosarcoma cell lines MG63 and U2OS than in hFOB1.19.
These data revealed that PROSER2 served as an oncogene in the
progression of osteosarcoma.
PROSER2 shRNA lentivirus was used to infect MG63 and
U2OS cells. The role of PROSER2 in phenotypic effects was
investigated; interference with PROSER2 significantly decreased
viability, migration and invasion of MG63 and U2OS cells. GSEA was
performed on the Xiantao and the ‘GPCR ligand binding’, ‘calcium
signaling pathway’, and ‘Wnt signaling pathway’ were found to be
enriched in the PROSER2 high-expression group of patients with
osteosarcoma from TCGA database. Next, western blot analysis to was
used to investigate whether PROSER2 regulates proliferation,
migration and invasion of MG63 cells via the Wnt/Ca2+
signaling pathway. It was revealed that levels of Wnt5a, NFATc1,
COX2, MMP-2 and MMP-9 were significantly decreased in PROSER2 shRNA
lentivirus-infected MG63 cells. Wnt5a has previously been found to
promote migration and invasion of osteosarcoma cells via the
Wnt5a/receptor tyrosine kinase-like orphan receptor (ROR)
2(27),
Wnt5a/ROR1/disheveled-associated activator of morphogenesis
1(28) and SRC/ERK/MMP-14 pathway
(29), which was consistent with
the results from the present study indicating that Wnt5a exerted a
tumorigenic role in progression, migration and invasion of
osteosarcoma. Overexpression of COX2 has been detected in various
types of human malignancy and inhibition of COX2 has been found to
suppress tumor cell proliferation and osteosarcoma metastasis
(30,31). MMP-2 and MMP-9 participate in
degrading the extracellular matrix and are involved in the invasion
and migration of cancer cells (32,33).
In the present study, PROSER2 knockdown significantly decreased
expression of NFATc1, COX2, MMP-2 and MMP-9 in osteosarcoma cells.
Notably, nuclear NFATc1 was detected in PROSER2 shRNA
lentivirus-infected MG63 cells using western blot analysis. The
results revealed that interference with PROSER2 significantly
decreased translocation of NFATc1 in MG63 cells, which increased
the inhibitory role of CsA in suppressing apoptosis, migration and
invasion of MG63 cells.
However, the present study did not collect enough
clinical specimens to prove the role of PROSER2 in the progression
of osteosarcoma. Future studies should verify its role and test
whether it could be used as the prognostic or diagnostic biomarker
for clinical therapy of osteosarcoma.
In conclusion, PROSER2 was associated with poor
prognosis in patients with osteosarcoma and served an oncogenic
role in promoting proliferation, migration and invasion of
osteosarcoma cells via the Wnt/Ca2+/NFATc1 signaling
pathway by increasing nuclear localization of NFATc1.
Supplementary Material
Differentially enriched terms by GSEA
analysis
GO/KEGG analysis
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Xinglin Project
of Chengdu University of Traditional Chinese Medicine (grant no.
YYZX2021045).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZB designed the experiments, performed statistical
analysis and wrote the manuscript draft. ZL performed western
blotting analysis and the flow cytometry assay. YZ and YL performed
bioinformatics and the MTT assay. SX performed the lentivirus
transfection assay and helped with the western blotting. SL and JL
collected the clinical samples and performed cell culture. All
authors have read and approved the final manuscript. ZB and ZL
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
This study was approved by the Medical Ethics
Committee of the Chengdu Fifth People's Hospital (approval no.
2020A-00173).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang G, Wu Y, Wan R, Sang H, Liu H and
Huang W: The role of noncoding RNAs in the regulation, diagnosis,
prognosis and treatment of osteosarcoma (review). Int J Oncol.
59(69)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gazouli I, Kyriazoglou A, Kotsantis I,
Anastasiou M, Pantazopoulos A, Prevezanou M, Chatzidakis I,
Kavourakis G, Economopoulou P, Kontogeorgakos V, et al: Systematic
review of recurrent osteosarcoma systemic therapy. Cancers (Basel).
13(1757)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Assi T, Kattan J, Nassereddine H, Rassy E,
Briand S, Court C, Verret B, Le Cesne A and Mir O: Chemotherapy in
the management of periosteal osteosarcoma: A narrative review. J
Bone Oncol. 30(100389)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sugito W and Kamal AF: Clinical outcome
following prolonged neoadjuvant chemotherapy and delayed surgery in
osteosarcoma patients: An evidence-based clinical review. Acta Med
Indones. 54:142–150. 2022.PubMed/NCBI
|
|
5
|
Zhao X, Wu Q, Gong X, Liu J and Ma Y:
Osteosarcoma: A review of current and future therapeutic
approaches. Biomed Eng Online. 20(24)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang T, Yu Q, Zhang W and Gao L:
Comprehensive analysis of the PROSER2-AS1-related ceRNA network and
immune cell infiltration in papillary thyroid carcinoma. Int J Gen
Med. 15:1647–1663. 2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hamada H, Okae H, Toh H, Chiba H, Hiura H,
Shirane K, Sato T, Suyama M, Yaegashi N, Sasaki H and Arima T:
Allele-specific methylome and transcriptome analysis reveals
widespread imprinting in the human placenta. Am J Hum Genet.
99:1045–1058. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Parviainen R, Skarp S, Korhonen L, Serlo
W, Mannikko M and Sinikumpu JJ: A single genetic locus associated
with pediatric fractures: A genome-wide association study on 3,230
patients. Exp Ther Med. 20:1716–1724. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sui Y, Ju C and Shao B: A lymph node
metastasis-related protein-coding genes combining with long
noncoding RNA signature for breast cancer survival prediction. J
Cell Physiol. 234:20036–20045. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen J, Guo X, Zeng G, Liu J and Zhao B:
Transcriptome analysis identifies novel prognostic genes in
osteosarcoma. Comput Math Methods Med. 2020(8081973)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jiang X, Liu J, Guan Y, Zhao Z, Meng F and
Wang X, Gao X, Zhou F, Chen Y and Wang X: The mechanism of the
WNT5A and FZD4 receptor mediated WNT/β-catenin pathway in the
degeneration of ALS spinal cord motor neurons. Biochem Biophys Res
Commun. 609:23–30. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Choi JH, Kim YM, Park HJ, Nam MH and Seo
YK: Extremely low-frequency electromagnetic fields increase
cytokines in human hair follicles through Wnt/β-catenin signaling.
Biomedicines. 10(924)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tong XK, Royea J and Hamel E: Simvastatin
rescues memory and granule cell maturation through the
Wnt/β-catenin signaling pathway in a mouse model of Alzheimer's
disease. Cell Death Dis. 13(325)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Daniels JR, Ma JZ, Cao Z, Beger RD, Sun J,
Schnackenberg L, Pence L, Choudhury D, Palevsky PM, Portilla D and
Yu LR: Discovery of novel proteomic biomarkers for the prediction
of kidney recovery from dialysis-dependent AKI patients. Kidney360.
2:1716–1727. 2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chu CY, Wang R and Liu XL: Roles of
Wnt/β-catenin signaling pathway related microRNAs in esophageal
cancer. World J Clin Cases. 10:2678–2686. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen Y, Chen Z, Tang Y and Xiao Q: The
involvement of noncanonical Wnt signaling in cancers. Biomed
Pharmacother. 133(110946)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vargas JY, Loria F, Wu YJ, Córdova G,
Nonaka T, Bellow S, Syan S, Hasegawa M, van Woerden GM, Trollet C
and Zurzolo C: The Wnt/Ca2+ pathway is involved in
interneuronal communication mediated by tunneling nanotubes. EMBO
J. 38(e101230)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang J, Chandrasekaran G, Li W, Kim DY,
Jeong IY, Lee SH, Liang T, Bae JY, Choi I, Kang H, et al:
Wnt-PLC-IP3-Connexin-Ca2+ axis maintains ependymal
motile cilia in zebrafish spinal cord. Nat Commun.
11(1860)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Farrera-Hernández A, Marín-Llera JC and
Chimal-Monroy J: WNT5A-Ca2+-CaN-NFAT signalling plays a
permissive role during cartilage differentiation in embryonic chick
digit development. Dev Biol. 469:86–95. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Huybrechts Y, Mortier G, Boudin E and Van
Hul W: WNT signaling and bone: Lessons from skeletal dysplasias and
disorders. Front Endocrinol (Lausanne). 11(165)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yuan J, Yuan Z, Ye A, Wu T, Jia J, Guo J,
Zhang J, Li T and Cheng X: Low GNG12 expression predicts adverse
outcomes: A potential therapeutic target for osteosarcoma. Front
Immunol. 12(758845)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang J, Liu W, Li JC, Li M, Li B and Zhu
R: Hepcidin downregulation correlates with disease aggressiveness
and immune infiltration in liver cancers. Front Oncol.
11(714756)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Du Y, Cao J, Jiang X, Cai X, Wang B, Wang
Y, Wang X and Xue B: Comprehensive analysis of CXCL12 expression
reveals the significance of inflammatory fibroblasts in bladder
cancer carcinogenesis and progression. Cancer Cell Int.
21(613)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Feng Z, Li L, Tu Y, Shu X, Zhang Y, Zeng
Q, Luo L, Wu A, Chen W, Cao Y and Li Z: Identification of circular
RNA-based immunomodulatory networks in colorectal cancer. Front
Oncol. 11(779706)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang J, Zhang A, Luo H and Ma C:
Construction and validation of a novel gene signature for
predicting the prognosis of osteosarcoma. Sci Rep.
12(1279)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wan J, Liu Y, Long F, Tian J and Zhang C:
circPVT1 promotes osteosarcoma glycolysis and metastasis by
sponging miR-423-5p to activate Wnt5a/Ror2 signaling. Cancer Sci.
112:1707–1722. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dai B, Shen Y, Yan T and Zhang A:
Wnt5a/ROR1 activates DAAM1 and promotes the migration in
osteosarcoma cells. Oncol Rep. 43:601–608. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang X, Zhao X, Yi Z, Ma B, Wang H, Pu Y,
Wang J and Wang S: WNT5A promotes migration and invasion of human
osteosarcoma cells via SRC/ERK/MMP-14 pathway. Cell Biol Int.
42:598–607. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Duan N, Zhang W, Song T, Li Z, Chen X and
Ma W: A naturally derived small molecule PSM0537 targets the
AF1Q-TCF4 interaction to suppress COX2 expression and inhibit cell
proliferation and metastasis in osteosarcoma. Am J Cancer Res.
11:2637–2653. 2021.PubMed/NCBI
|
|
31
|
Zhang X, Qu P, Zhao H, Zhao T and Cao N:
COX2 promotes epithelialmesenchymal transition and migration in
osteosarcoma MG63 cells via PI3K/AKT/NF-κB signaling. Mol Med Rep.
20:3811–3819. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Duan H, Ding X and Luo H: KISS-1, mediated
by promoter methylation, suppresses esophageal squamous cell
carcinoma metastasis via MMP2/9/MAPK axis. Dig Dis Sci.
67:4780–4796. 2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu J, Ding D, Liu F and Chen Y: Rhein
inhibits the progression of chemoresistant lung cancer cell lines
via the Stat3/Snail/MMP2/MMP9 pathway. Biomed Res Int.
2022(7184871)2022.PubMed/NCBI View Article : Google Scholar
|